CXCL12 forms a complex with HMGB1 that binds to the chemokine receptor CXCR4 and increases inflammatory cell migration.
Abstract
After tissue damage, inflammatory cells infiltrate the tissue and release proinflammatory cytokines. HMGB1 (high mobility group box 1), a nuclear protein released by necrotic and severely stressed cells, promotes cytokine release via its interaction with the TLR4 (Toll-like receptor 4) receptor and cell migration via an unknown mechanism. We show that HMGB1-induced recruitment of inflammatory cells depends on CXCL12. HMGB1 and CXCL12 form a heterocomplex, which we characterized by nuclear magnetic resonance and surface plasmon resonance, that acts exclusively through CXCR4 and not through other HMGB1 receptors. Fluorescence resonance energy transfer data show that the HMGB1–CXCL12 heterocomplex promotes different conformational rearrangements of CXCR4 from that of CXCL12 alone. Mononuclear cell recruitment in vivo into air pouches and injured muscles depends on the heterocomplex and is inhibited by AMD3100 and glycyrrhizin. Thus, inflammatory cell recruitment and activation both depend on HMGB1 via different mechanisms.
A variety of chemokines and inflammatory molecules are concomitantly produced at sites of inflammation and tissue damage and are responsible for leukocyte trafficking and homing. Although we understand reasonably well the effects of these molecules individually, much less is known about the consequences of the combined activity of chemokines with other chemokines or inflammatory molecules.
Chemokines control integrin function and cell locomotion (Murphy, 2002) by binding to seven transmembrane domain receptors coupled to heterotrimeric GTP-binding proteins (GPCRs [G-protein–coupled receptors]), which are differentially expressed in a wide range of cell types. The resulting combinatorial diversity in responsiveness to chemokines guarantees the proper tissue distribution of distinct leukocyte subsets under normal and pathological conditions. The chemokine CXCL12 binds to the chemokine receptor CXCR4 and plays an essential and unique role in homeostatic regulation of leukocyte traffic, hematopoiesis, organogenesis, cell differentiation and tissue regeneration (Murphy, 2002).
Many molecules involved in triggering inflammation have chemoattractant activities. These include pathogen-associated molecular patterns, such as LPS and N-formylated peptides (Chen and Pan, 2009), and damage-associated molecular patterns, which are endogenous molecules that signal cell distress and traumatic cell death and include IL-1α, S100 proteins, defensins, and HMGB1 (high mobility group box 1; Bianchi, 2009).
HMGB1 is a ubiquitously expressed, highly conserved nuclear protein that plays important roles in chromatin organization and transcriptional regulation (Bianchi, 2009). HMGB1 acts as a damage-associated molecular pattern after its release, which can occur passively from dead cells (Bianchi, 2009) or actively by secretion from activated immune cells, enterocytes, hepatocytes, and possibly several other types of cells under distress (Tsung et al., 2007). Secretion requires the translocation of HMGB1 from the nucleus to the cytoplasm and does not involve the endoplasmic reticulum and the Golgi apparatus (Wang et al., 1999; Bianchi, 2009). Stress conditions that induce HMGB1 secretion include hypoxia (Andrassy et al., 2008), treatment with specific antitumor drugs (Ditsworth et al., 2007), or lethal irradiation (Apetoh et al., 2007). Recently, it has emerged that the relocation of HMGB1 from the nucleus to the cytoplasm also controls autophagy and in turn autophagic cells can secrete HMGB1 (Livesey et al., 2009; Skinner, 2010; Tang et al., 2010).
Extracellular HMGB1 induces several responses, including the release of proinflammatory cytokines, cell proliferation, and cell migration (Bianchi, 2009). Several receptors have been implicated in HMGB1-mediated functions, including RAGE (receptor for advanced glycation end products) and TLR2 (Toll-like receptor 2), TLR4, and TLR9 (Lotze and Tracey, 2005; Tian et al., 2007). Downstream signaling is not completely understood, but involves Src, MAPK (mitogen-activated protein kinase), and NF-κB activation (Palumbo et al., 2009; Penzo et al., 2010). Moreover, the migration of immature DCs and smooth muscle cells in response to HMGB1 is sensitive to Bordetella pertussis toxin (PTX), indicating the involvement of GPCRs (Degryse et al., 2001; Yang et al., 2007), coupled to Gi/0 proteins.
It is currently not known whether all receptors and all signaling pathways are required for the different responses to HMGB1; recent evidence suggests that TLR4 but not RAGE is required for cytokine release (Yang et al., 2010) and that RAGE is involved in cell migration (Penzo et al., 2010). Surprisingly, HMGB1-induced cell migration requires activation of both the canonical and noncanonical NF-κB pathways, which lead to the transcription of the Cxcl12 gene (Penzo et al., 2010). In addition, HMGB1 protects CXCL12 from degradation, suggesting both a functional and physical interaction between the molecules (Campana et al., 2009). We show here by nuclear magnetic resonance (NMR) and surface plasmon resonance (SPR) analysis that HMGB1 and CXCL12 form a complex. Importantly, HMGB1 induces changes in residues 3–12 of CXCL12, which are fundamental for the triggering of CXCR4, the CXCL12 receptor. The conformational rearrangements of CXCR4 homodimers differ when triggered by CXCL12 alone or in complex with HMGB1, as determined by fluorescence resonance energy transfer (FRET).
We then investigated how HMGB1 and CXCL12 cooperate in promoting cell migration in vitro and in vivo. We show evidence that in the initial phase of tissue injury, the recruitment of mononuclear cells is mediated by the CXCL12–HMGB1 complex, which signals via CXCR4, whereas both RAGE- and TLR-mediated signaling are dispensable. Thus, the recruitment of inflammatory cells to damaged tissues appears distinct from the induction of cytokine release from the same cells, which depends on the TLR4 receptor, although both processes involve HMGB1.
RESULTS
HMGB1-induced migration of fibroblasts is inhibited by blocking CXCR4 or CXCL12
HMGB1 is a chemoattractant for a variety of cells; however, recent work has demonstrated that the redox state of HMGB1 affects its properties (Yang et al., 2012). The recombinant HMGB1 used throughout this work is fully reduced: it was purified and stored in the presence of DTT, and mass spectrometry confirmed that its three cysteine residues are in the thiol state (Fig. 1 A).
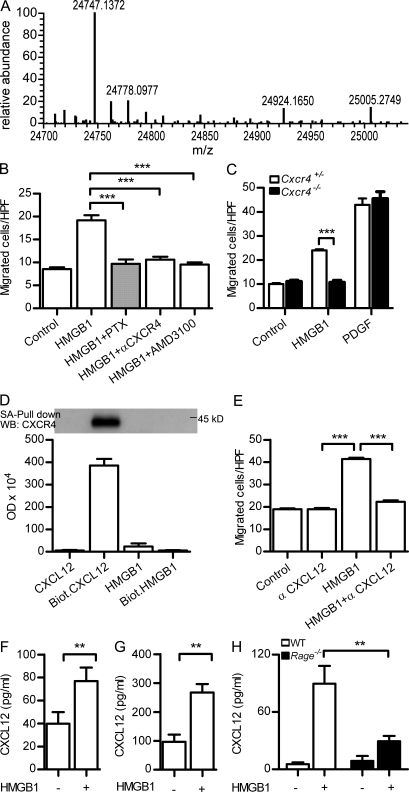
Figure 1.
Migration of mouse fibroblasts in the presence of HMGB1 requires CXCR4/CXCL12. (A) Deconvoluted mass spectra of recombinant HMGB1 used throughout our work. The mass of 24,747 corresponds to the fully reduced state of HMGB1 (amino acids 2–215; the first methionine is absent both in mammalian and in bacterially produced HMGB1). (B) Migration of murine NIH/3T3 in the presence of 1 nM HMGB1 is 100 nM PTX–sensitive, and it is blocked by 2 µg/ml anti-CXCR4 antibody or by 1 µM of the CXCR4 antagonist AMD3100. Migrated cells were counted per high-power field (HPF) and are shown as mean ± SEM of three independent experiments (***, P < 0.005 vs. HMGB1 using ANOVA plus Dunnett’s test). (C) Migration of Cxcr4+/− and Cxcr4−/− embryonic fibroblasts (MEFs) in the presence of 1 nM HMGB1 or 10 ng/ml PDGF. Migrated cells were counted per high-power field and are shown as mean ± SEM of three independent experiments (***, P < 0.001 using ANOVA plus Bonferroni posttest). (D) Pull-down of CXCR4 from cell lysates of pre-B 300.19–CXCR4+ was performed with 500 nM of biotinylated CXCL12 or 500 nM of biotinylated HMGB1 and analyzed by Western blot. Nonbiotinylated CXCL12 or HMGB1 was used at 500 nM as control. Densitometric analysis of the bands obtained in three independent experiments (mean ± SEM) is shown. (E) Migration in the presence of HMGB1 is blocked by 1 µg/ml anti-CXCL12. Migrated cells were counted per high-power field and are shown as mean ± SEM of three independent experiments (***, P < 0.005 vs. HMGB1 using ANOVA plus Dunnett’s test). (F–H) CXCL12 as detected by ELISA in the supernatant of mouse 3T3 (F), human monocytes (G), or Rage−/− MEFs (H) stimulated with 4 nM HMGB1 for 2 h. Mean ± SEM of three independent experiments is shown (**, P < 0.01).
