Introduction
Phosphatidylinositol Phosphate (PIP) kinases are responsible for the production of the lipid signaling molecule phosphatidylinositol 4,5-bisphosphate, PI4,5P2. PI4,5P2 can directly affect the function of an array of signal transduction pathways by interactions with PI4,5P2 effectors. PIP kinases modulate PI4,5P2 sensitive pathways by controlling the generation of PI4,5P2 through interactions between PIP kinases and protein partners which target the PIP kinases to specific sub-cellular compartments. In addition, the protein partners are often themselves PI4,5P2 regulated proteins. PIP kinase targeting in this manner allows for the spatial and temporal generation of PI4,5P2 to affect specific signaling pathways. Identification of these protein partners has allowed for the determination of many molecular mechanisms of PI4,5P2 signaling. Recently, a nuclear speckle targeted non-canonical poly(A) polymerase, Star-PAP, has been defined to have a functional interaction with the type Iα PIP kinase to process select mRNAs for their 3’ end formation. Star-PAP contains a poly(A) polymerase catalytic and core domains (PAP) though differs from the canonical PAP due to its unique domain arrangement and phosphoinositide regulation. Star-PAP is a duel specificity polymerase that harbors in vitro poly(A) polymerase activity that is stimulated by PI4,5P2, and also embodies features of Terminal Uridylyl Transferase (TUTase) in both of its domain arrangement and its in vitro ability to transfer UMP to cellular RNA including the small nuclear RNA U6. The Star-PAP complex of proteins contains a number of cleavage and polyadenylation components, an active PIPKIα capable of generating de novo PI4,5P2, and the PI4,5P2 sensitive protein kinase CKIα. CKIα can directly phosphorylate Star-PAP and in conjunction with PIPKIα, is required for expression and maintenance of the Star-PAP target mRNA HO-1. HO-1 mRNA encodes the cytoprotective enzyme heme oxygenase-1, which is an important detoxifying enzyme involved in protection from reactive oxygen species and cellular oxidative stresses. HO-1 is upregulated in response to oxidative stress through increase in transcription, placing Star-PAP, PIPKIα and CKIα as mediators of oxidative cellular stress response. Taken together, the Star-PAP complex represents a focal point for nuclear phosphoinositide signaling where Star-PAP, PIPKIα and CKIα can synergize to regulate the 3’ end formation of select mRNAs.
Type I PIP kinases generate PI4,5P2 to regulate signaling events
Phosphatidylinositol phosphate (PIP) kinases are lipid kinases that function to generate phosphoinositide signaling molecules, which play critical roles throughout the life cycle of metazoans (Gardocki et al., 2005). The type I PIP kinases, PIPKIα, -β, -γ are PI-4’-phosphate 5’-kinases that synthesize the signaling molecule PI4,5P2 utilizing PI4P as a substrate (Heck et al., 2007). Over the last decade a wealth of accumulated evidence has indicated that the different phosphorylated PIs serve not only as intermediates in the synthesis of the higher phosphorylated phosphoinositides, but also as regulators of different protein targets in their own right. Type I PIP kinases generate the lion’s share of PI4,5P2 in the cell, though these three isoforms display distinct sub-cellular targeting and function (Fig. 1). PIPKIα targets to membrane ruffles and nuclear speckles, PIPKIβ targets to a perinuclear region, and PIPKIγ targets to focal adhesions. This mechanism of synthesis is a mode of regulating where and when PI4,5P2 is generated and acts to tightly control PI4,5P2 regulated signal transduction(Doughman et al., 2003).
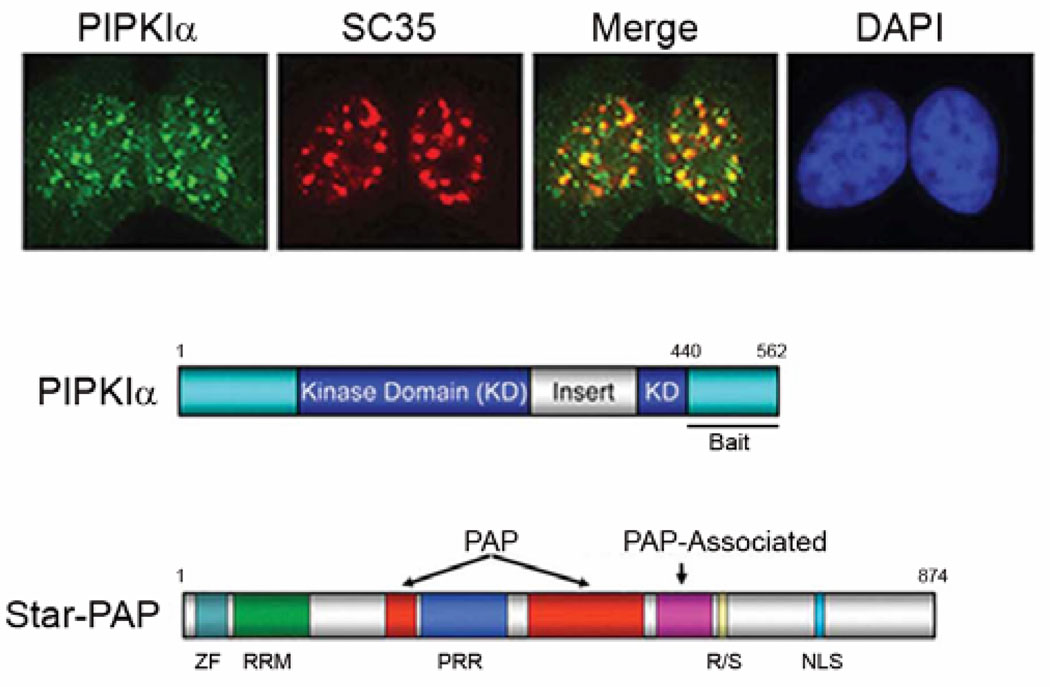
Fig. 1.
Depicted above are the three members of the type I phosphatidylinositol 4-phosphate 5’-kinase family,PIPKIα, -β and -γ. Each kinase produces the same phosphoinositide product but at distinct sub-cellular sites thereby spatially restricting activation of phosphoinositide sensitive pathways. (ref. Heck et.al., 2008)
Of the vast assortment of lipids present in eukaryotic cells, phosphoinositides play highly specialized roles to regulate a diverse set of signaling events. The large number of specialized membrane compartments in eukaryotes results in organized and polarized structural arrangements within the cell. To regulate this organization and maintain integrity, cells require a pool of adaptable lipid messages that can undergo rapid modifications for which soluble messages are not suitable. In this model, phosphoinositide lipid messengers are specifically labeled for delivery to a particular membrane compartment where they can orchestrate targeting and fusion with other compartments, or regulate compartmental assembly of specific components through direct regulation of their functions. When the sub-cellular destination has been reached by the signaling lipid and function carried out, the phosphorylated species undergoes dynamic dephosphorylation and re-phosphorylation events that generate a variety of phosphorylated isoforms of the lipid messenger, which can be re-routed to alternative sub-cellular locations to carry out specialized functions.
Phosphoinositide signaling through PI4,5P2
Phosphoinositides function in signal transduction pathways to regulate a number of processes in eukaryotic cells. Traditionally, phosphatidylinositol (PI) which constitutes a minor component ~10% of total membrane lipid (Cohn et al., 1988), is anchored in a membrane lipid-bilayer via its acyl chains leaving its inositol head group exposed and accessible for phosphorylation by the family of PIP kinases. Phosphorylation of PI can generate seven different phosphorylated PIPn species, each with their own signaling capacity (Clarke, 2003). Receptor stimulation of the plasma membrane PI cycle is well characterized (Alb et al., 1996) and leads to the production of lipid second messengers, such as PI4,5P2. PI4,5P2 is a versatile molecule in that it has potent signaling strength to initiate downstream events(McLaughlin and Murray, 2005; McLaughlin et al., 2002), as well as bind to activate or repress enzymes(Brockman and Anderson, 1991; Sciorra et al., 1999). Additionally, PI4,5P2 can be converted to PI3,4,5P3, which is a potent signaling molecule(Insall and Weiner, 2001). PI4,5P2 can be dephosphorylated at the 4’ or ‘5 position of the inositol ring(Wiradjaja et al., 2007) to generate pools of PI4P or PI5P(D'Angelo et al., 2008). Furthermore, PI4,5P2 can be metabolized by PI-specific lipases, PLC enzymes, to generate diacylglycerol (DAG) and soluble second messengers such as inositol-1,4,5-trisphosphate, (IP3) that triggers calcium fluxes(Chu and Stefani, 1991; Wang et al., 2004), which exerts a strong influence on signaling events(Irvine, 1992). IP3 can be phosphorylated by a family of inositol multi-kinases that can generate up to ten different soluble inositol polyphosphate second messengers, which can modulate a number of signaling pathways in the cell(York et al., 2001). Phosphoinositide-based signaling cascades via PI4,5P2 has been shown to mediate not only transmission of hormones and neurotransmitters but also other cellular mechanisms including assembly of the actin cytoskeleton (Janmey and Stossel, 1987; Lassing and Lindberg, 1985), regulating interactions between cytoskeletal proteins and the plasma membrane(Anderson and Marchesi, 1985), vesicular trafficking(Czech, 2000; Downes et al., 2005; Huijbregts et al., 2000; Wenk and De Camilli, 2004), secretion (Hay et al., 1995; Martin, 2001), cell motility/cytoskeletal assembly(Janmey, 1994; Niggli, 2005; Yin and Janmey, 2003), regulation of ion channels (Delmas et al., 2005; Li et al., 2005), apoptosis(Mejillano et al., 2001), and the regulation of nuclear events (Boronenkov et al., 1998; Cocco et al., 1987; Gonzales and Anderson, 2006; Gozani et al., 2003; Irvine, 2003; Irvine, 2006; Macbeth et al., 2005; Mellman et al., 2008; Osborne et al., 2001; York and Majerus, 1994; Zhao et al., 1998).
Roles for nuclear phosphoinositides
The understanding of events regulated by nuclear phosphoinositides has lagged behind those in the cytosol and at the plasma membrane. Nuclear PI signaling was initially proposed when it was shown that nuclear PI and PI4P kinase activities were seen in preparations that were enriched with nuclear membranes (Smith and Wells, 1983; Irvine, 2002). Direct evidence of a nuclear phosphoinositide cycle was first demonstrated in MEL cells, mouse erythroleukaemia cells, when a nuclear pool of PI4P and PI4,5P2 were metabolized differently from lipids in the cytosol (Cocco et al., 1987). It was later shown that differential cellular stimuli with agents such as bombesin, a mitogenic stimulus(Irvine, 2003; Watt et al., 1991) or IGF-1(Divecha et al., 1991), Insulin Growth Factor-1, stimulated the generation of the PI cycle within different compartments(Divecha et al., 1991). Bombesin only activated the characterized plasma membrane inositol lipid cycle whereas IGF-1 stimulated a distinct nuclear polyphosphoinositol lipid metabolism(Divecha et al., 1991).
