Abstract
An understanding of heart development is critical in any systems biology approach to cardiovascular disease. The interpretation of data generated from high-throughput technologies (such as microarray and proteomics) is also essential to this approach. However, characterizing the role of genes in the processes underlying heart development and cardiovascular disease involves the non-trivial task of data analysis and integration of previous knowledge. The Gene Ontology (GO) Consortium provides structured controlled biological vocabularies that are used to summarize previous functional knowledge for gene products across all species. One aspect of GO describes biological processes, such as development and signaling.
In order to support high-throughput cardiovascular research, we have initiated an effort to fully describe heart development in GO; expanding the number of GO terms describing heart development from 12 to over 280. This new ontology describes heart morphogenesis, the differentiation of specific cardiac cell types, and the involvement of signaling pathways in heart development and aligns GO with the current views of the heart development research community and its representation in the literature. This extension of GO allows gene product annotators to comprehensively capture the genetic program leading to the developmental progression of the heart. This will enable users to integrate heart development data across species, resulting in the comprehensive retrieval of information about this subject.
The revised GO structure, combined with gene product annotations, should improve the interpretation of data from high-throughput methods in a variety of cardiovascular research areas, including heart development, congenital cardiac disease, and cardiac stem cell research. Additionally, we invite the heart development community to contribute to the expansion of this important dataset for the benefit of future research in this area.
Keywords: annotation, cardiovascular, development, Gene Ontology, heart
Introduction
Forming as the result of an elegant coordination of integrated processes, the heart is one of the first organs to develop in a vertebrate embryo. Understanding this developmental process is critical to the understanding of cardiovascular disease (CVD), a leading cause of mortality worldwide (Batsis and Lopez-Jimenez, 2010). One aspect of CVD is damaged heart tissue, and the possibility of using stem cells to repair a heart, is an active area of research (Bollini et al., 2011). However, to fully understand how to repair a heart we must first understand the processes by which a heart is formed. Heart development is a complex process controlled by a multitude of coordinated cellular processes resulting in proper patterns of cell differentiation and tissue morphogenesis (Abu-Issa and Kirby, 2007; Dyer and Kirby, 2009). The events in the process are still being identified, thus, the study of genes and proteins involved in cardiovascular development is an important research area. As with any multi-genic process, there is an important role for high-throughput methods to characterize the genes and proteins involved in both developmental and disease processes. However, interpretation of high-throughput data with regard to previously established work is a non-trivial task, associated with a need for categorization of gene functions to enable data analysis. To assist with the interpretation of these data, the Gene Ontology Consortium (GOC) provides a robust hierarchical controlled vocabulary, the Gene Ontology (GO) (Ashburner et al., 2000). Individually each GO term can be applied to gene products across all species to summarize individual experiments. However, the power of GO lies in the fact that it is a categorization of gene products characteristics, rather than a categorization of gene products themselves. Thus, large numbers of gene products can be grouped on the basis of their characteristics, as defined by GO terms. Many high-throughput analysis tools have been developed for this purpose (Hendrickson et al., 2008; Malik et al., 2010; Werner, 2008). GO currently contains over 33,000 terms and is routinely used for the analysis of large datasets (Colak et al., 2009; Herbert et al., 2009; Mace et al., 2009).
The GOC comprises of GO editors who develop the ontology, and GO annotators who read the primary literature and create annotations using the ontology (Gene Ontology Consortium, 2009). The two groups of curators work closely together. As a result GO terms are continuously examined, evaluated and tested during the annotation process based on the experimental literature. The GOC has set up several mechanisms for handling ontology issues. We utilize a SourceForge tracker for ontology questions, have a ‘help’ resource available from the GO web site and provide contact information for curator interest groups who coordinate biological areas of ontology development (see box). There are more than a dozen model organism databases (MOD) represented in the GOC, which use GO to annotate the gene products of their specific organism. Additionally there are other annotation groups focusing on specific areas of biology. The BHF-UCL (British Heart Foundation-University College London) GO team focuses on cardiovascular biology and has the specific remit of annotating human proteins involved in cardiovascular processes and disease (Lovering et al., 2008). An advantage to having a specialized annotation group like the BHF-UCL group is that it allows for development of a whole branch of the ontology alongside the creation of gene annotations using the new ontology terms. GO annotators read the primary literature and create GO annotations using a variety of evidence codes that describe the nature of the experiments that support the annotation. In practice annotations to developmental processes are mostly derived from direct assays, mutational studies or gene interaction studies. Annotators are cautious about using expression data for GO annotations since expression may correlate with a process, but not necessarily be actively involved in the process.
In general, there are two ways in which ontology development can be coordinated with annotation: 1) terms can be requested individually as annotators find need for them; 2) large-scale development of the ontology can be undertaken in anticipation of the terms needed for annotation. In practice, we have found that the latter method is very efficient for developing focused areas of the ontology (Diehl et al., 2007; Feltrin et al., 2009; Maccagnan et al., 2010). Specifically, we find that collaboration between expert annotators, ontology developers and experimental biologists working in the relevant field is a productive way to create an accurate and complete representation of an area of biology in the shortest amount of time.