Already in the first description of the chemoattractant activity of HMGB1 it was noted that the response of primary rat aortic smooth muscle cells to HMGB1 is sensitive to PTX pretreatment (Degryse et al., 2001). In mouse NIH/3T3 fibroblasts, HMGB1-induced migration was also blocked by PTX, as well as by a selective antibody to the chemokine receptor CXCR4 and by the specific CXCR4 antagonist, AMD3100 (Fig. 1 B). In addition, HMGB1 did not induce migration of embryonic fibroblasts lacking CXCR4 (Fig. 1 C). A direct interaction of HMGB1 with CXCR4 was excluded by pull-down experiments performed on a lysate of mouse pre-B 300.19 cells transfected with CXCR4: CXCR4 was pulled down with 500 nM of biotinylated CXCL12 but not with 500 nM of biotinylated HMGB1 (Fig. 1 D).
To test the possibility that HMGB1-induced cell migration involves the interaction of CXCL12 with CXCR4, we tested the migration toward HMGB1 in the presence of a neutralizing anti-CXCL12 antibody. Indeed, the antibody blocked HMGB1-induced migration (Fig. 1 E). These experiments suggested that CXCL12 is necessary for HMGB1 to induce cell migration. They also indicate that CXCL12 is already available in the medium in which the cells are cultured or suspended or is made available after the exposure of the cells to HMGB1. In fact, both 3T3 cells (Fig. 1 F) and freshly isolated human monocytes (Fig. 1 G) released a basal level of CXCL12; this level was significantly increased after the exposure of the cells to HMGB1, in accordance with the recent observation that HMGB1 induces Cxcl12 transcription (Penzo et al., 2010). The release of CXCL12 from HMGB1-stimulated cells depends on the presence of RAGE: embryonic fibroblasts from C57BL/6 mice lacking RAGE (Rage−/−) secreted a reduced amount of CXCL12 in response to HMGB1 compared with cells from WT animals (Fig. 1 H).
HMGB1 enhances CXCL12-induced migration, extracellular signal-regulated kinase (ERK) phosphorylation, and intracellular Ca2+ mobilization in human monocytes and CXCR4-transfected cells
We further investigated in vitro whether HMGB1 and CXCL12 cooperate in modulating cell migration or whether the migration observed in the presence of HMGB1 is exclusively induced by CXCL12. As shown in Fig. 2 A, 1 nM CXCL12 did not induce migration of human monocytes, whereas optimal migration was observed at 100 nM. HMGB1 alone did not induce migration up to 300 nM (not depicted). In the presence of 300 nM HMGB1, CXCL12 induced strong monocyte migration already at 1 and 10 nM. We determined, by dose finding experiments, that 150 nM is the lowest HMGB1 concentration that can enhance migration in the presence of a suboptimal CXCL12 concentration (10 nM; Fig. 2 B). HMGB1 does not enhance the activity of other inflammatory chemokines, such as CXCL8, CCL2, and CCL7, and only marginally enhances CCL19- and CCL21-induced migration (not depicted).
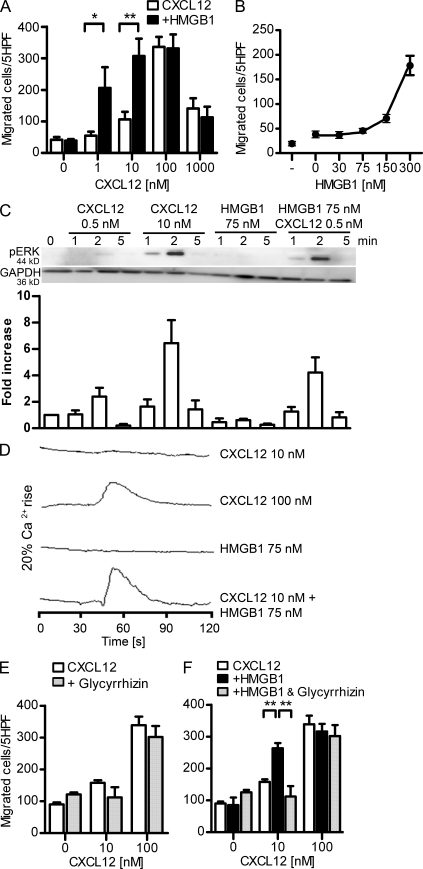
Figure 2.
HMGB1 increases CXCL12-induced activities in human monocytes and CXCR4-transfected cells and is sensitive to glycyrrhizin. (A) Chemotaxis toward increasing concentrations of CXCL12 ± 300 nM HMGB1. Migrated cells were counted per five high-power fields (HPF) and are shown as mean ± SEM of three independent experiments performed with cells from different donors (*, P < 0.05; **, P < 0.01, ANOVA plus Bonferroni posttest). (B) Dose–response curve of HMGB1 in the presence of 10 nM CXCL12. Migrated cells were counted per five high-power fields and are shown as mean ± SEM of three independent experiments. (C) Time course of ERK1&2 phosphorylation induced by CXCL12 alone or in the presence of HMGB1. The intensity of pERK1&2 bands was analyzed by densitometry, normalized with matching GAPDH, and compared with the unstimulated control. One representative blot is shown; the bars and error bars represent mean ± SEM of four independent experiments with cells from different donors. (D) Changes in [Ca2+]i were monitored in cells loaded with 50 nM Fura-2-AM and stimulated with CXCL12 and HMGB1. One representative set of measurements out of three independent experiments is shown. (E and F) Chemotaxis toward increasing concentrations of CXCL12 ± 200 µM glycyrrhizin ± 300 nM HMGB1. In all these experiments, migrated monocytes were counted per five high-power fields. Bars and error bars represent mean ± SEM of three different experiments performed with monocytes from different donors (**, P < 0.01, ANOVA plus Bonferroni posttest).
Chemokines, upon activation of their selective receptors, induce intracellular signaling cascades involved in cell activation and motility, such as ERK phosphorylation and Ca2+ release from stores. In the presence of a suboptimal CXCL12 concentration (0.5 nM), we could detect ERK phosphorylation in human monocytes in the presence of 75 nM HMGB1 (Fig. 2 C) but not in the presence of CXCL12 or HMGB1 alone. As a control, phosphorylation of ERK was induced by an optimal CXCL12 concentration (10 nM). Similar results were obtained when assessing CXCL12-induced intracellular Ca2+ mobilization in pre-B 300.19/CXCR4+ cells in the presence or absence of HMGB1. A rapid increase of intracellular Ca2+ was observed in cells stimulated with a high concentration of CXCL12 (100 nM) but not in cells stimulated with a suboptimal CXCL12 concentration (10 nM) or with 75 nM HMGB1 alone. However, a suboptimal CXCL12 concentration (10 nM) induced intracellular Ca2+ rise in the presence of 75 nM HMGB1, similar to the one observed with 100 nM CXCL12 (Fig. 2 D).
Glycyrrhizin, the glycoconjugated triterpene produced by the licorice plant Glycyrrhiza glabra, inhibits the chemoattractant and mitogenic activities of HMGB1 on 3T3 fibroblasts and binds to both HMG-box domains (BoxA and BoxB; Mollica et al., 2007). Glycyrrhizin did not affect CXCL12-induced migration (Fig. 2 E) but abrogated the synergistic effect exerted by HMGB1 (Fig. 2 F).
HMGB1 enhances CXCL12-induced migration exclusively via CXCR4 and in a RAGE- and TLR-independent manner
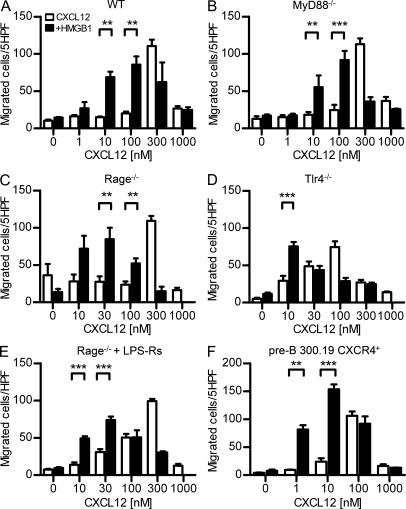
The synergy of HMGB1 and CXCL12 might require HMGB1 receptors (RAGE, TLR2, or TLR4), CXCL12 receptors (CXCR4), or both. Freshly isolated human monocytes express CXCR4, TLR2, and TLR4 on their surface, whereas RAGE expression can be detected only at the RNA level (not depicted). To test which receptors are required, we used BM cells from C57BL/6 mice, either WT or lacking RAGE (Rage−/−), the proximal interactor of TLR receptors MyD88 (MyD88−/−), or TLR4 (Tlr4−/−). WT BM cells migrated with a typical bell-shaped dose response to CXCL12 alone, and HMGB1 shifted the bell-shaped curve to lower CXCL12 concentrations (Fig. 3 A). BM cells from MyD88−/−, Rage−/−, and Tlr4−/− mice responded to CXCL12 and to the presence of HMGB1 similarly to WT BM cells (Fig. 3, B–D). Additionally, Rage−/− BM cells in the presence of the inhibitory TLR4 molecule, LPS-Rs (Maroso et al., 2010), did not modify the response to CXCL12 in the presence of HMGB1 (Fig. 3 E), indicating that neither RAGE nor TLR signaling is involved in the synergy between HMGB1 and CXCL12 and suggesting that the effect of HMGB1 on cell migration is mediated by CXCR4 alone. In support of this hypothesis, we used pre-B 300.19 cells, in which RAGE, TLR2, and TLR4 are not present and which do not respond to CXCL12 (not depicted). When we stably transfected pre-B 300.19 cells with human CXCR4, we observed a 100-fold increase in the chemoattractant potency of CXCL12 in the presence of 300 nM HMGB1 (Fig. 3 F).
Figure 3.
HMGB1 enhances CXCL12-induced migration via CXCR4 and is independent from RAGE or TLR signaling. (A–F) Migration induced by CXCL12 ± 300 nM HMGB1 on murine BM cells from C57BL/6 WT (A), MyD88−/− (B), Rage−/− (C), Tlr4−/− (D), and Rage−/− in the presence of 10 µg/ml LPS-Rs (E) or murine pre-B 300.19 cells transfected with human CXCR4 (F). Migrated cells were counted in five high-power fields (HPF) and are shown as mean ± SEM of four independent experiments performed with cells from different mice (**, P < 0.01; ***, P < 0.005, ANOVA plus Bonferroni posttest).
