Abstract
Laminin 332 (LN332) is a basally expressed extracellular matrix protein that enhances the migration and invasion of breast carcinoma cells. The goal of this study was to examine LN332 expression breast carcinoma. Triple negative breast carcinomas lack estrogen receptor (ER), progesterone receptor (PR) expression and HER2 positivity. Immunohistochemistry for ER, PR, HER2, and dual silver in situ hybridization for the HER2 gene were used to define the phenotype of 243 breast cancers in biopsies or arrays. Immunohistochemistry for LN332 revealed that 70 % of triple negative carcinomas stained for LN332. Cytokeratin 5/6 (CK5/6), epidermal growth factor receptor (EGFR) and p63 alone stained fewer triple negative breast carcinomas each, but the combination of LN332 and CK 5/6 or EGFR identified 92% of triple negative breast carcinoma.. Of the 163 non- triple negative cases, LN332 was expressed in only 15%. The identification of LN332 in triple negative breast carcinomas is consistent with gene profiling studies showing its expression among breast carcinomas with a basal phenotype. The observation that a pro-invasive protein such as LN332 is expressed in breast cancer suggests another mechanism by which the triple negative phenotype could be aggressive.
Keywords: Laminin 332, Breast carcinoma, Basal-like phenotype, Triple negative, Immunohistochemistry, HER2 dual in situ hybridization
Introduction
Laminins are large extracellular glycoproteins that are expressed by basal epithelium and are important components of basement membranes (1). Laminins are heterotrimers containing α, β and γ chains forming a cross-shaped structure (1, 2). Laminin 332 (LN332), consisting of α3, β3 and γ2 laminin chains, has been formerly known as laminin 5, kalinin, nicein, ladsin and epiligrin (3). LN332 is of particular interest among the laminin isoforms for its tumor promoting properties (4). The major functions of LN332 include binding of epithelial cells to the basement membrane through the formation of hemidesmosomes (5), and the migration of epithelial cells during wound repair (6–8). The main role of LN332 in normal tissues is in the maintenance of epithelial-mesenchymal cohesion in tissues exposed to external disruptive forces (9).
LN332 seems to have a more complex role in cell migration and tumor invasion (3). LN332 is known to stimulate the migration of various cells including carcinoma cells (8, 10–14). Since these properties are required for metastasis (15–18), LN332 has been implicated in tumor progression (19–23). LN332 expression has been shown to correlate with tumor invasiveness and poor patient prognosis (3). LN332 also is noted to accumulate at the interface of the tumor with the surrounding stroma or in the cytoplasm of cancer cells (24, 25). Besides a few exceptions, tumors derived from tissues that normally express LN332 show high levels of LN332 expression, whereas tissues that do not normally express LN332 give rise to tumors that likewise do not express LN332. (13, 26, 27). The role of LN332 in breast carcinoma, however has been less clear. Although the earliest studies reported LN332 at the invasive edge of breast carcinomas (25), other investigations concluded that LN332 expression was lost as breast carcinoma progressed (28). In vitro examinations by our group (12) and others (26) indicated that LN332 induces migratory and invasive properties to breast carcinoma cell lines, suggesting that LN332 may have a role in breast carcinoma progression. As an initial step to further dissect the relationship between LN332 and breast carcinoma, we tested the expression of LN332 in a series of surgically excised breast carcinomas by western blot and immunohistochemistry (IHC), with particular emphasis of relationships between staining and breast cancer subtype. Since our primary objective was to examine patterns of expression rather than clinical significance, we started with de-identified frozen tumor bank specimens and tissue arrays.
A molecular classification of breast cancers which includes the luminal A, B and C, HER2 over expressing, normal like and basal- like subgroups has been proposed, based on the molecular expression profile of genes expressed by breast carcinomas in several data sets (29). The basal-like phenotype, defined by expression of specific cytokeratins (CK) and other genes found in normal basal breast epithelium, is of particular interest because of the significantly worse prognosis initially observed in this group of patients (29), and subsequently confirmed (30–34) although some studies using different patient groups and definitions did not always find a significantly worse prognosis (35–37). The genetic profiling methods that currently define this phenotype, however, are not widely available, so a surrogate for the basal like phenotype based on proteins measureable by immunohistochemistry (IHC) such as epidermal growth factor receptor (EGFR) and CK 5/6 has been proposed (30,31,34). Triple negative (TN) breast cancer is defined by a lack of expression of estrogen receptor (ER), progesterone receptor (PR) and the absence of HER2 gene amplification or over-expression. Since many, although not all TN breast cancer equate roughly to those of the basal-like phenotype, the TN phenotype has also been used as a surrogate for basal-like breast cancers (38) in daily practice. Among the breast carcinomas we examined, LN332 was primarily expressed in the TN tumors.
