Abstract
LL-23 is a natural peptide corresponding to the N-terminal 23 amino acid residues of human host defense cathelicidin LL-37. LL-23 demonstrated, compared to LL-37, a conserved ability to induce the chemokine MCP-1 in human peripheral blood mononuclear cells, a lack of ability to suppress induction of the pro-inflammatory cytokine TNF-α in response to bacterial lipopolysaccharides (LPS), and reduced antimicrobial activity. Heteronuclear multidimensional NMR characterization of LL-23 revealed similar secondary structures and backbone dynamics in three membrane-mimetic micelles: SDS, dodecylphosphocholine (DPC), and dioctanoylphosphatidylglycerol. The NMR structure of LL-23 determined in perdeuterated DPC contained a unique serine that segregated the hydrophobic surface of the amphipathic helix into two domains. To gain further understanding, Ser9 of LL-23 was changed to either Ala or Val based on homologous primate cathelicidins. These changes made the hydrophobic surface of LL-23 continuous and enhanced antibacterial activity. While identical helical structures did not explain the altered activities, a reduced hydrogen-deuterium exchange from LL-23, LL-23A9, to LL-23V9 suggested a deeper penetration of LL-23V9 into the interior of the micelles, which correlated with enhanced activities. Moreover, these LL-23 variants had discrete immunomodulatory activities. Both restored the TNF-α dampening activity to the level of LL-37. Furthermore, LL-23A9, like LL-23, maintained superior protective MCP-1 production, while LL-23V9 was strongly immunosuppressive, preventing baseline MCP-1 induction and substantially reducing LPS-stimulated MCP-1 production. Thus, these LL-23 variants, designed based on a structural hot spot, are promising immune modulators that are easier to synthesize and less toxic to mammalian cells than the parent peptide LL-37.
Keywords: Antimicrobial peptides, Drug design, immune modulating peptides, micelles, NMR, protein dynamics, structure-activity relationship (SAR)
Naturally occurring antimicrobial peptides (AMPs) are key components of the innate immune systems of all life forms (1–6). The current version of the antimicrobial peptide database (http://aps.unmc.edu/AP/main.html/) contained 1800 AMPs as of October 2011. Among them, 151 entries originated from bacteria, 250 from plants, and 1283 from animals (7, 8). In the case of animal AMPs, 665 were discovered from amphibians, primarily frog skin. Likewise, AMPs such as LL-37, β-defensins, dermcidin, psoriasin (S100A7), and RNase 7 were detected in human skin (9, 10) and some of them can be produced upon UV irradiation (11). LL-37 exists in infant skin as well (12). The protective role of human host defense cathelicidin LL-37 against infection has been demonstrated in animal models (13, 14). In human skin, the activity of LL-37 is regulated by proteases at two levels: generation from its precursor protein hCAP-18 and degradation to smaller fragments, which may, or may not, be biologically active. Several active fragments of LL-37 have been detected in healthy human skin, including LL-23, LL-29, and KS-27 (15). The protease that cleaves hCAP-18 into LL-37 in neutrophils is proteinase 3, while other proteases such as kallikreins are proposed to cut LL-37 into smaller fragments in the skin. Importantly, the peptide cleavage pattern in healthy skin differs from that of diseased skin, suggesting that AMP profiles have potential diagnostic value and/or functional importance (16).
In order to study the structure-activity relationships of the natural fragments of human LL-37, which are poorly understood, we established bacterial expression systems for LL-23, LL-29, and KS-30. While LL-29 and KS-30 were found to be moderately antibacterial, LL-23 was poorly active (MIC > 150 μM) against Escherichia coli K12 (17). The structural basis for the poor antimicrobial activity of LL-23 has not yet been explained. In addition, the poor antibacterial activity of LL-23 suggests that it may have other functional roles. To provide insight into different membrane-mimetic models, we compared the NMR structure and dynamics of LL-23 in the micelles of dodecylphosphocholine (DPC), sodium dodecylsulfate (SDS), and dioctanoyl phosphatidylglycerol (D8PG). Because the results in DPC and D8PG are more similar, and deuterated D8PG is not yet commercially available, we determined the three-dimensional (3D) structure of LL-23 in complex with deuterated DPC micelles by NMR spectroscopy. A unique hydrophilic residue, Ser9, was found on the hydrophobic surface of the 3D structure of micelle-bound LL-23. Interestingly, the same position in the sequences of homologous cathelicidins from primates is occupied by an Ala or Val residue (18). To substantiate the essential role of Ser9, single amino acid residue variants of LL-23 were obtained by changing Ser9 to Ala or Val (Table 1). Because the N-terminal region of LL-37 is implicated in chemotaxis, we also evaluated the immunomodulatory activities of LL-23. Our studies revealed that position 9 of LL-23 is essential in regulating the immune and antimicrobial roles of the peptide. We observed that these natural amino acid variations at residue 9 of LL-23 had a dramatic effect on immunomodulatory properties, opening the door to engineering novel immune modulating peptides using a short fragment of LL-37 as a template. Here we report the structure, dynamics, antimicrobial activity, and immune regulation of LL-23 and its single amino acid variants generated based on non-human primate cathelicidin homologs.
TABLE 1.
Antibacterial Activity of the LL-23 Family against Two E. coli Strains
| Peptide | Amino acid sequencea | Purity (%) | MIC (μM) | |
|---|---|---|---|---|
| E. coli DC2b | E. coli K12c | |||
| LL-23 | LLGDFFRKSKEKIGKEFKRIVQR | 96.27 | 9 | 160 |
| LL23A9 | LLGDFFRKAKEKIGKEFKRIVQR | 96.79 | 9 | 80 |
| LL-23V9 | LLGDFFRKVKEKIGKEFKRIVQR | 95.20 | 2 | 40 |
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | 97.86 | 3 | 5 |
| LL-37A9 | LLGDFFRKAKEKIGKEFKRIVQRIKDFLRNLVPRTES | 99.02 | 6 | 15 |
| LL-37V9 | LLGDFFRKVKEKIGKEFKRIVQRIKDFLRNLVPRTES | 99.67 | 12 | 25 |
Materials and Methods
Peptides
For biological studies, all the peptides (Table 1) were chemically synthesized at isotopic natural abundance and purified to homogeneity (>95% pure) by reverse-phase HPLC (Genemed Synthesis, Inc., TX). The C-termini of these peptides were not amidated. In addition, 15N-labeled LL-23 was expressed in E. coli and purified using the established protocol (17). The recombinant form of LL-23 contained an additional Pro residue at the N-terminus of the peptide, which resulted from formic acid cleavage of the Asp-Pro site in the fusion protein created to release the peptide. Previous studies showed that this additional Pro had no effect on antibacterial activity against E. coli (17).
Antimicrobial Activity Assays
The antibacterial activities of LL-23 and its variants were determined using the standard broth microdilution method as described (19). In brief, a 5 ml culture was grown overnight. A fresh culture (5 mL) was inoculated with a small aliquot of the overnight culture and incubated at 37°C (250 rpm) until the optical density was approximately 0.5. The culture was then diluted to ~106 colony forming unit (CFU) per ml and 90 μL aliquots were placed into a 96-well microplate. The bacteria were treated with a series of peptide solutions (10 μL) with two-fold dilution. The assays were performed in triplet for each peptide. The plate was incubated at 37°C overnight (~16 hours) and read on a ChroMate 4300 Microplate Reader at 630 nm (GMI, Ramsey, MN). The minimal inhibitory concentration (MIC) was defined as the lowest peptide concentration that fully inhibits the bacterial growth (19). In addition, the effect of carbonate anion on peptide activity was evaluated to better mimic physiological conditions (20). Finally, the modified CLSI protocol for antibacterial assays used in initial testing was described elsewhere (21). The method was modified to prevent peptide binding to plastic.
The minimal bactericidal concentration (MBC) was determined by taking the wells just above the MIC (i.e. those that showed inhibition) and plating them for viable colonies. The MBC is the concentration at which no viable colonies can be obtained.
Immunomodulatory Assays
Venous blood from healthy volunteers was collected in Vacutainer collection tubes containing sodium heparin as an anticoagulant (BD Biosciences) in accordance with University of British Columbia ethical approval and guidelines. Isolation of peripheral blood mononuclear cells (PBMC), assessment of potential cytotoxicity by lactate dehydrogenase (LDH) release, stimulation of cells with peptides and bacterial lipopolysaccharide (LPS) and measurement of cytokine release by ELISA were all performed exactly as described previously (22).
Membrane Potential Measurements
Cytoplasmic membrane permeabilization was determined by using the membrane potential sensitive cyanine dye diSC35 (23). The mutant E. coli DC2 with increased outer membrane permeability was used so that diSC35 could reach the cytoplasmic membrane.
