Abstract
Exome sequencing of human breast cancers has revealed a substantial number of candidate cancer genes with recurring but infrequent somatic mutations. To determine more accurately their mutation prevalence, we performed a mutation analysis of 36 novel candidate cancer genes in 96 human breast cancers. Somatic mutations with potential impact on protein function were observed in the genes ADAM12, CENTB1, CENTG1, DIP2C, GLI1, GRIN2D, HDLBP, IKBKB, KPNA5, NFKB1, NOTCH1, and OTOF. These findings strengthen the evidence for involvement of the Notch, Hedgehog, NF-KB, and PIK3CA pathways in breast cancer development, and point to novel processes that likely are involved.
INTRODUCTION
It is widely accepted that cancer is caused by constitutional and somatic mutations in genes that control cell growth or genome stability (Vogelstein and Kinzler, 2004). Classical genetic techniques were used to discover frequently mutated cancer genes, such as TP53 and ERBB2, which subsequently guided genetic and functional characterization of the pathways in which they reside. However, the majority of recently discovered candidate cancer genes in adult solid tumors are mutated in <10% of patient tumors. Thus, the hunt for breast cancer genes mutated in a low fraction of patient tumors has necessitated unbiased mutational analyses at the gene family, exome or genome levels. Examples of such studies include (1) re-sequencing of genes encoding kinases, which uncovered a higher ratio of non-synonymous to synonymous mutations than expected by chance indicating accumulation of driver mutations in this gene set (Stephens et al, 2005); (2) exome-wide somatic mutation analyses (Sjöblom et al, 2006; Wood et al, 2007; Leary et al, 2008); (3) rearrangement analyses by paired-end sequencing, which have revealed an average of 90 chromosomal breakpoints per receptor-negative tumor (Stephens et al, 2009); and (4) whole genome sequencing, which revealed 50 somatic point mutations and small indels in coding sequences as well as 28 large deletions, 6 inversions and 7 translocations in the breast cancer metastasis studied (Ding et al, 2010). In breast cancers, exome sequencing revealed 140 candidate breast cancer genes (Sjöblom et al, 2006; Wood et al, 2007). The average receptor-negative breast cancer had point mutations or small insertions or deletions in 101 protein coding genes, 11 focal amplifications, and 7 focal deletions. In breast cancers, as well as in other tumor types, it is currently believed that the majority of somatic mutations are passengers, i.e. mutations which do not directly alter the net rate of cell growth or other phenotypes of essence to the tumor cell. However, the multitude of novel recurring but infrequent gene mutations discovered by exome or genome sequencing poses a challenge in distinguishing driver from passenger genes (Ali and Sjöblom, 2009). In the present study, we investigate 36 previously identified candidate breast cancer genes by mutational analysis of 96 additional tumors. Through bioinformatic analyses to predict the effect of specific mutations on protein function and the analysis of the pathways in which these mutated genes reside, we identify likely driver genes and pathways in breast tumorigenesis.
MATERIALS AND METHODS
Sample Collection and Handling
Ninety-six fresh frozen tumor samples were obtained from the Johns Hopkins Medical Institutions, the Dana-Farber Cancer Institute and the South Carolina Biorepository System and either macrodissected or laser capture microdissected to increase tumor cell fraction (Table S1). The patients have an average age at diagnosis of 54 years (range 30–89). Tumors were categorized into 3 subtypes, namely luminal, HER2+ and basal, according to the expression status of estrogen receptor (ER), progesterone receptor (PR) and HER2 by immunohistochemistry (Brenton et al, 2005). Among the 96 samples investigated in this study, 19 cases do not have sufficient expression information for classification, while the remaining 77 tumors consist of 54 luminal breast cancers (70%), 10 HER2+ breast cancers (13%) and 13 basal breast cancers (17%).
Tumor DNA was extracted from frozen tissue or purified cell lines, and whole genome amplification (REPLI-g WGA, Qiagen) was used to provide sufficient quantity of DNA for mutational analyses.
Sequencing Strategy and Mutational Analyses
Protein coding sequences of the 36 selected candidate cancer genes were amplified and sequenced from 96 breast tumor samples using previously described approaches (Sjöblom et al, 2006). PCR primers used to amplify targeted regions are listed in Table S2. DNA sequences were analyzed by Mutation Surveyor (SoftGenetics) followed by visual inspection to identify potential mutations. Sequence variants present in SNP databases (International HapMap Project and the 1000 Genomes Project, release 20100804) were removed. Putative mutations were sequenced de novo in the tumor DNA that had the mutation along with the patient-matched normal DNA. Two prediction tools, Cancer-Specific High-Throughput Annotation of Somatic Mutations (CHASM) (Carter et al, 2009) and MutationTaster (Schwarz et al, 2010), were used to predict functional effects of validated somatic mutations.
In order to calculate the mutation prevalence on a larger panel of samples, we combined the mutational data from Sjöblom et al. (2006) and Wood et al. (2007) with the data presented here. Since the tumor samples used in the validation screen in these studies varied across different genes, only the samples in which the gene was successfully sequenced were included in calculating mutation rate for certain genes (or in which all genes from the pathway were successfully sequenced in the case of pathway mutation prevalence calculation). To determine whether differences in mutation rates exist between different breast cancer subtypes, Fisher’s exact test was applied to the mutational data from samples that had subtype information. False discovery rate was controlled at 0.02 using the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995) to minimize false positives caused by multiple comparisons.