Materials and Methods
Cases for immunohistochemistry
We investigated 243 cases of breast carcinoma, which were obtained from the surgical pathology archives of the University of California, Irvine Medical Center and the City of Hope Medical Center between 1994 and 2007. One case was from a male, and the other patients were all female. For 201 cases, breast cancer tissue from mastectomy or lumpectomy cases was removed from the paraffin blocks and arranged into arrays of 9–12 cases per block. Cases on the arrays were from completely de-identified specimens, thus follow up information was not available. The remaining 42 cases were from biopsy specimens, and sections from the original paraffin blocks of those cases were used from the study. Cases that were placed in the array but did not have sufficient tissue to measure the markers were excluded from analysis. The hormone receptor (ER, PR) status and HER2 gene amplification result from fluorescence in situ hybridization (FISH) studies of the 42 non-arrayed cases was obtained from the surgical pathology reports. The 201 cases in the arrays were stained with immunohistochemistry for ER, PR, and HER2, as described below.
The study was approved by the Institutional Review Boards of the University of California, Irvine and the City of Hope Medical Center.
Immunohistochemistry
Breast cancer tissue arrays were prepared and stained with hematoxylin and eosin (H&E) and IHC for ER, PR, HER2, p63, CK5/6, and LN332. Formalin-fixed and and paraffin embedded (FFPE) sections of breast specimens were placed on positively charged slides (X-tra Slides, Surgipath Inc. Richmond IL) and deparaffinized with xylene substitute (Clear Rite 3 Cardinal Health, Dublin OH) and rehydrated through decreasing concentrations of 90:5:5 ethyl: methyl: isopropyl alcohol. Pretreatment was performed using Dako Target Retrieval Solution, pH 6 (Dako, Inc. Carpinteria CA) in a pressure cooker for 5 minutes. Antibodies and dilutions were as follows: LN332 β3 chain, (marketed as kalinin B1 by BD Transduction Laboratories, Lexington KY) clone 17 at 1:250, p63 clone 4A4 (Dako, Inc.) at 1:200, cytokeratin 5/6 clones D5/16B4 (Dako Inc.) at 1:200. ER clone1D5 (Dako, Inc) 1:100, PR clone PgR636 (Dako, Inc) at 1:20, EGFR clone 31G7 (Invitrogen, Carlsbad CA) at 1:50 and HER2 clone A0485 (Dako, Inc) at 1:500 dilution. Immunoperoxidase reactions were performed using a Dako Autostainer Plus automated immunostainer according to the manufacturer's instructions. Briefly, the automated steps included blockage of endogenous peroxidase with DakoCytomation Dual Endogenous Enzyme Block and reaction with the primary antibody. Incubation with DakoCytomation EnVision+ Dual Link System-HRP followed the primary antibody reaction. The chromogen was diaminobenzadine for all reactions. Negative controls were performed in the same fashion, except that the primary antibody was substituted with mouse immunoglobulin. Sections of HER2 over expressing breast carcinoma and myometrium provided the receptor antibody positive controls and breast tissue that included normal myoepithelial elements provided the positive control for every other antibody.