NMR Sample Preparations
For structure and dynamics characterization by heteronuclear NMR spectroscopy, 15N-labeled LL-23 and lipids were solubilized in 300 μl water containing 10% D2O as the field-locking signal. In the final sample, the peptide concentration was ~0.5 mM. The LL-23/lipid molar ratios were 1:20, 1:60, and 1:60 for protonated D8PG (Avanti Polar Lipids, Alabaster, AL), deuterated SDS, and deuterated DPC (Cambridge Isotope Laboratories, Andover, MA), respectively. The solution was transferred to a SHIGEMI tube (Tokyo, Japan) for NMR measurements. For determination of three-dimensional structure, NMR samples in 300 μl of a 90% H2O and 10% D2O solution contained 2 mM synthetic peptide (LL-23, LL-23A9 or LL-23V9) and 120 mM deuterated DPC at pH 5.4. These unlabeled samples were used to collect both 2D proton NMR spectra and HSQC spectra at natural abundance. For proton-deuterium (H-D) exchange experiments, the above samples of LL-23, LL-23A9 and LL-23V9 bound to DPC micelles were dried and re-solubilized in 99.9% D2O followed by immediate NMR measurements.
Translational Diffusion Coefficient Measurements
Translational diffusion coefficients (Df) are inversely proportional to the size of the particle. In particular, Df of a peptide changes substantially upon association with membrane-mimetic micelles (24). The Df values for synthetic LL-23 and its variants in complex with micelles were measured at 25°C on a 400-MHz Bruker NMR spectrometer using the 2D DOSY technique (25).
Multidimensional NMR Spectroscopy
For 2D homonuclear 1H NMR studies of unlabeled peptides, a set of spectra, including NOESY, TOCSY, and DQF-COSY (26–28), was recorded for signal assignments at 25°C. To confirm the assignments, data were also collected at 35°C. The 1H spectral width in both dimensions was 8510.6 Hz. To verify the proton assignments and to further refine the structure (29), gradient-enhanced HSQC spectra (30), between 1H and 15N, or 1H and 13C, were also recorded for 2 mM synthetic LL-23 or its variants at natural abundance. The 1H, 15N, and 13C carriers were set at 4.77, 118.27, and 36.37 ppm, respectively. Typically 30 increments (128 scans) and 80 increments (256 scans) were collected for the 15N (spectral width 2,200 Hz) and aliphatic 13C (spectral width 12,000 Hz) dimensions, respectively.
For multidimensional heteronuclear NMR studies, 15N-labeled recombinant LL-23 was utilized. 2D HSQC spectra were collected at a sweep width of 2200 Hz with 100 increments. For signal assignments, 3D 15N-edited NOESY-HSQC and TOCSY-HSQC were collected (31). Residue-specific heteronuclear 15N{1H} NOE values for LL-23 in complex with different micelles were obtained from 2D (1H, 15N) HSQC spectra with and without proton saturation. The 15N longitudinal and transverse relaxation times (T1 and T2) of LL-23 were measured by integrating the proton peaks in the entire amide region. The relaxation delays used for measuring T1 were 0, 0.05, 0.13, 0.25, 0.51, 1, and 2 s, while the relaxation delays used for measuring T2 were 0.01, 0.03, 0.09, 0.19, 0.35, 0.41, and 0.49 s. Chemical shifts were referenced as described (32). Data were recorded on a 600-MHz Varian INOVA NMR spectrometer equipped with a triple-resonance cryogenic probe with a z-axis gradient capability. These data were then transferred to and processed offline on a Silicon Graphics Octane workstation using NMRPipe and analyzed by PIPP (33, 34).
Three-dimensional Structures of the Peptides in Membrane-mimetic Micelles
For structural calculations of LL-23, LL-23A9, and LL-23V9, the major restraints were derived from 2D NOESY spectra (35). For LL-23, additional distance restraints were obtained from 3D 15N-separated NOESY. The cross peaks were integrated by PIPP and converted to distance restraints 1.8–2.8, 1.8–3.8, 1.8–5.0, and 1.8–6.0 Å corresponding to strong, medium, weak, and very weak types of NOE peaks, respectively. Based on 1Hα, natural abundance 15N, 13Cα, and 13Cβ chemical shifts, backbone angles were predicted by using an updated version of TALOS (36). Hydrogen bond restraints were introduced based on H/D exchange experiments, temperature coefficients, and the NOE-derived structures (18). An extended covalent structure was used as starting coordinates. An ensemble of 100 structures was calculated by using the simulated annealing protocol in the Xplor-NIH program (37). Twenty structures were accepted based on the following criteria: no NOE-derived distance violations greater than 0.20 Å, back dihedral angle violations less than 2°, rmsd for bond deviations from ideality less than 0.01 Å, and rmsd for angle deviations from ideality less than 5°. The structures were viewed and analyzed using MOLMOL (38) and PROCHECK (39). The 20 structures and 1H, 13C, and 15N chemical shifts of human LL-23 bound to DPC micelles were deposited with the Protein Data Bank (PDB entry: 2LMF) and BioMagResBank (BMRB entry: 18114).
Results
Antibacterial Activity of LL-23 and Its Variants
Initially, antimicrobial activities were evaluated as MIC against wild type E. coli K12 using a previously described protocol (18). The MIC decreased from LL-23, LL-23A9, to LL-23V9, indicating that the type of residues at position 9 influenced peptide activity (Table 1). To substantiate these small differences in MIC, we utilized a more sensitive E. coli strain DC2, which has a decreased outer membrane permeability barrier and is consequently supersusceptible to peptides (23). LL-23 was found to inhibit this supersusceptible strain at 9 μM, which was only 3-fold increased over the MIC of LL-37 against this same strain (Table 1), indicating that the outer membrane permeability barrier explained in part the very poor ability of LL-23 to kill wild type E. coli. When residue Ser9 was altered to Ala9 (i.e. LL-23A9), a similar MIC was found. A substantial increase in MIC was observed for the LL-23V9 variant (2 μM). For comparison, we also determined the MICs of LL-37 and the corresponding single residue variants using both supersensitive and wild type E. coli strains. Interestingly, the MIC increased slightly from LL-37, LL-37A9 to LL-37V9 (Table 1).
We also measured the antimicrobial activities of LL-23 and its single residue variants against the Gram-positive Staphylococcus aureus UAMS-1, a clinical isolate, and found it to be inactive (MIC > 160 μM). Consistent with this observation, LL-23 and its variants (Table 2) displayed no activity against a wild type S. aureus Rosenbach strain. In the presence of carbonate, only LL-23V9 showed a slightly reduced MIC (12 μM). To better delineate the activity differences of these peptides, we tested a cationic peptide-susceptible S. aureus strain, with an mprF mutation (Table 2). The MICs of both LL-23A9 and LL-23V9 reduced by four fold compared to LL-23 (24 μM). In the presence of physiologically relevant carbonate (20), the bacterium became even more susceptible with the MICs dropped to the 0.35–0.7 μM range for the Ala and Val variants of LL-23. We also measured the minimal bactericidal concentrations (MBC). While the wild type S. aureus strain was not killed at an MBC > 24 μM, the LL-23 variants were able to kill the mprF strain of S. aureus at 3–12 μM. Although the antimicrobial activity of these peptides depended on both bacterial strains and medium conditions (Tables 1 & 2), it is evident that LL-23V9 was more active than LL-23 while LL-23A9 was equivalent vs. E. coli DC2 and more active vs. E. coli K12 or S. aureus.
TABLE 2.
Activity of LL-23 and Its Variants against Staphylococcus aureus
| Peptide | MIC (μM) | MBC (μM) | ||||||
|---|---|---|---|---|---|---|---|---|
| WTa | mprF mutant | WT | mprF mutant | |||||
| −Cb | +C | −C | +C | −C | +C | −C | +C | |
| LL23 | >24 | >24 | 24 | 3 | >24 | >24 | >24 | 12 |
| LL23A9 | >24 | >24 | 6 | 0.35–0.7 | >24 | >24 | 6–12 | 6 |
| LL23V9 | >24 | 12 | 6 | 0.35–0.7 | >24 | >24 | 3–6 | 6–12 |
WT, wild type S. aureus; mprF mutant with multiple peptide resistance gene knocked out (62).
The assay culture contains physiological concentrations of carbonate that generally enhances susceptibility to cationic AMPs (20). In the table, +C denotes the presence of carbonate and −C indicates the absence of that salt.