IGF-1 induction decreases in nuclear pools of PI4P and PI4,5P2 with a concomitant increase in nuclear DAG with a simultaneous increase in nuclear translocation of PKC, an effector of DAG(Divecha et al., 1991; Martelli et al., 1991), providing evidence that nuclear DAG acts as a chemoattractant for PKC nuclear translocation. If the function of nuclear DAG (Hodgkin et al., 1998; Wakelam, 1998) is to attract DAG-dependent PKC isoforms to the nucleus, then a mechanism to turn off the signal should conceivably exist. This role could be fulfilled by the DAG Kinase DGK, which phosphorylates DAG to produce Phosphatidic Acid (PA)(Topham and Prescott, 1999). This interconversion of signaling molecules sets up an intriguing scenario, whereby, in the nucleus PI4,5P2 generated by a PIP Kinase elicits a signal which can be turned off by a PLC isoform to rapidly generate IP3 and DAG. IP3 elicits calcium signaling which can affect a myriad of functions(Alonso et al., 2006; Bucki and Gorski, 2001; Choe and Ehrlich, 2006; Ehrlich et al., 1994; Irvine, 1982; MacDonald, 1998; Malviya and Klein, 2006; Miyazaki, 1993; Miyazaki, 1995). IP3 can be converted to higher order inositides IP4, IP5, IP6 , as well as PP-IP molecules, which could all carry out distinct nuclear processes(Macbeth et al., 2005; Seeds et al., 2007; York, 2006). DAG is converted to PA by DGK(Goto et al., 2006) and PA in turn stimulates the PIP Kinase to generate PI4,5P2 driving the cycle back to the “beginning”, propagating the phosphoinositide signals to regulate nuclear events. An example of this scenario from the cytosol is seen with the Phospholipase D isoenzyme, PLD2(Billah, 1993; Cazzolli et al., 2006; Cummings et al., 2002; Jenkins et al., 1994; Jenkins and Frohman, 2005; Metz and Dunlop, 1991; Morris et al., 1997). PLD2 can bind PIPKIα(Divecha et al., 2000) whereby PLD2 activity is regulated by PI4,5P2 (Pertile et al., 1995; Sciorra et al., 1999). PIPKIα bound PLD2 can hydrolyze phosphatidylcholine into PA and choline (Ktistakis et al., 2003), stimulating PIPKIα to generate PI4,5P2 activating PLD2 and thus providing a potential bifurcated route to control the feedback within the PI cycle, and regulation of PLD2 activity and downstream events.
Not only has PIPKIα and its product PI4,5P2 been reported to be localized to nuclei (Boronenkov et al., 1998), but a repertoire of phosphoinositide metabolizing enzymes such as multiple forms of PI-PLCs, PI3K-C2α and PI3,4,5P3 and PTEN have been reported to localize to nuclei (Cocco et al., 2006; Cocco et al., 2004a; Cocco et al., 2004b; Cocco et al., 2001; Martelli et al., 2005a; Martelli et al., 2005b; Planchon et al., 2008). It has been demonstrated that PI-PLCβ1 showed increased activity in response to IGF-1 stimulation and nuclear localization (Martelli et al., 1992), providing evidence that this isoform of PLC operates in the nucleus. In contrast, PLC-γ1 was confined to the cytosol(Divecha et al., 1993), demonstrating that there are indeed differentially localized isoforms of PLC. Taken together, the nuclear PI cycle operates distinctly from the PI cycle in the cytosol, though there is great similarity in the molecules that are generated and enzymes utilized by both of these autonomous PI cycles. This implies that either structural components and/or upstream and downstream signaling must be pivotal in the selective activation and repression of these distinct PI cycles.
Phosphoinositide signaling pathways in nuclei of mammals have been shown to be key for numerous events such as the cell cycle, chromatin structure, DNA repair, tumor progression and cellular proliferation, transcription, RNA editing and mRNA metabolism (Gozani et al., 2003; Irvine, 2003; Lo Vasco et al., 2004; Macbeth et al., 2005; Osborne et al., 2001; York and Majerus, 1994; Yu et al., 1998; Zhao et al., 1998). PI4,5P2 co-immunoprecipitates with snRNPs and the hyperphosphorylated form of RNA Pol II, providing an intriguing suggestion for the involvement of PI4,5P2 as a structural or functional regulator in aspects of transcription and pre-mRNA processing(Osborne et al., 2001). Supporting this hypothesis is evidence that both PIPKIα and PI4,5P2 co-localize with SC35, a component of interchromatin granule clusters or nuclear speckles(Boronenkov et al., 1998; Lamond and Spector, 2003; Spector, 2001) which are membrane-less nuclear bodies enriched in factors required for splicing and 3’ end formation of pre-mRNAs, as well as being sites of phosphoinositide metabolism(Boronenkov et al., 1998; Bunce et al., 2008; Didichenko and Thelen, 2001; Evangelisti et al., 2006; Mortier et al., 2005). It is interesting to note that PI4,5P2 is enriched at nuclear speckles although speckles are devoid of detectable lipid bilayer membranes(Handwerger and Gall, 2006; Lamond and Spector, 2003), which raises the intriguing question of whether there are specific PI carrier proteins(Cunningham et al., 1995; Wirtz, 1997) that store PI4,5P2 to assure its precise synthesis and localization at speckles. However phosphoinositides are sequestered in the nucleus, the presence of phosphoinositide metabolism occurring at nuclear speckles strongly implicates functional roles for nuclear phosphoinositide signaling in pre-mRNA processing.
PIPKIα directly interacts with a nuclear speckle localized non-canonical poly(A) polymerase, Star-PAP, and resides in complex with 3’end formation machinery components
Based on the model of PI4,5P2 signaling specificity being dependent upon its interactions with protein partners and targeting factors (Doughman et al., 2003), a yeast two-hybrid screen was established to identify PIPKIα interacting proteins to define functional roles for PIPKIα and its product PI4,5P2 nuclear signaling events. The region of PIPKIα that was responsible for its nuclear targeting was determined to be in the carboxy-terminus, amino acids 440–562, therefore this portion of the molecule was used as bait in the yeast two-hybrid screen(Mellman et al.,2008) (Fig. 2). The screen was carried out in human B cell, breast, prostate, and placenta, as well as mouse pre B-cell and embryonic cDNA libraries. This screen yielded forty-two positive clones and sequence analysis indicated that fourteen of these clones contained full or partial ORFs in the correct orientation. A majority of these proteins are known or suspected of being present in the nucleus and interestingly, almost half of the proteins contained C2H2 zinc finger domains, which are known to be involved in protein-protein interactions as well as protein-nucleic acid interactions (Matthews and Sunde, 2002).
Fig. 2.
Endogenous PIPKIα localizes at nuclear speckles (top panel) evidenced by the co-localization of PIPKIα (green) with the SR protein SC35 (red). Merge shows co-localization. 4′,6-diamidino-2-phenylindole (blue). Amino acids 440–562 of PIPKIα was used as bait in a yeast two-hybrid screen to identify PIPKIα interacting proteins that function together in nuclear events (middle panel). Star-PAP was one of numerous nuclear localized proteins isolate din the screen and has been shown to function with PIPKIα in the 3’ end formation of select mRNAs (bottom panel) (ref. Mellman et. al., 2008).
At the time of the screen, many of the identified proteins had no known function. Therefore, to try and identify roles for these proteins in nuclear phosphoinositide signaling pathways a protein domain data base search was carried out. One hypothetical protein that at the time was termed RNA Binding Motif protein 21 (RBM21) (NM_022830.1) contained a particularly interesting domain architecture and became a focus of our studies from this screen. Data base analysis of RBM21 amino acid sequence determined it contained the conserved pol β nucleotidyl transferase motif, GSX10DXD, placing it as a member of pol β super family of nucleotidyl transferases(Martin and Keller, 2007). Specifically, RBM21 contained a poly(A) polymerase catalytic and core domains (PAP) as well as a PAP associated domain. In confirmation of the yeast two-hybrid screen, RBM21 can directly interact with full length PIPKIα as well as the carboxy-terminus of PIPKIα (amino acids 440–562) in vitro. In vivo, antibodies specific for RBM21 co-immunoprecipitates endogenous PIPKIα from RNase A treated fractions, confirming that in vivo these two enzymes are in association together. Endogenous RBM21 is found primarily in the nucleus at nuclear speckles, co-localizing with PIPKIα(Mellman et al., 2008) and PI4,5P2(Boronenkov et al., 1998). The presence of all three molecules in the same nuclear sub-compartment suggests that they are positioned to directly interact and function with each other in vivo.
Despite the conserved nucleotidyl transferase sequence motif and PAP domain, the domain arrangement of RBM21 showed clear differences when compared to both the canonical mammalian poly(A) polymerase PAPα(Raabe et al., 1991) and the non-canonical regulatory poly(A) polymerase, Germ Line Development 2 (GLD2)(Wang et al., 2002) (Fig. 2). RBM21 contains a C2H2 zinc finger motif followed by an RNA Recognition Motif (RRM) that differs both in sequence and location from the RNA binding domain of PAPα. Another distinguishing feature in RBM21 is its split PAP domain that is linked by a proline rich region (PRR). This is a unique characteristic of RBM21 compared to all other reported poly(A) polymerases. The carboxy-terminus of RBM21 contains R/S peptide repeats, characteristic of speckle targeting proteins(Lamond and Spector, 2003) followed by an Nuclear Localization Sequence (NLS). These unique domains of Star-PAP may be important for interactions with molecular partners and targeting to sub-cellular compartments, as well as for its biological function in pre-mRNA processing. Based upon the nuclear speckle localization, its in vitro poly(A) polymerase activity and the functional interaction with PIPKIα, RBM21 was subsequently named nuclear Speckle Targeted PIPKIα Regulated-Poly(A) Polymerase, Star-PAP.
When recombinant His-Star-PAP expressed and purified from E.coli BL 21 was subjected to a standard poly(A) polymerase assay it showed robust PAP activity(Mellman et al., 2008) towards a generic A15 poly(A) RNA oligo primer, the RNA oligo primer L1(Rouhana et al., 2005) as well as a 45mer RNA oligo primer of the sequence (UAGGGA5)A15, which was designed to bind the RRM of Star-PAP with high affinity(Ding et al., 1999). This RNA oligo primer was designed based upon Star-PAP similarity to the RRMs within hnRNPA1, which preferentially binds multiple repeats of the sequence UAGGGAn where n=2 or more. This suggests that the endogenous RNA target of Star-PAP harbors this motif or a motif with sequence similarity. Most interestingly, recombinant Star-PAP poly(A) polymerase activity is sensitive to and stimulated by exogenous PI4,5P2. The regulatory affect of PI4,5P2 controlling Star-PAP activity defines a mode of regulation for Star-PAP function in vivo. Based upon the principal of function following targeting and interaction, the association between Star-PAP and PIPKIα as well as their co-localization with PI4,5P2 in the nucleus strongly suggests that Star-PAP is an effector PI4,5P2 and thus at least one mode of Star-PAP function can operate under the regulation of PIPKIα.