In order to establish the groundwork for an expansion of the representation of GO biological processes involved in heart development, an initial meeting was held between gene annotators (from the BHF-UCL GO team and several model-organism databases), GOC ontology developers and cardiac development experts (all authors of this paper). At the start of the work, there were 12 terms in GO that represented all of heart development. Work during the meeting, as well as subsequent revisions and discussions have resulted in the addition of 281 new terms to date (see Supplemental Table for full list of new terms). These new terms have been added to GO and are fully integrated with relationships to other developmental processes in the existing biological processes ontology, and all additional parent terms. The new terms are publically available as part of the current version of GO. GO now includes terms describing an anatomical representation of heart development (such as the valves and the heart chambers), as well as terms that describe specific types of processes that contribute to heart development (such as cell differentiation and signaling pathways). In this article we introduce the new heart development Gene Ontology, demonstrate how it is used for annotations and invite the heart development community to contribute to this important resource.
A Gene Ontology primer
Gene Ontology (GO) is a controlled vocabulary that is used to classify the biological characteristics of gene products. GO terms describe three characteristics, Biological Process (BP), Molecular Function (MF) and Cellular Component (CC). BP terms describe the general process a gene product is involved in, MF terms describe the specific molecular function of a gene product and CC terms describe the subcellular compartment in which a gene product is found. This work focused on the expansion of the BP part of GO, which encompasses all developmental processes.
Gene annotators use experimentally supported data from published literature to associate specific GO terms with the genes that have been shown to bear the attributes described by the GO term. Taken as a whole, the set of annotations to a given gene product aims to describe the totality of what is currently known about that gene product’s role in biology, while an individual annotation describes the results of a single experiment. Annotators use their biological knowledge alongside information presented in the paper to judge the most specific term possible for each annotation. For example, individual experiments have shown that the homeobox NKX2-5 gene product takes part in the process of cardiac muscle cell differentiation (GO:0055007; BP) (Tanaka et al., 1999), has transcription factor activity (GO:0003700; MF) (Kasahara and Izumo, 1999) and is found in the nucleus (GO:0005634; CC) (Zhu et al., 2000). Thus the NKX2-5 gene product has been annotated to each of those terms. Additionally gene products may have multiple functions, often take part in more than one process and can be found in multiple subcellular compartments. GO allows a single gene product to be annotated to any number of terms from each of the three ontologies.
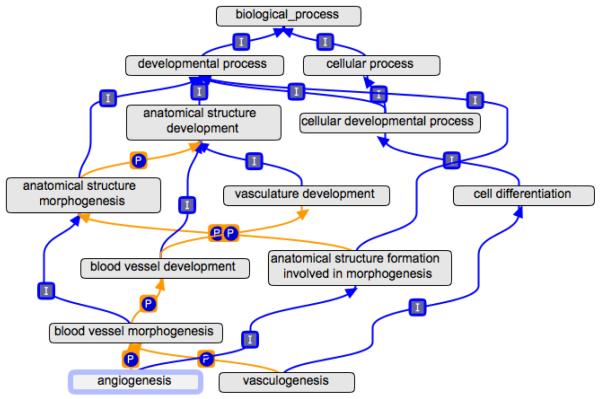
GO terms are structured in a directed acyclic graphs (DAG), where each term can have multiple relationships to broader ‘parent’ and more specific ‘child’ terms. The parent and child terms have specific relationships with each other. In GO, there are seven types of relationships (Gene Ontology Consortium, 2009; Smith et al., 2005), of which five are relevant to the heart development ontology. The ‘is_a’ relationship means that a child term is always a type of its parent term; for example heart development (GO:0007507) is a type of organ development (GO:0048513). The ‘part_of’ relationship means that the child term is always a part of the parent term; for example cell migration involved in vasculogenesis (GO:0035441) is part of vasculogenesis (GO:0001570). The ‘regulates’, ‘positively_regulates’, and ‘negatively_regulates’ relationships signify that the children have a regulatory effect on the parent; for example the term negative regulation of cardiac muscle tissue development (GO:0055026) has a ‘negatively_regulates’ relationship to the term cardiac muscle tissue development (GO:0048738). To illustrate how terms and relationships are used in GO, Figure 1 shows that the term vasculogenesis (GO:0001570) has two direct ancestors, cell differentiation (GO:0030154) and blood vessel morphogenesis (GO:0048514). Vasculogenesis ‘is a’ type of cell differentiation and is also ‘part of’ the process of blood vessel morphogenesis.
Figure 1.
The ancestor chart for GO terms vasculogenesis and angiogenesis. The graph shows the terms vasculogenesis (GO:0001570), angiogenesis (GO:0001525) and all of their ancestor terms. The lines marked with I indicate an ‘is_a’ relationship and those marked with P indicate a ‘part_of’ relationship. The root of this ontology is the ‘biological process’ term, thus all the terms shown here are biological processes. A gene product annotated to vasculogenesis (GO:0001570) is automatically associated with the term blood vessel morphogenesis, since vasculogenesis is a child of blood vessel morphogenesis. Drawn with the OBO-Edit tool.
An important benefit of building a DAG, rather than a flat-list of controlled vocabulary terms, is that relationships can be used to make inferences from one term to another. For example, vasculogenesis (GO:0001570) is part of blood vessel morphogenesis (GO:0048514) and blood vessel morphogenesis (GO:0048514) is part of blood vessel development (GO:0001568). Therefore, because the ‘part_of’ relationship is transitive, vasculogenesis (GO:0001570) can also be considered to be a part of blood vessel development (GO:0001568), and does not require a direct link between the two terms. This transitive nature is very useful when using GO to find gene products annotated as being involved in a particular process; for example a search for gene products annotated to blood vessel morphogenesis (GO:0048514) will include those annotated directly to the more specific processes of vasculogenesis (GO:0001570) and angiogenesis (GO:0001525), both of which are a part of blood vessel morphogenesis (GO:0048514).