RESULTS AND DISCUSSION
We selected 36 candidate breast cancer genes to investigate from Sjöblom et al. (2006) and Wood et al. (2007) fulfilling the criteria (1) having somatic mutations in at least one tumor in a discovery set of 11 breast cancers, and subsequently found mutated in at least one tumor when reassessed in a validation set of 24 breast cancers; (2) not previously demonstrated to be breast cancer genes by functional studies; (3) having a mutation prevalence >3-fold higher than the estimated background somatic mutation prevalence of 1–3 mutations/Mb; (4) having mutations with predicted effects on gene function. In addition, BAP1, IKBKB, NFKB1, NFKBIA, and NFKBIE were included as they had putative loss of function mutations in discovery set tumors and were implicated in known cancer pathways.
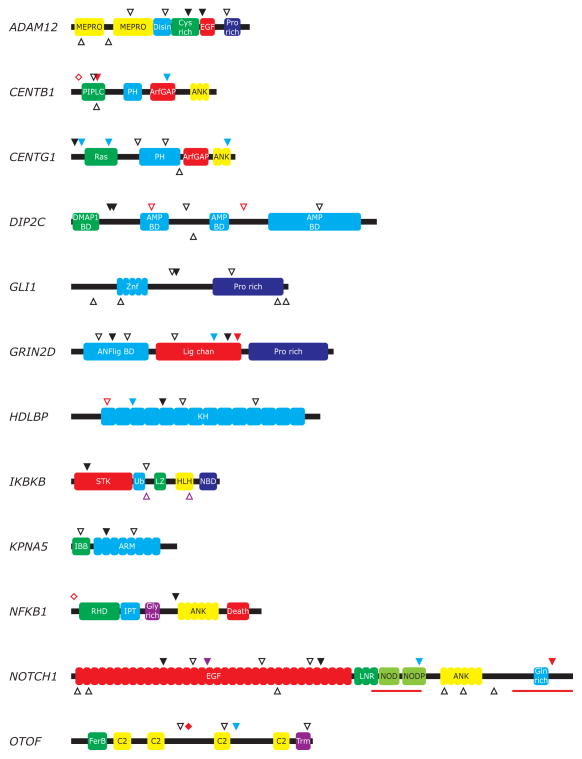
We assessed ~130 kb of protein-encoding sequences from the 36 genes in each of 96 breast cancers for a total of 12.5 MB sequence. We observed 28 somatic mutations, comprising 13 non-synonymous, 3 frameshift, 1 truncating, 1 splice site, and 10 synonymous mutations (Table 1). Novel non-synonymous somatic mutations were observed in one-third of the genes, namely ADAM12, CENTB1, CENTG1, DIP2C, GLI1, GRIN2D, HDLBP, IKBKB, KPNA5, NFKB1, NOTCH1, and OTOF (Fig. 1).
TABLE 1.
Mutational Analysis of Candidate Breast Cancer Genes
| Symbol | Chra | CDS lengthb |
Study | Sample IDc | gDNAd | cDNA | Protein | Classe | CHASM FDRf |
MutationTasterg |
|---|---|---|---|---|---|---|---|---|---|---|
| ABCA3 | 16p13.3 | 5355 | Sjöblom et al. | BB1T | chr16:2309588C>A | c.868C>A | L290M | NS | 0.9 | Disease causing |
| Sjöblom et al. | B7C | chr16:2285603G>C | c.2403G>C | E801D | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB16T | chr16:2276767C>G | c.3207C>G | H1069Q | NS | 0.9 | Polymorphism | |||
| ADAM12 | 10q26.2 | 2914 | Present | BP03TA | chr10:127727952A>G | c.1786A>G | T596A | NS | 0.4 | Polymorphism |
| Present | BB48T | chr10:127724615G>C | c.2003G>C | G668A | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | B2C | chr10:127779650G>C | c.901G>C | D301H | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB5T | chr10:127743547G>A | c.1436G>A | G479E | NS | 0.5 | Disease causing | |||
| Sjöblom et al. | B8C | chr10:127714867G>T | c.2376G>T | L792F | NS | 1 | Polymorphism | |||
| AIM1 | 6q21 | 3972 | Present | BB37T | chr6:107074968A>T | c.1968A>T | G656G | S | Polymorphism | |
| Wood et al. | BB32T | chr6:107067066G>T | c.157G>T | A53S | NS | 1 | Polymorphism | |||
| Wood et al. | B5C | chr6:107067369G>T | c.460G>T | E154X | STOP | Disease causing | ||||
| Wood et al. | BB24T | chr6:107074493A>G | c.1493A>G | E498G | NS | 1 | Polymorphism | |||
| Wood et al. | BB18T | chr6:107118333T>C | c.4916T>C | I1639T | NS | 0.9 | Disease causing | |||
| AMFR | 16q13 | 2044 | Sjöblom et al. | B7C | chr16:54958853A>G | IVS12+4A>G | sp | SP | ||
| Sjöblom et al. | BB10T | chr16:54954440A>T | c.1814A>T | D605V | NS | 1 | Disease causing | |||
| ATP8B1 | 18q21.31 | 3831 | Present | BB94T | chr18:53510096C>T | c.1161C>T | L387L | S | Polymorphism | |
| Sjöblom et al. | B8C | chr18:53513397C>A | IVS9+4C>A | sp | SP | |||||
| Sjöblom et al. | BB9T | chr18:53486774C>A | IVS17-4C>A | sp | SP | |||||
| Sjöblom et al. | BB14T | chr18:53479454C>T | c.2657C>T | A886V | NS | 0.3 | Disease causing | |||
| Sjöblom et al. | BB1T | chr18:53466940C>G | c.3534C>G | I1178M | NS | 1 | Disease causing | |||