Chemokine Release and Cytotoxicity
The immunomodulatory activities of the LL-23 variants were tested using PBMC. Previous studies have demonstrated that there are two major immunomodulatory activities of cationic peptides that correlate with protection in animal models, namely their ability to attract immune cells through the production of chemokines like MCP-1 (CCL2) and their ability to suppress the production of pro-inflammatory cytokines like TNF-α in response to bacterial signature molecules like LPS (40, 41). This selective modulation of innate immunity does not per se result in cytotoxicity, and we confirmed here that none of the LL-23 peptides or LPS utilized led to more than 4% of maximal release of cytosolic lactate dehydrogenase in PBMC.
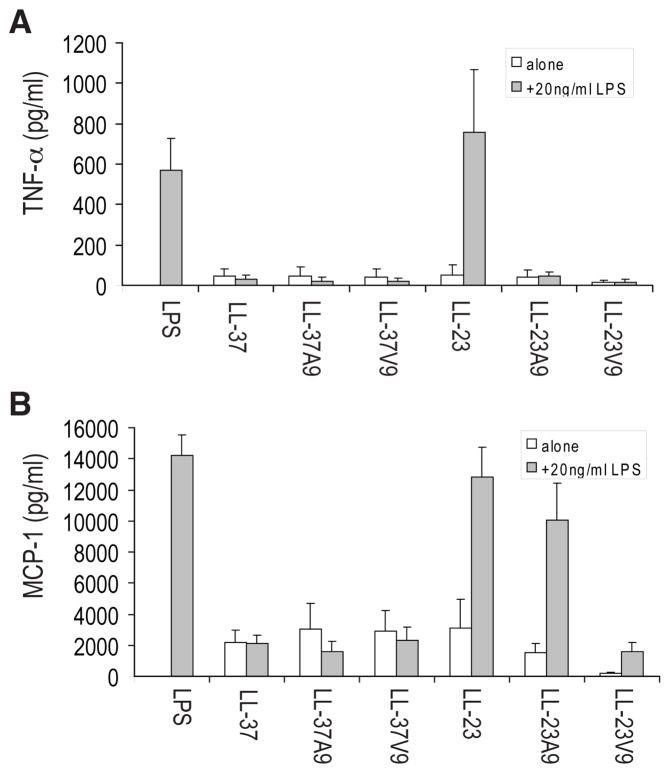
To assess their immunomodulatory activities, cytokine and chemokine release in the presence of LL-23 and its variants was evaluated and compared to LL-37 and its single amino acid substitution variants (Fig. 1). The LL-37 variants were identical to LL-37 in demonstrating no induction of the pro-inflammatory cytokine TNF-α, a moderate ability to induce chemokine MCP-1 (cf. treatment with LPS) and a strong ability to suppress LPS-induced TNFα while maintaining the level of induced MCP-1 (22, 42). In contrast, LL-23 was able to induce moderate levels of MCP-1 but lacked the ability to suppress the release of LPS-induced MCP-1 or TNF-α. However, alteration of Ser9 of LL-23 to either Ala or Val restored the ability to reduce LPS-mediated TNF-α release but had contrasting effects on MCP-1 production. Thus LL-23V9 failed to induce any MCP-1 by itself and strongly suppressed LPS-induced MCP-1 production, indicating that it was a potent anti-inflammatory peptide, while LL-23A9 by itself induced MCP-1 induction but, unlike LL-37 or LL-23, strongly maintained LPS-induced chemokine production.
FIGURE 1. Production of the pro-inflammatory cytokine TNF-α (A) or the chemokine MCP-1 (CCL2) (B) in response to peptides in the presence or absence of bacterial.
LPS. Human PBMC were treated with 20 μg/ml of peptide in the presence or absence of 20 ng/ml of LPS and after 24 h the production of TNF-α or MCP-1 was assessed by ELISA. The data in panel (A) demonstrated that none of the peptides was intrinsically pro-inflammatory and only LL-23 failed to suppress LPS-mediated TNF-α production. The data in panel (B) demonstrated that all of the peptides except LL-23V9 were able to induce substantial levels of baseline MCP-1 but differed in their ability to suppress LPS mediated MCP-1 production. Results represent the means ± SD of 5 experiments.
Membrane Depolarization
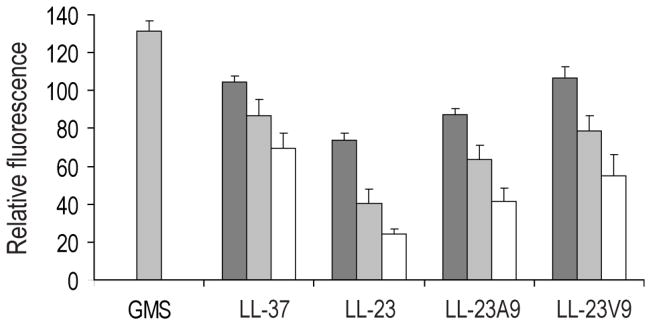
To provide insight into the antimicrobial mechanism of action, the effects of the peptides on bacterial membrane potential was measured in the supersusceptible E. coli DC2 mutant (23). The fluorophore diSC35 is a caged cation that concentrates within the cytoplasmic membrane of bacteria under the influence of the bacterial membrane potential gradient (Δψ); at these high concentrations it self quenches leading to a suppression of fluorescence. When the membrane becomes leaky for cations, including protons, the Δψ is dissipated leading to release of diSC35. As shown for other cationic peptides, LL-37 showed a concentration-dependent fluorescence increase from 3 to 12 μM (Fig. 2), indicating that this peptide was permeabilizing the membrane. At the same peptide concentrations, similar effects were observed with LL-23 and its variants in this supersusceptible strain, indicating that LL-23 did have the potential to act on bacterial membranes and reinforcing the idea that poor permeability across the outer membrane was the basis for the poor activity of this peptide against E. coli. Interestingly, LL-23A9 and LL-23V9 displayed increased permeability and the membrane permeability of LL-23V9 was comparable to LL-37 in this model bacterium (Fig. 2).
FIGURE 2. Depolarizing activity of the LL-37 derived peptides.
E. coli DC2 susceptible strain was used in these experiments (23). The percent dequenching of fluorescence of the membrane-potential sensitive fluorophore diSC35 was assessed as a function of peptide concentrations of 8 (white bars), 16 (light gray bars) and 32 (dark gray bars) μg/ml. (These concentrations correspond to 3, 6, and 12 μM for the LL-23 series and 2, 4, and 8 μM in the case of the LL-37). As a positive control gramicidin S (GMS) was used at 16 μg/ml.
Effects of the Micelle Type on the Structure and Backbone Dynamics of 15N-Labeled LL-23
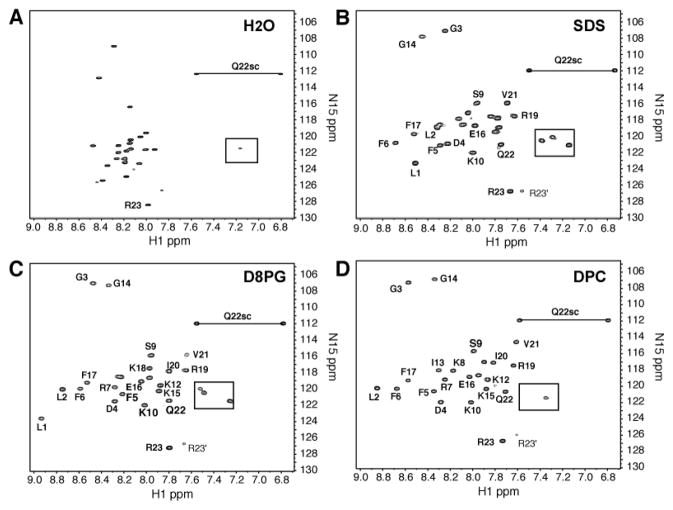
To provide atomic insight into the interactions of the peptide with different membranes, NMR studies of 15N-labeled LL-23 were conducted in three membrane-mimetic micelles: SDS, DPC, and D8PG. While deuterated SDS and DPC are widely utilized (35), D8PG is a newly established model (18, 43). D8PG possesses a phosphatidylglycerol head group, identical to the native anionic lipids in bacterial membranes. In water, the cross peaks of 15N-labeled LL-23 occupied a narrow range of 7.9–8.5 ppm in the proton dimension, indicating a random coiled conformation (Fig. 3A). The cross peaks of the peptide expanded to a wider range of approximately 7.6–8.9 ppm in the presence of micelles (Fig. 3, B–D). In addition, the cross peaks of the peptide became broader. These observations indicated the binding of the peptide to membrane-mimetic micelles and folding into an ordered structure (44). To provide additional evidence for complex formation, the T1 and T2 relaxation times of the peptide were measured in the absence or presence of micelles (18). Based on the T1/T2 ratios, the correlation times (τc) of the peptide with and without micelles were calculated (equation in Table 3). The τc is related to the size of the molecular system under investigation. In water, LL-23 had a τc of 3.5 ns. In the presence of micelles, the correlation times doubled (6.3–7.0 ns). The τc values of the peptide in the bound state were comparable to those previously measured for small proteins (45), indicating a complex formation between the peptide and micelle in each case. While the τc values of the LL-23 peptide measured in SDS and DPC micelles were essentially identical, the value was slightly larger in D8PG micelles (Table 3). It appeared that the peptide/D8PG complex was slightly larger than the complex of the peptide with either SDS or DPC.