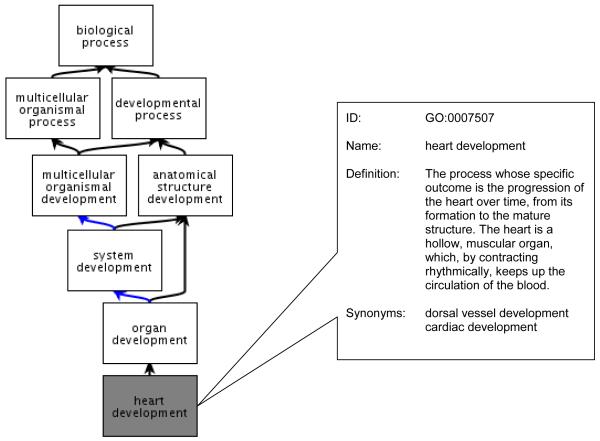
Each GO term has several different components (Figure 2). The GO ID is unique to each term. The definition is a textual description of what the term means and in many cases disambiguates the use of identical terms that are used to mean different things. Additionally the placement of the term within the ontology also provides a necessary definition through the terms relationships with its parents. For example the term lateral ventricle development (GO:0021670) may at first glance be ambiguous, but upon viewing the ontology it is clear that this term is a descendant of central nervous system development (GO:0007417), and thus ‘ventricle’ in this context refers to a brain ventricle and not a heart ventricle.
Figure 2.
Anatomy of a GO term. QuickGO view of GO (www.ebi.ac.uk/QuickGO) illustrates how each GO term has a unique GO ID number. The term name describes the overall concept, which is supplemented by what is often a very detailed definition. Some terms also have synonyms to aid searching.
GO allows for the description of processes that occur at multiple levels of biology: i) the organ level, ii) the multicellular (tissue) level and iii) the level of the single cell. It also allows for the description of generic processes that are used in multiple ways to accomplish a given objective. The developmental process section of the biological process ontology allows for the description of developmental events either from an anatomical perspective or from a process perspective. Standard developmental terms such as cell differentiation (GO:0030154) are defined generically and are then used consistently to describe the process in the context of all of the different processes in which it is involved (Hill et al., 2010). The following sections illustrate how the newly created ontology can be used to describe the heart developmental processes that occur at each of these three levels; heart morphogenesis at the organ level, cell differentiation in the heart at the cellular level, and signaling pathways to describe interactions between cells that make up tissues.
The terms and relationships in the ontology are carefully chosen so that regardless of the species the ontology is always correct. Species-neutrality enables experimentally supported annotations in model organisms, to be transferred to human gene products, if appropriate. For example, based on the observation that expression of mouse Mesp1 in embryonic stem cells (ESCs) results in transcriptional regulation of key genes controlling early mesoderm and endoderm cell fates and promotes the progression of cells toward a cardiac fate (Bondue et al., 2008), one of the terms that was used to annotate the mouse Mesp1 gene product is cardiac cell fate determination (GO:0060913). At the time of writing there are 26 experimentally supported annotations associated with the mouse Mesp1 gene product, whereas there are 10 such annotations associated to the human MESP1 gene product. The unique mouse annotations have been transferred to the orthologous human protein, thus enhancing our knowledge about how the orthologous gene products might be involved in human biology or disease.
Heart morphogenesis
In GO, morphogenesis is used to describe the initial formation of an anatomical structure and its subsequent shaping in an anatomical context. Because the ontology does not contain temporal relationships, GO does not describe lineage relationships between anatomical structures. Although often embedded in the definition of a term, lineage relationships themselves are not specified in the ontology. An important aspect in the creation of the development domain of GO is identifying when a structure begins to exist and what processes lead to its formation. In addition, GO developers need to reflect the prevailing thought of the scientific community of a given field. This approach allows GO to be used in a practical manner as it is aligned with current scientific thought. The heart is formed from a distinct group of cells (distinguishable from the surrounding cell population only by the proteins expressed) that make up the heart fields. These cells proliferate and migrate to form distinct subpopulations of cells, which then form the discrete substructures of the heart, for example the valves and the chambers. GO reflects that the specification of the heart field is one of the initial processes of heart development and contains terms for primary heart field specification (GO:0003138) and secondary heart field specification (GO:0003139). Both of these terms are types of a more generic heart field specification (GO:0003128). The induction process whereby the mesoderm, endoderm, and ectoderm interact are not part of the formation of the heart per se, but rather positively regulate its formation. The rationale behind this decision is that when this inductive process is occurring, the heart has not formed and the delineation of the field is the first step in the formation of the actual structure. This decision is consistent with all inductive interactions in the development domain of the ontology (Hill et al., 2010). Other terms that are part of the morphogenesis of the heart reflect both the morphogenesis of anatomical parts of the heart such as cardiogenic plate morphogenesis (GO:0003142) and endocardial cushion morphogenesis (GO:0003203), or conserved processes that contribute to the shaping of the structure itself such as apoptosis involved in heart morphogenesis (GO:0003278) and growth involved in heart morphogenesis (GO:0003241). The latter category of terms allows users of GO to search for gene products that might be conserved in apoptosis or growth involved morphogenesis of other anatomical structures.
