A patient with a homozygous premature stop codon in PIK3R1 showed an early developmental block in B cell development but minimal effects in other organ systems.
Abstract
Whole exome sequencing was used to determine the causative gene in patients with B cell defects of unknown etiology. A homozygous premature stop codon in exon 6 of PIK3R1 was identified in a young woman with colitis and absent B cells. The mutation results in the absence of p85α but normal expression of the p50α and p55α regulatory subunits of PI3K. Bone marrow aspirates from the patient showed <0.1% CD19+ B cells with normal percentages of TdT+VpreB+CD19− B cell precursors. This developmental block is earlier than that seen in patients with defects in the B cell receptor signaling pathway or in a strain of engineered mice with a similar defect in p85α. The number and function of the patient’s T cells were normal. However, Western blot showed markedly decreased p110δ, as well as absent p85α, in patient T cells, neutrophils, and dendritic cells. The patient had normal growth and development and normal fasting glucose and insulin. Mice with p85α deficiency have insulin hypersensitivity, defective platelet function, and abnormal mast cell development. In contrast, the absence of p85α in the patient results in an early and severe defect in B cell development but minimal findings in other organ systems.
Approximately 85% of patients with the early onset of infections, panhypogammaglobulinemia, and markedly reduced or absent B cells have mutations in the gene encoding BTK (Conley et al., 1998), a hematopoietic-specific tyrosine kinase that is phosphorylated and activated by signaling through the pre-BCR and BCR (de Weers et al., 1994; Guo et al., 2000). Another 5–7% of patients with isolated defects in B cell development have mutations in components of the pre-BCR, including μ heavy chain, Igα, Igβ, or λ5 (Conley et al., 2009). Mutations in BLNK, a scaffold protein which assembles BTK with other downstream molecules involved in response to BCR activation, account for another small percentage of patients. The bone marrow from patients with all of these disorders shows a similar block in B cell development. Although patients have normal numbers of pro-B cells, they have markedly reduced or absent pre-B cells; this block coincides with the earliest expression of the pre-BCR (Conley et al., 2009).
Mice with null mutations in μ heavy chain, Igα, or Igβ demonstrate a block in B cell differentiation that is similar to that seen in patients with mutations in the same gene; however, defects in Btk (Rawlings et al., 1993), λ5 (Kitamura et al., 1992), and Blnk (Pappu et al., 1999) result in a milder phenotype in mice than in humans. Btk-deficient mice have reduced serum IgM and IgG3 but normal concentrations of IgG1, IgG2a, and IgG2b. These mice fail to make antibody to some T cell–independent antigens but respond normally to most T cell–dependent antigens. The number of circulating and splenic B cells is ∼50% of normal; however, there are reduced numbers of marginal zone B cells in the spleen and peritoneal B1 cells in these mice. The number of pre-B cells in the bone marrow is normal. In contrast, patients with mutations in BTK have reduced serum concentrations of all immunoglobulin isotypes; they fail to make antibodies to all vaccine antigens, and they have <1% of the normal number of B cells in the peripheral circulation. Mice that are null for λ5 or BLNK have a block in B cell differentiation that is more severe than that seen in Btk-deficient mice, but they have easily detected peripheral B cells (Kitamura et al., 1992; Pappu et al., 1999).