Based upon Star-PAP in vivo interactions and localization, as well as its in vitro activity, we hypothesized that Star-PAP is acting as a poly(A) polymerase and is required for the 3’ end formation of pre-mRNAs. 3’ end formation of pre-mRNA requires a large set of interacting factors which coordinate to assemble on the pre-mRNA in a sequence specific manner(Sheets et al., 1990) and process pre-mRNA in a highly orchestrated fashion(Wahle, 1995). The factors required includes though is not limited to the presence of a PAP, Cleavage and Polyadenylation Specificity Factor (CPSF160, -100, -73 and -30) subunits, Cleavage Factors I (CFI68, -59 and -25) and Cleavage Factors II subunits, Cleavage Stimulatory Factor Subunits (CstF77,-64,-50), the scaffolding protein symplekin, SM proteins, Poly(A) Binding Proteins (PABPs) and the largest subunit of the carboxy terminal domain of RNA Polymerase II (CTD of RNA Pol II)(Colgan and Manley, 1997). This megadalton complex binds in the 3’ UTR of pre-mRNA at the poly(A) site, AAUAAA, whereby CPSF binds in a sequence dependent fashion, positioning the machinery for efficient cleavage and polyadenylation (Colgan and Manley, 1997).
3’ end formation of pre-mRNA requires the splicing of the last intron-exon boundary as well as cleavage and polyadenylation. In vivo, all three of these steps are coupled and all three steps requires the presence of a poly(A) polymerase(Christofori and Keller, 1988; Kyburz et al., 2006).
Based on the hypothesis that Star-PAP is functioning as a PAP, we performed a proteomic analysis of the endogenous Star-PAP complex from HEK293 cells which revealed that PIPKIα and Star-PAP together reside in a megadalton, pre-mRNA 3’ end formation complex devoid of detectable PAPα(Mellman et al., 2008). Star-PAP and PIPKIα were found to be in complex with a large number of the cleavage and polyadenylation complex members such as CPSF, CstF, SM proteins, symplekin and the CTD of RNA Pol II. To directly compare the canonical PAPα complex to the Star-PAP complex, PAPα and Star-PAP, respectively, were cloned into a pCMV-Flag expression vector (Sigma), expressed and purified from HEK293 cells and the purified complexes were subjected to Western blot analysis. Both Flag-tagged PAP complexes contained CPSF, CstF, symplekin, and SM proteins. Most interestingly, Flag-Star-PAP purified from HEK293 cells was found to contain PIP Kinase activity. Incubation of Flag-purified Star-PAP complexes with PI4P micelles and ATP resulted in the production of PI4,5P2. The PIP Kinase activity found associated with purified Flag-PAPα as a direct comparison of the two PAP complexes showed no PIP Kinase activity above background. This strongly supports the hypothesis that PIPKIα in the Flag-Star-PAP complex is capable of producing de novo PI4,5P2 in proximity to Star-PAP in vivo to regulate the function of Star-PAP and possibly other unique Star-PAP interacting proteins for the efficient 3’ end formation of pre-mRNAs.
When PI3P or PI5P micelles (poor type I PIPK substrates) were used in the kinase assay there was very little PI4,5P2 production observed with either the Star-PAP or PAPα complexes. Likewise, when PI or PI4,5P2 micelles were used, there was almost no detectable kinase activity associated with either complex(Gonzales et al., 2008). These results indicate that type I PIP Kinase activity is specifically associated with Star-PAP and not PAPα. Furthermore, this was the only phosphoinositide kinase activity that co-purifies with Flag-tagged Star-PAP or PAPα. PIPKIα was the only type I PIP kinase detectable by Western blotting of the Star-PAP complex, indicating that PIPKIα is the only phosphoinositide kinase associated with Star-PAP.
Like other pre-mRNA processing proteins, Star-PAP contains a zinc finger module. A single zinc finger binds only a short stretch of nucleotides and is not sufficient to provide an interaction with a specific DNA or RNA sequence, as zinc finger proteins that interact with nucleic acids usually contain multiple zinc finger motifs in order to provide sufficient stringency to achieve specific binding(Hall, 2005). Therefore, it will be of interest to determine whether the single zinc finger found in Star-PAP is mediating protein-protein interactions or targeting a specific RNA sequence. It is interesting to note that a number of single zinc finger containing proteins were identified in the PIPKIα yeast two-hybrid screen. It may be that zinc fingers are the common motif required for mediating interactions with carboxy-terminus of PIPKIα. Further investigations awaits to determine the physiological significance of zinc finger mediated interactions between PIPKIα and its suite of interacting proteins and how this affects cellular signaling pathways and gene expression.
PIPKIα synergizes with Star-PAP to target select mRNAs for 3’ end formation
Based on the association of Star-PAP with the mRNA cleavage and polyadenylation machinery we hypothesized that Star-PAP is involved in the 3’ processing and polyadenylation of mRNAs. To define a role for Star-PAP in modulation of mRNA, we set out to determine in vivo Star-PAP RNA substrates (Mellman et. al., 2008). Because correct polyadenylation of mRNA is critical for the stability of mRNAs(Colgan and Manley, 1997), we therefore predicted that siRNA knock down of Star-PAP would result in a decrease of cellular mRNAs that require Star-PAP for their 3’ end processing and polyadenylation. We hypothesized that if PIPKIα acts to modulate Star-PAP function, the loss of PIPKIα by siRNA will cause a decrease in the pool of target mRNAs that require both Star-PAP and PIPKIα for their maturation. Therefore, we knocked down Star-PAP or PIPKIα, respectively and performed a microarray analysis of total polyadenylated mRNAs from each group. A significant (conditional false discovery rate ≤ 0.01) change in mRNA level compared with control cells (n = 3) was detected for 4,481 genes with Star-PAP RNAi knockdown and 4,542 genes with PIPKIα RNAi knockdown (Mellman et. al., 2008). There was an overlap of 2,350 significant gene changes in both conditions, of which 2,262 were in the same direction (Mellman et. al., 2008).
A large group of the identified genes encode proteins involved in detoxification and/or oxidative stress response (Mellman et. al., 2008). Of these biomedically relevant potential Star-PAP mRNA targets a small group were chosen for validation by quantitative real-time RT-PCR (qRT-PCR), including the mRNA encoding heme oxygenase-1 (HO-1) (NM_002133.1). The expression levels of the chosen candidate mRNAs were consistent with the microarray analysis, demonstrating that Star-PAP is required for the expression of these mRNAs (Mellman et. al., 2008). PIPKIα RNAi knockdown also significantly decreased the expression levels of these same mRNAs, indicating a biological relationship between PIPKIα and the expression of select Star-PAP-dependent mRNAs. Knockdown of both Star-PAP and PIPKIα showed no additive effect on the loss of HO-1 mRNA, providing evidence that Star-PAP and PIPKIα function in a common pathway to control its expression (Mellman et. al., 2008).
To determine direct targets of Star-PAP, RNA immunoprecipitation (RIP)(Gilbert et al., 2004) was used. Star-PAP was associated with HO-1 mRNA but not with the non-target mRNAs encoding glutamate cysteine ligase, catalytic subunit (GCLC) (NM_001498.2) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (NM_002046.3). Because HO-1 is a direct target of Star-PAP, it was selected for use in exploring the mechanism by which Star-PAP controls the expression of its select target mRNA.
HO-1 enzyme catalyzes the rate limiting step in the conversion of heme to potent signalling molecules, including biliverdin and carbon monoxide, which possesses antioxidant and other protective properties. Regulation of HO-1 is achieved primarily through regulation of its mRNA levels and induction of HO-1 mRNA is a key cellular response to reactive oxygen species and other cellular stresses(Takahashi et al., 2004). HO-1 expression is induced in response to cellular stresses and the end products of HO-1 metabolism exhibit antioxidant and other cytoprotective functions. Because of this, HO-1 is thought to be an important component of the cellular response to stress. In cell culture, HO-1 mRNA expression can be induced by quinone compounds such as tert-butyl hydroquinone (tBHQ) which induces an antioxidant response in cells(Keum et al., 2006). To test the requirements for PIPKIα and Star-PAP in HO-1 mRNA induction, HEK293 cells were treated with control, PIPKIα or Star-PAP siRNA oligos. After siRNA treatment, cells were treated for 4 hours with 100µM tBHQ or DMSO only. Total RNA was isolated and HO-1 levels were analyzed by qRT–PCR. tBHQ can induce HO-1 expression in HEK293 cells by ~7–10 fold and siRNA knock down of PIPKIα as well as siRNA knock down of Star-PAP not only reduced basal levels of HO-1 mRNA, but also the maximal expression upon induction as compared to control cells (Mellman et. al., 2008).
In order to determine whether PIPKIα and Star-PAP are functioning together in the 3’ end formation of HO-1 mRNA, a functional in vivo 3’ end formation assay was established(Mellman et al., 2008). We examined a step in 3’ processing that is functionally coupled to polyadenylation which is 3’ end cleavage. Poly(A) polymerases are not only required for the generation of polyadenylation but they are also required for the pre-mRNA cleavage reaction that precedes polyadenylation(Zhao et al., 1999). We had predicted that if Star-PAP is acting as a poly(A) polymerase towards HO-1 mRNA then it must be playing a direct role in the 3’ end cleavage of HO-1 mRNA. Hence, in the absence of Star-PAP, 3’ end cleavage of HO-1 should be inhibited.
We examined whether Star-PAP and/or PIPKIα siRNA knock down affected cleavage of HO-1 mRNA by directly measuring the amount of uncleaved mRNA relative to total mRNA levels. To assess mRNA cleavage, total RNA from control, Star-PAP or PIPKIα knock down cells was reverse transcribed with random hexamer primers and the level of uncleaved mRNA was measured by qRT–PCR using primers sets that span the 3’ cleavage site of HO-1 mRNA. The level of uncleaved mRNA was normalized to the total mRNA levels for each mRNA examined, control versus experimental. Total target RNA levels measured using the same primer sets used to validate the microarray discussed above were therefore specific for the mRNA and the RNA samples were DNaseI treated prior to the reverse transcription reaction.