Dual Silver in situ Hybridization (SISH)
In order to determine whether cases on the arrays stained with HER2 and scored as 2+ had HER2 gene amplification, dual SISH, which was also used in routine clinical practice in our laboratory, was performed on all the arrays. Since most of the arrays had at least one 2+ staining case and to provide consistency, SISH was performed on all cases in each array. SSH was also necessary to determine if some of the 2+ staining cases were negative for HER2 gene amplification and therefore potentially TN. Because of its use under bright field conditions for FFPE tissues, dual SISH (39) provides a useful alternative to FISH, and was performed using a fully automated protocol of the BenchMark Ultra System (Ventana Medical, Tucson AZ). All reagents, including DNP HER2 and Chromosome 17 centromere (Chr17) probes, and the ultraView SISH and ultraView Alkaline Phosphatase Red ISH detection kits were obtained from Ventana.. The specific hybridization of the HER2 probe to its target was visualized by an insoluble precipitate of silver chromogen using Silver C, a horse radish peroxidase-conjugated multimer and Silver chromogen. The Chr17 probe was sequentially applied and visualized by the soluble precipitate of the alkaline phosphatase-based Fast Red chromogenic system. For visualizing the complete morphology of the tissue, the slides were counterstained with hematoxylin. The black HER2 SISH copies and the Chr17 Red copies were enumerated in the tumor nuclei using brightfield microscopy under a 40x objective. A ratio of HER2 to Chr17 signals or 2.0 was considered amplification. In cases of discordance between IHC and SISH or FISH ( 9 cases, 3.7%) the SISH or FISH result was used to determine the true HER2 status. The reproducibility and concordance of the silver HER2 in situ hybridization with FISH has been previously reported (40).
Evaluation
The TN phenotype was defined as less than 1% of cells in the tumor staining for ER and PR, and by 0 or 1+ staining of HER2 by IHC, or absence of HER2 gene amplification byFISH or Dual SISH. Staining 5% or more of tumor cells for p63, CK5/6, and LN332 was considered as a positive result for each of these markers. Only nuclear staining for p63 and cytoplasmic staining for CK 5/6 and LN332 was considered a positive reaction. Peri-tumoral or stromal staining of LN332 was not evaluated. For EGFR IHC, positivity was defined as any staining of tumor cell membranes or cytoplasm (31). Nottingham scores were evaluated (41). For all assessments, cases were reviewed independently by at least two observers. In cases of initial disagreement, the cases were reviewed together with a double-headed microscope, and a consensus was reached.
Results
Immunohistochemistry
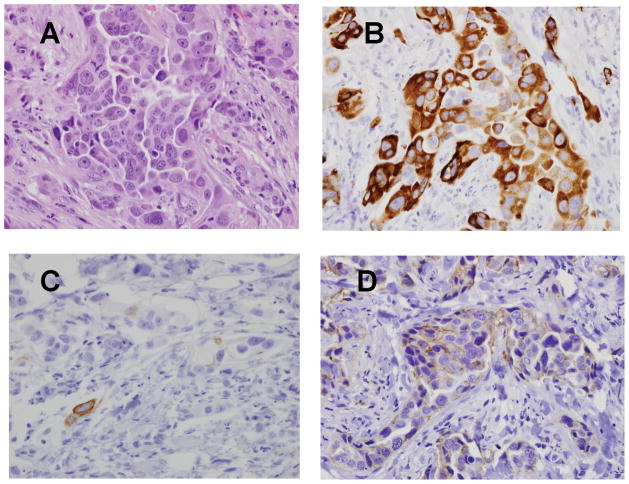
80 cases among 243 were categorized into the TN group. The mean and median Nottingham scores in the TN group were 8.05 and 8 respectively, and the mean and, median scores of in the non- TN group were 6.84 and 7 respectively. In addition to ER, PR and HER2 expression, LN332, CK5/6, EGFR and p63, proteins normally expressed in the basal layer of normal glands (24, 42) were examined. Morphologically, all TN cases were high grade infiltrating ductal adenocarcinomas. Special subtypes of TN carcinomas, such as metaplastic carcinomas were not evaluated in this study, but the expression these markers, including LN332, in metaplastic carcinomas has been reported previously (24). Examples of the pattern of expression of these proteins are illustrated in Fig 1. The numbers of cases in each group expressing each of these histologic markers are listed in the Table. While positive expression of LN332 was identified in the tumor cells of 56 cases (70%) of the 80 TN cases, expression was identified in only 15.2% of the non-TN cases. Cytoplasmic staining was specifically evaluated because it was more likely to be a product of the tumor itself rather than a secretory protein of stromal cells or residual basal epithelium. As we have noted previously (12, 24), LN332 often is more prominent in the cells at the tumor stroma interface, reminiscent of its basal location in normal breast. Among the TN tumors, 65 out of 80 (81.3%) met a commonly used IHC definition of basal phenotype, namely EGFR and CK5/6 expression. This percentage is comparable to the 76% percent of EGFR or CK5/6 staining tumors in a group of basal tumors defined by gene profile analysis (31). Of these 65 basal tumors, 47 (72.3%) were positive for LN332.