Cell differentiation in the heart
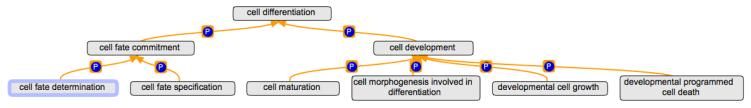
Much of development at the cellular level can be considered to be successive cell differentiation events that eventually lead to terminally differentiated cells that contribute to the functioning of a mature anatomical structure. GO describes cell differentiation in two parts. First an undifferentiated cell somehow decides what it will become and then in the second step the cell undergoes the actual process of becoming differentiated. In GO, the term cell differentiation (GO:0030154) is the parent of the terms cell fate commitment (GO:0045165) that describes how a cell decides what to become, and cell development (GO:0048468) that describes how a cell undergoes the actual differentiation process. The relationship between these terms and further descendants of these terms are shown in Figure 3. The way that GO defines cell differentiation, and in particular the phrases used in the definitions of these terms, is discussed in greater depth in a review of Gene Ontology and developmental biology (Hill et al.).
Figure 3.
Cell differentiation ontology. Gene products specify what a cell is going to be through the process of cell fate commitment (GO:0045165) and subsequently expressed gene products allow the cell to become so through the process of cell development (GO:0048468). The lines are marked with P to indicate a ‘part_of’ relationship. Drawn with the OBO-Edit tool.
This structure allows GO to describe the differentiation of any cell type in the heart. As a result, we have included 26 cell type terms specific to the heart in the revised ontology (Table 1). These range from very general cell type terms like cardiac fibroblast cell differentiation (GO:0060935) to very specific cell type terms like atrioventricular bundle cell differentiation (GO:0003167).
Table 1.
The 26 newly created heart-specific cell type terms currently in GO. This is not an exhaustive list, and other cell types will be added when needed for new annotations.
| Gene Ontology Term Name | Gene Ontology ID |
|---|---|
| atrial cardiac muscle cell differentiation | GO:0003167 |
| atrioventricular bundle cell differentiation | GO:0003168 |
| atrioventricular node cell differentiation | GO:0003255 |
| cardiac blood vessel endothelial cell differentiation | GO:0003292 |
| cardiac endothelial cell differentiation | GO:0003293 |
| cardiac fibroblast cell differentiation | GO:0003348 |
| cardiac glial cell differentiation | GO:0003349 |
| cardiac muscle cell differentiation | GO:0007513 |
| cardiac muscle cell myoblast differentiation | GO:0010002 |
| cardiac neuron differentiation | GO:0051890 |
| cardiac Purkinje fiber cell differentiation | GO:0051891 |
| cardiac septum cell differentiation | GO:0051892 |
| cardiac vascular smooth muscle cell differentiation | GO:0055007 |
| cardioblast differentiation | GO:0055011 |
| endocardial cell differentiation | GO:0055012 |
| endocardial precursor cell differentiation | GO:0060379 |
| epicardium-derived cardiac endothelial cell differentiation | GO:0060920 |
| epicardium-derived cardiac fibroblast cell differentiation | GO:0060921 |
| epicardium-derived cardiac vascular smooth muscle cell differentiation |
GO:0060922 |
| heart valve cell differentiation | GO:0060932 |
| His-Purkinje system cell differentiation | GO:0060935 |
| neural crest-derived cardiac fibroblast cell differentiation | GO:0060938 |
| neural crest-derived cardiac glial cell differentiation | GO:0060942 |
| pacemaker cell differentiation | GO:0060945 |
| pericardial cell differentiation | GO:0060946 |
| sinoatrial node cell differentiation | GO:0060947 |
| ventricular cardiac muscle cell differentiation | GO:0060950 |
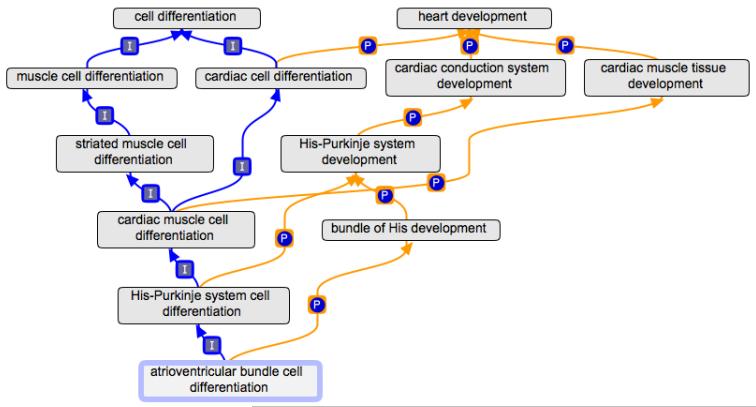
The structure that is now in place will permit gene product annotations to be integrated into knowledge about not only the generic types of cells that cardiac cells represent but also integrated into the way they play specific roles in the development of the heart. For example, atrioventricular bundle cell differentiation is related to generalized cardiac muscle cell differentiation as well as to the development of the cardiac conduction system (Figure 4).
Figure 4.
An example of the relationship between specific cell types and the role the cell plays in heart development. GO allows atrioventricular bundle cells to have a direct ‘is_a’ relationship to generalized cardiac cells, as well a ‘part_of’ relationship to cardiac conduction. I indicates an ‘is_a’ relationship, P indicates a ‘part_of’ relationship. Drawn with the OBO-Edit tool.