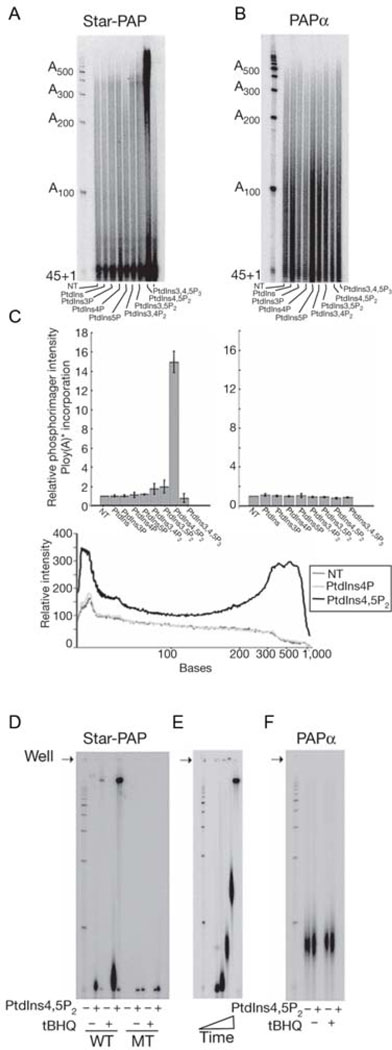
Star-PAP knock down resulted in a dramatic accumulation of uncleaved HO-1 mRNA relative to the total amount of HO-1 mRNA present. Significantly, PIPKIα knock down did not dramatically affect the amount of HO-1 mRNA cleavage even though PIPKIα is required for its expression, consistent with PIPKIα acting as a modulator of Star-PAP function. While Star-PAP knock down may be inhibiting HO-1 expression by causing defects in cleavage, PIPKIα knock down may be reducing HO-1 mRNA levels by affecting other aspects of 3’ processing such as causing reduced Star-PAP function by loss of de novo PI4,5P2 generation. The amount of non-Star-PAP target mRNA GCLC that was uncleaved was unchanged by either Star-PAP or PIPKIα knock down. This requirement of Star-PAP and PIPKIα for 3’ end cleavage demonstrates that the Star-PAP complex is playing a direct and functional role in the 3’ end formation of HO-1 mRNA. This is a specialized function, as the 3’ end processing of the non-Star-PAP mRNA target GCLC was unaffected by loss of Star-PAP as well as loss of PIPKIα. Oxidative stress induction by incubation of HEK293 cells with 100µM tBHQ for 4 hours stimulated the assembly of the endogenous Star-PAP complex with large fold increases in the association of PIPKIα, CPSF subunits as well as the CTD of RNA Pol II, demonstrating that the Star-PAP complex is subject to extra-cellular signaling dependent assembly with its protein partners into a highly stable complex. The enzyme activity and sensitivity to PI4,5P2 of Flag-Star-PAP was enhanced in complexes purified from tBHQ-treated cells as compared to Flag-Star-PAP purified from resting cells, suggesting that the Star-PAP enzyme may undergo post-translational modifications to activate the enzyme under oxidative stress conditions (Fig. 3). Star-PAP expressed and purified in E.coli BL21 displays PAP activity that is sensitive to PI4,5P2, which demonstrates that PI4,5P2 can bind Star-PAP to stimulate its polymerase activity. How PI4,5P2 is binding Star-PAP to affect its activity is of paramount importance to understand how the Star-PAP complex is activated, stabilized and binds its RNA and/or rNTP substrate for 3’ end formation.
Fig. 3.
Star-PAP has poly(A) polymerase activity that is stimulated by PI4,5P2.
A, B, Effects of 50µM inositol phospholipid micelles on recombinant His-Star-PAP as compared to PAPα. C, Incorporation of [α-32P]ATP into poly(A) RNA products larger than A200 in the presence of phospholipid micelles by Star-PAP and PAPα from A and B. Poly(A) tail tracings measuring RNA extension with [α-32P]ATP into poly(A) RNA products comparing Star-PAP incubated with PI4,5P2, PI4P or a non-treated His-Star-PAP. D, Affinity-purified Flag-Star-PAP (WT) or catalytic mutant Flag-Star-PAP (MT) from stably expressing HEK293 cells subsequent to treatment with tBHQ and /or PI4,5P2. E, Time course subsequent to treatment with tBHQ in D, in the presence of PI4,5P2. F, Flag-PAPα activity after treatment with 100µM tBHQ and/or the presence of PI4,5P2. (ref. Mellman et. al., 2008).
The nature of the Star-PAP complex is that it contains a wealth of associated protein components, some unique components while some are shared with other 3’ end formation machineries such as PAPα and GLD2 poly(A) polymerase. Star-PAP complex is a phosphoinositide sensitive complex that is highly regulated and acts to target select mRNAs for their 3’ end formation. HO-1 mRNA is one of potentially many mRNAs that require the Star-PAP complex for their maturation. How the Star-PAP complex targets its RNA substrate, what the required signals are for such targeting and whether there is an RNA or DNA Star-PAP consensus sequence in Star-PAP targets are areas of active investigation.
Star-PAP complex contains the PI4,5P2 sensitive protein kinase Casein Kinase Iα, CKIα
The Star-PAP complex contains a number of shared associated proteins that also assemble in the canonical PAPα complex including SM proteins, CPSF, CstF, symplekin and RNA Pol II (Mellman et. al., 2008). A distinguishing feature of the Star-PAP complex is that it contains unique associated proteins as compared to the canonical PAPα complex, including PIPKIα and the PI4,5P2 sensitive protein kinase Casein Kinase Iα, CKIα(Gonzales et. al., 2008). CKIα is a serine/threonine protein kinase which phosphorylates acidic peptides in a variety of substrates, and was one of the earliest protein kinase activities to be purified and characterized biochemically (Fu et. al., 2001). The characterization of substrate specificity of CKI isoforms initially led to the identification of the canonical sequence S/T(P)X1-2S/T indicating that modification of serine or threonine residues by CKI requires the preceding phosphorylation of amino acid residues amino-terminal of the target site; additionally non-canonical CKI motifs have been found (Knippschild et. al., 2005). However, phosphorylation by CKI of a substrate does not seem to be strictly dependent upon the consensus sequence, as it was shown that phosphorylation also depends on the tertiary structure of the substrate(Cegiekska et. al., 1998).
Several reports have demonstrated that CKI activity can be isolated from a soluble cytosolic protein fraction from many different mammalian and plant tissue types with the pure and active species being monomeric of molecular weight in the ~30kDa range (Fu et. al., 2001). CKI activity can also be isolated from nuclei and yielded a similar or slightly larger sized protein than from the cytosol suggesting that a second nuclear form of CKI may exist(Tuazon et. al., 1991). Later cDNA cloning studies revealed that CKI actually constitutes an entire protein kinase family including at least 7 related vertebrate genes named α, β, γ1–3, δ and ε (Rowles et. al., 1991). The α and γ3 genes are subject to alternative splicing, leading potentially up to 11 different CKI proteins(Fish et. al., 1995, Green and Bennett 1998, Zhai et. al., 1995). ckIα has been reported to be alternatively spliced into four splice variants named CKIα, CKIαS, CKIαL and CKIαSL. The four isoforms differ only in the presence of two inserts, L and S (Fu et. al., 2001). The L insert contains 28 amino acids which are located within the kinase catalytic domain, while the S insert contains 12 amino acids and is located near the end of the short carboxy-terminal extension beyond the catalytic domain.
The functions of each CKI protein kinase are not completely understood, although in vitro studies suggest several potential substrates. These include the cytoplasmic domain of the insulin receptor β subunit in the plasma membrane(Rapuano and Rosen 1991), the cyclic AMP response element modulator in the nucleus(de Groot et. al., 1993), cytosolic muscle glycogen synthase(Flowtow and Roach 1989), neuronal cytoskeletal proteins such as microtubule-associated protein tau and neurofilaments(Floyd et. al., 1991; Singh et. al., 1994), a subset of synaptic vesicle proteins(Gross et. al., 1995) and the non-canonical poly(A) polymerase Star-PAP(Gonzales et. al., 2008). It was reported that the CKI protein kinase isoform that directly phosphorylates Star-PAP is CKIα, and these two enzymes co-localize in the nucleus at speckles where they function in a large phosphoinositide-regulated protein complex regulating the 3’ end processing of select mRNAs (Gonzales et. al., 2008).
The Star-PAP complex includes PIPKIα and the activity of Star-PAP is regulated by the PIPKIα product PI4,5P2. PI4,5P2 tends to regulate multiple proteins at its site of generation (Bairstow et. al., 2006), thus it was exciting to find recently that there are at least two protein kinase activities associated with the Star-PAP complex and that one of these activities is inhibited by PI4,5P2(Gonzales et al., 2008). Subsequently, it was identified that one of the protein kinase activities associated with the Star-PAP complex was the PI4,5P2 sensitive protein kinase CKIα. Western blot analysis of Flag-purified complexes with antibodies specific for CKIα revealed that CKIα co-purifies specifically with Star-PAP and not PAPα. CKIα is capable of phosphorylating Star-PAP in its PRR domain and in vivo CKIα along with PIPKIα are required for the maintenance of a subset of Star-PAP target mRNAs (Gonzales et al., 2008). Protein kinase activities have been shown to regulate the activity of canonical PAP. For example PAPα can be inhibited in a cell cycle-dependent fashion by cyclin-dependent kinases (Colgan et. al., 1998). Additionally, PAPα was identified as a target of ERK. PAPα phosphorylation of serine 537 by ERK increased its non-specific polyadenylation activity in vitro. This PAP activity was also activated by stimulation of ERK with phorbol-12-myristate-13-acetate in vivo. These data suggest that ERK is a novel regulatory kinase for PAPα and further, that PAP activity could be regulated by extra-cellular stimuli through an ERK-dependent signaling pathway(s). It will be of interest to determine how phosphorylation can modulate Star-PAP function and what the cellular signals are which promote the regulatory aspects of CKIα in the Star-PAP complex.
The protein kinase activity of CKIα is specifically inhibited by PI4,5P2 and it was shown that concentrations as low as 12.5µM PI4,5P2 can potently inhibit the Flag-Star-PAP associated CKIα protein kinase activity(Gonzales et al., 2008). Phosphorylation is important for the function of many of the proteins found in nuclear speckles, including the SR family of splicing factors. CKIα localizes at nuclear speckles in vivo and is capable of directly phosphorylating SR proteins in vitro and is therefore implicated in the regulation of pre-mRNA splicing(Gross et al., 1999). The presence of a PI4,5P2 sensitive protein kinase at nuclear speckles which is capable of phosphorylating splicing factors and functionally interacts with the Star-PAP complex provides a link between splicing, and cleavage and polyadenylation for 3’ formation of Star-PAP target mRNA. Interestingly, RIP revealed that CKIα is associated with Star-PAP target mRNA HO-1 and the use of CKI inhibitors CKI-7 and IC261 caused a significant decrease in the expression of HO-1 mRNA upon induction by tBHQ, demonstrating a role for CKIα in a stress response pathway with PIPKIα and Star-PAP to efficiently process the 3’ end of Star-PAP target mRNAs. Interestingly, CKIα does not RIP with the non-stress response Star-PAP target mRNA cationic transport regulator 1(CHAC1) (NM_219270) and the loss of CKIα via siRNA or by aforementioned CKIα inhibitors does not affect CHAC1 mRNA levels, suggesting that the Star-PAP complex may vary in composition in dependence of the RNA that is targeted for processing(Gonzales et al., 2008) .
The unexpected and new nuclear substrate and function for PI4,5P2 sensitive protein kinase CKIα reinforces the understanding of phosphoinositide sensitive protein complexes which contain multiple components that are regulated by PI4,5P2 (Bairstow et al., 2006). CKIα functions with PIPKIα and both are required for the expression of the Star-PAP target mRNA HO-1, which encodes a critical cytoprotective enzyme. The involvement of Star-PAP, PIPKIα and CKIα in regulating the expression of a pivotal stress response gene demonstrates a clear role for a nuclear phosphoinositide signaling pathway in response to oxidative stress.