| BAP1 | 3p21.1 | 2326 | Wood et al. | B6C | chr3:52415311C>T (homozygous) | c.781C>T | Q261X | STOP | Disease causing | |
| CENTB1 | 17p13.1 | 2399 | Present | BB73T | chr17:7187475_7187475insA | c.398_398insA | fs | FS | Disease causing | |
| Present | BS43T | chr17:7192220C>T | c.1464C>T | R488R | S | Polymorphism | ||||
| Sjöblom et al. | BB3T | chr17:7185990G>C | IVS2-1G>C | sp | SP | |||||
| Sjöblom et al. | B3C | chr17:7186514A>G | c.341A>G | K114R | NS | 0.9 | Disease causing | |||
| CENTG1 | 12q14.1 | 2655 | Present | BP15T | chr12:56422069G>C | c.53G>C | R18P | NS | 0.9 | Disease causing |
| Present | BB69T | chr12:56421965C>A | c.157C>A | R53R | S | Disease causing | ||||
| Present | BS43T | chr12:56412998G>A | c.573G>A | V191V | S | Polymorphism | ||||
| Present | BS43T | chr12:56406725C>T | c.2388C>T | G796G | S | Polymorphism | ||||
| Sjöblom et al. | BB40T | chr12:56411623G>A | c.1015G>A | A339T | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | B8C | chr12:56409800G>T | c.1438G>T | D480Y | NS | 1 | Disease causing | |||
| CYP1A1 | 15q24.1 | 1587 | Sjöblom et al. | BB29T | chr15:72802018C>A | c.474C>A | Y158X | STOP | Disease causing | |
| Sjöblom et al. | B2C | chr15:72799993C>T | c.1429C>T | R477W | NS | 0.1 | Disease causing | |||
| DBN1 | 5q35.3 | 2062 | Sjöblom et al. | B7C | chr5:176820163G>T | IVS9-1G>T | sp | SP | ||
| Sjöblom et al. | B7C | chr5:176820162G>A | c.832G>A | E278K | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB16T | chr5:176817072G>C | c.1918G>C | E640Q | NS | 0.9 | Disease causing | |||
| DIP2C | 10p15.3 | 4967 | Present | BP26L | chr10:458776C>G | c.592C>G | Q198E | NS | 1 | Disease causing |
| Present | BB98T | chr10:455128G>A | c.616G>A | A206T | NS | 1 | Disease causing | |||
| Sjöblom et al. | BB27T | chr10:435084_435083insAGCACAGCTTGCTTTGGGGTC AAACGTGGATCAGCAGCCTCTTGGTCAGTAAA |
c.1225_1226insAGCACAGCTTGCTTTGGGGTCAAA CGTGGATCAGCAGCCTCTTGGTCAGTAAA |
fs | FS | |||||
| Sjöblom et al. | B8C | chr10:420086C>A | c.1757C>A | A586E | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB12T | chr10:398592delT | c.2632delT | fs | FS | Disease causing | ||||
| Sjöblom et al. | B6C | chr10:363080G>A | c.3790G>A | V1264M | NS | 0.9 | Disease causing | |||
| FLJ13479 | 16p11.2 | 1876 | Sjöblom et al. | BB5T | chr16:30983086G>A | c.196G>A | A66T | NS | 0.9 | Disease causing |
| Sjöblom et al. | B8C | chr16:30980894G>A | c.856G>A | G286S | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB12T | chr16:30980758C>G | c.992C>G | T331R | NS | 0.9 | Polymorphism | |||
| Sjöblom et al. | BB30T | chr16:30980083G>A | c.1667G>A | R556Q | NS | 0.9 | Polymorphism | |||
| GEN1 | 2p24.2 | 2831 | Sjöblom et al. | BB9T | chr2:17874166_17874165delC | c.785_786delC | fs | FS | Disease causing | |
| Wood et al. | B10C | chr2:17875550_17875553delGTAA | c.824_827delGTAA | fs | FS | Disease causing | ||||
| GAB1 | 4q31.21 | 2165 | Sjöblom et al. | BB22T | chr4:144694410A>G | c.248A>G | Y83C | NS | 0.9 | Disease causing |
| Sjöblom et al. | B3C | chr4:144717323C>A | c.1160C>A | T387N | NS | 1 | Disease causing | |||
| GLI1 | 12q13.3 | 3409 | Present | BS43T | chr12:56150393C>T | c.1603C>T | P535S | NS | 0.9 | Polymorphism |
| Sjöblom et al. | BB9T | chr12:56149713C>T | c.1541C>T | T514I | NS | 0.9 | Polymorphism | |||
| Sjöblom et al. | B2C | chr12:56151239G>C | c.2449G>C | E817Q | NS | 1 | Polymorphism | |||
| GRIN2D | 19q13.32 | 4107 | Present | BS45T | chr19:53616943C>T | c.2181C>T | P727P | S | Polymorphism | |