The arrangement of the cell differentiation terms allows for the placement of new terms easily into the ontology. It also allows for the detailed annotation of gene products at a level of detail that is supported by the experimental data. For example, in the context of ESC differentiation, when ESCs are transfected with POU5F1 cDNA and stimulated with BMP2, pluripotency is lost, the cells express SOX17 and adopt a cardiovascular fate (Stefanovic et al., 2009). However, without the BMP2 signal, transfection with POU5F1 cDNA maintains the pluripotency of the ESC. Thus, POU5F1 specifies a cardiac cell fate only if the cell is subject to a specific environment, and so POU5F1 was annotated with cardiac cell fate specification (GO:0060912). (The mouse Pou5f1 and human POU5F1 gene products respectively have 68 and 18 additional experimentally supported annotations). On the other hand, once the human ESC expresses SOX17, it is committed to a cardiac cell fate regardless of the environment (Stefanovic et al., 2009); thus SOX17 was annotated with cardiac cell fate determination (GO:0060913). (The mouse Sox17 and human SOX17 gene products respectively have 15 and 11 other experimentally supported annotations).
Signaling pathways in heart development
The revised heart development ontology allows for the annotation of gene products that describe signaling processes between different types of heart cells. Terms like endodermal-mesodermal cell signaling involved in heart induction (GO:0003134) describe signaling processes where it is known that the cells of the endoderm signal to cells of the mesoderm, but the molecular nature of the signaling is unknown. Where the molecular nature of the signaling is known, we are able to create terms for the specific pathways. So far we have created terms for the BMP, hedgehog, FGF, Notch, TGF-beta and WNT signaling pathways as children of cell surface receptor linked signaling pathway involved in heart development (GO:0061311). By creating these high-level generic terms, we have built the framework for the addition of more specific terms, involving these and other signaling pathways, as needed to annotate new data. For example, it has shown that heart induction is a result of several signaling pathways, the BMP signaling pathway (Dyer and Kirby, 2009), the Notch pathway (MacGrogan et al., 2010) and the FGF receptor signaling pathway (Samuel and Latinkic, 2009); whilst the canonical Wnt receptor signaling pathway plays a negative role (Palpant et al., 2007; van de Schans et al., 2008). As an example of the types of signaling pathway terms GO can support, we have created terms such as BMP signaling pathway involved in heart induction (GO:0003130), fibroblast growth factor receptor signaling pathway involved in heart induction (GO:0003135) and Notch signaling pathway involved in heart induction (GO:0003137), all as ‘part of’ heart induction (GO:0003129) (Bondue et al., 2008; Lough et al., 1996; Schlange et al., 2000).
We have also created the term negative regulation of heart induction by canonical Wnt receptor signaling pathway (GO:0003136) to describe the role of that pathway in the process. Annotations are made to gene products involved in each of the pathways, and these can then be used during analysis of high-throughput data to identify members of signaling pathways that play roles in heart induction. For example, BMP2 induces a cardiac fate (Stefanovic et al., 2009) and has therefore been annotated to BMP signaling pathway involved in heart induction (GO:003130) while the Mesp1 gene product promotes cardiovascular differentiation by inhibiting the canonical Wnt signaling pathway (David et al., 2008), hence Mesp1 was annotated to the term positive regulation of heart induction by negative regulation of canonical Wnt receptor signaling pathway (GO:0090082). This term positively regulates heart induction and therefore describes the role of the Mesp1 gene product. As an example of how we plan to handle the complexity of signaling pathways in heart development, we have expanded the Wnt-receptor signaling portion of the ontology in detail.
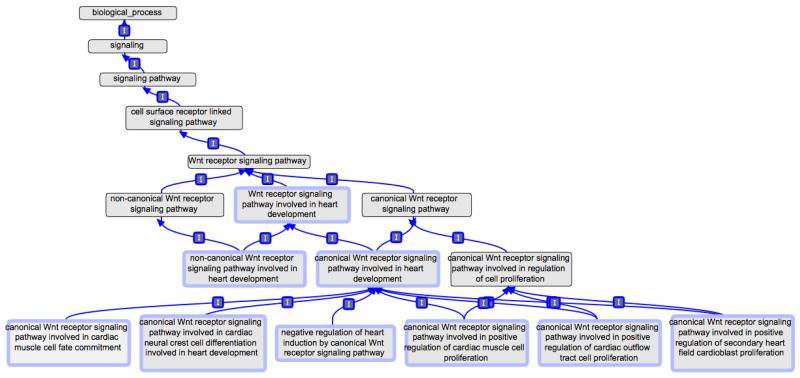
As shown in Figure 5, Wnt receptor signaling pathway involved in heart development (GO:0003306) has two child terms, canonical Wnt receptor signaling pathway involved in heart development (GO:0061316) and non-canonical Wnt receptor signaling pathway involved in heart development (GO:0061341). Based on the literature, the canonical Wnt receptor signaling pathway involved in heart development (GO:0061316) term has been given six child terms to describe the role of canonical Wnt signaling in cardiac muscle cell fate commitment (GO:0061317), cardiac neural crest cell differentiation (GO:0061310), cardiac muscle cell proliferation (GO:0061315), cardiac outflow tract cell proliferation (GO:0061324), secondary heart field cardioblast proliferation (GO:0003267), and heart induction (Brade et al., 2006; Kwon et al., 2007; Song et al., 2010).
Figure 5.
The Wnt signaling ontology illustrates the way in which GO can be used to represent signaling pathways involved in heart development. I indicates an ‘is_a’ relationship. Drawn with the OBO-Edit tool.
We have begun using these terms for the annotation of gene products. As detailed annotations accumulate, it will become increasingly easy to identify all proteins that act in signaling pathways resulting in specific developmental events in the heart. As more experimental evidence becomes available to support the role of a particular signaling pathway in a particular developmental event, we will continue to expand the ontology and the associated annotations.