Star-PAP is a novel member of the CID1 family of non-canonical poly(A) polymerases and embodies features of TUTase
RNA-specific nucleotidyl transferases (rNTs) that act by covalently attaching ribonucleotides to the 3’ end of their RNA substrates have been studied for over forty years and are customarily divided into canonical and non-canonical type enzymes(Martin and Keller, 2007). Canonical rNTs include the CCA-adding enzyme which is essential for the function of tRNAs as acceptors of amino acids(Xiong and Steitz, 2004), TUTases involved in mitochondrial mRNA editing in Trypanosomal Brucei (T. brucei), Leishmania major (L. major) and Leishmania tarentolae (L. tarentolae) (Simpson et al., 2004), members of the PAPOLA gene family PAPa, -β and –γ, and all of the splice variants therein (Kashiwabara et al., 2002; Kyriakopoulou et al., 2001; Raabe et al., 1991) as well as certain antibiotic resistance enzymes. Non-canonical rNTPs are varied and include the many members of the CID1 family of non-canonical poly(A) polymerases, such as CID1, and the “CID-1like proteins” such as the regulatory cytoplasmic PAP GLD2 and the exosome-related PAP Trf4/5p (LaCava et al., 2005; Stevenson and Norbury, 2006; Wang et al., 2002), and Star-PAP.
The cid1 gene was isolated in a genetic screen as a suppressor of “checkpoint Rad” mutants in fission yeast. The mutants were sensitive to hydroxyurea (HU) which is an inhibitor of ribonucleotide reductase, though these cidIΔ were also sensitive to a combination of HU and caffeine, hence the name CID1 for caffeine induced death 1. cidIΔ mutants affect S-M checkpoints when they are crossed with mutants of DNA Pol δ and ε, DNA polymerases required for chromosomal replication(Wang et al., 2000). CID1 was initially characterized as a regulatory cytoplasmic poly(A) polymerase but has now been defined to be a regulatory poly(U) polymerase, (PUP) an enzyme which uridylates its RNA substrate rather than adenylating.
The gld2 gene, germline development 2, was isolated as a regulator of mitosis/meiosis decision in C. elegans (Kadyk and Kimble, 1998), though the discovery of GLD2 being a regulatory poly(A) polymerase was not revealed until years later (Wang et. al., 2002). gld2Δ fail to complete spermatogenesis and oogenesis in the nematode. In mouse, GLD2 was proposed to play a role in a positive feedback during the progression of metaphase I to metaphase II during oocyte maturation(Nakanishi et al., 2006). GLD2 in humans is a regulator of germline development and has been proposed to play functional roles in Long Term Potentiation (LTP). GLD2 is abundant in anatomical regions of the brain required for long term cognitive and emotional learning in humans and mouse(Rouhana et al., 2005). In vivo, GLD2 is dependent upon its interacting protein partner GLD3, which contains an RRM and is required for GLD2 to bind its mRNA substrate and stimulates the PAP activity of GLD2(Wang et. al., 2002).
Trf4/5p poly(A) polymerase is a component of the TRAMP complex characterized in yeast. RNA targets of the TRAMP, Trf4/5p, Air1/2, Mtr41 Polyadenylation complex include tRNAs, U14 snoRNA, U5 snRNA and pre-ribosomal RNA(LaCava et al., 2005). Air1/2 contain multiple zinc knuckle motifs to bind its RNA target and Mtr41 acts as a helicase to unwind the RNA and expose the 3’ end to make it accessible for Trf4/5p to polyadenylate the RNA. The oligo(A) tract allows recruitment of the exosome, which degrades its RNA substrate in a 3’ to 5’ direction. The RNA bound in the TRAMP complex can undergo multiple rounds of re-adenylation until the RNA is completely degraded. The fact that TRAMP polyadenylation complex acts to degrade RNA is in contrary to role of polyadenylation of mRNA, which is believed to be a stabilizing modification. This function is reminiscent of bacterial poly(A) polymerases where polyadenylation marks an RNA for degradation(Grunberg-Manago, 1999).
There are two striking properties that the members of the CID1-like family of non-canonical poly(A) polymerase feature that is distinct from canonical poly(A) polymerase and every known member embodies at least one of these properties. The first is that many of these enzymes can not bind their RNA substrate on their own and require an interacting RNA binding protein, as in the cases of GLD2/GLD3 and the TRAMP complex. A second feature is that there are a number of CID1-like non-canonical poly(A) polymerases that have the capacity to transfer UMP to its RNA substrate in addition to, or rather than, AMP. Hence, there are non-canonical poly(A) polymerases that display biological traits of being a PUP, or as being a TUTase(Stevenson and Norbury, 2006). Intriguingly, Star-PAP falls into the latter category.
PUP activity was first demonstrated in tobacco leaves over thirty years ago(Brishammar and Juntti, 1975), and the first reported evidence of TUTase came around the same time period, which suggested that a host TUTase was involved in poliovirus replicase initiation(Dasgupta et al., 1980). It was not until the advent of high throughput sequencing that is was established that tracts of poly(U) could be added post-transcriptionally rather than simply being encoded in the DNA of the gene. Analysis of cytoplasmic mRNA provided solid evidence of post-transcriptionally added oligo(U) tracts to RNA. Poly(U) tracts ~30–40 nucleotides in length were reported for about one-fifth of all poly(A)-containing mRNA(Korwek et al., 1976). Poly(U) tracts were subsequently found in cytoplasmic and nuclear RNA, in both polyadenylated and non-polyadenylated fractions. It was later reasoned that cellular RNA could be separated into separate categories according to poly(A) and poly(U) content. First, those containing both poly(A) and poly(U) sequences, which was estimated to be ~ 5–15% of nuclear RNA. Second, those containing only poly(A) tracts, was estimated to be~ 50%. Third, those RNAs solely with poly(U) tracts, which was estimated to be ~2%, and lastly RNAs which contain neither poly(A) or poly(U) sequences, which was estimated to be ~ 30% (Molloy, 1980).
The question that comes to mind when examining poly(U) mRNA is, why does the cell require 3’ end U addition? There are at least two separate potential reasons why this mechanism evolved. First, U-addition and deletion is a means for expanding the gene expression repertoire. By de novo creation of sequence specific sites, mRNA editing can change an ORF to give rise to diverse proteins from a single gene, additionally, mRNA editing can add alternative poly(A) sites, AAUAAA, again expanding the possible protein products from a single gene. The second reason is a structural argument. Oligo(U) tracts would be able to base pair with existing poly(A), forming a hairpin. The formation of such structures could affect the stability, export and/or translatability of an mRNA, ultimately affecting mRNA half-life. Although, it is known that uridylation occurs not only in mRNAs such as Actin mRNA in S.pombe(Rissland et. al., 2007), but in non-coding small RNAs as well.
A “hybrid” 3’ end on a small RNA composed of AMP and UMP could act as signal, or act as a sequence specific platform for RNA binding proteins to interact and modulate the function of small nuclear RNAs in the processing of mRNAs. Many small RNAs are uridylated such as microRNAs and endogenous siRNAs, whereby 3’ end uridylation appears to be destabilizing to these RNAs (Chen et al., 2000; Shen and Goodman, 2004). Additionally, histone mRNA has been recently reported to be uridylated and this acts to destabilize the message(Mullen and Marzluff, 2008). The small nuclear RNA U6, U6 snRNA, is uridylated and this modification stabilizes this RNA; U6 snRNA is also adenylated though the functional significance of this modification remains unknown (Chen et al., 2000).
It was reported that Star-PAP has the capacity to transfer UMP to the 3’ end of U6 snRNA and acts as the U6 TUTase(Trippe et al., 2006). Indeed, Star-PAP has the capacity to transfer UMP to total cellular RNA and can uridylate U6 in vitro. U6 snRNA is a spliceosome component and transcribed by RNA Pol III and accordingly transcriptional termination is signaled by UUUUU(Kiss, 2004). The 3’ end of U6 snRNA is stabilized by the formation of a 2,3’-cyclic phosphate and the majority of U6 molecules contains this modification(Lund and Dahlberg, 1992). The remaining population of U6 contains either a 3’ tail containing a short stretch of poly(U) or a 3’ tail containing poly(U) which is followed by adenylation(Chen et al., 2000). Presumably without maintaining the 3’end of U6 snRNA, U6 snRNA is exonucleolytically trimmed, becoming a poor substrate for the 3’ terminal cyclase. Consequently, U6 snRNA would suffer complete degradation resulting in a vast decrease in the amount of competent spliceosomes to process the nascent mRNA, and mRNA metabolism would be altered globally resulting in cellular death(Trippe et al., 2006). If Star-PAP plays a functional role in the assembly of spliceosomes through U6 snRNA 3’ end modification for global splicing of bulk mRNA, then a functional in vivo assay could potentially be established to demonstrate this. Since spliceosome assembly occurs in a step wise manner, it is conceivable to isolate intermediates (Bessonov et al., 2008) that harbor an active Star-PAP complex. This could provide insights into the U6 snRNP domain and define a role for Star-PAP function in sustaining catalytic activity of the spliceosome to couple with 3’ end formation of mRNA.
Finally, it must be emphasized that included in the U6 TUTase report(Trippe et al., 2006) was the statement that U6 is the only snRNA that has its own modifying enzyme, and the question was poised asking why is U6 snRNA afforded the luxury of having its own 3’ end modifying enzyme while the other snRNAs that make up the spliceosome, U1, U2, U4 and U5 do not? If Star-PAP is indeed the sole U6 TUTase and U6 is the sole RNA substrate for Star-PAP in vivo, then the answer will most likely be centered directly on phosphoinositide based signaling through the Star-PAP interacting protein, PIPKIα, its product PI4,5P2 and most probably the breadth of nuclear phosphoinositide signaling molecules that control gene expression.
Star-PAP harbors features of TUTase in regards to its domain arrangement and its in vitro transferase activity. As featured in Star-PAP, canonical TUTase described in T.brucei has a poly(A) polymerase domain that is split by a linker domain(Aphasizhev et al., 2002). Although, the linker domain, the PRR domain in Star-PAP, is highly divergent and shows no identity to any reported TUTase(Stagno et al., 2007). Attempts to detect sequence similarity to any known protein or secondary structural elements in Star-PAP PRR domain have been unsuccessful(Stagno et al., 2007). This strongly supports the hypothesis that the PRR domain in Star-PAP is unique to Star-PAP and that this domain provides a three-dimensional requirement for the phosphoinositide-dependent nature of Star-PAP function as a duel specificity non-canonical poly(A) polymerase.
Summary
Star-PAP is a recently identified nuclear speckle localized non-canonical poly(A) polymerase that has a functional interaction with PIPKIα, and whose activity is modulated by the PIPKIα product, PI4,5P2. Similar to other poly(A) polymerases, such as the canonical PAPα and the non-canonical GLD2 PAP, Star-PAP resides in a large complex of proteins involved in the 3’ end formation of mRNAs (Fig. 4). The Star-PAP complex shares components with the canonical PAPα complex though contains unique associated proteins such as PIPKIα and CKIα. The Star-PAP complex assembles into a highly stable 3’ end processing machine upon oxidative stress induction. This assembled complex shows enhanced enzyme activity and hypersensitivity to exogenous PI4,5P2, implying that an activated Star-PAP is distinctly modified and/or contains unique factors as compared to Star-PAP purified from resting cells.