FIGURE 3.
HSQC spectra of 15N-labeled LL-23 (A) in water, in complex with (B) SDS, (C) D8PG, or (D) DPC at pH 6 and 35°C. The peptide concentration was ~0.5 mM (1.5 mg/ml). For peptide/lipid ratios and other data, see Table 3. For clarity peaks are partially labeled. There are two sets of peaks for R23. Side-chain signals are boxed or connected by a line.
TABLE 3.
NMR Characterization of 15N-labeled LL-23 in Water or Bound to Micelles
| Detergent/lipid | P/L Ratioa | T1 (ms)b | T2 (ms)b | τc (ns)c |
|---|---|---|---|---|
| Waterd | 1:0 | 804 | 427 | 3.5 |
| SDS | 1:60 | 645±6 | 165±3 | 6.3±0.1 |
| DPC | 1:60 | 669±21 | 169±11 | 6.4±0.1 |
| D8PG | 1:20 | 711 | 154 | 7.0 |
Peptide/lipid molar ratio. All the measurements were conducted at pH 6, 35°C.
Exponential fits to obtain T1 and T2 for each case are of high quality as reflected by a correlation coefficient of 0.99 or better.
The correlation time (τc) was calcualted based on the following relationship: T1/T2 ≈ 0.5ωo2τc2 for proteins (< 25 kDa), where the angular velocity ωo = 2πυo and υo is the precession frequency of 15N (60.785 MHz on our 14.1 Tesla magnet) (63).
Measurements were duplicated in SDS or DPC, but not in water or D8PG.
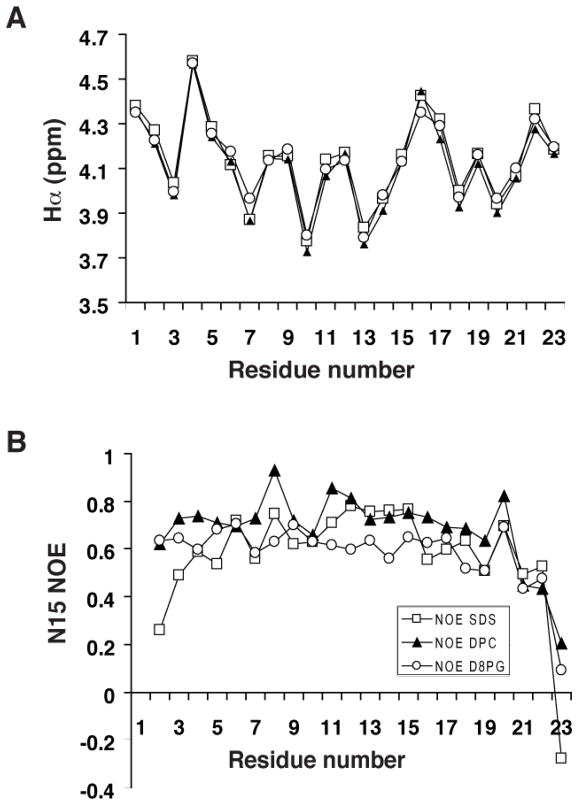
Chemical shifts are sensitive to protein conformation. Therefore, the Hα chemical shifts of LL-23 in SDS, DPC, or D8PG micelles were compared in Fig. 4A. Nearly identical Hα chemical shifts supported similar backbone conformations of the peptide in these micelles. To provide site-specific insight into the backbone dynamics of LL-23 in these model systems, heteronuclear 15N NOEs were measured for each residue of LL-23 in these micelles. In both DPC and D8PG micelles, Fig. 4B indicates that LL-23 was rigid from residues 2 to 20 (15N NOE ~0.6–0.8). There were increased motions for residues 21–22, and residue 23 appeared to be more mobile as indicated by a small or negative 15N NOE. In SDS, residues 2–3 were moderately mobile (15N NOE 0.2–0.5) followed by a rigid helical region between residues 4–20. Nevertheless, the motion pattern for residues 21–23 of the peptide in SDS was similar to those observed in DPC or D8PG micelles. Note that the C-terminal backbone 15N nucleus of Arg23 showed an increased motion from DPC, D8PG, to SDS micelles (Fig. 4B). This was likely due to increased charge repulsions between the negatively charged C-terminal end of LL-23 and the head group of the micelles from DPC (neutral), D8PG (anionic), to SDS (strongly anionic) (46). A different micelle-dependent phenomenon was demonstrated for the N-terminus of the peptide. The strong head group of SDS made the micelle surface more acidic (47), leading to a strong cross peak for Leu1 (visible due to a precedent Pro residue in the recombinant construct) in SDS, moderate in D8PG, and too weak to be detected in DPC owing to rapid exchange at pH 6 (Fig. 3). Meanwhile, the N-terminal amide protons for residues L1, L2, and G3 shifted upfield in SDS (Fig. 3B) compared to those in D8PG or DPC (Fig. 3 C&D), leading to an improved spectral dispersion for those protons in the latter two systems. Dynamically, however, the peptide behaved more similarly in DPC and D8PG micelles (Fig. 4B). In addition, the HSQC spectral dispersions and peak positions of LL-23 in DPC and D8PG were also more comparable (Fig. 3). These results indicated that DPC might be more similar to D8PG than SDS. Because deuterated D8PG is not yet commercially available, we utilized deuterated DPC micelles below as a model for high-quality structural determination of LL-23 and its variants in the membrane-bound state.
FIGURE 4. Structure and dynamics of 15N-labeled LL-23 in complex with SDS, D8PG, or DPC micelles.
(A) The Hα chemical shift plot of LL-23 in SDS (open squares), D8PG (open circles), or DPC (filled triangles). (B) The 15N NOE values of LL-23 measured in complex with SDS, D8PG, or DPC micelles under the conditions described in Fig. 3.
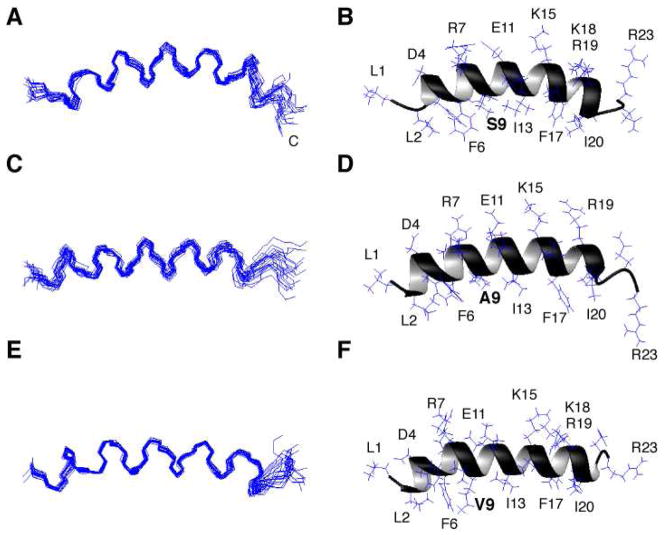
Three-dimensional Structures of LL-23, LL-23A9, and LL-23V9 in Complex with DPC Micelles
The above T1/T2 ratio-based method for calculating correlation times requires isotope labeling of AMPs. In the absence of isotope labeling, translational diffusion coefficients (Df) can be measured as an indication of complex formation whereby the larger the complex, the slower its diffusion (24). The diffusion coefficient for the peptide alone in solution was 1.6 × 10−6 cm2/s, whereas LL-23, LL-23A9, and LL-23V9 in the presence of DPC micelles all had diffusion coefficients around 1.0 × 10−6 cm2/s, indicating a complex formation in all the cases (24).