HMGB1 forms a heterocomplex with CXCL12
Synergy between HMGB1 and CXCL12 depends on CXCR4, but we could not show a direct interaction of HMGB1 with CXCR4 (Fig. 1 D). The synergy does not depend on RAGE, TLR2, or TLR4, suggesting that HMGB1 and CXCL12 might form a physical complex that acts on CXCR4. Modulation of chemokine activities has been shown to occur via the formation of heterocomplexes in vitro (Paoletti et al., 2005; von Hundelshausen et al., 2005) and in vivo (Koenen et al., 2009).
We confirmed that purified HMGB1 and CXCL12 can form a complex upon mixing: two different anti-CXCL12 antibodies coimmunoprecipitated HMGB1 (not depicted). To observe and characterize the interaction between CXCL12 and HMGB1, we then used NMR chemical shift mapping. By comparing 15N-labeled heteronuclear single-quantum coherence (HSQC) spectra before and after addition of the partner, it is possible to confirm an interaction even between low-affinity binding partners (Varani et al., 2007) and also to deduce which residues are affected.
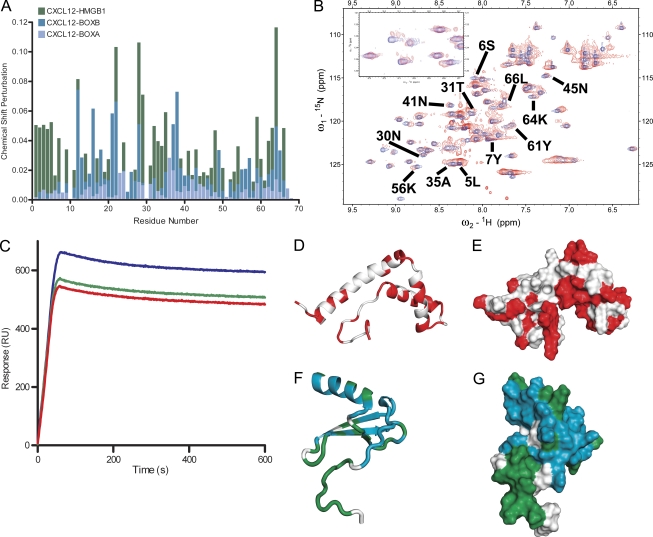
We first recorded the NMR spectra of free 15N-labeled CXCL12 and subsequently added either unlabeled HMGB1 or one of its HMG-box domains, BoxA or BoxB. The NMR signal of 38 out of 68 CXCL12 residues shifted upon addition of an equimolar amount of BoxB, proving that the two molecules interact (Fig. 4 A). The majority of the changes in CXCL12 involve residues 13–25, 40–45, and the C terminus, whereas the N terminus of CXCL12, in particular the flexible first 8 aa required for CXCR4 triggering (Crump et al., 1997; Wu et al., 2010), are clearly not affected by BoxB. Most CXCL12 residues involved in the interaction with BoxB were also affected by the addition of BoxA, but the chemical shift changes were slightly different and generally smaller in magnitude (Fig. 4 A), probably reflecting interactions with residues not conserved between BoxA and BoxB (which have a very similar structure but are only 20% identical at the amino acid level). In the reverse experiments, we added CXCL12 to 15N-labeled BoxB: 44 out of 80 BoxB residues were affected by CXCL12 binding, confirming the interaction and pinpointing it to the concave surface formed by the BoxB helices (Fig. 4, D and E). Intriguingly, this is the site where glycyrrhizin binds to HMGB1 (Mollica et al., 2007).
Figure 4.
NMR analysis of CXCL12 interactions with HMGB1, BoxA, and BoxB. (A) Chemical shift perturbation analysis of CXCL12 in complex with HMGB1, BoxA, or BoxB, calculated as described in Materials and methods. (B) 15N-HSQC spectra of CXCL12 free (blue) and in complex with HMGB1 (red); some of the residues showing chemical shift changes upon complex formation are labeled. (C) SPR sensograms of the interaction between HMGB1 and CXCL12 in the presence of increasing concentrations of glycyrrhizin. HMGB1 was immobilized on the sensor surface according to standard techniques, and 500 nM CXCL12 was then passed over the surface. CXCL12 (blue) shows a typical response curve with the signal increasing while the protein is flowed over the surface and decreasing after it stops flowing. In the presence of 500 nM glycyrrhizin (green), the signal intensity decreases and decreases further in the presence of 5 µM glycyrrhizin (red), which is indicative of decreased CXCL12 binding. The experiment was reproduced three times with the same results. RU, response units. (D and E) Cartoon (D) and surface (E) representation of BoxB: residues with significant chemical shift changes upon the addition of CXCL12 are in red. (F and G) Similar representation of CXCL12 residues showing NMR chemical shift changes upon binding of BoxA or BoxB are in blue; residues that change only upon the addition of HMGB1 are in green.
The addition of HMGB1 to 15N-CXCL12 at a 1:2 ratio caused more extensive (57 of 68 residues) and profound changes in the NMR spectrum of CXCL12 (Fig. 4, A, B, and F–G) than those induced by individual HMG-boxes. Only one set of peaks was present, and no signal corresponding to free CXCL12 was observed, suggesting a (CXCL12)2(HMGB1) stoichiometry. CXCL12 can bind to both HMG-box domains independently but displays different chemical shifts; because a single set of peaks was observed, this suggests that the heterocomplex is dynamic and the CXCL12 molecules exchange between the free state, BoxA binding, and BoxB binding.
Residues 3–12 in the N terminus of CXCL12, which are directly involved in CXCR4 recognition and triggering (Crump et al., 1997), were affected by the binding of full-length HMGB1 but not of either BoxA or BoxB alone (Fig. 4, A, B, and F–G). HMGB1 also affected the 31–35 loop, which is spatially close to the N terminus. This suggests that HMGB1 might induce a rearrangement of the CXCL12 conformation capable of enhancing the CXCL12-induced responses.
SPR experiments show that CXCL12 can bind to HMGB1 immobilized on the sensor surface, with signal intensity increasing with increasing concentrations of CXCL12. The intensity decreased, instead, in the presence of increasing concentrations of glycyrrhizin (Fig. 4 C), indicating that this small molecule inhibits the HMGB1–CXCL12 interaction. This supports and strengthens our NMR data because glycyrrhizin is known to bind HMGB1 (Mollica et al., 2007) in the same region where CXCL12 binds according to our mapping experiments.
CXCL12 and HMGB1–CXCL12 complexes interact differently with CXCR4 homodimers
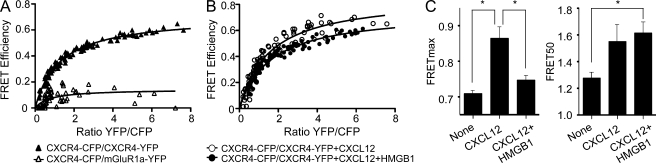
To test whether CXCL12 and the HMGB1–CXCL12 complex interact similarly or differently with CXCR4, we used sensitized FRET. HEK293 cells were transiently cotransfected with constant amounts of donor (CXCR4 fused to CFP; CXCR4-C) and increasing amounts of acceptor (CXCR4 fused to YFP; CXCR4-Y), and FRET saturation curves were determined. The results indicate that CXCR4 homodimerizes in the absence of ligands (FRETmax, 0.708 ± 0.008; FRET50, 1.274 ± 0.042); this process is specific, as in HEK293 cells coexpressing constant amounts of CXCR4-CFP and increasing amounts of mGluR1α-YFP, a seven-transmembrane receptor used as negative control, the FRET signal was negligible (FRETmax, 0.144 ± 0.024; FRET50, ND; Fig. 5 A). To determine the effect of ligand binding on CXCR4 homodimers, HEK293 cells transiently cotransfected with CXCR4-C/CXCR4-Y, as above, were stimulated with CXCL12 alone or in combination with HMGB1, and sensitized FRET was determined as above (Fig. 5 B). The maximal FRET signal (FRETmax) depends on FRET efficiency, which is affected by the distance and the relative orientation between the fluorescent proteins. The apparent affinity between CXCR4 monomers is defined by FRET50, the number of CXCR4 complexes at which half-maximal FRET signal is observed. In the presence of CXCL12 alone, FRET50 did not change significantly (1.548 ± 0.122 vs. 1.274 ± 0.042 in untreated controls; P > 0.05), indicating that both CXCR4 partners are bound apparently with similar affinity, but FRETmax increased significantly (0.8636 ± 0.029 vs. 0.708 ± 0.008 in controls; P < 0.05), indicating a change in conformation of CXCR4 dimers (Fig. 5 C). In contrast, stimulation with HMGB1–CXCL12 did not cause a significant modification of FRETmax (0.7462 ± 0.013 vs. 0.708 ± 0.008 in controls; P > 0.05) but significantly increased FRET50 (1.612 ± 0.08 vs. 1.274 ± 0.042 in controls; P < 0.05), indicating that HMGB1–CXCL12 increases the number of CXCR4 dimers formed and does not cause the same rearrangement in CXCR4 complexes as CXCL12 alone. This observation was confirmed, as we detected a significant difference in FRETmax (P < 0.05) between cells activated with CXCL12 alone and those stimulated with HMGB1–CXCL12 (Fig. 5 C).
Figure 5.