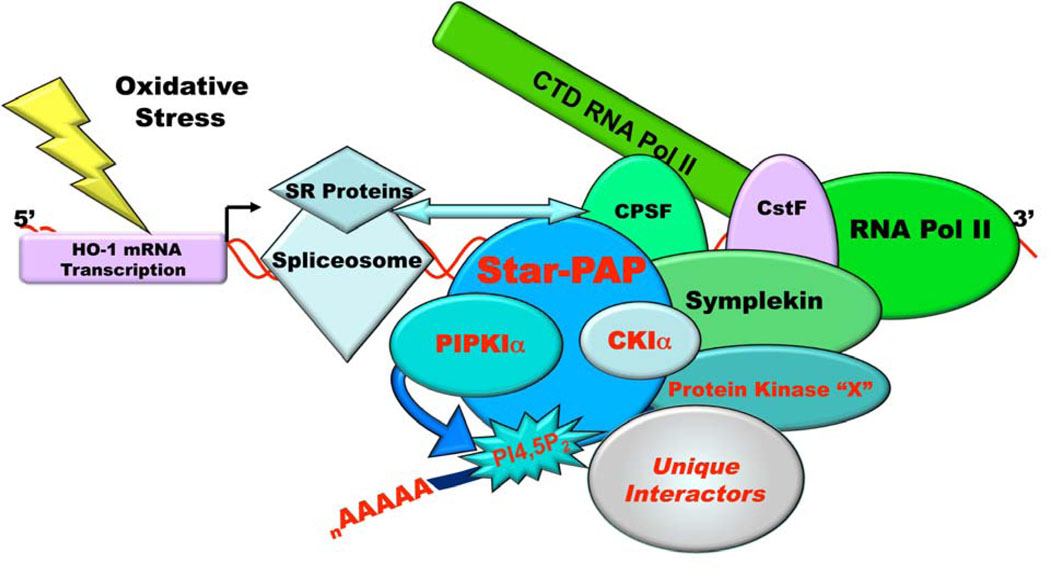
Fig. 4.
Model of the Star-PAP complex induced to assemble on its target gene ho-1 for the 3’ end processing of HO-1 mRNA. A stimuli such as oxidative stress induction can drive the inclusion of a phosphoinositide signaling components PIPKIα and CKIα into a stable Star-PAP complex that targets the stress response gene heme oxygenase-1, ho-1.
The association between a poly(A) polymerase and the polyadenylation complex is vital for its proper function, accordingly, Star-PAP is required for the expression and 3’ end formation of select mRNAs. In addition to poly(A) adding activity, Star-PAP embodies features of Terminal Uridylyl Transferase activity, TUTase, and can transfer UMP to cellular RNA such as the small nuclear RNA U6, signifying that Star-PAP is a duel specificity RNA nucleotidyl transferase. Additionally, the Star-PAP complex harbors lipid kinase activity capable of generating de novo PI4,5P2 and protein kinase activity that can be inhibited by PI4,5P2. It was demonstrated that the PI4,5P2 sensitive kinase CKIα is at least one of the kinases responsible for this activity and that CKIα can directly phosphorylate Star-PAP in its Proline Rich Region domain suggesting that multiple aspects of Star-PAP function can be regulated by phosphoinositide signaling. The Star-PAP complex therefore represents a site where multiple phosphoinositide signaling pathways converge to control the synthesis of select mRNAs.
Consistent with this, CKIα as well as PIPKIα are required for the synthesis of the Star-PAP target mRNA HO-1, which encodes the cytoprotective enzyme heme oxygenase -1, HO-1. CKIα and Star-PAP are associated with HO-1 mRNA in vivo, suggesting that Star-PAP, CKIα and PIPKIα work together to modulate the production of this and other select mRNAs. It will be useful to identify more uniquely associated Star-PAP proteins to begin defining additional signal transduction pathways that converge on Star-PAP functions and may allow for the discovery of additional Star-PAP target mRNAs. It will also be helpful to define signaling components in the tBHQ-induced oxidative stress response pathway that work upstream of Star-PAP, PIPKIα and CKIα. This may provide information about how nuclear phosphoinositide generation and signaling is regulated by various stimuli.
Acknowledgements
We would like to acknowledge Dr. C.A. Barlow for critical conversations in the writing process. We acknowledge the National Institutes of Health, NIH, for funding. Additionally, DLM acknowledges the American Heart Association, AHA, for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Alb JG, Jr, Kearns MA, Bankaitis VA. Phospholipid metabolism and membrane dynamics. Curr Opin Cell Biol. 1996;8:534–541. doi: 10.1016/s0955-0674(96)80032-9. [DOI] [PubMed] [Google Scholar]
- Alonso MT, Villalobos C, Chamero P, Alvarez J, Garcia-Sancho J. Calcium microdomains in mitochondria and nucleus. Cell Calcium. 2006;40:513–525. doi: 10.1016/j.ceca.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Anderson RA, Marchesi VT. Regulation of the association of membrane skeletal protein 4.1 with glycophorin by a polyphosphoinositide. Nature. 1985;318:295–298. doi: 10.1038/318295a0. [DOI] [PubMed] [Google Scholar]
- Aphasizhev R, Sbicego S, Peris M, Jang SH, Aphasizheva I, Simpson AM, Rivlin A, Simpson L. Trypanosome mitochondrial 3' terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Bairstow SF, Ling K, Su X, Firestone AJ, Carbonara C, Anderson RA. Type Igamma 661 phosphatidylinositol phosphate kinase directly interacts with AP2 and regulates endocytosis. J Biol Chem. 2006;281:20632–20642. doi: 10.1074/jbc.M601465200. [DOI] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lurhmann R. Isolation of an active step one spliceosome and composition of its RNP core. Nature. 2008;452:846850. doi: 10.1038/nature06842. [DOI] [PubMed] [Google Scholar]
- Billah MM. Phospholipase D and cell signaling. Curr Opin Immunol. 1993;5:114–123. doi: 10.1016/0952-7915(93)90090-f. [DOI] [PubMed] [Google Scholar]
- Boronenkov IV, Loijens JC, Umeda M, Anderson RA. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol Biol Cell. 1998;9:3547–3560. doi: 10.1091/mbc.9.12.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brishammar S, Juntti N. A poly(U) polymerase in tobacco leaves. Biochim Biophys Acta. 1975;383:351–358. doi: 10.1016/0005-2787(75)90304-4. [DOI] [PubMed] [Google Scholar]
- Brockman JL, Anderson RA. Casein kinase I is regulated by phosphatidylinositol 4,5-bisphosphate in native membranes. J Biol Chem. 1991;266:2508–2512. [PubMed] [Google Scholar]
- Bucki R, Gorski J. Current views on function and regulation of Ca2+ levels in cell nuclei. Postepy Hig Med Dosw. 2001;55:157–175. [PubMed] [Google Scholar]
- Bunce MW, Boronenkov IV, Anderson RA. Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J Biol Chem. 2008;283:8678–8686. doi: 10.1074/jbc.M710222200. [DOI] [PubMed] [Google Scholar]
- Cazzolli R, Shemon AN, Fang MQ, Hughes WE. Phospholipid signalling through phospholipase D and phosphatidic acid. IUBMB Life. 2006;58:457–461. doi: 10.1080/15216540600871142. [DOI] [PubMed] [Google Scholar]
- Cegielska A, Gietzen KF, Rivers A, Virshup DM. Autoinhibition of casein kinase I epsilon (CKI epsilon) is relieved by protein phosphatases and limited proteolysis. J Biol Chem. 1998;273:1357–1364. doi: 10.1074/jbc.273.3.1357. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sinha K, Perumal K, Reddy R. Effect of 3' terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. Rna. 2000;6:1277–1288. doi: 10.1017/s1355838200000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CU, Ehrlich BE. The inositol 1,4,5-trisphosphate receptor (IP3R) and its regulators: sometimes good and sometimes bad teamwork. Sci STKE. 2006:re15. doi: 10.1126/stke.3632006re15. [DOI] [PubMed] [Google Scholar]
- Christofori G, Keller W. 3' cleavage and polyadenylation of mRNA precursors in vitro requires a poly(A) polymerase, a cleavage factor, and a snRNP. Cell. 1988;54:875–889. doi: 10.1016/s0092-8674(88)91263-9. [DOI] [PubMed] [Google Scholar]
- Chu A, Stefani E. Phosphatidylinositol 4,5-bisphosphate-induced Ca2+ release from skeletal muscle sarcoplasmic reticulum terminal cisternal membranes. Ca2+ flux and single channel studies. J Biol Chem. 1991;266:7699–7705. [PubMed] [Google Scholar]
- Clarke JH. Lipid signalling: Picking out the PIPs. Curr Biol. 2003;13:R815–R817. doi: 10.1016/j.cub.2003.09.054. [DOI] [PubMed] [Google Scholar]
- Cocco L, Faenza I, Fiume R, Maria Billi A, Gilmour RS, Manzoli FA. Phosphoinositide-specific phospholipase C (PI-PLC) beta1 and nuclear lipid-dependent signaling. Biochim Biophys Acta. 2006;1761:509–521. doi: 10.1016/j.bbalip.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Cocco L, Gilmour RS, Ognibene A, Letcher AJ, Manzoli FA, Irvine RF. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem J. 1987;248:765–770. doi: 10.1042/bj2480765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocco L, Manzoli L, Barnabei O, Martelli AM. Significance of subnuclear localization of key players of inositol lipid cycle. Adv Enzyme Regul. 2004a;44:51–60. doi: 10.1016/j.advenzreg.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Cocco L, Maraldi NM, Manzoli FA. New frontiers of inositide-specific phospholipase C in nuclear signalling. Eur J Histochem. 2004b;48:83–88. [PubMed] [Google Scholar]
- Cocco L, Martelli AM, Gilmour RS, Rhee SG, Manzoli FA. Nuclear phospholipase C and signaling. Biochim Biophys Acta. 2001;1530:1–14. doi: 10.1016/s1388-1981(00)00169-4. [DOI] [PubMed] [Google Scholar]
- Cohn RC, Poncz L, Waller RL, Dearborn DG. Phosphoinositide content of erythrocyte membranes in cystic fibrosis. J Lab Clin Med. 1988;111:336–340. [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Zhao W, Prives C, Manley JL. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. Embo J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R, Parinandi N, Wang L, Usatyuk P, Natarajan V. Phospholipase D/phosphatidic acid signal transduction: role and physiological significance in lung. Mol Cell Biochem. 2002;234–235:99–109. [PubMed] [Google Scholar]
- Cunningham E, Thomas GM, Ball A, Hiles I, Cockcroft S. Phosphatidylinositol transfer protein dictates the rate of inositol trisphosphate production by promoting the synthesis of PIP2. Curr Biol. 1995;5:775–783. doi: 10.1016/s0960-9822(95)00154-0. [DOI] [PubMed] [Google Scholar]
- Czech MP. PIP2 and PIP3: complex roles at the cell surface. Cell. 2000;100:603–606. doi: 10.1016/s0092-8674(00)80696-0. [DOI] [PubMed] [Google Scholar]
- D'Angelo G, Vicinanza M, Di Campli A, De Matteis MA. The multiple roles of PtdIns(4)P -not just the precursor of PtdIns(4,5)P2. J Cell Sci. 2008;121:1955–1963. doi: 10.1242/jcs.023630. [DOI] [PubMed] [Google Scholar]
- Dasgupta A, Zabel P, Baltimore D. Dependence of the activity of the poliovirus replicase on the host cell protein. Cell. 1980;19:423–429. doi: 10.1016/0092-8674(80)90516-4. [DOI] [PubMed] [Google Scholar]
- de Groot RP, den Hertog J, Vandenheede JR, Goris J, Sassone-Corsi P. Multiple and cooperative phosphorylation events regulate the CREM activator function. Embo J. 1993;12:3903–3911. doi: 10.1002/j.1460-2075.1993.tb06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P, Coste B, Gamper N, Shapiro MS. Phosphoinositide lipid second messengers: new paradigms for calcium channel modulation. Neuron. 2005;47:179–182. doi: 10.1016/j.neuron.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Didichenko SA, Thelen M. Phosphatidylinositol 3-kinase c2alpha contains a nuclear localization sequence and associates with nuclear speckles. J Biol Chem. 2001;276:48135–48142. doi: 10.1074/jbc.M104610200. [DOI] [PubMed] [Google Scholar]