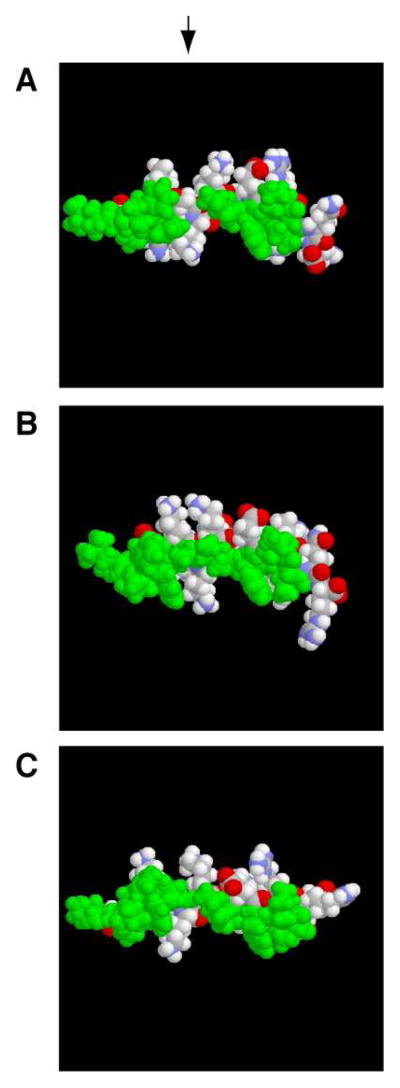
Sequential NMR signal assignments for LL-23 or its variants in complex with DPC micelles were achieved by using the established 2D NMR method (35). In brief, amino acid spin systems were identified in the TOCSY spectrum followed by the establishment of their connectivities via the NOESY spectrum. In each case, the NOE patterns in the fingerprint region contained (i, i+3) and (i, i+4) types of connectivities, typical of helical structures. TALOS (36) analysis of 1Hα, 15N, 13Cα, and 13Cβ chemical shifts of micelle-bound peptides identified a helical region spanning residues 2–21 for LL-23 and its single-residue variants, respectively. The NMR restraints used for determining the 3D structures of LL-23 and its variants are summarized in Table 4. Fig. 5 presents an ensemble of 20 structures for each peptide accepted using the criteria defined in Methods. The root mean square deviations (rmsd) were 0.39, 0.62, and 0.36 Å for LL-23 (A), LL-23A9 (C), and LL-23V9 (E), respectively, when the backbone atoms of residues 2–20 were superimposed. A Procheck analysis (39) of the ensemble of structures showed that 89.5 to 94.7% of the residues of these peptides are located in the most favored region of the Ramachandran plot (Table 4). Thus, these structures were determined to high quality. Detailed statistics for each structural ensemble is summarized in Table 4. As shown in the ribbon diagram of the peptide structure, the hydrophobic surface of LL-23 was segregated by a hydrophilic residue Ser9 (Fig. 5B), whereas both LL-23A9 and LL-23V9 possessed continuous hydrophobic surfaces (Fig. 5, D and F). The continuity of the hydrophobic surfaces (in green) in the case of the LL-23 variants was evident in the colored space-filling models in Fig. 6. Interestingly, a similar hydrophobic gap exists in the 3D structure of intact LL-37 bound to SDS micelles (Fig. 7A).
TABLE 4.
Structural Statistics of Human LL-23 and Its Mutants in DPC Micelles
| Structural restraints | LL-23 | LL-23A9 | LL-23V9 |
|---|---|---|---|
| NOE restraints (total) | 255 | 217 | 257 |
| Intra-residue | 74 | 73 | 54 |
| Sequential | 89 | 76 | 86 |
| Short range | 89 | 68 | 117 |
| Backbone angles (φ/ψ)a | 38 | 42 | 42 |
| Structure and quality | |||
| Helical region | 2–20 | 2–20 | 2–21 |
| Helicity based on structure (%) | 83 | 83 | 87 |
| Helicity based on Ha chemical shifts (%) | 78 | 77 | 79 |
| Backbone rmsd (Å)b | 0.39 | 0.62 | 0.36 |
| NOE-derived distance violations (Å) | < 0.5 | < 0.5 | < 0.5 |
| TALOS-derived angle violations (°) | < 5 | < 5 | < 5 |
| Ramachandran plotc | |||
| Residues in the most favored region (%) | 89.5 | 89.5 | 94.7 |
| Residues in the additional allowed region (%) | 10.5 | 10.5 | 5.3 |
FIGURE 5. Solution structures of human LL-23 and its single residue variants in complex with deuterated DPC at pH 5.4 and 25°C.
Shown are ensembles of 20 backbone structures of LL-23 (A), LL-23A9 (C), and LL-23V9 (E) with amino acid residues 2–20 superimposed, and ribbon diagrams of LL-23 (B), LL-23A9 (D), and LL-23V9 (F) with side chains labeled. In panels B, D, and F, the residue at position 9 is bolded.
FIGURE 6.
Space-filling models of LL-23 (A), LL-23A9 (B) and LL-23V9 (C) in complex with membrane-mimetic DPC micelles. Color code: oxygen, red; nitrogen, blue; carbon and hydrogen, white; and hydrophobic side chains, green. The hydrophobic defect at residue Ser9 of LL-23 (pointed by an arrow) is a hot spot, which formed the basis for structure-based peptide design in panels (B) and (C).
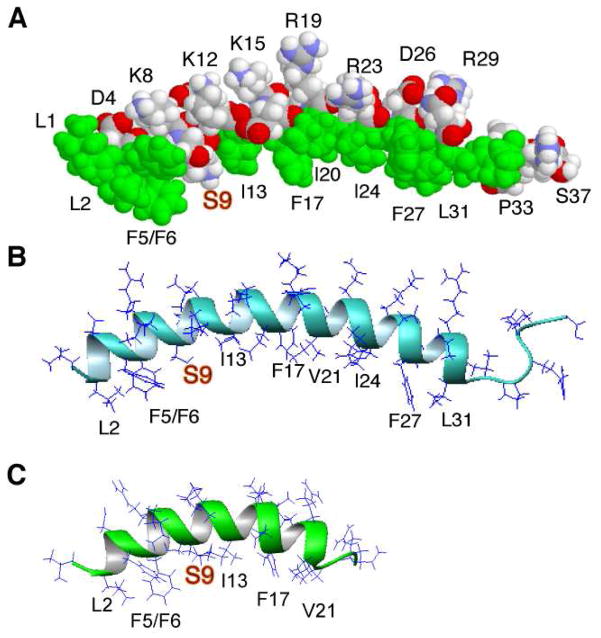
FIGURE 7.
Structural comparison of human LL-37 with LL-23 in complex with micelles. The structure of LL-37 (PDB entry: 2K6O) was reported previously (18). The space-filling and ribbon diagram models of LL-37 are shown in panels (A) and (B), while a ribbon diagram of LL-23 (PDB entry: 2LMF) is given in panel (C). Both LL-37 and LL-23 consist of an amphipathic helix followed by a short disordered tail at the C-terminus. Note that only the amphipathic helix portion is required to associate with bacterial membranes. Also in both structures, a hydrophilic Ser9 (in gold) is located on the hydrophobic surface, leading to two hydrophobic domains in each. The two-domain structure (18) explains the cooperative LPS binding of LL-37 (56) and weak LPS binding of LL-23 (see the text).
Hydrogen-deuterium Exchange Dynamics Explains Peptide Activity Differences
Since the 3D structures of LL-23, LL-23A9, and LL-23V9 were similar, we also investigated the dynamic aspect of these peptides bound to DPC micelles based on H-D exchange experiments. This was performed by removing the water in the NMR samples followed by adding the same volume of deuterium oxide. As a consequence of H-D exchanges, those exchangeable proton signals of the peptides became weaker and eventually disappeared from proton-detected NMR spectra. In the case of LL-23, the amide proton signals of terminal residues 2–5 and 22–23 disappeared in 20 min. After 21 h, only the backbone amide proton signals for residues I13, F17, and I20 of the peptide remained (Table 5). All the backbone amide signals of LL-23 disappeared within 6 days. After 21 h, for LL-23A9, only the amide proton signals of A9, K10, I13, and F17 were detected, while in the case of LL-23V9, only the peaks for V9, K10, K12, I13, and F17 were detected. At 72 h, only the peaks of I13 and V9 of LL-23V9 remained. The signal of I13 was even observed for LL-23A9 and LL-23V9 after two weeks. Overall, there was a trend of slowed exchange of increasing residues from LL-23, LL-23A9, to LL-23V9 after 21 h (Table 5). The slowed exchange of the amide protons of the LL-23 variants might reflect their deeper penetration into the DPC micelles. The increased membrane penetration ability from LL-23, LL-23A9 to LL-23V9 was in line with their increased peptide hydrophobicity according to HPLC retention times (Table 5) (49). Remarkably, the exchange dynamics of LL-23 and its variants was fully consistent with their antimicrobial activities. We propose that deeper penetration of LL-23V9 into the bacterial cytoplasmic membrane was important for its enhanced antibacterial activity against both the supersusceptible and normal strains of Gram-negative E. coli (Table 1).