CXCL12 and the CXCL12–HMGB1 heterocomplex trigger different conformational changes in CXCR4 homodimers. (A and B) FRET saturation curves were generated in HEK293T cells transiently cotransfected with a constant amount of CXCR4-CFP (CXCR4-C; 2.0 µg; ∼300,000 FU) and increasing quantities of CXCR4-YFP (CXCR4-Y; 0.25–4.25 µg; ∼80,000-2,000,000 FU) or mGluR1α-Y (0.5–6.0 µg; ∼110,000-2,100,000 FU) as negative control. The curves represent data obtained in eight independent experiments. (B) Effect of CXCL12 (100 nM, 30 min) or CXCL12 + HMGB1 (100 nM and 300 nM, 30 min) on CXCR4 homodimers. The curves represent data obtained in 12 independent experiments. (C) FRETmax and FRET50 values shown were deduced using a nonlinear regression equation applied to a single binding site model and are representative of 8–12 independent experiments. FRETmax signals for CXCL12-treated and CXCL12–HMGB1-treated cells increase significantly compared with untreated cells (*, P < 0.05). Furthermore, the FRETmax value for CXCL12-treated cells is significantly higher (*, P < 0.05) than the FRETmax for HMGB1–CXCL12-treated cells. Error bars indicate SEM.
HMGB1 and CXCL12 cooperate to induce cell migration in vivo via CXCR4 and in a RAGE-independent manner
CXCL12 is one of the most abundant chemokines and is expressed on the endothelial vessels, ensuring trafficking of blood cells expressing CXCR4 in basal (homeostatic) conditions. Based on our in vitro experiments, the HMGB1–CXCL12 heterocomplex is much more active than CXCL12 alone. To test the in vivo relevance of the HMGB1–CXCL12 heterocomplex, air pouches were established in C57BL/6 mice, and 10 pmol CXCL12 was injected alone or together with HMGB1. As shown in Fig. 6 A, the complex induced a massive influx of WBCs 6 h after injection and was much more potent than CXCL12 alone.
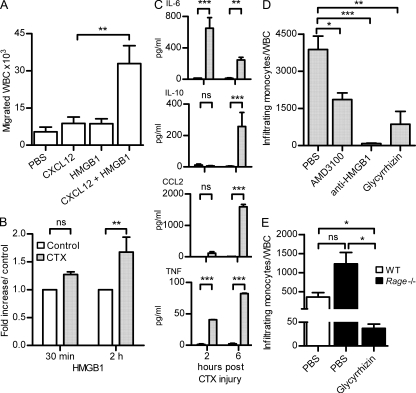
Figure 6.
Migration of WBCs in vivo depends on the HMGB1–CXCL12 heterocomplex. (A) WBCs were collected from air pouches 6 h after injection of PBS, 10 pmol CXCL12, 300 pmol HMGB1, or CXCL12 + HMGB1. Bars and error bars represent mean ± SEM of cell influx from at least six mice per condition (**, P < 0.01, ANOVA plus Dunnett’s test). (B) HMGB1–CXCL12 heterocomplex detected by hybrid ELISA in the muscles injured with CTX and in the untreated contralateral ones. Results are expressed as fold increase of the complex in CTX-treated muscle compared with the untreated controls and normalized to the total weight of muscle. Mean ± SEM of three independent experiments is shown (**, P < 0.01, ANOVA plus Bonferroni posttest). (C) Mouse cytokines were measured at 2 and 6 h after injury in muscles treated with CTX and in the untreated contralateral ones (control). Results are expressed as picogram/milliliter of cytokines detected with cytometric bead assay and normalized to the total weight of tissue. Mean ± SEM of three independent experiments is shown (**, P < 0.01; ***, P < 0.001, ANOVA plus Bonferroni posttest). (D) Mononuclear CD11bhigh+ cells infiltrating the muscle of WT BALB/c mice were counted 6 h after CTX injury and normalized against circulating total leukocytes (WBC). Mice were treated with PBS, AMD3100, α-HMGB1 antibody, or glycyrrhizin as described in Materials and methods. Mean ± SEM of infiltrating cells from at least three mice per condition is shown (*, P < 0.05; **, P < 0.01; ***, P < 0.005, ANOVA plus Dunnett’s posttest). (E) Mononuclear CD11bhigh+ cells infiltrating the muscles of Rage−/− mice were counted 6 h after CTX injury and normalized against circulating total leukocytes (WBC). Mice were treated with PBS or glycyrrhizin before injury. Mean ± SEM of infiltrating cells from three mice is shown (*, P < 0.05, ANOVA plus Dunnett’s posttest).
The results reported in the previous sections suggest that HMGB1 released as a result of tissue damage promotes monocyte recruitment to the injured tissue by using CXCR4 as a receptor. To test this hypothesis, we studied the initial phase of mononuclear cell migration in a model of muscle injury induced by cardiotoxin (CTX) injection. A single dose of CTX was injected in the tibialis anterior (TA) muscle of BALB/c WT mice. CTX injury induced the infiltration of a mononuclear cell population positive for CD11b and Ly6C already at 3 h after injection (not depicted). The amount of transcripts specific for mononuclear cells (CD11b, CD11c, Ly6c, CCL2, and CCR2) in the entire TA muscle increased as early as 3 h after CTX damage and remained elevated up to 48 h later (not depicted). HMGB1 was present in both myocytes and inflammatory cells and was also colocalized with CXCL12 on endothelial cells (not depicted). To measure the amount of circulating HMGB1–CXCL12 heterocomplex, we set up a hybrid ELISA in which the surface-bound antibody captured CXCL12 and the detection antibody recognized HMGB1 (see Materials and methods). Using this hybrid ELISA, we confirmed the presence of the HMGB1–CXCL12 heterocomplex in the muscles 2 h after injury (Fig. 6 B). We also assessed the level of cytokines that are involved in inflammatory reactions 2 and 6 h after CTX injury (Fig. 6 C). IL-6 was already released at 2 h, indicating an influx of monocytes. IL-10 and CCL2 were released at a later time (6 h), and low concentrations of TNF were detected during the first hours after injury. These results suggest that the activity of the complex HMGB1–CXCL12 induces the recruitment of monocytes soon after the injury, whereas the release of CCL2, a potent monocyte chemoattractant, supports cell influx at later times.
To verify that the initial monocyte/macrophage recruitment after muscle injury is mainly caused by the CXCL12/CXCR4 axis, we recovered and counted monocytes/macrophages by flow cytometry after CXCR4 blockage with its specific antagonist AMD3100 (DiPersio et al., 2009). AMD3100 (430 µg per day) or PBS was infused continuously (Jujo et al., 2010) over 3 d from an implanted osmotic pump. At day 3, mice were treated with CTX, and the injured and contralateral TA muscles were collected after 6 h. Although AMD3100 induced higher levels of circulating BM cells, as expected (DiPersio et al., 2009), the influx of monocytes/macrophages into the injured TA muscle was significantly decreased compared with the PBS-treated mice (Fig. 6 D), confirming that the initial cell recruitment is mainly mediated by CXCR4.
To fully characterize the role of HMGB1 in CXCL12-dependent migration in vivo, we used an anti-HMGB1 monoclonal antibody known to block HMGB1 activities in vitro and in vivo (Sitia et al., 2011) and glycyrrhizin, which abrogates the synergy between HMGB1 and CXCL12 in vitro (Fig. 2 F). A single dose of either glycyrrhizin (200 µg) or anti-HMGB1 antibody (200 µg) was injected intravenously 3 h before CTX-induced injury, and cellular infiltration was assessed at 6 h. Both treatments strongly inhibited monocyte infiltration in the injured muscle, confirming the relevance in vivo of the synergy between HMGB1 and CXCL12 (Fig. 6 D).
RAGE has been indicated, so far, as the receptor responsible for HMGB1-induced migration (Orlova et al., 2007), whereas TLR4 is responsible for HMGB1-dependent cytokine release (Yang et al., 2012). We investigated the influx of mononuclear cells after CTX injury in Rage−/− mice (Fig. 6 E). In C57BL/6 mice, the total number of circulating mononuclear cells was different than in BALB/c mice, as was the proportion of cells recruited to damaged tissue (compare PBS-treated control mice in Fig. 6 [D and E]). The absence of RAGE in C57BL/6 mice did not prevent, but actually promoted the recruitment of monocytes into the injured muscle, excluding the possibility that inflammatory cells are recruited through RAGE signaling. Glycyrrhizin (200 µg per mouse) also significantly inhibited monocyte/macrophage recruitment in this mouse strain, as well as in BALB/c mice, confirming that cellular influx into the injured area is mediated by the HMGB1–CXCL12 complex.
DISCUSSION
The present study shows that HMGB1 promotes the recruitment of inflammatory cells to injured tissues by forming a heterocomplex with the chemokine CXCL12 that acts exclusively via CXCR4, the receptor for CXCL12. The CXCL12/CXCR4 axis induces several cellular responses in vitro and in vivo, including cell migration and recruitment of hematopoietic stem cells to ischemic heart (Jujo et al., 2010). We show that suboptimal concentrations of CXCL12, which per se do not trigger in vitro migration or signaling events such as ERK phosphorylation and intracellular Ca2+ rise, induce robust responses in the presence of HMGB1. This synergy might in principle be caused by the convergence and nonlinear response of the signaling pathways, to the formation of a complex between ligands, or of a complex between receptors. Experiments on BM cells from MyD88−/−, Tlr4−/−, Rage−/− mice in the presence of LPS-Rs and on pre-B 300.19 cells transfected only with human CXCR4 exclude both the possibilities that CXCR4 interacts with RAGE or TLR4 or that the downstream signaling pathways converge. That leaves open only the interaction of the ligands, HMGB1 and CXCL12.