GO is applicable across species
GO is designed so that it can be used for the annotation of all gene products across all species. This allows GO to be used to examine orthologous gene products from a variety of species and to make conclusions about conserved or diverse biological processes based on annotations to similar or dissimilar GO terms. It also allows us to transfer annotations from one species to another when genes appear to have conserved functions across species (Mi et al., 2009). For example, orthologs of TBX5 have been shown to be key players in the patterning of the ventricle. These gene products have differential expression patterns in the 3-chambered amphibian heart compared to the 4-chambered mammalian and avian hearts. In the red-eared slider turtle (Trachemys scripta elegans), the expression of Tbx5 was homogenous throughout its single ventricle, whereas in the chick it was restricted to the left ventricle (Koshiba-Takeuchi et al., 2009). Furthermore, the loss of Tbx5 in the mouse ventricle resulted in the development of a single ventricle, similar to that of the turtle (Koshiba-Takeuchi et al., 2009). In this relatively simple scenario, the mouse and chick Tbx5 gene products are annotated to ventricular septum development (GO:0003281) and cardiac left ventricle formation (GO:0003218). The turtle Tbx5 gene product would be annotated to the parent of the latter term, cardiac ventricle formation (GO:0003211). Thus the structure of the ontology allows for the differential annotation of orthologous gene products that take part in related but distinct processes in different species.
However, there are also cases where different species have specific structures. In the zebrafish (Danio rerio), the bulbus arteriosus is an elastic heart chamber that receives blood from the heart, and maintains its flow to the gill arches. There is no orthologous structure in mammals, so we created the term bulbus arteriosus development (GO:0003232) to capture this fish-specific process.
There are also some processes that appear to be species specific. In the zebrafish, the establishment of left/right asymmetry of the heart tube has been observed to occur through a two step process of heart tube displacement and rotation called ‘heart jogging’ and ‘heart looping’ (Chen et al., 1997). The resulting left/right asymmetry of the heart tube is reversed or otherwise disrupted in a number of zebrafish mutants affecting genes including the chd, acvr1l, smad5, bmp7a, ndr2, foxh1, pkd2, lrrc6, spaw, and cha genes (Chen et al., 1997; Hashimoto et al., 2004; Long et al., 2003). Experimentally supported data from such mutants has been curated to a number of GO BP terms relating to heart development, including heart jogging (GO:0003146), heart looping (GO:00011947), cell migration involved in heart jogging (GO:0003305), and determination of left/right symmetry (GO:0007368). The process of heart jogging has not been observed in mammals, in addition there does not appear to be an orthologous process. Therefore, restrictions in the ontology have been created to ensure that any annotations made to zebrafish gene products to heart jogging (GO:0003146) and its child terms cannot be transferred to mammalian orthologs. The GOC has mechanisms in place to ensure that inappropriate annotations are not made (Deegan et al., 2010).
Community Annotation
With the new ontology structure for heart development in place, the challenge now is to utilize the new terms in annotations and to continue to refine and add to the ontology. This is a daunting task for MOD curators since the existing experimental literature is voluminous. Manual GO annotation is a painstaking process, with a large volume of literature to sort through and relatively few curators at each MOD dedicated to GO annotation. One way to increase the amount of information about how gene products play a role in heart development is to take advantage of the orthology strategies described above. However, the annotation effort can also be greatly helped by input from research scientists working in collaboration with professional curators. This exercise in ontology development is proof that input from experts in the field can have dramatic effects on a bioinformatics resource such as GO. Since experimental biologists are necessarily much more familiar with their field of research, they know the details and location of the experimental evidence to support a GO annotation or a new ontology term better than a GO curator, who does not necessarily specialize in the field.
Research scientists can help to improve the GO annotation dataset for heart development in a number of ways. 1) Review the information we have captured about a gene in which they specialize, informing us of inaccurate or missing annotations. 2) Help identify key papers in a field or research, resulting in the manual curation representing the most comprehensive and accurate information. 3) Review the ontology to be sure that it is complete and aligns with the current thought in the field. 4) Provide feedback about how they have used GO in their analyses, reporting about what they liked, didn’t like or features they would like to see available. 5) Researchers could also provide GO annotations from their own papers, this has been working well with the Arabidopsis community (Ort and Grennan, 2008). Input can be provided to the GOC in multiple ways (see box). Once suggestions are made ontology curators will address issues that are raised, with help from domain experts in the field. We suggest that a good time for researchers to provide GO annotations would be when proofreading accepted papers. This would ensure the latest data would be included in the GO annotation dataset. Annotated papers may be cited more often, since GO is widely used in bioinformatics analyses.
Although the GOC consists of curators working at several different MODs in multiple different locations around the world, GO curators communicate regularly and utilize each other’s expertise. For example, a curator working on human gene products may contact a curator working for another MOD as needed, to annotate a paper on an organism which is unfamiliar to them. Therefore the GO curators are able to call upon a wide range of species-specific expertise as needed. Additionally, GO curators are proactive about involving expert researchers as needed to work on a particular area of the ontology, as evidenced by the work described here and by previous ontology development projects (Feltrin et al., 2009; Maccagnan et al., 2010).
Conclusions
In this article we have described the work that we have done to refine and expand the representation of heart development in GO. We have expanded the heart development section of the ontology from 12 to over 250 terms. In the process, we have integrated the representation of heart development with other cellular and developmental processes in GO. Biological Process GO terms can now be used to interpret all aspects of heart development processes, from an anatomical perspective, to a cell differentiation perspective, through to a cellular perspective such as signaling. Heart development is a complex process underpinned by multiple gene regulatory networks and signaling pathways, and the use of GO terms can help to simplify the categorization of gene products which are important to this process. Additionally, the fact that GO terms and their associated annotations are freely available means that this dataset is a vitally important resource for the heart research community, from those wishing to analyze high-throughput data (Marques et al., 2010; Tranter et al., 2010) to those wishing to use the heart as a model for understanding how organogenesis initiates and unfolds (Abdulla et al., 2010).