TABLE 5.
Summary of H-D Exchange Results of Amide Protons in Three Peptide/DPC Micelles after 21 Hours
| Peptide | Sequence pattern of slowly exchanged amides (bold and underlined) | Amides detected | HPLC retention time (min)a |
|---|---|---|---|
| LL-23 | LLGDFFRKSKEKIGKEFKRIVQR | 3 | 9.702 |
| LL-23A9 | LLGDFFRKAKEKIGKEFKRIVQR | 4 | 10.331 |
| LL-23V9 | LLGDFFRKVKEKIGKEFKRIVQR | 5 | 10.817 |
The retention time of the peptide was measured on a Waters HPLC system equipped with an analytical reverse-phase Waters symmetry C8 column (150 × 3.9 mm). The peptide detected at 215 nm was eluted with a gradient of acetonitrile (containing 1% TFA) from 5% to 95% at a flow rate of 1 ml/min.
Discussion
This study provided unique insight into the structure, dynamics, and functions of LL-23, a natural derivative of human cathelicidin LL-37, and of the differential activities of hypothesized primate variants. The structure and dynamics of LL-23 were similar in SDS, DPC, and D8PG micelles, indicating that these membrane-mimetic micelles had little impact on the peptide conformation. A helical structure was found between residues 2 and 20 of the peptide with the three C-terminal residues disordered. The increased motions at the C-terminus of LL-23 could be attributed to peptide chain truncation, as these residues are structured in intact LL-37 (Fig. 7B). Mobile residues were also observed at the C-terminus of LL-37 and they were not required for membrane binding (Fig. 7). Except for the disordered C-terminus, the LL-23 structure in DPC micelles was identical to that in the high-resolution structure of intact LL-37 bound to SDS micelles (Fig. 7). The fact that both the structures of LL-23 (this study) and LL-37 (18) were independent of micelle types led us to conclude that the differences between the two LL-37 structures, one determined in DPC micelles by 2D NMR (48) and the other in SDS micelles determined by 3D NMR (18), might stem from the insufficient spectral resolution in the 2D NMR case as initially pointed out by Li et al. (49). In the high-resolution structures of both the SDS-bound LL-37 and DPC-bound LL-23 (Fig. 7), the helix started from residue 2 and the aromatic rings of F5 and F6 were perpendicular to each other as a result of aromatic-aromatic interactions.
Interestingly, structural determination revealed a hydrophobic defect in the structure of LL-23, whereby a hydrophilic Ser9 segregated the hydrophobic surface into two clusters (Fig. 6). However, there was no increased motion at Ser9 based on heteronuclear 15N NOE measurements (Fig 4B). Therefore, the poor activity of LL-23 was not due to increased motion at that position on the ps-ns time scale. Similar results were observed in the high-quality structure of intact LL-37 (Fig. 7A) (18). Zelezetsky and colleagues sequenced cathelicidins from more than a dozen primates (50). Sequence comparison revealed that at position 9, human LL-37 possesses a Ser residue, while the same position in non-human primate cathelicidins is an Ala or Val residue (18). Our results indicated that a change of hydrophilic Ser9 on the hydrophobic surface of LL-23 to hydrophobic Ala9 or Val9 did not change the backbone structure of the peptide (Fig. 5). Thus, the activity differences of LL-23 and its variants could not be explained at the backbone level of the 3D structure in this case. This observation revealed the challenge in correlating 3D structure of AMPs with antimicrobial activity. What then was responsible for the activity difference? We observed that the H-D exchange rates were reduced when Ser9 of LL-23 was substituted by Ala9 or Val9. Therefore, the existence of hydrophilic Ser9 on the hydrophobic surface did not allow the LL-23 to penetrate as deeply into the membrane environment, consistent with its poor ability to kill even supersusceptible or carbonate treated bacteria (Table 1) or polarize the bacterial membranes. In contrast, when hydrophilic Ser9 was substituted with hydrophobic Val9, the reduced exchange rate corresponded to higher antimicrobial activity as well as improved membrane polarizing ability (Fig. 2). Against wild type E. coli K12, the negligible antimicrobial activity of LL-23 was clearly a function of its poor ability to penetrate the outer membrane and possibly its sensitivity to rapid efflux from the cell. When this barrier was overcome by using an outer membrane altered supersusceptible strain DC2, the residual antimicrobial activity of LL-23 could be attributed to a large extent to the hydrophobic gap in the 3D structure. This hydrophobic gap, an inherent structural defect, split the hydrophobic surface of LL-23 into two portions (Fig. 6A), thereby weakening its capability of membrane penetration and depolarization.
Besides direct microbial killing, LL-37 is known to regulate immune response. It is now well understood that in addition to surface receptors, LL-37 is able to rapidly traverse into mammalian cells in a manner reminiscent of cell penetrating peptides (51) and interact with cytoplasmic interaction partners in a manner that determines its immunomodulatory properties (52). In particular, the N-terminal region of LL-37 is implicated in direct chemokine activity (53). Unlike the potent anti-inflammatory activity of LL-37, this study showed that LL-23 had a weak ability in suppressing the LPS-induced release of either TNF-α or MCP-1, which was restored in the case of the primate-specific sequence variant peptides. This suppression activity of LL-37 is related to its LPS-binding ability (54, 55). Our previous NMR characterization mapped the LPS binding region of LL-37 to residues 2–31 (18). Since Ser9 separates the hydrophobic surface of LL-37 into two domains (Fig. 7A), our 3D structure explains the cooperative LPS binding observed earlier (56). One such domain was located between residues 15–32 of LL-37 (57). Because LL-23 contained the short N-terminal LPS-binding domain and part of the strong LPS-binding domain (Fig. 5B), it is not surprising that this peptide was unable to effectively suppress LPS-induced TNF-α release (Fig. 1A). However, a change of Ser9 of LL-23 to Ala9 or Val9 reduced TNF-α release, presumably by enhancing its LPS binding ability. The continuous hydrophobic surface of the LL-23 variants (Fig. 6) might also explain in part the increased suppression of LPS-induced release of chemokine MCP-1 (Fig. 1B). However, the effects of LL-37 in antagonizing LPS-stimulation of pro-inflammatory cytokine expression are only partly dependent on LPS binding (22, 54). Thus we assume that these effects reflected both relative ability to bind to LPS and direct influence on signaling.
Importantly, this series of peptide variants offered very different immunomodulatory activities with LL-23 being able to induce chemokine production but no anti-inflammatory effect, LL-23V9 having a potent anti-inflammatory activity but no chemokine production ability, and LL-23A9 having not only the anti-inflammatory activity and chemokine induction abilities of LL-37 but being able to additionally maintain the protective chemokine response to LPS. Our studies indicated that these substantially shorter variant peptides not only shared properties with their parent peptide LL-37, but also provided the starting points for optimizing peptides with rather distinct and useful immunomodulatory activities. These important findings opened the door to peptide engineering using LL-23 as a model, which clearly possessed favorable properties compared to LL-37 in several aspects. First, LL-23 has a shortened sequence, which is less costly to synthesize. Second, LL-23 showed low toxicity to mammalian cells due to sequence truncation that disrupted the strong antimicrobial region of LL-37 (49, 58). Third, the short sequence of LL-23 also facilitated detailed immune modulation studies of additional peptide variants. Since these LL-23 variants interacted well with both anionic and neutral membrane mimics, it was tempting to speculate that their differences in membrane-interaction dynamics and surface properties are responsible for these properties by influencing their ability to be taken up into host cells and their interaction with discrete receptors.
In summary, our structure-activity relationship studies provided important insight into the functional roles of LL-23. It is a peptide with weak antimicrobial activity as well as weak suppression of pro-inflammatory cytokine induced by LPS. These poor activities, to a large extent, might be attributable to the segregation of the hydrophobic surface of LL-23 into two small hydrophobic clusters by Ser9 (i.e. a structural hot spot) (Fig. 6). It is understandable now that mutations at that hot spot of LL-23 drastically affected peptide activities. Interestingly, similar single-residue variants of LL-37, unlike LL-23, did not show distinct differences in immune modulations (Fig. 1). Such differences between LL-23 and LL-37 in response to single residue changes were not determined by the 3D structures (Fig. 7), because the structures for residues 2–21 in both peptides were identical, including the location of hydrophilic Ser9 on the hydrophobic surface (Figs. 6A & 7A). Rather, we might attribute the difference to the truncation of the LL-37 sequence that disrupted the strong LPS-binding region (57). In the presence of that region, the effect of the Ser9 to Val9 substitution on the immune modulating function of LL-37 could have been masked. In the case of LL-23, the same replacement drastically altered the properties of the molecule as a consequence of the merging of the two isolated and small hydrophobic moieties into a continuous and stronger membrane, endotoxin (LPS), or receptor-binding surface (see Fig. 6). Such a sequence difference between LL-23 and LL-37 underscored peptide-dependent functional outcomes even though they share the same structure in the corresponding region.