We indeed show that HMGB1 and CXCL12 form a complex, which we characterized by NMR and SPR experiments. CXCL12 can interact with the individual HMGB-boxes of HMGB1, giving similar but distinguishable NMR spectra. When CXCL12 is mixed in twofold excess with full-length HMGB1, a single set of peaks is recorded that is distinguishable from the spectra emerging from the interaction with the single HMG-boxes. We thus infer that the HMGB1–CXCL12 complex is formed by one molecule of HMGB1 and two molecules of CXCL12, each interacting with one HMG-box domain and exchanging rapidly within the heterocomplex. The vast majority of CXCL12 residues are affected by the interaction with HMGB1. Notably, the N-terminal residues, which have been shown to be very flexible and responsible for CXCR4 recognition and triggering (Crump et al., 1997; Wu et al., 2010), are only affected by formation of the functional complex between HMGB1 and CXCL12 molecules. Heterocomplexes forming between chemokines or with HMGB1 could be responsible for maintaining the agonist in an optimal conformation for triggering the receptor. We can also speculate that HMGB1 might bring together two molecules of CXCL12 and present them with the optimal spatial arrangement to a dimer of CXCR4, thus increasing the effective activity of CXCL12 in inducing cellular responses.
Our FRET data show that CXCR4 homodimers exist in the absence of ligands and that CXCL12 and HMGB1–CXCL12 complexes trigger different CXCR4 homodimer rearrangements. In accordance with previous FRET studies (Percherancier et al., 2005; Levoye et al., 2009), we observed that addition of CXCL12 increased the maximal FRET signal, whereas the FRET50 value was not significantly affected. These data suggest that CXCL12 binding alters the conformation rather than the number of preformed CXCR4 dimers. In contrast, HMGB1–CXCL12 addition did not modify FRETmax but increased FRET50 values, indicating the formation of a larger number of CXCR4 dimers. These data and the significant difference in FRETmax detected between cells activated with CXCL12 alone or with HMGB1–CXCL12 indicated that CXCL12 and the heterocomplex differ in promoting rearrangements within the CXCR4 dimers. These effects might explain the differences observed in the activation of signaling pathways.
Our findings explain why the chemoattractant activities of CXCL12 and HMGB1 appear to use the same signaling pathways in fibroblasts, including NF-κB canonical and noncanonical pathways (Penzo et al., 2010). They also account for the inhibition of HMGB1-induced migration by PTX, which inhibits the α subunit of the Gi protein typically associated with chemokine receptors (Murphy, 2002), and update our understanding of the mechanism of action of glycyrrhizin, which does not only prevent the binding of HMGB1 to RAGE as previously proposed, but rather appears to prevent the formation of the HMGB1–CXC12 heterodimer.
The use of CXCR4 for HMGB1 signaling conforms to a general pattern whereby HMGB1 forms complexes with partner molecules that then act via the partner’s receptor (Bianchi, 2009). HMGB1 forms a heterocomplex with IL-1β, which acts via the IL-1R receptor enhancing the activity of IL-1β alone (Sha et al., 2008). HMGB1 also binds DNA and promotes its interaction with the DNA-sensing TRL9 receptor (Ivanov et al., 2007).
Modulation of human leukocyte migration occurring via the formation of heterocomplexes between chemokines has been widely studied in vitro (Paoletti et al., 2005; von Hundelshausen et al., 2005; Venetz et al., 2010). It was recently shown in vivo that monocyte recruitment into the atherosclerotic plaque is promoted by the formation of a heterodimer between the CCL5 and CXCL4 chemokines, which can be prevented by a peptide rationally designed to mimic the heterodimer interface (Koenen et al., 2009). Our in vitro and in vivo results show that the complex HMGB1–CXCL12 enhances cell migration and that glycyrrhizin inhibits this enhancement. Of note, an NMR study showed that CXCL12 and glycyrrhizin (Mollica et al., 2007) interact with the same region of HMGB1, and SPR experiments (Fig. 4) indicate that glycyrrhizin interferes with the formation of the HMGB1–CXCL12 heterocomplex.
HMGB1 induces monocytes/macrophages to secrete TNF, among other cytokines. The HMGB1 dependence of the recruitment of inflammatory cells into damaged tissues was already shown in several animal models, most notably in heart ischemia/reperfusion (Andrassy et al., 2008), in peritonitis (Orlova et al., 2007), and in hepatitis (Sitia et al., 2007, 2011). Indeed, we show here that an HMGB1 blocking antibody abolishes the recruitment of monocytes/macrophages in muscle tissue damaged by CTX injection and that AMD3100, a selective CXCR4 antagonist, interferes with recruitment although a higher number of inflammatory cells egress from the BM. We highlight that the recruitment of inflammatory cells to damaged tissue and the activation of the same cells both require HMGB1 but different receptors: CXCR4 for recruitment and TLR4 for activation (Yang et al., 2010, 2012). TLR4 is triggered by oxidized HMGB1, containing one disulfide bond in BoxA (Yang et al., 2012), whereas in our work we consistently used fully reduced HMGB1, in which all cysteines are in the thiol form.
In our model, RAGE does not appear to be required for inflammatory cell recruitment. Both in vitro (Degryse et al., 2001) and in a mouse model heart ischemia/reperfusion (Andrassy et al., 2008), the chemoattractant activity of HMGB1 appears to involve RAGE because it is inhibited by anti-RAGE antibodies, and the migration of Rage−/− cells is severely impaired. In contrast, we propose that the role of RAGE in cell migration is to trigger Cxcl12 transcription (Penzo et al., 2010) so that CXCL12 production is increased and sustained over time. Additional CXCL12 production is unnecessary in vitro if enough CXCL12 is already present in the medium, or in our model of CTX-induced muscle damage where CXCL12 is presented by endothelial or epithelial cells (Agace et al., 2000; Venetz et al., 2010).
Blocking the HMGB1–CXCL12 interaction might be beneficial in the early phase of tissue damage, such as in ischemic and trauma patients, and in chronic diseases like rheumatoid arthritis, where an excessive recruitment of leukocytes is supported by HMGB1 (Andersson and Harris, 2010). Notably, other activities of HMGB1 appear completely independent from the formation of the complex with CXCL12; indeed, HMGB1-induced TNF secretion proceeds via TLR4 (Yang et al., 2010), and maturation of DCs and proliferation of several cell types proceed via RAGE (Yang et al., 2007; unpublished data). Drugs that prevent the formation of the HMGB1–CXCL12 heterocomplex might be rationally designed and provide a very specific tool to selectively block some HMGB1 effects, such as migration, without affecting effects induced through other receptors.
MATERIALS AND METHODS
Cells.
PBMCs were isolated from buffy coats of donor blood (Central Laboratory of the Swiss Red Cross, Basel, Switzerland) by Ficoll-Paque density centrifugation. Monocytes, CD14+, were isolated by a positive immunoselection procedure (CD14 MicroBeads; Miltenyi Biotec), according to the manufacturer’s instructions, or by Percoll gradient. Stable transfection of human CXCR4 in murine pre-B 300.19 was performed as described previously (Loetscher et al., 1994). Experiments performed with human monocytes and cells transfected with human CXCR4 were approved by Swiss federal authority for Biotechnology and Public Health (N.A000208/1). NIH/3T3 mouse fibroblasts were grown in DME supplemented with 10% FCS. BM cells from C57BL/6 mice (WT, Rage−/−, MyD88−/−, or Tlr4−/−) were isolated by flushing the medullary cavities of tibias and femurs with saline phosphate buffer. Rage−/− BM cells were incubated for 30 min in the presence or absence of 10 µg/ml LPS-Rs before performing chemotaxis assay. HEK293T cells were from the American Type Culture Collection. WT and mutant mouse embryonic fibroblast (MEF) matched pairs were derived from same-litter embryos obtained from Rage+/− or Cxcr4+/− crosses.
Constructs.
The human CXCR4 cDNA was amplified by PCR from pcDNA3-CXCR4 construct using the following primers and cloned into pECFP-N1 and pEYFP-N1 (Takara Bio Inc.): 5′ HindIII (5′-ATAAGCTTATGGAGGGGATCAGTATATACATTC-3′) and 3′ AgeI (5′-GACCGGTGGATCCCGTAAGCTGGAGTGAAAACTTGAAG-3′). mGluR1α cloned into pEYFP-N1 vector (mGluR1α-YFP) was provided by R. Franco (Universidad Autònoma de Barcelona, Barcelona, Spain).
Mice and treatments.
C57BL/6 and BALB/c mice were purchased from Harlan and the Jackson Laboratory, respectively, and C57BL/6 MyD88−/− mice (Adachi et al., 1998) were provided by S. Akira (Osaka University, Suita, Osaka, Japan). C57BL/6 Rage−/− mice (Liliensiek et al., 2004) were provided by A. Bierhaus (University of Heidelberg, Heidelberg, Germany). C57BL/6 Tlr4−/− mice were provided by M. Manz (University of Zurich, Zurich, Switzerland). Cxcr4+/− mice (Zou et al., 1998) were provided by D. Littman (New York University School of Medicine, New York, NY). Sterile injury was induced by injection of 15 µM CTX in the TA muscle. Air pouches were established in 8-wk-old male C57BL/6 mice by injecting subcutaneously 5 ml and 3 ml of air at day 0 and day 3, respectively. At day 6, mice were injected with 200 µl PBS, 10 pmol CXCL12, 300 pmol HMGB1, or 10 pmol CXCL12 + 300 pmol HMGB1 in the air pouch. After 6 h, cells were collected from the air pouch and stained with α-Ly6C (BD) and α-CD11b (BioLegend) antibodies. For AMD3100 pretreatment, mice were implanted with mini-osmotic pumps (DURECT Corporation) that infused 1.3 mg AMD3100 or an equivalent volume of saline over a period of 3 d before CTX injury (Jujo et al., 2010). For glycyrrhizin or α-HMGB1 antibody pretreatment, mice were injected intravenously with 200 µg glycyrrhizin (Minophagen Pharmaceutical Co.) or α-HMGB1 monoclonal antibody DPH1.1 3 h before CTX-induced injury. After 6 h, infiltrating cells were isolated from muscles at 37°C in a buffer containing Collagenase P and Collagenase D (Roche). Cells were stained with α-Ly6C (BD), α-CD11b (BioLegend), and α-CCR2 (FAB5538A; R&D Systems) antibodies, and the population of CD11b+ was enriched using anti-PE microbeads (Miltenyi Biotech) and analyzed by flow cytometry per time unit (120 s) at constant flow. Results were normalized against the total circulating leucocytes.