| Present | BB85T | chr19:53636974T>C | c.2389T>C | S797P | NS | 0.9 | Disease causing | |||
| Present | BP35- | 53637319_53637328insACATGGCGGG | c.2541_2550insACATGGCGGG | fs | FS | Disease causing | ||||
| Sjöblom et al. | B2C | chr19:53593879C>T | c.418C>T | P140S | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | B4C | chr19:53600193G>A | c.856G>A | G286R | NS | 0.9 | Polymorphism | |||
| Sjöblom et al. | BB28T | chr19:53610100A>G | c.1580A>G | E527G | NS | 0.9 | Disease causing | |||
| HDLBP | 2q37.3 | 4015 | Present | BB91T | chr2:241914920C>T | c.939C>T | P313P | S | Polymorphism | |
| Present | BB54T | chr2:241909360G>C | c.1398G>C | K466N | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB31T | chr2:241916116_241916107delCAAAATCCAG | c.546_555delCAAAATCCAG | fs | FS | Disease causing | ||||
| Sjöblom et al. | BB18T | chr2:241907435A>C | c.1704A>C | K568N | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | B2C | chr2:241896108A>T | c.2816A>T | D939V | NS | 0.9 | Disease causing | |||
| HOXA3 | 7p15.2 | 1348 | Sjöblom et al. | B11C | chr7:26923376G>A | c.124G>A | D42N | NS | 1 | Polymorphism |
| Sjöblom et al. | BB28T | chr7:26923109G>A | c.391G>A | A131T | NS | 0.9 | Polymorphism | |||
| IKBKB | 8p11.21 | 2439 | Present | BB48T | chr8:42266871G>C | c.241G>C | E81Q | NS | 0.3 | Disease causing |
| Sjöblom et al. | B4C | chr8:42293532G>T | c.1078G>T | A360S | NS | 0.9 | Disease causing | |||
| KEAP1 | 19p13.2 | 1915 | Sjöblom et al. | B2C | chr19:10471642G>A | c.68G>A | C23Y | NS | 0.9 | Polymorphism |
| Sjöblom et al. | BB20T | chr19:10461011C>T | c.1565C>T | A522V | NS | 0.9 | Polymorphism | |||
| KIAA1946 | 2q32.1 | 2545 | Wood et al. | B4C | chr2:187441459G>T | c.817G>T | V273F | NS | 0.9 | Polymorphism |
| Wood et al. | B2C | chr2:187452229G>C | c.1654G>C | E552Q | NS | 1 | Polymorphism | |||
| Wood et al. | BB34T | chr2:187452859C>G | c.2284C>G | H762D | NS | 1 | Disease causing | |||
| KPNA5 | 6q22.2 | 1732 | Present | BP26L | chr6:117129974C>G | c.535C>G | L179V | NS | 0.9 | Disease causing |
| Sjöblom et al. | B11C | chr6:117119916C>G | c.144C>G | F48L | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB5T | chr6:117152189G>T | c.957G>T | R319S | NS | 0.9 | Disease causing | |||
| LOC340156 | 6p25.2 | 1255 | Wood et al. | B11C | chr6:2694440G>C | c.88G>C | E30Q | NS | 0.4 | Polymorphism |
| Wood et al. | B3C | chr6:2638020G>T | c.232G>T | A78S | NS | 1 | Polymorphism | |||
| Wood et al. | BB5T | chr6:2624573A>G | c.827A>G | N276S | NS | 0.1 | Polymorphism | |||
| LRRFIP1 | 2q37.3 | 2435 | Sjöblom et al. | BB5T | chr2:238399276A>G | IVS2+3A>G | sp | SP | ||
| Sjöblom et al. | B11C | chr2:238411419C>G | c.203C>G | S68C | NS | 1 | Disease causing | |||
| Sjöblom et al. | BB35T | chr2:238454328C>T | c.1900C>T | Q634X | STOP | Disease causing | ||||
| MRE11A | 11q21 | 2187 | Sjöblom et al. | BB7T | chr11:93844523T>G | c.710T>G | F237C | NS | 0.9 | Disease causing |
| Sjöblom et al. | B11C | chr11:93843398C>T | c.904C>T | H302Y | NS | 0.9 | Polymorphism | |||
| NCOA6 | 20q11.22 | 6296 | Sjöblom et al. | B3C | chr20:32794543C>T | c.3178C>T | P1060S | NS | 0.9 | Disease causing |
| Sjöblom et al. | BB15T | chr20:32794148C>A | c.3573C>A | S1191R | NS | 0.9 | Polymorphism | |||
| NFKB1 | 4q24 | 3094 | Present | BS72T | chr4:103875968G>A | c.1594G>A | V532I | NS | 0.9 | Polymorphism |
| Sjöblom et al. | B11C | chr4:103808177G>A | IVS1-1G>A | sp | SP | |||||