The structure we have established not only places heart development in alignment with other developmental processes in GO, but also allows for the interpretation of gene product annotations using the variety of ontology structures and the detail of terms. This new structure should prove useful for researchers in the field who are performing genomic studies or searching for genes that could be involved in a given process. As the terms are used for annotation and as researchers generate new biological knowledge, we anticipate additional terms will need to be created. The existing mechanisms established by the GOC for revising the ontology will continue to be used. However, the ontology foundations presented here will permit easy additions of new terms in the future as children of existing heart development terms. For example the differentiation of a new cell type can be added as a differentiation of an existing generic cell type and a part of the development of the structure to which it contributes.
With the structure of the heart development ontology now in place, the rate-limiting step for using GO to its full potential is the annotation of gene products to the new terms in the ontology. Curators from the relevant model organism databases are working alongside the BHF-UCL GO team to screen the literature and identify papers containing gene products that are involved in heart development. It is at this step, and in the continued development of the ontology where experts in the field could help us to streamline the process. By helping us to identify key genes, literature and areas of the ontology on which to focus, researchers could make certain that the limited curatorial resources available are utilized in the most efficient manner. In the end this will add to the value of our resource for the benefit of the entire cardiovascular research community.
Supplementary Material
Research highlights.
The usefulness of high-throughput data depends on the ability to interpret it.
Gene Ontology (GO) annotations are used to interpret high-throughput data.
Understanding heart development is important in understanding heart disease.
We have expanded the available GO terms for heart development from 12 to over 280.
This dataset could improve interpretation of cardiovascular high-throughput data.
Acknowledgements
The BHF-UCL team is funded by the British Heart Foundation (SP/07/007/23671), as part of the Cardiovascular GO Annotation Initiative. The Gene Ontology Consortium is funded by NHGRI grant HG002273 to J. Blake, J. M. Cherry, S. Lewis and M. Ashburner. Dr. Constance Smith and Dr. Cynthia Smith for their critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdulla T, Imms R, Schleich JM, Summers R. Multiscale information modelling for heart morphogenesis. Journal of Physics: Conference Series. 2010;238 [Google Scholar]
- Abu-Issa R, Kirby ML. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsis JA, Lopez-Jimenez F. Cardiovascular risk assessment--from individual risk prediction to estimation of global risk and change in risk in the population. BMC Med. 2010;8:29. doi: 10.1186/1741-7015-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollini S, Smart N, Riley PR. Resident cardiac progenitor cells: At the heart of regeneration. J Mol Cell Cardiol. 2011 doi: 10.1016/j.yjmcc.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Brade T, Manner J, Kuhl M. The role of Wnt signalling in cardiac development and tissue remodelling in the mature heart. Cardiovasc Res. 2006;72:198–209. doi: 10.1016/j.cardiores.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Chen JN, van Eeden FJ, Warren KS, Chin A, Nusslein-Volhard C, Haffter P, Fishman MC. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124:4373–82. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- Colak D, Kaya N, Al-Zahrani J, Al Bakheet A, Muiya P, Andres E, Quackenbush J, Dzimiri N. Left ventricular global transcriptional profiling in human end-stage dilated cardiomyopathy. Genomics. 2009;94:20–31. doi: 10.1016/j.ygeno.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Muller-Hocker J, Kitajima S, Lickert H, Rupp R, Franz WM. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–45. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- Deegan JINC, Dimmer EC, Mungall CJ. Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development. BMC Bioinformatics. 2010;11:530. doi: 10.1186/1471-2105-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl AD, Lee JA, Scheuermann RH, Blake JA. Ontology development for biological systems: immunology. Bioinformatics. 2007;23:913–5. doi: 10.1093/bioinformatics/btm029. [DOI] [PubMed] [Google Scholar]
- Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol. 2009;336:137–44. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltrin E, Campanaro S, Diehl AD, Ehler E, Faulkner G, Fordham J, Gardin C, Harris M, Hill D, Knoell R, Laveder P, Mittempergher L, Nori A, Reggiani C, Sorrentino V, Volpe P, Zara I, Valle G, Deegan J. Muscle Research and Gene Ontology: New standards for improved data integration. BMC Med Genomics. 2009;2:6. doi: 10.1186/1755-8794-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2009;38:D331–5. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Rebagliati M, Ahmad N, Muraoka O, Kurokawa T, Hibi M, Suzuki T. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development. 2004;131:1741–53. doi: 10.1242/dev.01070. [DOI] [PubMed] [Google Scholar]