- Ding J, Hayashi MK, Zhang Y, Manche L, Krainer AR, Xu RM. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Banfic H, Irvine RF. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. Embo J. 1991;10:3207–3214. doi: 10.1002/j.1460-2075.1991.tb04883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Rhee SG, Letcher AJ, Irvine RF. Phosphoinositide signalling enzymes in rat liver nuclei: phosphoinositidase C isoform beta 1 is specifically, but not predominantly, located in the nucleus. Biochem J. 1993;289(Pt 3):617–620. doi: 10.1042/bj2890617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divecha N, Roefs M, Halstead JR, D'Andrea S, Fernandez-Borga M, Oomen L, Saqib KM, Wakelam MJ, D'Santos C. Interaction of the type Ialpha PIPkinase with phospholipase D: a role for the local generation of phosphatidylinositol 4, 5-bisphosphate in the regulation of PLD2 activity. Embo J. 2000;19:5440–5449. doi: 10.1093/emboj/19.20.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughman RL, Firestone AJ, Anderson RA. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J Membr Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- Downes CP, Gray A, Lucocq JM. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005;15:259–268. doi: 10.1016/j.tcb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Ehrlich BE, Kaftan E, Bezprozvannaya S, Bezprozvanny I. The pharmacology of intracellular Ca(2+)-release channels. Trends Pharmacol Sci. 1994;15:145–149. doi: 10.1016/0165-6147(94)90074-4. [DOI] [PubMed] [Google Scholar]
- Evangelisti C, Riccio M, Faenza I, Zini N, Hozumi Y, Goto K, Cocco L, Martelli AM. Subnuclear localization and differentiation-dependent increased expression of DGK-zeta in C2C12 mouse myoblasts. J Cell Physiol. 2006;209:370–378. doi: 10.1002/jcp.20744. [DOI] [PubMed] [Google Scholar]
- Fish KJ, Cegielska A, Getman ME, Landes GM, Virshup DM. Isolation and characterization of human casein kinse I epsilon (CKI), a novel member of the CKI gene family. J Biol Chem. 1995;270:14875–14883. doi: 10.1074/jbc.270.25.14875. [DOI] [PubMed] [Google Scholar]
- Flowtow H, Roach PJ. Synergistic phosphorylation of rabbit muscle glycogen synthase by cyclic AMP-dependent protein kinase and casein kinase; Implications for hormonal regulation of glycogen synthase. J Biol Chem. 1989;264:9126–9128. [PubMed] [Google Scholar]
- Flotow H, Roach PJ. Role of acidic residues as substrate determinants for casein kinase I. J Biol Chem. 1991;266:3724–3727. [PubMed] [Google Scholar]
- Floyd CC, Grant P, Gallant PE, Pant HC. Principal neurofilament-associated protein kinase in squid axoplasm is related to casein kinase I. J Biol Chem. 1991;266:4987–4994. [PubMed] [Google Scholar]
- Fu Z, Chakraborti T, Morse S, Bennett GS, Shaw G. Four casein kinase I isoforms are differentially partitioned between the nucleus and the cytoplasm. Exp Cell Res. 2001;269:275–286. doi: 10.1006/excr.2001.5324. [DOI] [PubMed] [Google Scholar]
- Gardocki ME, Jani N, Lopes JM. Phosphatidylinositol biosynthesis: biochemistry and regulation. Biochim Biophys Acta. 2005;1735:89–100. doi: 10.1016/j.bbalip.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Gonzales ML, Anderson RA. Nuclear phosphoinositide kinases and inositol phospholipids. J Cell Biochem. 2006;97:252–260. doi: 10.1002/jcb.20655. [DOI] [PubMed] [Google Scholar]
- Gonzales ML, Mellman DL, Anderson RA. CKIalpha is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J Biol Chem. 2008;283:12665–12673. doi: 10.1074/jbc.M800656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Hozumi Y, Kondo H. Diacylglycerol, phosphatidic acid, and the converting enzyme, diacylglycerol kinase, in the nucleus. Biochim Biophys Acta. 2006;1761:535–541. doi: 10.1016/j.bbalip.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Green CL, Bennett GS. Identification of four alternatively spliced isoforms of chicken casein kinase I alpha that are all expressed in diverse cell types. Gene. 1998;216:189–195. doi: 10.1016/s0378-1119(98)00291-1. [DOI] [PubMed] [Google Scholar]
- Gross SD, Hoffmanv DP, Fisette PL, Baas P, Anderson RA. A phosphatidylinositol 4,5-bisphosphate -sensitive casein kinase I alpha associates with synaptic vesicles and phosphorylates a subset of vesicle proteins. J Cell Biol. 1995;130:711–724. doi: 10.1083/jcb.130.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Gross SD, Loijens JC, Anderson RA. The casein kinase Ialpha isoform is both physically positioned and functionally competent to regulate multiple events of mRNA metabolism. J Cell Sci. 1999;112(Pt 16):2647–2656. doi: 10.1242/jcs.112.16.2647. [DOI] [PubMed] [Google Scholar]
- Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu Rev Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Hansen LK, Mooney DJ, Vacanti JP, Ingber DE. Integrin binding and cell spreading on extracellular matrix act at different points in the cell cycle to promote hepatocyte growth. Mol Biol Cell. 1994;5:967–975. doi: 10.1091/mbc.5.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Fisette PL, Jenkins GH, Fukami K, Takenawa T, Anderson RA, Martin TF. ATP-dependent inositide phosphorylation required for Ca(2+)-activated secretion. Nature. 1995;374:173–177. doi: 10.1038/374173a0. [DOI] [PubMed] [Google Scholar]
- Heck JN, Mellman DL, Ling K, Sun Y, Wagoner MP, Schill NJ, Anderson RA. A conspicuous connection: structure defines function for the phosphatidylinositol-phosphate kinase family. Crit Rev Biochem Mol Biol. 2007;42:15–39. doi: 10.1080/10409230601162752. [DOI] [PubMed] [Google Scholar]
- Hodgkin MN, Pettitt TR, Martin A, Michell RH, Pemberton AJ, Wakelam MJ. Diacylglycerols and phosphatidates: which molecular species are intracellular messengers? Trends Biochem Sci. 1998;23:200–204. doi: 10.1016/s0968-0004(98)01200-6. [DOI] [PubMed] [Google Scholar]
- Huijbregts RP, Topalof L, Bankaitis VA. Lipid metabolism and regulation of membrane trafficking. Traffic. 2000;1:195–202. doi: 10.1034/j.1600-0854.2000.010301.x. [DOI] [PubMed] [Google Scholar]
- Insall RH, Weiner OD. PIP3, PIP2, and cell movement--similar messages, different meanings? Dev Cell. 2001;1:743–747. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF. The enzymology of stimulated inositol lipid turnover. Cell Calcium. 1982;3:295–309. doi: 10.1016/0143-4160(82)90018-5. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Inositol lipids in cell signalling. Curr Opin Cell Biol. 1992;4:212–219. doi: 10.1016/0955-0674(92)90035-b. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Nuclear lipid signaling. Sci STKE. 2002:RE13. doi: 10.1126/stke.2002.150.re13. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Nuclear inositide signalling -- expansion, structures and clarification. Biochim Biophys Acta. 2006;1761:505–508. doi: 10.1016/j.bbalip.2006.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- Jenkins GH, Fisette PL, Anderson RA. Type I phosphatidylinositol 4-phosphate 5-kinase isoforms are specifically stimulated by phosphatidic acid. J Biol Chem. 1994;269:11547–11554. [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kashiwabara S, Noguchi J, Zhuang T, Ohmura K, Honda A, Sugiura S, Miyamoto K, Takahashi S, Inoue K, Ogura A, Baba T. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science. 2002;298:1999–2002. doi: 10.1126/science.1074632. [DOI] [PubMed] [Google Scholar]
- Keum YS, Han YH, Liew C, Kim JH, Xu C, Yuan X, Shakarjian MP, Chong S, Kong AN. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm Res. 2006;23:2586–2594. doi: 10.1007/s11095-006-9094-2. [DOI] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Korwek EL, Nakazato N, Edmonds M, Venkatesan S. Poly(uridylic acid) sequences in messenger ribonucleic acid of HeLa cells. Biochemistry. 1976;15:4643–4649. doi: 10.1021/bi00666a015. [DOI] [PubMed] [Google Scholar]
- Knippschild U, et al. The casein kinase I family: participation in multiple cellular processes in eukaryotes. Cell Signal. 2005;17:675–689. doi: 10.1016/j.cellsig.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Ktistakis NT, Delon C, Manifava M, Wood E, Ganley I, Sugars JM. Phospholipase D1 and potential targets of its hydrolysis product, phosphatidic acid. Biochem Soc Trans. 2003;31:94–97. doi: 10.1042/bst0310094. [DOI] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3' end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Kyriakopoulou CB, Nordvarg H, Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J Biol Chem. 2001;276:33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Lee SH, Choi HS, Kim H, Lee Y. ERK is a novel regulatory kinase for poly(A) polymerase. Nucleic Acids Res. 2008;36:803–813. doi: 10.1093/nar/gkm1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Gamper N, Hilgemann DW, Shapiro MS. Regulation of Kv7 (KCNQ) K+ channel open probability by phosphatidylinositol 4,5-bisphosphate. J Neurosci. 2005;25:9825–9835. doi: 10.1523/JNEUROSCI.2597-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Vasco VR, Calabrese G, Manzoli L, Palka G, Spadano A, Morizio E, Guanciali-Franchi P, Fantasia D, Cocco L. Inositide-specific phospholipase c beta1 gene deletion in the progression of myelodysplastic syndrome to acute myeloid leukemia. Leukemia. 2004;18:1122–1126. doi: 10.1038/sj.leu.2403368. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Cyclic 2',3'-phosphates and nontemplated nucleotides at the 3' end of spliceosomal U6 small nuclear RNA's. Science. 1992;255:327–330. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JR. Nuclear calcium: transfer to and from the cytosol. Biol Signals Recept. 1998;7:137–147. doi: 10.1159/000014540. [DOI] [PubMed] [Google Scholar]
- Malviya AN, Klein C. Mechanism regulating nuclear calcium signaling. Can J Physiol Pharmacol. 2006;84:403–422. doi: 10.1139/y05-130. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Fiume R, Faenza I, Tabellini G, Evangelista C, Bortul R, Follo MY, Fala F, Cocco L. Nuclear phosphoinositide specific phospholipase C (PI-PLC)-beta 1: a central intermediary in nuclear lipid-dependent signal transduction. Histol Histopathol. 2005a;20:1251–1260. doi: 10.14670/HH-20.1251. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Follo MY, Evangelisti C, Fala F, Fiume R, Billi AM, Cocco L. Nuclear inositol lipid metabolism: more than just second messenger generation? J Cell Biochem. 2005b;96:285–292. doi: 10.1002/jcb.20527. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Gilmour RS, Bertagnolo V, Neri LM, Manzoli L, Cocco L. Nuclear localization and signalling activity of phosphoinositidase C beta in Swiss 3T3 cells. Nature. 1992;358:242–245. doi: 10.1038/358242a0. [DOI] [PubMed] [Google Scholar]