Conclusions
The significance of host defense peptide LL-37 to human health is now firmly established. Differential expression of this peptide is associated with a variety of diseases, including cancer (13, 59). The 3D structure of human LL-37 provides insight into its antimicrobial and cooperative LPS-binding properties (18, 58). The biology of human LL-37 is further complicated by protease-generated fragments for which structure and function are poorly understood. This heteronuclear NMR study of the structure and dynamics of LL-23 is an important step toward the understanding of its functional roles. Our results of LL-23 in membrane-mimetic micelles (SDS, DPC, or D8PG) made the trend in NMR chemical shifts, peptide structure, and dynamics more understandable. For example, the magnitude of motion of the C-terminal residue on the ps-ns time scale was in the following order: SDS > D8PG > DPC (Fig. 4B), which might be due to increased negative charge repulsions between the peptide and micelles.
In DPC micelles, LL-23, LL-23A9, and LL-23V9 shared the same helical structure (Fig. 5), which made a direct structure-activity correlation unsuccessful at the backbone level. Nevertheless, these peptides differed at a critical side chain, which was sufficient to cause them to interact with membranes differently (Table 5 and Fig. 2). In particular, they displayed different exchange dynamics, which provided a means for correlation with antimicrobial activity. LL-23 with a defect on the hydrophobic surface (Fig. 7) was unable to effectively penetrate bacterial outer membranes and interacted poorly with outer membrane defective bacteria. However, a substitution of Ser9 of LL-23 by a hydrophobic residue Val, as observed in homologous primate cathelicidins, led to a continuous hydrophobic surface on LL-23V9, enabling a deeper penetration into the bacterial membranes of outer membrane defective bacteria to exert its damaging effects. Our H-D exchange data also indicate that residue alteration was directly responsible for this enhanced ability and caused a shift of the slowed exchanged residues from the peptide C-terminus (Val 21 disappeared) to the hot spot (residue 9 remained) (Table 5). In addition, while LL-23 had modest chemokine inducing activity, its variants showed distinct and potent immunomodulatory activities (Fig. 1). Thus, a very minor sequence change on the common structural scaffold of LL-23 led to peptides with dramatically different and potent immunomodulatory properties as a consequence of their differences in interactions with cells dynamically. In conclusion, a single amino acid change on the hydrophobic surface of LL-23 substantially modulated its antimicrobial as well as immune modulating activities. In contrast, a similar modification on LL-37 did not alter its immune modulating activities but only reduced its antimicrobial activity slightly (Fig. 1 and Table 1). We propose that the LL-23 family investigated here represents interesting templates for designing distinct and new immune modulating peptides. Because these peptides primarily modulate the human immune system, there is little chance for bacteria to develop resistance. In particular, our observation that LL-23V9 by itself was unable to induce MCP-1 might be of use in designing anti-inflammatory peptides to treat inflammatory diseases such as asthma, rheumatoid arthritis, inflammatory bowel disease, and atherosclerosis (3, 60, 61).
Acknowledgments
We thank Dr. Nicole Kruse (Bruker Biospin) for assistance in implementation of the DOSY technique, and Dr. Biswajit Mishra (UNMC) for HPLC measurements.
Abbreviations
- AMPs
antimicrobial peptides
- CLSI
Clinical and Laboratory Standards Institute
- DPC
dodecylphosphocholine
- D8PG
dioctanoyl phosphatidylglycerol
- ELISA
enzyme-linked immunoabsorbent assay
- HSQC
heteronuclear single quantum coherence spectroscopy
- LDH
lactate dehydrogenase
- LPS
lipopolysaccharides
- MCP-1
monocyte chemotactic protein-1
- MBC
minimal bactericidal concentration
- MIC
minimal inhibitory concentration
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser enhancement
- PBMC
human peripheral blood mononuclear cells
- rmsd
root mean square deviations
- SDS
sodium dodecylsulfate
- TNF-α
tumor necrosis factor alpha
- TOCSY
total correlated spectroscopy
Footnotes
Research grants from the NIH to GW and CIHR to REWH are gratefully acknowledged. In addition, the Nebraska Research Initiative funding to GW allowed for the purchase of a new Bruker Avance III 400-MHz NMR spectrometer and a HPLC system. REWH holds a Canada Research Chair.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Boman HG. Antibacterial peptides: basic facts and emerging concepts. J Inter Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- 3.Easton DM, Nijnik A, Mayer ML, Hancock REW. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009;27:582–590. doi: 10.1016/j.tibtech.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opinion Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 5.Zanetti M. The role of cathelicidins in the innate host defenses of mammals. Curr Issues Mol Biol. 2005;7:179–196. [PubMed] [Google Scholar]
- 6.Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z, Wang G. APD: the antimicrobial peptide database. Nucleic Acids Res. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci. 2006;63:469–486. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamasaki K, Gallo RL. Antimicrobial peptides in human skin disease. Eur J Dermatol. 2008;18:1–11. doi: 10.1684/ejd.2008.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser R. Research in practice: Antimicrobial peptides of the skin. J Dtsch Dermatol Ges. 2011;9:678–680. doi: 10.1111/j.1610-0387.2011.07708.x. [DOI] [PubMed] [Google Scholar]
- 12.Marchini G, Lindow S, Brismar H, Stabi B, Berggren V, Ulfgren AK, Lonne-Rahm S, Agerberth B, Gudmundsson GH. The newborn infant is protected by an innate antimicrobial barrier: peptide antibiotics are present in the skin and vernix caseosa. Br J Dermatol. 2002;147:1127–1134. doi: 10.1046/j.1365-2133.2002.05014.x. [DOI] [PubMed] [Google Scholar]
- 13.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 14.Lee PH, Ohtake T, Zaiou M, Murakami M, Rudisill JA, Lin KH, Gallo RL. Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proc Natl Acad Sci USA. 2005;102:3750–3755. doi: 10.1073/pnas.0500268102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamasaki K, Schauber J, Coda A, Lin H, Dorschner RA, Schechter NM, Bonnart C, Descargues P, Hovnanian A, Gallo RL. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 2006;20:2068–2080. doi: 10.1096/fj.06-6075com. [DOI] [PubMed] [Google Scholar]
- 16.Yamasaki K, Di Nardo A, Bardan A, Murakami M, Ohtake T, Coda A, Dorschner RA, Bonnart C, Descargues P, Hovnanian A, Norhenn VB, Gallo RL. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nature Med. 2007;13:975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Li X, Wang G. On-resin cleavage of bacterially expressed fusion proteins for purification of active recombinant peptides SK-29, KR-20, LL-29, and LL-23 from human sweat or skin. Protein Expr Purif. 2007;55:395–405. doi: 10.1016/j.pep.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 18.Wang G. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J Biol Chem. 2008;283:32637–32643. doi: 10.1074/jbc.M805533200. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Li Y, Li X. Correlation of three-dimensional structures with the antibacterial activity of a group of peptides designed based on a nontoxic bacterial membrane anchor. J Biol Chem. 2005;280:5803–5811. doi: 10.1074/jbc.M410116200. [DOI] [PubMed] [Google Scholar]
- 20.Dorschner RA, Lopez-Garcia B, Peschel A, Kraus D, Morikawa K, Nizet V, Gallo RL. The mammalian ionic environment dictates microbial susceptibility to antimicrobial defense peptides. FASEB J. 2006;20:35–42. doi: 10.1096/fj.05-4406com. [DOI] [PubMed] [Google Scholar]
- 21.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 22.Mookherjee N, Brown KL, Bowdish DME, Doria S, Falsafi R, Hokamp K, Roche FM, Mu R, Doho GH, Pistolic J, Powers J, Bryan J, Brinkman FSL, Hancock REW. Modulation of the TLR-mediated inflammatory response by the endogenous human host defense peptide LL-37. J Immunol. 2006;176:2455–2464. doi: 10.4049/jimmunol.176.4.2455. [DOI] [PubMed] [Google Scholar]