The animal experimentation performed at the San Raffaele Science Institute was approved by Comitato Istituzionale per la Buona Sperimetazione Animale della Fondazione San Raffaele del Monte Tabor on July 15, 2011. The animal experimentation performed at the Institute for Research in Biomedicine was approved by the Dipartimento della Sanitá e della Socialitá (Authorization N. 19/2010).
CXCL12 and HMGB1.
CXCL12 was synthesized using tBoc solid-phase chemistry (Clark-Lewis et al., 1997). Full-length HMGB1, produced and stored in buffers containing DTT (Knapp et al., 2004), was provided by HMGBiotech S.r.l. Using the Cambrex Limulus Amoebocyte Assay QCL-1000 (Lonza), we detected ∼1.1 pg LPS in 1 mg HMGB1 and 2.2 pg LPS after 1 mg HMGB1 was terminally digested with trypsin (therefore excluding any interference of HMGB1 with the limulus assay). The amount of LPS present in the assays in which HMGB1 was used is totally ineffective when administered alone. For NMR experiments, 15N-labeled CXCL12 or 15N-labeled BoxB were obtained by growing Escherichia coli Rosetta cells (EMD) transfected with pET30 vector (EMD) containing the respective sequences in M9 minimal media containing 15N-H4Cl. CXCL12 was renatured from inclusion bodies. BoxB and CXCL12 were purified according to standard techniques (Proudfoot and Borlat, 2000). CXCL12 was eluted in 20 mM NaCl and 20 mM Na phosphate buffer, pH 6.
nSpray-ESI-MS analysis of intact proteins.
Protein samples in solution were acidified with 10% formic acid and desalted with ZipTip C4 (Millipore) according to the manufacturer’s protocol for direct nano-ESI-MS analysis on an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems). MS spectra were acquired in positive ion mode in the Orbitrap from m/z 1,000–2,500 with a resolution set to 60,000 or 100,000 at m/z 400. Peak deconvolution with the Bayesian Protein Reconstruct algorithm was performed using Xtract from QualBrowser software (version 2.07; Thermo Fisher Scientific) for molecular mass determination.
ELISA.
The concentration of CXCL12 in the supernatant of 2 × 106 human monocytes, murine 3T3 fibroblasts, or Rage−/− MEFs per milliliter stimulated for 2 h with HMGB1 was determined with sandwich ELISA kit (R&D Systems) according to the manufacturer’s protocol. HMGB1 bound to CXCL12 in muscle from CTX-treated and untreated mice, obtained by muscle fragmentation and tissue disaggregation in buffer containing Collagenase D (Roche), was determined by hybrid ELISA. In summary, the plate was coated with capture anti-CXCL12 antibody from ELISA kit (R&D Systems), and the detection of HMGB1 bound to CXCL12 in muscle was assessed with anti-HMGB1 antibody from HMGB1 sandwich ELISA kit (IBL International). The hybrid ELISA did not detect HMGB1 alone or CXCL12 alone.
Cytometric bead array (CBA).
IL-6, IL-10, CCL2, and TNF in muscle supernatant, obtained by muscle fragmentation and tissue disaggregation in buffer containing Collagenase D (Roche), from CTX-treated and untreated mice, were detected using the CBA inflammatory kit (BD) according to the manufacturer’s protocol.
Chemotaxis assays.
Chemotaxis was assayed in 48-well Boyden microchambers (Neuro Probe) as previously described (Uguccioni et al., 1995). In brief, CXCR4-transfected cells and freshly isolated monocytes (5 × 104) were diluted in RPMI 1640 supplemented with 20 mM Hepes, pH 7.4, and 1% pasteurized plasma protein solution (5% PPL SRK; Swiss Red Cross Laboratory). Cells were then added to the upper wells. After 90 min of incubation for monocytes and 120 min for the CXCR4-transfected cells, the membrane was removed, washed on the upper side with PBS, fixed, and stained. All assays were performed in triplicate, and for each well, the migrated cells were counted at 1,000-fold magnification in randomly selected fields. The following reagents were used to inhibit cell migration: PTX (EMD), anti-CXCR4 (Abcam), anti-CXCL12 (K15C; provided by F. Arenzana-Seisdedos, Institut Pasteur, Paris, France), and AMD3100 (Sigma-Aldrich).
Intracellular Ca2+ rise.
Pre-B 300.19–CXCR4+ cells (0.3 × 106) in 100 µl were loaded with Fura-2-AM (final concentration 50 nM) on poly-lysine–coated slides. Loaded cells were washed with buffer containing 155 mM NaCl, 4.5 mM KCl, 5 mM Hepes, 1 mM MgCl2, 2 mM CaCl2, and 10 mM glucose, and imaging was recorded with a 40× oil-immersion objective on an inverted microscope (Axiovert 200; Carl Zeiss) with excitation at 340 nm and 380 nm using the Polychrom V illumination system from TILL Photonics GmbH. Chemokines were injected after 60 s of recording, and recording was continued up to 3 min. The 340/380 ratio provides a relative measure of cytoplasmic-free Ca2+ concentration.
ERK phosphorylation.
Freshly isolated monocytes (106 cells/time point) were incubated in RPMI/Hepes for 10 min at 37°C and stimulated with CXCL12 ± HMGB1. The reaction was stopped by the addition of 10% trichloroacetic acid. Whole cell lysates were separated on 11% SDS-PAGE and transferred to polyvinylidene difluoride membranes. Activated ERK was detected with an anti–diphospho-ERK antibody (1:10,000; Sigma-Aldrich). Enhanced chemiluminescence was used for detection of horseradish peroxidase–conjugated secondary antibody (Bio-Rad Laboratories, Hercules, CA). Equal loading was confirmed by reprobing with an anti-GAPDH antibody (AbD Serotec). Densitometric analysis of the bands obtained by Western blotting was performed with AIDA software (Aida Image analyzer version 3.28).
Pull-down of CXCR4.
Pre-B 300.19–CXCR4+ cells were incubated with biotinylated or nonbiotinylated CXCL12 or HMGB1 at 500 nM for 10 min on ice. Cells were lysed in 30 mM Hepes, 100 mM KCl, 20 mM NaCl, 2 mM MgCl2, 5% glycerol, 5 µM GDP, 2% Heptanetriol, 0.4% lauryl maltoside, 0.08% cholesteryl hemisuccinate, 1 µM microcystin, 1 mM NaVO4, and Complete protease inhibitors (Roche). Receptor–ligand complexes were pulled down using streptavidin-Sepharose (SA; GE Healthcare) and eluted using sample buffer containing 50 mM Tris-HCl, pH 6.8, 2% glycerol, 100 mM Na dodecyl-sulfate, and bromophenol blue. CXCR4 was analyzed by Western blotting using anti-CXCR4 (SZ1567; custom prepared; Eurogentec) and horseradish peroxidase–conjugated goat anti–rabbit antibody (Santa Cruz Biotechnology, Inc.). Enhanced chemiluminescence was used for detection according to the manufacturer’s instructions (Thermo Fisher Scientific).
NMR experiments.
15N-HSQC experiments were performed on a Bruker 700-MHz spectrometer with CryoProbe. Acquisition time varied between 30 min (free CXCL12 or BoxB) and 16 h (complexes). An HSQC spectrum was recorded on the free, labeled component; a second HSQC was recorded after addition of the unlabeled partner (1:1 ration for the CXCL12–BoxB or –BoxA complexes; 2:1 for CXCL12–HMGB1). Backbone assignments were obtained by comparison to previously published data (Biological Magnetic Resonance Bank accession numbers 15148, 15149, and 16145).
Chemical shift mapping.
Residues whose NMR signal changes position upon complex formation were identified by visual comparison of free and bound spectra of CXCL12 or BoxB in complex with partners. For quantitative analysis, the amount of chemical shift change upon complex formation was calculated for each residue according to the formula where ΔN and ΔH are the chemical shift difference between free and bound spectrum in the nitrogen and proton dimension.
SPR.
SPR experiments were performed on a Proteon-XPR36 instrument (Bio-Rad Laboratories). HMGB1 was immobilized on a GLM sensor surface through amine coupling according to standard protocols. In a first experiment, CXCL12 was added at concentrations of 250 and 500 nM in different sensor channels, showing increased binding to HMGB1 at increasing concentration. In a second experiment, glycyrrhizin was flowed over the surface at concentrations of 0, 500, or 5,000 nM, followed by the addition of 500 nM CXCL12. CXCL12 binding to HMGB1 was detected in all cases, but the signal intensity decreased at increasing concentrations of glycyrrhizin. All experiments were conducted at pH 6.0 in 20 mM Na phosphate buffer and 20 mM NaCl. Several reference channels were used, either flowing CXCL12 and glycyrrhizin over a sensor surface in the absence of HMGB1 or flowing buffer without CXCL12 or glycyrrhizin over a surface with immobilized HMGB1.
FRET experiments.