Figure 1.
TN Breast carcinoma staining. A. H&E. B. LN332 β3 chain. C. CK5/6 D. EGFR Example of strong LN332 expression but less EGFR or CK5/6 staining. Original magnifications 200X
Table.
The expression of p63, CK5/6, EGFR and LN332 in TN and non-TN breast carcinomas.
| Triple Negative group | Non-TN group | Total Cases | ||
|---|---|---|---|---|
| LN332 | positive | 56 | 25 | 81 |
| negative | 24 | 138 | 162 | |
| EGFR | positive | 51 | 17 | 68 |
| negative | 28 | 117 | 145 | |
| p63 | positive | 17 | 14 | 31 |
| negative | 63 | 149 | 212 | |
| CK5/6 | positive | 38 | 14 | 52 |
| negative | 42 | 149 | 191 | |
| LN332 +CK5/6 +EGFR | positive | 74 | 31 | 105 |
| negative | 6 | 103 | 109 | |
| EGFR +CK5/6 | positive | 65 | 22 | 87 |
| negative | 15 | 112 | 127 |
TN: Triple Negative, CK5/6: cytokeratins 5 and 6, EGFR: epidermal growth factor receptor, LN332: laminin 332
Although only a minority of the non-TN cases were positive for LN332, we examined whether there was any differences between the non TN tumors that were of luminal (ER and or PR positive) and HER2 over-expressing types. Of the 163 non-TN cases, 51 were HER2 over-expressing and 131 expressed ER, PR or both. LN332 was expressed in 14 (27.5%) HER2 expressing carcinomas but only 10 (7.6%) ER and/or PR positive tumors. Of the ER and/or PR positive tumors expressing LN332, 6 (60%) were high grade (Nottingham score 8 or 9), and these higher grade tumors included 3 that over expressed HER2, while only 32%, of the non-LN332 expressing ER/PR positive tumors had a score of 8 or 9.
Discussion
Using an anti-LN332 β chain antibody on a small group of breast carcinomas and by its co-expression with the γ chain in a previous study (24), LN332 expression was noted in 70% of TN breast carcinomas but only 14.7% of non-TN carcinomas.
Although LN 332 expression was identified in a high proportion of TN carcinoma, this association has not been clearly described in some prior studies and a range of conclusions regarding the role of LN332 in breast carcinoma have been proposed. For example, LN332 has been identified in immortalized human breast epithelial cell lines, but is either markedly decreased or not present in mammary carcinoma cell lines (22, 26, 43, 44). A possible explanation for the discrepancy is that many of the cell lines used for these in vitro studies were ER or HER2 expressing cell lines rather than TN lines, or in the case for MDA-MB-435, of melanoma rather than breast origin (45). In earlier reports using surgical specimens of mammary adenocarcinomas studied by immunohistochemistry, LN332 was identified in the basal cell layer of normal mammary glands and in the peritumor stroma in a minority of mammary adenocarcinoma cases (46, 28). In a few small series (22, 26, 28, 43), observations such as these were cited as evidence that LN332 is down-regulated during mammary tumorigenesis, and a potential marker of benign breast disease. In contrast, our study provides evidence that LN332 is often present in one of the more aggressive subsets of mammary adenocarcinoma. The likely reasons for this discrepancy include the small study size and no specification of the receptor status of any of the samples in those investigations. In two other studies of 212 (47) and 220 (46) cases respectively evaluated for LN332 expression by IHC, both reported LN 332 expression in approximately 25% of cases, but one study specified that the majority of those cases had only peritumoral LN332 staining, with cytoplasmic staining in only 5.7% of cases (46). In that series, LN332 expression was identified in a greater proportion of either ER negative, PR negative or HER2 negative carcinomas, but the proportion of tumors negative for all three markers but expressing LN332 was not specified. On the other hand, using gene array analysis, LN332 chain-specific transcripts were identified in a subset of carcinomas identified as having a basal-like gene expression profile (29). We have also previously described that LN332 was highly expressed in 96% of metaplastic carcinomas of the breast (24). Complementary to our study, unspecified laminin reacting to the Novocastra LAM-89 antibody was detected in 42.3% of cases of sporadic basal like breast carcinoma and 80% of tumors from patients with BRCA1 mutations (48). In conclusion, based on the results of this and our previous study, breast carcinoma is among the malignancies that express LN332, and non-expression in some cases is more a reflection of the luminal phenotype rather than the tissue type.