- 23.Wu M, Hancock REW. Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J Biol Chem. 1999;274:29–35. doi: 10.1074/jbc.274.1.29. [DOI] [PubMed] [Google Scholar]
- 24.Keifer PA, Peterkofsky AP, Wang G. Effects of detergent alkyl chain length and chemical structure on the properties of a micelle-bound bacterial membrane targeting peptide. Anal Biochem. 2004;331:33–39. doi: 10.1016/j.ab.2004.03.074. [DOI] [PubMed] [Google Scholar]
- 25.Morris KF, Johnson CS. Diffusion-ordered two-dimensional nuclear magnetic resonance spectroscopy. J Am Chem Soc. 1992;114:3139–3141. [Google Scholar]
- 26.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. J Chem Phys. 1979;71:4546–4553. [Google Scholar]
- 27.Bax A, Davis DG. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 28.Rance M, Sørensen OW, Bodenhausen G, Wagner G, Enrst RR, Wüthrich K. Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem Biophys Res Commun. 1983;117:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 29.Wang G. Structural biology of antimicrobial peptides by NMR spectroscopy. Curr Org Chem. 2006;10:569–581. [Google Scholar]
- 30.Kay LE, Keifer PA, Saarinen T. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc. 1992;114:10663–10665. [Google Scholar]
- 31.Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 32.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J Biomol NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J Magn Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York: 1986. [Google Scholar]
- 36.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 37.Schwieters CD, Kuszewski J, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 38.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 40.Scott MG, Dullaghan E, Mookherjee N, Glavas N, Waldbrook M, Thompson A, Wang A, Lee K, Doria S, Hamill P, Yu J, Li Y, Donini O, Guarna MM, Finlay BB, North JR, Hancock REW. An anti-infective peptide that selectively modulates the innate immune response. Nature Biotech. 2007;25:465–472. doi: 10.1038/nbt1288. [DOI] [PubMed] [Google Scholar]
- 41.Nijnik A, Madera L, Ma S, Waldbrook M, Elliott MR, Easton DM, Mayer ML, Mullaly SC, Kindrachuk J, Jenssen H, Hancock REW. Synthetic cationic peptide IDR-1002 provides protection against bacterial infections through chemokine induction and enhanced leukocyte recruitment. J Immunol. 2010;184:2539–2550. doi: 10.4049/jimmunol.0901813. [DOI] [PubMed] [Google Scholar]
- 42.Bowdish DM, Davidson DJ, Speert DP, Hancock REW. The human cationic peptide LL-37 induces activation of the extracellular signal-regulated kinase and p38 kinase pathways in primary human monocytes. J Immunol. 2004;172:3758–3765. doi: 10.4049/jimmunol.172.6.3758. [DOI] [PubMed] [Google Scholar]
- 43.Wang G, Keifer PA, Peterkofsky A. Solution structure of the N-terminal amphitropic domain of Escherichia coli glucose-specific enzyme IIA in membrane-mimetic micelles. Protein Sci. 2003;12:1087–1096. doi: 10.1110/ps.0301503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Keifer PA, Peterkofsky A. Short-chain diacyl phosphatidylglycerols: which one to choose for NMR structural determination of a membrane-associated peptide from Escherichia coli? Spectroscopy. 2004;18:257–264. [Google Scholar]
- 45.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Foeman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homolog 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 46.Wang G. NMR studies of a model antimicrobial peptide in the micelles of SDS, dodecylphosphocholine, or dioctanoylphosphatidylglycerol. Open Magn Reson J. 2008;1:9–15. [Google Scholar]
- 47.Wang G, Treleaven D, Cushley RJ. Conformation of human serum apolipoprotein A-I(166–185) in the presence of sodium dodecyl sulfate or dodecylphosphocholine by 1H-NMR and CD. Evidence for specific peptide-SDS interactions. Biochim Biophys Acta. 1996;1301:174–184. doi: 10.1016/0005-2760(96)00037-9. [DOI] [PubMed] [Google Scholar]
- 48.Porcelli F, Verardi R, Shi L, Henzler-Wilderman KA, Ramamoorthy A, Veglia G. NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry. 2008;47:5565–5572. doi: 10.1021/bi702036s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li X, Li Y, Han H, Miller DW, Wang G. Solution structures of human LL-37 fragments and NMR-based identification of a minimal membrane-targeting antimicrobial and anticancer region. J Am Chem Soc. 2006;128:5776–5785. doi: 10.1021/ja0584875. [DOI] [PubMed] [Google Scholar]
- 50.Zelezetsky I, Pontillo A, Puzzi L, Antcheva N, Segat L, Pacor S, Crovella S, Tossi A. Evolution of the primate cathelicidin. Correlation between structural variations and antimicrobial activity. J Biol Chem. 2006;281:19861–19871. doi: 10.1074/jbc.M511108200. [DOI] [PubMed] [Google Scholar]
- 51.Lau YE, Rozek A, Scott MG, Goosney DL, Davidson DJ, Hancock REW. Interaction and cellular localization of the human host defense peptide LL-37 with lung epithelial cells. Infect Immun. 2005;73:583–591. doi: 10.1128/IAI.73.1.583-591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mookherjee N, Lippert DND, Hamill P, Falsafi R, Nijnik A, Kindrachuk J, Pistolic J, Gardy J, Miri P, Naseer M, Foster LJ, Hancock REW. Intracellular receptor for human host defense peptide LL-37 in monocytes. J Immunol. 2009;183:2688–2696. doi: 10.4049/jimmunol.0802586. [DOI] [PubMed] [Google Scholar]
- 53.Braff MH, Hawkins MA, Di Nardo A, Lopez-Garcia B, Howell MD, Wong C, Lin K, Streib JE, Dorschner R, Leung DY, Gallo RL. Structure-function relationships among human cathelicidin peptides: dissociation of antimicrobial properties from host immunostimulatory activities. J Immunol. 2005;174:4271–4278. doi: 10.4049/jimmunol.174.7.4271. [DOI] [PubMed] [Google Scholar]
- 54.Mookherjee N, Wilson HL, Doria S, Popowych Y, Falsafi R, Yu J, Li Y, Veatch S, Roche FM, Brown KL, Brinkman FS, Hokamp K, Potter A, Babiuk L, Griebel PJ, Hancock REW. Bovine and human cathelicidin cationic host defense peptides similarly suppress transcriptional responses to bacterial lipopolysaccharide. J Leuko Biol. 2006;80:1563–1774. doi: 10.1189/jlb.0106048. [DOI] [PubMed] [Google Scholar]
- 55.Rosenfeld Y, Papo N, Shai Y. Endotoxin (lipopolysaccharide) neutralization by innate immunity host-defense peptides. J Biol Chem. 2006;281:1636–1643. doi: 10.1074/jbc.M504327200. [DOI] [PubMed] [Google Scholar]
- 56.Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI. Activities of LL-37, a cathelin-associated antimicrobial peptides of human neutrophils. Antimicrob Agents Chemother. 1998;42:2206–2214. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagaoka I, Hirota S, Niyonsaba F, Hirata M, Adachi Y, Tamura H, Tanaka S, Heumann D. Augmentation of the lipopolysaccharide-neutralizing activities of human cathelicidin CAP18/LL-37-derived antimicrobal peptides by replacement with hydrophbic and cationic amino acid residues. Clin Diagn Lab Immunol. 2002;9:972–982. doi: 10.1128/CDLI.9.5.972-982.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang G, Epand RF, Mishra B, Lushnikova T, Thomas VC, Bayles KW, Epand RM. Decoding the functional roles of cationic side chains of the major antimicrobial region of human cathelicidin LL-37. Antimicrob Agents Chemother. 2011 doi: 10.1128/AAC.05637-11. doi: 10.1128/AAC.05637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu WKK, Wang G, Coffelt SB, Betancourt AM, Lee CW, Yu J, Sung JJY, Cho CH. Emerging roles of the host defense peptide LL-37 in human cancer and its potential therapeutic applications. Int J Cancer. 2010;127:1741–1747. doi: 10.1002/ijc.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang G. Antimicrobial Peptides: Discovery, Design and Novel therapeutic Strategies. CABI; Wallingford, England: 2010. [Google Scholar]
- 61.Edfeldt K, Agerberth B, Rottenberg ME, Gudmundsson GH, Wang XB, Mandal K, Xu Q, Yan ZQ. Arterioscler Thromb Vasc Biol. 2006;26:1551–1557. doi: 10.1161/01.ATV.0000223901.08459.57. [DOI] [PubMed] [Google Scholar]
- 62.Staubiz P, Neumann H, Schneider T, Wiedemann I, Peschel A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol Lett. 2004;231:67–71. doi: 10.1016/S0378-1097(03)00921-2. [DOI] [PubMed] [Google Scholar]
- 63.Van de Ven FJM. Multidimensional NMR in Liquids. WILEY-VCH; N.Y: 1995. [Google Scholar]