FRET saturation curves were performed as described previously (Martínez Muñoz et al., 2009). Gain settings were identical for all experiments to maintain a constant relative contribution of fluorescent proteins to the detection channels for spectral imaging and linear unmixing (Zimmermann et al., 2002). The contribution of CFP and YFP alone was measured in each detection channel (spectral signature) and normalized to the sum of the signal obtained for both channels. For quantitation, the spectral signature was taken into consideration for linear unmixing to separate the two emission spectra. To determine the fluorescence emitted by each fluorescent protein, we applied the following general formulas: CFP = S/(1 + 1/R) and YFP = S/(1 + R), where S = ChCFP + ChYFP, R = ((YFP530Q) − YFP486)/(CFP486 − (CFP530Q)), and Q = ChCFP/ChYFP. ChCFP and ChYFP represent the signal in the 486- and 530-nm detection channels (Ch); CFP486, CFP530, YFP530, and YFP486 represent the normalized contributions of CFP and YFP to channels 486–530, as determined from spectral signatures of the fluorescent proteins.
Statistics.
Statistical analysis was performed using the two-tailed Student’s t test (unpaired) for means in the CXCL12 measurements by ELISA. Migration assays with CXCL12 and HMGB1 and CBA measurements of cytokines were analyzed by analysis of variance (ANOVA) plus Bonferroni posttest. ANOVA followed by Dunnett’s multiple comparison tests was used for the in vivo experiments and migration of 3T3 mouse fibroblasts. To determine FRET50 and FRETmax, values were extrapolated from data analysis using a nonlinear regression equation applied to a single binding site model with a 95% confidence interval (Prism 5.0; GraphPad Software).
Acknowledgments
We thank Gabriela Danelon for excellent technical assistance, Angela Cattaneo for mass spectrometry, Dr. Giovanni Sitia, Denise Bottinelli, and Tanja Rezzonico for assisting in the experiments with mice, Dr. Giovanna Musco for the constructs and protocols for the expression and purification of HMG-box domains, Dr. Maura Casalgrandi and Alessandro Catucci for the production of HMGB1, and Dr. Roberta Palumbo for discussion.
This work was funded by grants from European Union (EU) FP6 (INNOCHEM, LSHB-CT-2005-518167; and DEC-VAC, LSHP-CT-2005-018685 to M. Uguccioni) and FP7 (Endostem to HMGBiotech S.r.l. and ADITEC 280873 and TIMER 281608 to M. Uguccioni), the Swiss National Science Foundation (3100A0-118048/1 to M. Uguccioni), the San Salvatore Foundation, the Helmut Horten Foundation, the Institute for Arthritis Research, the Swiss Vaccine Research Institute, Associazione Italiana Ricerca sul Cancro (AIRC; to M.E. Bianchi), the Spanish Ministry of Science and Innovation (SAF 2008-03388 to M. Mellado and L. Martínez Muñoz), and the Instituto de Salud Carlos III (RD08/0075/0010; RIER to M. Mellado and L. Martínez Muñoz). A. Raucci was partially supported by a fellowship from the AIRC. NMR time was granted at the Biomolekulares Magnetresonanz Zentrum der Universität Frankfurt as part of the EU project Bio-NMR, contract no. 261863.
The authors have no conflicting financial interests. However, M.E. Bianchi is founder and part owner of HMGBiotech S.r.l., a company which provides goods and services related to HMGB proteins, and A. Raucci and B. Celona were partially supported by HMGBiotech S.r.l.
Author contributions: M. Schiraldi carried out most of the work. M. Schiraldi, F. De Marchis, and B. Celona performed migration assays and in vivo experiments. E. Venereau fully characterized the chemical identity of HMGB1. E. Livoti, L. Varani, M. Pedotti, and A. Proudfoot designed and carried out protein production and NMR analysis. L. Martínez Muñoz, M. Schiraldi, and M. Mellado performed the FRET experiments. M. Schiraldi, T. Apuzzo, and M. Thelen performed CXCR4 pull-down experiments and developed reagents. A. Bachi was responsible for mass spectrometry. M. Schiraldi, A. Raucci, M.E. Bianchi, and M. Uguccioni designed the experiments and discussed the data. M. Schiraldi, M.E. Binachi, and M. Uguccioni wrote the paper. All authors commented and contributed to the work.
Footnotes
Abbreviations used:
- ANOVA
- analysis of variance
- CBA
- cytometric bead array
- CTX
- cardiotoxin
- ERK
- extracellular signal-regulated kinase
- FRET
- fluorescence resonance energy transfer
- HSQC
- heteronuclear single-quantum coherence
- MEF
- mouse embryonic fibroblast
- NMR
- nuclear magnetic resonance
- PTX
- Bordetella pertussis toxin
- RAGE
- receptor for advanced glycation end products
- SPR
- surface plasmon resonance
- TA
- tibialis anterior
- TLR
- Toll-like receptor
References
- Adachi O., Kawai T., Takeda K., Matsumoto M., Tsutsui H., Sakagami M., Nakanishi K., Akira S. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 9:143–150 10.1016/S1074-7613(00)80596-8 [DOI] [PubMed] [Google Scholar]
- Agace W.W., Amara A., Roberts A.I., Pablos J.L., Thelen S., Uguccioni M., Li X.Y., Marsal J., Arenzana-Seisdedos F., Delaunay T., et al. 2000. Constitutive expression of stromal derived factor-1 by mucosal epithelia and its role in HIV transmission and propagation. Curr. Biol. 10:325–328 10.1016/S0960-9822(00)00380-8 [DOI] [PubMed] [Google Scholar]
- Andersson U., Harris H.E. 2010. The role of HMGB1 in the pathogenesis of rheumatic disease. Biochim. Biophys. Acta. 1799:141–148 [DOI] [PubMed] [Google Scholar]
- Andrassy M., Volz H.C., Igwe J.C., Funke B., Eichberger S.N., Kaya Z., Buss S., Autschbach F., Pleger S.T., Lukic I.K., et al. 2008. High-mobility group box-1 in ischemia-reperfusion injury of the heart. Circulation. 117:3216–3226 10.1161/CIRCULATIONAHA.108.769331 [DOI] [PubMed] [Google Scholar]
- Apetoh L., Ghiringhelli F., Tesniere A., Obeid M., Ortiz C., Criollo A., Mignot G., Maiuri M.C., Ullrich E., Saulnier P., et al. 2007. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med. 13:1050–1059 10.1038/nm1622 [DOI] [PubMed] [Google Scholar]
- Bianchi M.E. 2009. HMGB1 loves company. J. Leukoc. Biol. 86:573–576 10.1189/jlb.1008585 [DOI] [PubMed] [Google Scholar]
- Campana L., Bosurgi L., Bianchi M.E., Manfredi A.A., Rovere-Querini P. 2009. Requirement of HMGB1 for stromal cell-derived factor-1/CXCL12-dependent migration of macrophages and dendritic cells. J. Leukoc. Biol. 86:609–615 10.1189/jlb.0908576 [DOI] [PubMed] [Google Scholar]
- Chen L.Y., Pan Z.K. 2009. Synergistic activation of leukocytes by bacterial chemoattractants: potential drug targets. Endocr. Metab. Immune Disord. Drug Targets. 9:361–370 http://www.ncbi.nlm.nih.gov/pubmed/19601917?dopt=Abstract [DOI] [PubMed] [Google Scholar]
- Clark-Lewis I., Vo L., Owen P., Anderson J. 1997. Chemical synthesis, purification, and folding of C-X-C and C-C chemokines. Methods Enzymol. 287:233–250 10.1016/S0076-6879(97)87018-8 [DOI] [PubMed] [Google Scholar]
- Crump M.P., Gong J.H., Loetscher P., Rajarathnam K., Amara A., Arenzana-Seisdedos F., Virelizier J.L., Baggiolini M., Sykes B.D., Clark-Lewis I. 1997. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 16:6996–7007 10.1093/emboj/16.23.6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degryse B., Bonaldi T., Scaffidi P., Müller S., Resnati M., Sanvito F., Arrigoni G., Bianchi M.E. 2001. The high mobility group (HMG) boxes of the nuclear protein HMG1 induce chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. J. Cell Biol. 152:1197–1206 10.1083/jcb.152.6.1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiPersio J.F., Uy G.L., Yasothan U., Kirkpatrick P. 2009. Plerixafor. Nat. Rev. Drug Discov. 8:105–106 10.1038/nrd2819 [DOI] [PubMed] [Google Scholar]
- Ditsworth D., Zong W.X., Thompson C.B. 2007. Activation of poly(ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J. Biol. Chem. 282:17845–17854 10.1074/jbc.M701465200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S., Dragoi A.M., Wang X., Dallacosta C., Louten J., Musco G., Sitia G., Yap G.S., Wan Y., Biron C.A., et al. 2007. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood. 110:1970–1981 10.1182/blood-2006-09-044776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jujo K., Hamada H., Iwakura A., Thorne T., Sekiguchi H., Clarke T., Ito A., Misener S., Tanaka T., Klyachko E., et al. 2010. CXCR4 blockade augments bone marrow progenitor cell recruitment to the neovasculature and reduces mortality after myocardial infarction. Proc. Natl. Acad. Sci. USA. 107:11008–11013 10.1073/pnas.0914248107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S., Müller S., Digilio G., Bonaldi T., Bianchi M.E., Musco G. 2004. The long acidic tail of high mobility group box 1 (HMGB1) protein forms an extended and flexible structure that interacts with specific residues within and between the HMG boxes. Biochemistry. 43:11992–11997 10.1021/bi049364k [DOI] [PubMed] [Google Scholar]