The number of TN cases in the current study is the largest examined for LN332 expression to the best of our knowledge, however the proportion of TN cases was purposefully high since our focus was on this subset of carcinomas. In addition to LN332, we examined the basal markers CK 5/6 and EGFR, which showed that 92.5 % of TN carcinomas were positive for one or more of these three markers. Since LN332 is a basally expressed protein and since gene expression of LN332 chains was included in the gene expression profile definition of the basal phenotype (29), our results raise the possibility that expression of the basal marker LN332 as well as EGFR and CK 5/6 may help in identifying breast carcinoma cases having the basal phenotype. In order to identify the true proportion of LN332 staining tumors with a basal phenotype, future studies with LN332 should include a group of cases which have molecular profiles, with comparison to other IHC basal markers to obtain a panel that best identifies the basal phenotype.
Since the primary goal of this study was to obtain preliminary data on whether LN332 was expressed in breast carcinomas, the cases we used were primarily from tumor arrays without follow up information for the individual tumors on the array to allow us to determine whether LN332 expression had any correlation with prognosis. In an earlier study, a trend toward a worse prognosis was noted in LN332 expressing tumors, but the number of positive cases was too small to reach significance (46). Among metaplastic carcinomas, LN332 expression was greater in patients with lymph node metastases (12). Further more, the observation that about half as many of the non-LN332 expressing ER/PR positive tumors were high grade compared to the LN332 expressing ER/PR positive tumors raises the possibility that LN332 may have an effect on the biological properties of luminal tumors as well. The number in this series is too small, however, to draw definite conclusions. For all the above reasons, it will be worthwhile to examine LN332 expression in a larger cohort with long term follow up.
The higher proportion of HER2 positive tumors expressing LN332 compared with ER and PR positive tumors is reminiscent of earlier reports of HER2 and basal-like marker co-expression in breast carcinomas (35,49). HER2 over expressing breast carcinomas with basal like features may have particularly aggressive characteristics (35). Specifically, Liu et al noted that about one third of HER2 positive breast carcinomas co-expressed a basal marker, a slightly greater proportion than we observed. This group of patients had a poorer 5 year disease free survival and overall survival compared with patients whose tumors were HER2 positive but without basal markers or basal phenotype by IHC for basal cytokeratins or EGFR (35). Further study will be needed with a greater number of patients to determine if LN332 positivity and HER2 over expression show any difference in prognosis.
The question remains whether LN332 expression has any effect on the migratory or invasive phenotype. of breast carcinoma cells that over express LN332. Previous studies on the co-expression of LN332 and its receptors, which might result in autocrine stimulation of migration, suggest this possibility. For example, β4 integrin, one of the components of one of the LN332 receptors, is expressed in 55.6 % of basal breast carcinomas (55). Furthermore, the gene signature of β4-expressing breast carcinomas includes LAMB3 and LAMC2, which encode the β3 and γ2 chains of LN332, respectively. This observation raises the possibility that LN332 might provide an autocrine stimulus for motility or invasion in these cells, although co-expression of LN332 and β4 integrin was not always observed in another study (46). Our previous work showing that metaplastic breast carcinomas also express the α3 integrin, a LN332 receptor that mediates cell motility, is evidence that the co-expression of LN332 and α3 integrin in these cells provides an autocrine stimulus for motility (12). Additional experiments are needed to determine whether the expression of LN332 in TN breast carcinomas affects their migratory or invasive phenotype.
Acknowledgments
The authors thank Brook Bell Smith and colleagues at Ventana for their assistance with SISH protocol development, and Tracy Kingsley, Michelle Chang and Sofia Loera for their technical contributions. The study was funded by the National Cancer Institute Support Grant CA-62203.to the Chao Family Cancer Center, Dr. Frank Meyskens, Principal Investigator.