| NFKBIA | 14q13.2 | 1002 | Sjöblom et al. | B6C | chr14:34942227_34942226insC (homozygous) | c.427_428insC | fs | FS | Disease causing | |
| NFKBIE | 6p21.1 | 1551 | Wood et al. | B7C | chr6:44336225C>T | c.1138C>T | Q380X | STOP | Disease causing | |
| Wood et al. | B8C | chr6:44335809C>A | c.1386C>A | D462E | NS | 0.9 | Disease causing | |||
| NOTCH1 | 9q34.3 | 7943 | Present | BB60T | chr9:136688077G>A | c.1405G>A | D469N | NS | 0.9 | Disease causing |
| Present | BS48T | chr9:136684927C>A | c.2079C>A | C693X | STOP | Disease causing | ||||
| Present | BB77T | chr9:136677098G>A | c.3811G>A | E1271K | NS | 0.9 | Disease causing | |||
| Present | BP01T | chr9:136672719C>T | c.5229C>T | A1743A | S | Polymorphism | ||||
| Present | BS50T |
chr9:136666686_136666668delCCC AGCAGCCTGGCGGTGC |
c.7345_7363delCCCAGC AGCCTGGCGGTGC |
fs | FS | |||||
| Wood et al. | B10C | chr9:136685817G>T | c.1858G>T | D620Y | NS | 0.9 | Disease causing | |||
| Wood et al. | BB32T | chr9:136680082C>T | c.2912C>T | T971I | NS | 0.9 | ||||
| Wood et al. | B3C | chr9:136677262G>A | c.3647G>A | G1216D | NS | 0.5 | ||||
| OTOF | 2p23.3 | 3917 | Present | BB68T | chr2:26602024C>T | IVS1790-4C>T | sp | SP | ||
| Present | BB94T | chr2:26599529C>A | c.2518C>A | R840R | S | Polymorphism | ||||
| Sjöblom et al. | BB32T | chr2:26605168G>A | c.1666G>A | E556K | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | BB33T | chr2:26600351A>G | c.2338A>G | I780V | NS | 0.9 | Disease causing | |||
| Sjöblom et al. | B7C | chr2:26592647T>C (homozygous) | c.3605T>C | I1202T | NS | 0.9 | Polymorphism | |||
| PIK3R1 | 5q13.1 | 2295 | Sjöblom et al. | B1C | chr5:67625349_67625351dupTAA (homozygous) | c.1356_1358dupTAA | fs | FS | Polymorphism | |
| SIX4 | 14q23.1 | 2307 | Sjöblom et al. | B7C | chr14:60260479G>C (homozygous) | c.4G>C | E2Q | NS | 1 | |
| Sjöblom et al. | B3C | chr14:60256443G>A | c.1274G>A | G425D | NS | 0.9 | ||||
| Sjöblom et al. | BB28T | chr14:60249884C>A | c.2277C>A | D759E | NS | 0.9 | ||||
| TCF1 | 12q24.31 | 1976 | Sjöblom et al. | B1C | chr12:119894790A>G (homozygous) | c.817A>G | K273E | NS | 0.9 | Disease causing |
| Sjöblom et al. | BB12T | chr12:119900103G>A | c.1721G>A | S574N | NS | 0.9 | Polymorphism | |||
| TMEM123 | 11q22.2 | 602 | Wood et al. | BB31T | chr11:101777989A>C | c.259A>C | N87H | NS | 0.9 | |
| Wood et al. | B5C | chr11:101777516T>C | c.509T>C | M170T | NS | 0.9 | ||||
| VEPH1 | 3q25.31-32 | 2606 | Sjöblom et al. | BB9T | chr3:158581786delG | c.988delG | fs | FS | Disease causing | |
| Sjöblom et al. | BB5T | chr3:158581776G>T | c.998G>T | S333I | NS | 0.9 | Polymorphism | |||
| Sjöblom et al. | B8C | chr3:158461684_158461683delAA | c.2443_2444delAA | fs | FS | Disease causing |
Chr, chromosome band.
CDS length, nucleotides in protein-coding sequences including four flanking intronic bases (splice site) on each side of the exons.
Samples used in Sjöblom et al. (2006) and Wood et al. (2007) were referred to using sample IDs from Wood et al. (2007) instead of their ATCC IDs used in Sjöblom et al. (2006) (B1C =Hs 578T; B2C= HCC1008; B3C = HCC1954; B4C = HCC38; B5C = HCC1143; B6C = HCC1187; B7C = HCC1395; B8C = HCC1599; B9C = HCC1937; B10C = HCC2157; B11C = HCC2218).
gDNA: genomic positions, corresponding to UCSC hg17 build 35.1 release.
Class: NS, non-synonymous; S, synonymous; STOP, nonsense; SP, splice site; FS, frameshift.
CHASM FDR, false discovery rate of calling driver mutations determined by CHASM.
MutationTaster, prediction of mutation significance provided by MutationTaster.
Figure 1.