- Koenen R.R., von Hundelshausen P., Nesmelova I.V., Zernecke A., Liehn E.A., Sarabi A., Kramp B.K., Piccinini A.M., Paludan S.R., Kowalska M.A., et al. 2009. Disrupting functional interactions between platelet chemokines inhibits atherosclerosis in hyperlipidemic mice. Nat. Med. 15:97–103 10.1038/nm.1898 [DOI] [PubMed] [Google Scholar]
- Levoye A., Balabanian K., Baleux F., Bachelerie F., Lagane B. 2009. CXCR7 heterodimerizes with CXCR4 and regulates CXCL12-mediated G protein signaling. Blood. 113:6085–6093 10.1182/blood-2008-12-196618 [DOI] [PubMed] [Google Scholar]
- Liliensiek B., Weigand M.A., Bierhaus A., Nicklas W., Kasper M., Hofer S., Plachky J., Gröne H.J., Kurschus F.C., Schmidt A.M., et al. 2004. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J. Clin. Invest. 113:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey K.M., Tang D., Zeh H.J., Lotze M.T. 2009. Autophagy inhibition in combination cancer treatment. Curr. Opin. Investig. Drugs. 10:1269–1279 [PubMed] [Google Scholar]
- Loetscher M., Geiser T., O’Reilly T., Zwahlen R., Baggiolini M., Moser B. 1994. Cloning of a human seven-transmembrane domain receptor, LESTR, that is highly expressed in leukocytes. J. Biol. Chem. 269:232–237 [PubMed] [Google Scholar]
- Lotze M.T., Tracey K.J. 2005. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 5:331–342 10.1038/nri1594 [DOI] [PubMed] [Google Scholar]
- Maroso M., Balosso S., Ravizza T., Liu J., Aronica E., Iyer A.M., Rossetti C., Molteni M., Casalgrandi M., Manfredi A.A., et al. 2010. Toll-like receptor 4 and high-mobility group box-1 are involved in ictogenesis and can be targeted to reduce seizures. Nat. Med. 16:413–419 10.1038/nm.2127 [DOI] [PubMed] [Google Scholar]
- Martínez Muñoz L., Lucas P., Navarro G., Checa A.I., Franco R., Martínez-A C., Rodríguez-Frade J.M., Mellado M. 2009. Dynamic regulation of CXCR1 and CXCR2 homo- and heterodimers. J. Immunol. 183:7337–7346 10.4049/jimmunol.0901802 [DOI] [PubMed] [Google Scholar]
- Mollica L., De Marchis F., Spitaleri A., Dallacosta C., Pennacchini D., Zamai M., Agresti A., Trisciuoglio L., Musco G., Bianchi M.E. 2007. Glycyrrhizin binds to high-mobility group box 1 protein and inhibits its cytokine activities. Chem. Biol. 14:431–441 10.1016/j.chembiol.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Murphy P.M. 2002. International Union of Pharmacology. XXX. Update on chemokine receptor nomenclature. Pharmacol. Rev. 54:227–229 10.1124/pr.54.2.227 [DOI] [PubMed] [Google Scholar]
- Orlova V.V., Choi E.Y., Xie C., Chavakis E., Bierhaus A., Ihanus E., Ballantyne C.M., Gahmberg C.G., Bianchi M.E., Nawroth P.P., Chavakis T. 2007. A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. EMBO J. 26:1129–1139 10.1038/sj.emboj.7601552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo R., De Marchis F., Pusterla T., Conti A., Alessio M., Bianchi M.E. 2009. Src family kinases are necessary for cell migration induced by extracellular HMGB1. J. Leukoc. Biol. 86:617–623 10.1189/jlb.0908581 [DOI] [PubMed] [Google Scholar]
- Paoletti S., Petkovic V., Sebastiani S., Danelon M.G., Uguccioni M., Gerber B.O. 2005. A rich chemokine environment strongly enhances leukocyte migration and activities. Blood. 105:3405–3412 10.1182/blood-2004-04-1648 [DOI] [PubMed] [Google Scholar]
- Penzo M., Molteni R., Suda T., Samaniego S., Raucci A., Habiel D.M., Miller F., Jiang H.P., Li J., Pardi R., et al. 2010. Inhibitor of NF-kappa B kinases alpha and beta are both essential for high mobility group box 1-mediated chemotaxis [corrected]. J. Immunol. 184:4497–4509 10.4049/jimmunol.0903131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percherancier Y., Berchiche Y.A., Slight I., Volkmer-Engert R., Tamamura H., Fujii N., Bouvier M., Heveker N. 2005. Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J. Biol. Chem. 280:9895–9903 10.1074/jbc.M411151200 [DOI] [PubMed] [Google Scholar]
- Proudfoot A.E., Borlat F. 2000. Purification of recombinant chemokines from E. coli. Methods Mol. Biol. 138:75–87 [DOI] [PubMed] [Google Scholar]
- Sha Y., Zmijewski J., Xu Z., Abraham E. 2008. HMGB1 develops enhanced proinflammatory activity by binding to cytokines. J. Immunol. 180:2531–2537 [DOI] [PubMed] [Google Scholar]
- Sitia G., Iannacone M., Müller S., Bianchi M.E., Guidotti L.G. 2007. Treatment with HMGB1 inhibitors diminishes CTL-induced liver disease in HBV transgenic mice. J. Leukoc. Biol. 81:100–107 10.1189/jlb.0306173 [DOI] [PubMed] [Google Scholar]
- Sitia G., Iannacone M., Aiolfi R., Isogawa M., van Rooijen N., Scozzesi C., Bianchi M.E., von Andrian U.H., Chisari F.V., Guidotti L.G. 2011. Kupffer cells hasten resolution of liver immunopathology in mouse models of viral hepatitis. PLoS Pathog. 7:e1002061 10.1371/journal.ppat.1002061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner M. 2010. Autophagy: in the hands of HMGB1. Nat. Rev. Mol. Cell Biol. 11:756–757 10.1038/nrm2994 [DOI] [PubMed] [Google Scholar]
- Tang D., Kang R., Livesey K.M., Cheh C.W., Farkas A., Loughran P., Hoppe G., Bianchi M.E., Tracey K.J., Zeh H.J., III, Lotze M.T. 2010. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 190:881–892 10.1083/jcb.200911078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Avalos A.M., Mao S.Y., Chen B., Senthil K., Wu H., Parroche P., Drabic S., Golenbock D., Sirois C., et al. 2007. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat. Immunol. 8:487–496 10.1038/ni1457 [DOI] [PubMed] [Google Scholar]
- Tsung A., Klune J.R., Zhang X., Jeyabalan G., Cao Z., Peng X., Stolz D.B., Geller D.A., Rosengart M.R., Billiar T.R. 2007. HMGB1 release induced by liver ischemia involves Toll-like receptor 4–dependent reactive oxygen species production and calcium-mediated signaling. J. Exp. Med. 204:2913–2923 10.1084/jem.20070247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uguccioni M., D’Apuzzo M., Loetscher M., Dewald B., Baggiolini M. 1995. Actions of the chemotactic cytokines MCP-1, MCP-2, MCP-3, RANTES, MIP-1 α and MIP-1 β on human monocytes. Eur. J. Immunol. 25:64–68 10.1002/eji.1830250113 [DOI] [PubMed] [Google Scholar]
- Varani L., Bankovich A.J., Liu C.W., Colf L.A., Jones L.L., Kranz D.M., Puglisi J.D., Garcia K.C. 2007. Solution mapping of T cell receptor docking footprints on peptide-MHC. Proc. Natl. Acad. Sci. USA. 104:13080–13085 10.1073/pnas.0703702104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venetz D., Ponzoni M., Schiraldi M., Ferreri A.J., Bertoni F., Doglioni C., Uguccioni M. 2010. Perivascular expression of CXCL9 and CXCL12 in primary central nervous system lymphoma: T-cell infiltration and positioning of malignant B cells. Int. J. Cancer. 127:2300–2312 10.1002/ijc.25236 [DOI] [PubMed] [Google Scholar]
- von Hundelshausen P., Koenen R.R., Sack M., Mause S.F., Adriaens W., Proudfoot A.E., Hackeng T.M., Weber C. 2005. Heterophilic interactions of platelet factor 4 and RANTES promote monocyte arrest on endothelium. Blood. 105:924–930 10.1182/blood-2004-06-2475 [DOI] [PubMed] [Google Scholar]
- Wang H., Bloom O., Zhang M., Vishnubhakat J.M., Ombrellino M., Che J., Frazier A., Yang H., Ivanova S., Borovikova L., et al. 1999. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 285:248–251 10.1126/science.285.5425.248 [DOI] [PubMed] [Google Scholar]
- Wu B., Chien E.Y., Mol C.D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F.C., et al. 2010. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 330:1066–1071 10.1126/science.1194396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Chen Q., Yang H., Tracey K.J., Bustin M., Oppenheim J.J. 2007. High mobility group box-1 protein induces the migration and activation of human dendritic cells and acts as an alarmin. J. Leukoc. Biol. 81:59–66 10.1189/jlb.0306180 [DOI] [PubMed] [Google Scholar]
- Yang H., Hreggvidsdottir H.S., Palmblad K., Wang H., Ochani M., Li J., Lu B., Chavan S., Rosas-Ballina M., Al-Abed Y., et al. 2010. A critical cysteine is required for HMGB1 binding to Toll-like receptor 4 and activation of macrophage cytokine release. Proc. Natl. Acad. Sci. USA. 107:11942–11947 10.1073/pnas.1003893107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lundbäck P., Ottosson L., Erlandsson-Harris H., Venereau E., Bianchi M.E., Al-Abed Y., Andersson U., Tracey K.J., Antoine D.J. 2012. Redox modification of cysteine residues regulates the cytokine activity of HMGB1. Mol. Med. 10.2119/molmed.2011.00389 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zimmermann T., Rietdorf J., Girod A., Georget V., Pepperkok R. 2002. Spectral imaging and linear un-mixing enables improved FRET efficiency with a novel GFP2-YFP FRET pair. FEBS Lett. 531:245–249 10.1016/S0014-5793(02)03508-1 [DOI] [PubMed] [Google Scholar]
- Zou Y.R., Kottmann A.H., Kuroda M., Taniuchi I., Littman D.R. 1998. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 393:595–599 10.1038/31269 [DOI] [PubMed] [Google Scholar]