Grant Support:
The study was funded by the National Cancer Institute Support Grant CA-62203.to the Chao Family Cancer Center, Dr. Frank Meyskens, Principal Investigator.
References
- 1.Sasaki T, Fassler R, Hohenester E. Laminin: the crux of basement membrane assembly. J Cell Biol. 2004;164(7):959–963. doi: 10.1083/jcb.200401058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin GR, Timpl R. Laminin and other basement membrane components. Annu Rev Cell Biol. 1987;3:57–85. doi: 10.1146/annurev.cb.03.110187.000421. [DOI] [PubMed] [Google Scholar]
- 3.Marinkovich MP. Tumour microenvironment: laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 2007;7(5):370–380. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- 4.Patarroyo M, Tryggvason K, Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol. 2002;12(3):197–207. doi: 10.1016/S1044-579X(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 5.Sonnenberg A, de Melker AA, Martinez de Velasco AM, Janssen H, Calafat J, Niessen CM. Formation of hemidesmosomes in cells of a transformed murine mammary tumor cell line and mechanisms involved in adherence of these cells to laminin and kalinin. J Cell Sci. 1993;106 (Pt 4):1083–1102. doi: 10.1242/jcs.106.4.1083. [DOI] [PubMed] [Google Scholar]
- 6.Goldfinger LE, Hopkinson SB, deHart GW, Collawn S, Couchman JR, Jones JC. Theα3 laminin subunit, α6β4 and α3β1 integrin coordinately regulate wound healing in cultured epithelial cells and in the skin. J Cell Sci. 1999;112(Pt 16):2615–2629. doi: 10.1242/jcs.112.16.2615. OK. [DOI] [PubMed] [Google Scholar]
- 7.Kainulainen T, Hakkinen L, Hamidi S, et al. Laminin-5 expression is independent of the injury and the microenvironment during reepithelialization of wounds. J Histochem Cytochem. 1998;46(3):353–360. doi: 10.1177/002215549804600309. [DOI] [PubMed] [Google Scholar]
- 8.Zhang K, Kramer RH. Laminin 5 deposition promotes keratinocyte motility. Exp Cell Res. 1996;227(2):309–322. doi: 10.1006/excr.1996.0280. [DOI] [PubMed] [Google Scholar]
- 9.Ryan MC, Christiano AM, Engvall E, et al. The functions of laminins: lessons from in vivo studies. Matrix Biol. 1996;15(6):369–381. doi: 10.1016/s0945-053x(96)90157-2. [DOI] [PubMed] [Google Scholar]
- 10.Giannelli G, Fransvea E, Bergamini C, Marinosci F, Antonaci S. Laminin-5 chains are expressed differentially in metastatic and nonmetastatic hepatocellular carcinoma. Clin Cancer Res. 2003;9 (10 Pt 1):3684–3691. [PubMed] [Google Scholar]
- 11.Salo S, Haakana H, Kontusaari S, Hujanen E, Kallunki T, Tryggvason K. Laminin-5 promotes adhesion and migration of epithelial cells: identification of a migration-related element in the gamma2 chain gene (LAMC2) with activity in transgenic mice. Matrix Biol. 1999;18(2):197–210. doi: 10.1016/s0945-053x(99)00012-8. [DOI] [PubMed] [Google Scholar]
- 12.Carpenter PM, Dao AV, Arain ZS, et al. Motility induction in breast carcinoma by mammary epithelial laminin 332 (laminin 5) Mol Cancer Res. 2009;7(4):462–75. doi: 10.1158/1541-7786.MCR-08-0148. [DOI] [PubMed] [Google Scholar]
- 13.Tsuji T, Kawada Y, Kai-Murozono M, et al. Regulation of melanoma cell migration and invasion by laminin-5 and α3β1 integrin (VLA-3) Clin Exp Metastasis. 2002;19(2):127–34. doi: 10.1023/a:1014573204062. [DOI] [PubMed] [Google Scholar]
- 14.Fukushima Y, Ohnishi T, Arita N, Hayakawa T, Sekiguchi K. α3β1-mediated interaction with laminin-5 stimulates adhesion, migration and invasion of malignant glioma cells. Int J Cancer. 1998;76(1):63–72. doi: 10.1002/(sici)1097-0215(19980330)76:1<63::aid-ijc11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 15.Condeelis J, Segall JE. Intravital imaging of cell movement in tumours. Nat Rev Cancer. 2003;3(12):921–30. doi: 10.1038/nrc1231. [DOI] [PubMed] [Google Scholar]