- Martelli AM, Neri LM, Gilmour RS, Barker PJ, Huskisson NS, Manzoli FA, Cocco L. Temporal changes in intracellular distribution of protein kinase C in Swiss 3T3 cells during mitogenic stimulation with insulin-like growth factor I and bombesin: translocation to the nucleus follows rapid changes in nuclear polyphosphoinositides. Biochem Biophys Res Commun. 1991;177:480–487. doi: 10.1016/0006-291x(91)92009-9. [DOI] [PubMed] [Google Scholar]
- Martin G, Keller W. RNA-specific ribonucleotidyl transferases. Rna. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TF. PI(4,5)P(2) regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- Matthews JM, Sunde M. Zinc fingers--folds for many occasions. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- McLaughlin S, Wang J, Gambhir A, Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu Rev Biophys Biomol Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- Mejillano M, Yamamoto M, Rozelle AL, Sun HQ, Wang X, Yin HL. Regulation of apoptosis by phosphatidylinositol 4,5-bisphosphate inhibition of caspases, and caspase inactivation of phosphatidylinositol phosphate 5-kinases. J Biol Chem. 2001;276:1865–1872. doi: 10.1074/jbc.M007271200. [DOI] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Metz S, Dunlop M. Phospholipase D in the pancreatic islet: evidence suggesting the involvement of phosphatidic acid in signal transduction. Adv Prostaglandin Thromboxane Leukot Res. 1991;21A:287–290. [PubMed] [Google Scholar]
- Miyazaki S. IP3 receptor-mediated spatial and temporal Ca2+ signaling of the cell. Jpn J Physiol. 1993;43:409–434. doi: 10.2170/jjphysiol.43.409. [DOI] [PubMed] [Google Scholar]
- Miyazaki S. Inositol trisphosphate receptor mediated spatiotemporal calcium signalling. Curr Opin Cell Biol. 1995;7:190–196. doi: 10.1016/0955-0674(95)80027-1. [DOI] [PubMed] [Google Scholar]
- Molloy GR. The isolation and characterization of HeLa cell messenger RNA-like molecules containing uridylic acid-rich oligonucleotide sequences. J Biol Chem. 1980;255:10375–10381. [PubMed] [Google Scholar]
- Morris AJ, Frohman MA, Engebrecht J. Measurement of phospholipase D activity. Anal Biochem. 1997;252:1–9. doi: 10.1006/abio.1997.2299. [DOI] [PubMed] [Google Scholar]
- Mortier E, Wuytens G, Leenaerts I, Hannes F, Heung MY, Degeest G, David G, Zimmermann P. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. Embo J. 2005;24:2556–2565. doi: 10.1038/sj.emboj.7600722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5' to 3' and 3' to 5'. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S, Miyado K, Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Nebl T, Oh SW, Luna EJ. Membrane cytoskeleton: PIP(2) pulls the strings. Curr Biol. 2000;10:R351–R354. doi: 10.1016/s0960-9822(00)00465-6. [DOI] [PubMed] [Google Scholar]
- Niggli V. Regulation of protein activities by phosphoinositide phosphates. Annu Rev Cell Dev Biol. 2005;21:57–79. doi: 10.1146/annurev.cellbio.21.021704.102317. [DOI] [PubMed] [Google Scholar]
- Osborne SL, Thomas CL, Gschmeissner S, Schiavo G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J Cell Sci. 2001;114:2501–2511. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- Pertile P, Liscovitch M, Chalifa V, Cantley LC. Phosphatidylinositol 4,5-bisphosphate synthesis is required for activation of phospholipase D in U937 cells. J Biol Chem. 1995;270:5130–5135. doi: 10.1074/jbc.270.10.5130. [DOI] [PubMed] [Google Scholar]
- Planchon SM, Waite KA, Eng C. The nuclear affairs of PTEN. J Cell Sci. 2008;121:249–253. doi: 10.1242/jcs.022459. [DOI] [PubMed] [Google Scholar]
- Raabe T, Bollum FJ, Manley JL. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- Rapuano M, Rosen OM. Phosphorylation of the insulin receptor by a casein kinase I-like enzyme. 1991;266:12902–12907. [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by non-canonical poly(A) polymerases. Mol Cell Biol. 2007;207:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. Rna. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowles J, Slaughter C, Moomaw C, Hsu J, Cobb MH. Purification of casein kinase I and isolation of cDNAs encoding multiple casein kinase-like enzymes. Proc Natl Acad Sci U S A. 1991;88:9548–9552. doi: 10.1073/pnas.88.21.9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra VA, Rudge SA, Prestwich GD, Frohman MA, Engebrecht J, Morris AJ. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. Embo J. 1999;18:5911–5921. doi: 10.1093/emboj/18.21.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeds AM, Frederick JP, Tsui MM, York JD. Roles for inositol polyphosphate kinases in the regulation of nuclear processes and developmental biology. Adv Enzyme Regul. 2007;47:10–25. doi: 10.1016/j.advenzreg.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–57805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. Rna. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TJ, Grundke-Iqbal I, McDonald B, Iqbal K. Comparison of the phosphorylation of microtubule-associated protein tau by non-proline dependent protein kinases. Mol Cell Biochem. 1994;131:181–189. doi: 10.1007/BF00925955. [DOI] [PubMed] [Google Scholar]
- Smith CD, Wells WW. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J Biol Chem. 1983;258:9368–9373. [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson AL, Norbury CJ. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Morita K, Akagi R, Sassa S. Heme oxygenase-1: a novel therapeutic target in oxidative tissue injuries. Curr Med Chem. 2004;11:1545–1561. doi: 10.2174/0929867043365080. [DOI] [PubMed] [Google Scholar]
- Topham MK, Prescott SM. Mammalian diacylglycerol kinases, a family of lipid kinases with signaling functions. J Biol Chem. 1999;274:11447–11450. doi: 10.1074/jbc.274.17.11447. [DOI] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. Rna. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuazon PT, Traugh JA. Casein kinase I and II--multipotential serine protein kinases: structure, function and regulation. Adv Second Messenger Phosphoprotein Res. 1991;23:123–164. [PubMed] [Google Scholar]
- Wahle E. 3'-end cleavage and polyadenylation of mRNA precursors. Biochim Biophys Acta. 1995;1261:183–194. doi: 10.1016/0167-4781(94)00248-2. [DOI] [PubMed] [Google Scholar]
- Wakelam MJ. Diacylglycerol--when is it an intracellular messenger? Biochim Biophys Acta. 1998;1436:117–126. doi: 10.1016/s0005-2760(98)00123-4. [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble JA. regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Wang SW, Toda T, MacCallum R, Harris AL, Norbury C. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase delta or epsilon is inactivated. Mol Cell Biol. 2000;20:3234–3244. doi: 10.1128/mcb.20.9.3234-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Li WH, Wang J, Xu K, Dong P, Luo X, Yin HL. Critical role of PIP5KI{gamma}87 in InsP3-mediated Ca(2+) signaling. J Cell Biol. 2004;167:1005–1010. doi: 10.1083/jcb.200408008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Hudson DL, Lamb AG, Bolsover SR, Silver RA, Aitchison MJ, Whitaker M. Mitogens induce calcium transients in both dividing and terminally differentiating keratinocytes. J Cell Sci. 1991;99(Pt 2):397–405. doi: 10.1242/jcs.99.2.397. [DOI] [PubMed] [Google Scholar]
- Wenk MR, De Camilli P. Protein-lipid interactions and phosphoinositide metabolism in membrane traffic: insights from vesicle recycling in nerve terminals. Proc Natl Acad Sci U S A. 2004;101:8262–8269. doi: 10.1073/pnas.0401874101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiradjaja F, Ooms LM, Tahirovic S, Kuhne E, Devenish RJ, Munn AL, Piper RC, Mayinger P, Mitchell CA. Inactivation of the phosphoinositide phosphatases Sac1p and Inp54p leads to accumulation of phosphatidylinositol 4,5-bisphosphate on vacuole membranes and vacuolar fusion defects. J Biol Chem. 2007;282:16295–16307. doi: 10.1074/jbc.M701038200. [DOI] [PubMed] [Google Scholar]
- Wirtz KW. Phospholipid transfer proteins revisited. Biochem J. 1997;324(Pt 2):353–360. doi: 10.1042/bj3240353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Steitz TA. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- Yin HL, Janmey PA. Phosphoinositide regulation of the actin cytoskeleton. Annu Rev Physiol. 2003;65:761–789. doi: 10.1146/annurev.physiol.65.092101.142517. [DOI] [PubMed] [Google Scholar]
- York JD. Regulation of nuclear processes by inositol polyphosphates. Biochim Biophys Acta. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
- York JD, Guo S, Odom AR, Spiegelberg BD, Stolz LE. An expanded view of inositol signaling. Adv Enzyme Regul. 2001;41:57–71. doi: 10.1016/s0065-2571(00)00025-x. [DOI] [PubMed] [Google Scholar]
- York JD, Majerus PW. Nuclear phosphatidylinositols decrease during S-phase of the cell cycle in HeLa cells. J Biol Chem. 1994;269:7847–7850. [PubMed] [Google Scholar]
- Yu H, Fukami K, Watanabe Y, Ozaki C, Takenawa T. Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. Eur J Biochem. 1998;251:281–287. doi: 10.1046/j.1432-1327.1998.2510281.x. [DOI] [PubMed] [Google Scholar]
- Zander L, Bemark M. Immortalized mouse cell lines that lack a functional Rev3 gene are hypersensitive to UV irradiation and cisplatin treatment. DNA Repair (Amst) 2004;3:743–752. doi: 10.1016/j.dnarep.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Zhai L, et al. Casein kinase Igamma subfamily. Molecular cloning, expression, and characterization of three mammalian isoforms and complementation of defects in the Saccharomyces cerevisiae YCK genes. J Biol Chem. 1995;270:12717–12724. doi: 10.1074/jbc.270.21.12717. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]