Somatic mutations in candidate breast cancer genes. Known and novel somatic mutations in candidate breast cancer genes are indicated in their respective protein domain structures. Tumor DNA was extracted from 96 primary human breast tumors, whereas patient-matched normal DNA was derived from blood or adjacent normal tissues. Human genome sequences (hg17 build 35.1), transcript coordinates (RefSeq release 16, March 2006), and single nucleotide polymorphisms were obtained from the UCSC Santa Cruz Genome Bioinformatics Site (http://genome.ucsc.edu). The ~3.4 million single nucleotide polymorphisms (SNPs) of dbSNP (release 125) that were validated in the HapMap project were used to exclude known polymorphisms, as were rare constitutional variants previously observed by our group in other sequencing studies (Jones et al, 2008; Parsons et al, 2008). PCR, sequencing reactions, and mutational analyses using Mutation Surveyor software (SoftGenetics LLC) were performed as described (Sjöblom et al, 2006). Filled symbols, mutations observed in the present study. Open symbols above domain structure, mutations previously observed in breast cancers in Sjöblom et al. (2006) or Wood et al. (2007). Open symbols below domain structure, cancer-derived mutations present in COSMIC or previously observed in colorectal, pancreatic, or brain tumors (Jones et al, 2008; Parsons et al, 2008). Black triangles, missense mutations. Blue triangles, synonymous mutations. Purple triangles, nonsense mutations. Red triangles, frameshift mutations. Red diamonds, splice site mutations. Domain name abbreviations: AMP BD, AMP-binding enzyme; ANFlig BD, ANF receptor family ligand binding region; ANK, Ankyrin repeat; ArfGAP, ARF GTPase-activating proteins domain; ARM, Armadillo/beta-catenin-like repeat; Cys rich, Cysteine-rich domain; C2, C2 domain; Death, Death domain; Disin, Disintegrin domain; DMAP1 BD, DMAP1-binding domain; EGF, EGF-like domain; FerB, Central domain B in proteins of the Ferlin family; Gln rich, Glutamine-rich; Gly rich, Glycine-rich domain; HLH, Helix-loop-helix domain; IBB, Importin beta binding domain; IPT, Ig-like, plexins, transcription factors domain; KH, K homology RNA binding domain; Lig chan, Ligand-gated ion channel; LNR, Lin-12/Notch repeat; LZ, Leucine zipper; MEPRO, ADAM type metalloprotease domain; NBD, NEMO-binding domain; NOD, Notch protein; NODP, Notch protein; PH, Pleckstrin homology domain; PIPLC, Phosphatidylinositol-specific phospholipase X-box domain; Pro rich, Proline-rich domain; Ras, Ras family; RHD, Rel homology transcription factor domain; STK, Serine/Threonine protein kinase domain; Trm, Transmembrane helices; Ub, Ubiquitin-like domain; Znf, Zinc finger domain.
Mutations in NF-KB pathway components contribute to the development of hematological malignancies such as multiple myeloma (Annunziata et al, 2007). We have previously demonstrated truncating, frameshift, and splice site mutations, respectively, in NFKBIE, NFKBIA, and NFKB1 along with multiple non-synonymous mutations in IKBKB and KEAP1 in breast tumors (Sjöblom et al, 2006; Wood et al, 2007). We here identify additional non-synonymous mutations in NFKB1 and the kinase domain of IKBKB in breast cancers.
The novel IKBKB E81Q kinase domain mutation is a predicted driver missense mutation (CHASM, FDR = 0.3). At the crossroads of NF-KB and PI3K signaling are members of the Centaurin gene family. Downregulation of the GTPase-activating protein CENTB1 has been shown to enhance NF-KB signaling, which provides a plausible explanation for the early splice site and frameshift mutations observed in breast cancers (Yamamoto-Furusho et al, 2006). CENTG1 is a known proto-oncogene, amplified in ~10% of glioblastomas and an activator of PIK3CA pathway signaling, which should encourage further functional studies of the missense mutations observed in breast cancers (Liu et al, 2007). The non-synonymous mutations observed in CENTB1 and CENTG1 in the current study are predicted to be disease-causing by MutationTaster (Schwarz et al, 2010). The combined mutation prevalence of these NF-KB pathway components mentioned above is 8% of breast tumor cases. Mutations in genes of the NF-KB pathway (NFKB, NFKBIA, NFKBIE, IKBKB, KEAP1, CENTB1 and CENTG1) are non-randomly distributed among breast tumor subtypes (luminal, n=60; HER2+, n=13 and triple-negative, n=19 from this study and from Sjöblom et al. (2006); P =0.003). These genes are mutated in 22% (7 out of 32) HER2+ or triple-negative breast cancers, which is significantly higher than 1.7% (1 out of 60) in the luminal tumors (P=0.002, Fisher’s exact test, FDR=0.02). Mutation frequencies of other genes and pathways, including DIP2C (n=110, P=0.125), GLI1 (n=110, P=0.084), GRIN2D (n=110, P=0.429), HDLBP (n=110, P=0.429), KPNA5 (n=110, P=0.084), OTOF (n=110, P=1.000) and Notch pathway (NOTCH1 and ADAM12, n=87, P=0.853) were not significantly different among breast tumor subtypes.
Notch signaling has previously been implicated in human oncogenesis, and the NOTCH1 gene is a target for insertion and rearrangement by the mouse mammary tumor virus (MMTV) (Yanagawa et al, 2000). We here identify NOTCH1 as a human breast cancer gene, based on the detection of a frameshift mutation in its C-terminal regulatory region. Frameshift mutations near the carboxy-terminus, such as the one newly identified in this study, are known to activate NOTCH1 in T-cell acute lymphoblastic leukemia (Weng et al, 2004). Activating non-synonymous and frameshift mutations in NOTCH1 have been observed in ~10% of non-small cell lung cancers (Westhoff et al, 2009). Similarly, Notch signaling has been found aberrantly activated in human breast cancer cell lines and tissue samples, but not in normal breast tissues. Furthermore, induced Notch signaling can transform normal human breast epithelial cells, resulting in growth beyond confluence, remarked change in cell shape, loss of cell-cell adhesion and resistance to drug-induced apoptosis (Stylianou et al, 2006). The consequences of non-synonymous mutations in the extracellular domain of NOTCH1 have not previously been described and their putative functional roles merit further investigation.