- 16.Mareel MM, De Baetselier P, Van Roy FM. Mechanisms of Invasion and Metastasis. Boca Raton, FL: CRC Press; 1991. pp. 73–220. [Google Scholar]
- 17.Kantor JD, Zetter BR. Cell motility in breast cancer. In: Dickson R, Lippman M, editors. Mammary Tumor Cell Cycle, Differentiation and Metastasis. Boston, MA: Kluwer Academic Publishers; 1996. pp. 303–23. [Google Scholar]
- 18.Stracke ML, Aznavoorian SA, Beckner ME, Liotta LA, Schiffmann E. Cell motility, a principal requirement for metastasis. Exs. 1991;59:147–62. doi: 10.1007/978-3-0348-7494-6_10. [DOI] [PubMed] [Google Scholar]
- 19.Miyazaki K. Laminin-5 (laminin-332): Unique biological activity and role in tumor growth and invasion. Cancer Sci. 2006;97(2):91–98. doi: 10.1111/j.1349-7006.2006.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aumailley M, Bruckner-Tuderman L, Carter WG, et al. A simplified laminin nomenclature. Matrix Biol. 2005;24(5):326–332. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Giannelli G, Antonaci S. Biological and clinical relevance of Laminin-5 in cancer. Clin Exp Metastasis. 2000;18(6):439–443. doi: 10.1023/a:1011879900554. [DOI] [PubMed] [Google Scholar]
- 22.Katayama M, Sekiguchi K. Laminin-5 in epithelial tumour invasion. J Mol Histol. 2004;35(3):277–286. doi: 10.1023/b:hijo.0000032359.35698.fe. [DOI] [PubMed] [Google Scholar]
- 23.Guess CM, Quaranta V. Defining the role of laminin-332 in carcinoma. Matrix Biol. 2009;28(8):445–55. doi: 10.1016/j.matbio.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter PM, Wang-Rodriguez J, Chan OT, Wilczynski SP. Laminin 5 expression in metaplastic breast carcinomas. Am J Surg Pathol. 2008;32(3):345–353. doi: 10.1097/PAS.0b013e3181592201. [DOI] [PubMed] [Google Scholar]
- 25.Pyke C, Salo S, Ralfkiaer E, Rømer J, Danø K, Tryggvason K. Laminin-5 is a marker of invading cancer cells in some human carcinomas and is coexpressed with the receptor for urokinase plasminogen activator in budding cancer cells in colon adenocarcinomas. Cancer Res. 1995;55(18):4132–4139. [PubMed] [Google Scholar]
- 26.Martin KJ, Kwan CP, Nagasaki K, et al. Down-regulation of laminin-5 in breast carcinoma cells. Mol Med. 1998;4(9):602–613. [PMC free article] [PubMed] [Google Scholar]
- 27.Holler E. Laminin isoform expression in breast tumors. Breast Cancer Res. 2005;7(4):166–167. doi: 10.1186/bcr1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henning K, Berndt A, Katenkamp D, Kosmehl H. Loss of laminin-5 in the epithelium-stroma interface: an immunohistochemical marker of malignancy in epithelial lesions of the breast. Histopathology. 1999;34(4):305–309. doi: 10.1046/j.1365-2559.1999.00634.x. [DOI] [PubMed] [Google Scholar]
- 29.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van de Rijn M, Perou CM, Tibshirani R, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161(6):1991–6. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen TO, Hsu FD, Jensen K, et al. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 32.Abd El-Rehim DM, Pinder SE, Paish CE, et al. Expression of luminal and basal cytokeratins in human breast carcinoma. J Pathol. 2004;203(2):661–71. doi: 10.1002/path.1559. [DOI] [PubMed] [Google Scholar]