- Hendrickson EL, Lamont RJ, Hackett M. Tools for interpreting large-scale protein profiling in microbiology. J Dent Res. 2008;87:1004–15. doi: 10.1177/154405910808701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert JM, Buffa FM, Vorschmitt H, Egginton S, Bicknell R. A new procedure for determining the genetic basis of a physiological process in a non-model species, illustrated by cold induced angiogenesis in the carp. BMC Genomics. 2009;10:490. doi: 10.1186/1471-2164-10-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DP, Berardini TZ, Howe DG, Van Auken KM. Representing ontogeny through ontology: a developmental biologist’s guide to the gene ontology. Mol Reprod Dev. 2010;77:314–29. doi: 10.1002/mrd.21130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H, Izumo S. Identification of the in vivo casein kinase II phosphorylation site within the homeodomain of the cardiac tisue-specifying homeobox gene product Csx/Nkx2.5. Mol Cell Biol. 1999;19:526–36. doi: 10.1128/mcb.19.1.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Mori AD, Kaynak BL, Cebra-Thomas J, Sukonnik T, Georges RO, Latham S, Beck L, Henkelman RM, Black BL, Olson EN, Wade J, Takeuchi JK, Nemer M, Gilbert SF, Bruneau BG. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature. 2009;461:95–8. doi: 10.1038/nature08324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci U S A. 2007;104:10894–9. doi: 10.1073/pnas.0704044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–16. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Lough J, Barron M, Brogley M, Sugi Y, Bolender DL, Zhu X. Combined BMP-2 and FGF-4, but neither factor alone, induces cardiogenesis in non-precardiac embryonic mesoderm. Dev Biol. 1996;178:198–202. doi: 10.1006/dbio.1996.0211. [DOI] [PubMed] [Google Scholar]
- Lovering RC, Dimmer E, Khodiyar VK, Barrell DG, Scambler P, Hubank M, Apweiler R, Talmud PJ. Cardiovascular GO annotation initiative year 1 report: why cardiovascular GO? Proteomics. 2008;8:1950–3. doi: 10.1002/pmic.200800078. [DOI] [PubMed] [Google Scholar]
- Maccagnan A, Riva M, Feltrin E, Simionati B, Vardanega T, Valle G, Cannata N. Combining ontologies and workflows to design formal protocols for biological laboratories. Autom Exp. 2010;2:3. doi: 10.1186/1759-4499-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace LC, Yermalitskaya LV, Yi Y, Yang Z, Morgan AM, Murray KT. Transcriptional remodeling of rapidly stimulated HL-1 atrial myocytes exhibits concordance with human atrial fibrillation. J Mol Cell Cardiol. 2009;47:485–92. doi: 10.1016/j.yjmcc.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGrogan D, Nus M, de la Pompa JL. Notch signaling in cardiac development and disease. Curr Top Dev Biol. 2010;92:333–65. doi: 10.1016/S0070-2153(10)92011-5. [DOI] [PubMed] [Google Scholar]
- Malik R, Dulla K, Nigg EA, Korner R. From proteome lists to biological impact--tools and strategies for the analysis of large MS data sets. Proteomics. 2010;10:1270–83. doi: 10.1002/pmic.200900365. [DOI] [PubMed] [Google Scholar]
- Marques FZ, Campain AE, Yang YH, Morris BJ. Meta-analysis of genome-wide gene expression differences in onset and maintenance phases of genetic hypertension. Hypertension. 2010;56:319–24. doi: 10.1161/HYPERTENSIONAHA.110.155366. [DOI] [PubMed] [Google Scholar]
- Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD, PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium Nucleic Acids Res. 2009;38:D204–10. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Grennan AK. Plant Physiology and TAIR partnership. Plant Physiol. 2008;146:1022–3. doi: 10.1104/pp.104.900252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palpant NJ, Yasuda S, MacDougald O, Metzger JM. Non-canonical Wnt signaling enhances differentiation of Sca1+/c-kit+ adipose-derived murine stromal vascular cells into spontaneously beating cardiac myocytes. J Mol Cell Cardiol. 2007;43:362–70. doi: 10.1016/j.yjmcc.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel LJ, Latinkic BV. Early activation of FGF and nodal pathways mediates cardiac specification independently of Wnt/beta-catenin signaling. PLoS One. 2009;4:e7650. doi: 10.1371/journal.pone.0007650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlange T, Andree B, Arnold HH, Brand T. BMP2 is required for early heart development during a distinct time period. Mech Dev. 2000;91:259–70. doi: 10.1016/s0925-4773(99)00311-1. [DOI] [PubMed] [Google Scholar]
- Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Relations in biomedical ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Li Y, Wang K, Zhou CJ. Cardiac neural crest and outflow tract defects in Lrp6 mutant mice. Dev Dyn. 2010;239:200–10. doi: 10.1002/dvdy.22079. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Abboud N, Desilets S, Nury D, Cowan C, Puceat M. Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol. 2009;186:665–73. doi: 10.1083/jcb.200901040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–80. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tranter M, Ren X, Forde T, Wilhide ME, Chen J, Sartor MA, Medvedovic M, Jones WK. NF-kappaB driven cardioprotective gene programs; Hsp70.3 and cardioprotection after late ischemic preconditioning. J Mol Cell Cardiol. 2010;49:664–72. doi: 10.1016/j.yjmcc.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Schans VA, Smits JF, Blankesteijn WM. The Wnt/frizzled pathway in cardiovascular development and disease: friend or foe? Eur J Pharmacol. 2008;585:338–45. doi: 10.1016/j.ejphar.2008.02.093. [DOI] [PubMed] [Google Scholar]
- Werner T. Bioinformatics applications for pathway analysis of microarray data. Curr Opin Biotechnol. 2008;19:50–4. doi: 10.1016/j.copbio.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Zhu W, Shiojima I, Hiroi Y, Zou Y, Akazawa H, Mizukami M, Toko H, Yazaki Y, Nagai R, Komuro I. Functional analyses of three Csx/Nkx-2.5 mutations that cause human congenital heart disease. J Biol Chem. 2000;275:35291–6. doi: 10.1074/jbc.M000525200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.