We also identified two novel non-synonymous mutations in the protease gene ADAM12. Recently, two previously identified breast cancer derived mutations in ADAM12 (D301H and G479E) were shown to prevent its insertion in the plasma membrane in a dominant-negative fashion, thereby leading to decreased shedding of the Notch ligand Delta-like I (Dyczynska et al, 2008). The novel T596A substitution is located in a cysteine-rich domain and has characteristics of a likely driver mutation (CHASM, FDR = 0.4). The mutations in NOTCH1 or ADAM12 in 8.4% of breast tumors (9 of 107), along with functional data, collectively point to a role for Notch pathway aberration in the development of breast carcinomas.
The sonic hedgehog effector GLI1 (glioma-associated oncogene homolog 1) is known to undergo amplification in a fraction of patients with malignant glioma (Kinzler et al, 1987). Single somatic mutations of GLI1 have previously been reported in urinary tract tumors and skin cancers (COSMIC, http://www.sanger.ac.uk/genetics/CGP/cosmic/). We have observed three non-synonymous GLI1 mutations in breast cancers, which merit further functional studies as GLI1 is a proto-oncogene expressed in normal mammary epithelial cells as well as in breast cancers (http://www.proteinatlas.org).
Several genes involved in RNA metabolism have recently emerged as putative cancer genes (Sjöblom et al, 2006). The somatic mutation prevalence (5% of cases) along with multiple frameshift mutations links the human homologue of disco-interacting protein, DIP2C (KIAA0934), to the development of breast cancer. Further, the missense mutations observed are all located in regions strongly conserved throughout evolution (data not shown) and predicted to be disease-causing (Schwarz et al, 2010). The DIP class of RNA-binding nuclear genes, which interact with the D. melanogaster gene disco during the establishment of the nervous system, has been implicated in maintenance of cell fate specification (DeSousa et al, 2003). The ubiquitously expressed HDLBP/vigilin gene, which has been connected to mRNA metabolism and estrogen-mediated stabilization of mRNAs, is composed of 15 KH nucleic acid binding domains and is essential to human cells as evidenced by siRNA knockdown (Goolsby and Shapiro, 2003). However, relatively little is known about the function of these genes, and further investigations into their roles in normal and tumor tissues are required.
Previous studies have identified mutations in genes in the nuclear pore complex and nuclear transport processes, such as NUP133, NUP214, and KPNA5, in breast cancers and other malignancies (Sjöblom et al, 2006; Mitelman et al, 2007). We here identify an additional non-synonymous mutation in the second ARM domain of the importin subunit alpha-6, KPNA5, which is thought to be involved as an adaptor in nuclear localization signal (NLS)-dependent protein import into the nucleus (Yang et al, 2010). Intriguingly, the protein products that rely on KPNA5 for nuclear import are still unknown. Further, we have identified 5 non-synonymous mutations in the ligand binding and channel-forming domains, along with one truncating mutation at the end of the channel-forming domain, in GRIN2D. The N-methyl-D-aspartate (NMDA) receptor subunit epsilon 4, GRIN2D, forms a heterotetrameric ligand-gated cation channel together with GRIN1. Interestingly, GRIN2D expression is regulated by estrogen (Ikeda et al, 2010). Functional NMDA receptor complexes containing the GRIN2D gene product have been demonstrated in human breast cancer cells and tissues, and the in vitro and in vivo tumor growth can be inhibited by NMDA receptor antagonists (North et al, 2010). This raises the possibility that the GRIN2D mutations observed here are oncogenic. Mutations in OTOF, a calcium-sensing protein that triggers membrane fusion and exocytosis, may also provide a link between calcium signaling and cancer.
Taken together, we provide data to strengthen the role of mutations in a subset of novel candidate cancer genes in breast tumorigenesis. We have identified additional somatic mutations in genes of the Notch, Hedgehog, NFKB, and PIK3CA pathways as well as in processes not yet strongly linked to human cancer such as RNA processing and calcium signaling. The mutation prevalence of CAN genes in this study differs from previously published work (Sjöblom et al, 2006; Wood et al, 2007). Potential explanations include the inability of mutational screens based on a low number of samples to pinpoint the true mutation prevalence, and the sample cohort compositions in terms of subtypes of breast cancers used in the studies. We also noticed a difference in prediction of mutation significance provided by CHASM and MutationTaster, that among the 72 non-synonymous mutations in Table 1 which have predictions from both methods, only 6 were classified as disease causing mutations by CHASM with the FDR controlled at 0.4 while 46 were suggested causal by MutationTaster, and only 3 mutations were consistently identified as significant mutations by both methods. While computational tools predicting mutation significance can be applied to prioritize targets for subsequent studies, the functional significance of mutations has to be proven through experimental analyses. The observation of multiple mutations in genes outside established cancer pathways may indicate that our understanding of these pathways is incomplete, or that hitherto unknown pathways and phenotypes are involved in tumor formation.
Supplementary Material
Acknowledgments
Supported by: Young Investigator Award and project grants from the Swedish Cancer Foundation (T.S.), Virginia and D.K. Ludwig Fund for Cancer Research, and National Institutes of Health grants CA43460, CA573445, CA62924, and CA121113.