- 33.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 34.Cheang MC, Voduc D, Bajdik C, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008;14(5):1368–76. doi: 10.1158/1078-0432.CCR-07-1658. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Fan Q, Zhang Z, Li X, Yu H, Meng F. Basal-HER2 phenotype shows poorer survival than basal-like phenotype in hormone receptor-negative invasive breast cancers. Hum Pathol. 2008;39(2):167–174. doi: 10.1016/j.humpath.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Fulford LG, Reis-Filho JS, Ryder K, et al. Basal-like grade III invasive ductal carcinoma of the breast: patterns of metastasis and long-term survival. Breast Cancer Res. 2007;9(1):R41. doi: 10.1186/bcr1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jumppanen M, Gruvberger-Saal S, Kauraniemi P, et al. Basal-like phenotype is not associated with patient survival in estrogen-receptor-negative breast cancers. Breast Cancer Res. 2007;9(1):R16. doi: 10.1186/bcr1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang P, Skinner KA, Hicks DG. Molecular classification of breast carcinomas by immunohistochemical analysis: are we ready? Diagn Mol Pathol. 2009;18 (3):125–32. doi: 10.1097/PDM.0b013e31818d107b. [DOI] [PubMed] [Google Scholar]
- 39.Nitta H, Hauss-Wegrzyniak B, Lehrkamp M, et al. Development of automated brightfield double In Situ hybridization (BDISH) application for HER2 gene and chromosome 17 centromere (CEN 17) for breast carcinomas and an assay performance comparison to manual dual color HER2 fluorescence In Situ hybridization (FISH) Diagn Pathol. 2008;3:41. doi: 10.1186/1746-1596-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbone A, Botti G, Gloghini A, et al. Delineation of HER2 gene status in breast carcinoma by silver in situ hybridization is reproducible among laboratories and pathologists. J Mol Diagn. 2008;10(6):527–36. doi: 10.2353/jmoldx.2008.080052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002;41(3A):154–161. [PubMed] [Google Scholar]
- 42.Reis-Filho JS, Simpson PT, Martins A, Preto A, Gärtner F, Schmitt FC. Distribution of p63, cytokeratins 5/6 and cytokeratin 14 in 51 normal and 400 neoplastic human tissue samples using TARP-4 multi-tumor tissue microarray. Virchows Arch. 2003;443(2):122–32. doi: 10.1007/s00428-003-0859-2. [DOI] [PubMed] [Google Scholar]
- 43.Korah R, Das K, Lindy ME, Hameed M, Wieder R. Coordinate loss of fibroblast growth factor 2 and laminin 5 expression during neoplastic progression of mammary duct epithelium. Hum Pathol. 2007;38(1):154–160. doi: 10.1016/j.humpath.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Miller KA, Eklund EA, Peddinghaus ML, et al. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J Biol Chem. 2001;276(46):42863–8. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- 45.Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 melanoma cells--a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104(1):13–9. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- 46.Diaz LK, Cristofanilli M, Zhou X, et al. β4 integrin subunit gene expression correlates with tumor size and nuclear grade in early breast cancer. Mod Pathol. 2005;18(9):1165–75. doi: 10.1038/modpathol.3800411. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez-Angulo AM, Sahin A, Krishnamurthy S, et al. Biologic markers in axillary node-negative breast cancer: differential expression in invasive ductal carcinoma versus invasive lobular carcinoma. Clin Breast Cancer. 2006;7(5):396–400. doi: 10.3816/CBC.2006.n.056. [DOI] [PubMed] [Google Scholar]
- 48.Rodríguez-Pinilla SM, Sarrió D, Honrado E, et al. Vimentin and laminin expression is associated with basal-like phenotype in both sporadic and BRCA1-associated breast carcinomas. J Clin Pathol. 2007;60(9):1006–12. doi: 10.1136/jcp.2006.042143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertucci F, Finetti P, Cervera N, et al. How basal are triple-negative breast cancers? Int J Cancer. 2008;123(1):236–40. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- 50.Lu S, Simin K, Khan A, Mercurio AM. Analysis of integrin β4 expression in human breast cancer: association with basal-like tumors and prognostic significance. Clin Cancer Res. 2008;14(4):1050–8. doi: 10.1158/1078-0432.CCR-07-4116. [DOI] [PubMed] [Google Scholar]