References
- Ali MA, Sjöblom T. Molecular pathways in tumor progression: from discovery to functional understanding. Mol Biosyst. 2009;5:902–908. doi: 10.1039/b903502h. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, Dave S, Hurt EM, Tan B, Zhao H, Stephens O, Santra M, Williams DR, Dang L, Barlogie B, Shaughnessy JD, Jr, Kuehl WM, Staudt LM. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B. 1995;57:289–300. [Google Scholar]
- Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23:7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSousa D, Mukhopadhyay M, Pelka P, Zhao X, Dey BK, Robert V, Pelisson A, Bucheton A, Campos AR. A novel double-stranded RNA-binding protein, disco interacting protein 1 (DIP1), contributes to cell fate decisions during Drosophila development. J Biol Chem. 2003;278:38040–38050. doi: 10.1074/jbc.M303512200. [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, Abbott RM, Hoog J, Dooling DJ, Koboldt DC, Schmidt H, Kalicki J, Zhang Q, Chen L, Lin L, Wendl MC, McMichael JF, Magrini VJ, Cook L, McGrath SD, Vickery TL, Appelbaum E, Deschryver K, Davies S, Guintoli T, Crowder R, Tao Y, Snider JE, Smith SM, Dukes AF, Sanderson GE, Pohl CS, Delehaunty KD, Fronick CC, Pape KA, Reed JS, Robinson JS, Hodges JS, Schierding W, Dees ND, Shen D, Locke DP, Wiechert ME, Eldred JM, Peck JB, Oberkfell BJ, Lolofie JT, Du F, Hawkins AE, O’Laughlin MD, Bernard KE, Cunningham M, Elliott G, Mason MD, Thompson DM, Jr, Ivanovich JL, Goodfellow PJ, Perou CM, Weinstock GM, Aft R, Watson M, Ley TJ, Wilson RK, Mardis ER. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyczynska E, Syta E, Sun D, Zolkiewska A. Breast cancer-associated mutations in metalloprotease disintegrin ADAM12 interfere with the intracellular trafficking and processing of the protein. Int J Cancer. 2008;122:2634–2640. doi: 10.1002/ijc.23405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goolsby KM, Shapiro DJ. RNAi-mediated depletion of the 15 KH domain protein, vigilin, induces death of dividing and non-dividing human cells but does not initially inhibit protein synthesis. Nucleic Acids Res. 2003;31:5644–5653. doi: 10.1093/nar/gkg768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Fukushima T, Ogura H, Tsukui T, Mishina M, Muramatsu M, Inoue S. Estrogen regulates the expression of N-methyl-D-aspartate (NMDA) receptor subunit epsilon 4 (Grin2d), that is essential for the normal sexual behavior in female mice. FEBS Lett. 2010;584:806–810. doi: 10.1016/j.febslet.2009.12.054. [DOI] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O’Brien SJ, Wong AJ, Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70–73. doi: 10.1126/science.3563490. [DOI] [PubMed] [Google Scholar]
- Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, Parsons DW, Jones S, Sjöblom T, Park BH, Parsons R, Willis J, Dawson D, Willson JK, Nikolskaya T, Nikolsky Y, Kopelovich L, Papadopoulos N, Pennacchio LA, Wang TL, Markowitz SD, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hu Y, Hao C, Rempel SA, Ye K. PIKE-A is a proto-oncogene promoting cell growth, transformation and invasion. Oncogene. 2007;26:4918–4927. doi: 10.1038/sj.onc.1210290. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- North WG, Gao G, Memoli VA, Pang RH, Lynch L. Breast cancer expresses functional NMDA receptors. Breast Cancer Res Treat. 2010;122:307–314. doi: 10.1007/s10549-009-0556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C, O’Meara S, Parker A, Tarpey P, Avis T, Barthorpe A, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Edwards K, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Shepherd R, Small A, Solomon H, Stephens Y, Tofts C, Varian J, Webb A, West S, Widaa S, Yates A, Brasseur F, Cooper CS, Flanagan AM, Green A, Knowles M, Leung SY, Looijenga LH, Malkowicz B, Pierotti MA, Teh B, Yuen ST, Nicholson AG, Lakhani S, Easton DF, Weber BL, Stratton MR, Futreal PA, Wooster R. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, Jia M, Latimer C, Teague JW, Lau KW, Burton J, Quail MA, Swerdlow H, Churcher C, Natrajan R, Sieuwerts AM, Martens JW, Silver DP, Langerod A, Russnes HE, Foekens JA, Reis-Filho JS, van ‘t Veer L, Richardson AL, Borresen-Dale AL, Campbell PJ, Futreal PA, Stratton MR. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Research. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Westhoff B, Colaluca IN, D’Ario G, Donzelli M, Tosoni D, Volorio S, Pelosi G, Spaggiari L, Mazzarol G, Viale G, Pece S, Di Fiore PP. Alterations of the Notch pathway in lung cancer. Proc Natl Acad Sci U S A. 2009;106:22293–22298. doi: 10.1073/pnas.0907781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yamamoto-Furusho JK, Barnich N, Xavier R, Hisamatsu T, Podolsky DK. Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation. J Biol Chem. 2006;281:36060–36070. doi: 10.1074/jbc.M602383200. [DOI] [PubMed] [Google Scholar]
- Yanagawa S, Lee JS, Kakimi K, Matsuda Y, Honjo T, Ishimoto A. Identification of Notch1 as a frequent target for provirus insertional mutagenesis in T-cell lymphomas induced by leukemogenic mutants of mouse mammary tumor virus. J Virol. 2000;74:9786–9791. doi: 10.1128/jvi.74.20.9786-9791.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SN, Takeda AA, Fontes MR, Harris JM, Jans DA, Kobe B. Probing the specificity of binding to the major nuclear localization sequence-binding site of importin-alpha using oriented peptide library screening. J Biol Chem. 2010;285:19935–19946. doi: 10.1074/jbc.M109.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.