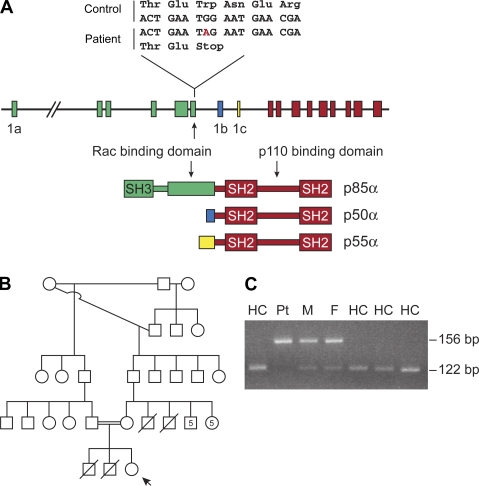
A B cell defect similar to that seen in Btk-deficient mice was reported in mice that are deficient in the p85α regulatory or p110δ catalytic subunit of class I PI3K (Fruman et al., 1999; Suzuki et al., 1999; Jou et al., 2002). PI3Ks are a broadly expressed group of enzymes that respond to a variety of extracellular signals to influence cell cycle progression, cell growth and survival, cell migration, and metabolic control (Engelman et al., 2006; Fruman, 2010; Vanhaesebroeck et al., 2010). There are multiple isoforms of PI3K, all of which function as heterodimers composed of a regulatory and a catalytic subunit. The gene that encodes p85α, PIK3R1, also encodes two additional regulatory isoforms, p55α and p50α (Fig. 1 A). The nine 3′ exons are shared by all three isoforms with two distinct promoters and two exon 1 sequences upstream of these nine exons controlling the production of p55α and p50α. The p85α subunit is ubiquitously expressed at relatively high concentrations in humans and mice, and somatic mutations in this gene have been identified in patients with malignancy (Engelman et al., 2006; Vanhaesebroeck et al., 2010).
Figure 1.
Mutation in PIK3R1 resulting in the absence of B lineage cells. (A) The intron/exon organization of PIK3R1 is shown, with the exons encoding p85α, p50α, and p55α color-coded to match the schematic drawing of the proteins. The specific mutation in exon 6 of PIK3R1 is shown above the gene. (B) The pedigree of the patient’s family is shown. Males who died of infection at <2 yr of age are indicated by a diagonal line through a square. The numbers within the square and circle indicate the number of males and females, respectively. The patient is indicated by an arrow. (C) PCR-based assay for mutation screening is shown. The lanes labeled HC contain digested PCR products from healthy controls. The lanes labeled Pt, M, and F contain digested PCR products from the patient, her mother, and her father, respectively.
Mice with deletions of the last five exons of Pik3r1 are null for all three isoforms and die in the perinatal period with hepatic necrosis and muscle and skeletal abnormalities (Fruman, 2010). However, lymphoid reconstitution of RAG-deficient mice with cells that lack all three isoforms results in normal T cell development, but the number of B cells in the spleen and lymph nodes is reduced to 10–30% of control. A second strain of knockout mice was constructed with a deletion of exon 1 of the p85α isoform (Suzuki et al., 1999). These mice, which are null for p85α but retain expression of p55α and p50α, are viable but have reduced numbers of B cells, enhanced responsiveness to insulin, and increased production of IL-12 by DCs (Fruman, 2010). Both strains of knockout mice have normal or elevated numbers of pro-B cells (B220+/CD43+) in the bone marrow and a B cell phenotype that is similar to that seen in Btk-deficient mice. In this manuscript, we report the identification of a patient with a premature stop codon in PIK3R1 resulting in absence of p85α but normal expression of p55α and p50α. The phenotype of this patient was restricted to colitis and a severe defect in B cell development with agammaglobulinemia, absent B cells, and markedly reduced or absent pro-B cells.
RESULTS AND DISCUSSION
Mutation detection
Whole exome sequencing was used to determine the genetic defect in patients with failure of B cell development of unknown etiology. A 19-yr-old female with a premature stop codon in PIK3R1 was identified. Direct sequencing of PCR products from this gene demonstrated that the patient was homozygous and both parents were heterozygous for a G to A substitution in codon 298, resulting in the replacement of the wild-type tryptophan with a premature stop codon in exon 6 (Fig. 1 A). To identify other patients with similar mutations, DNA from 55 patients with defects in B cell development of unknown etiology were screened for mutations in PIK3R1. No other mutations were found.
The patient, who has been previously reported (de la Morena et al., 1995), was from a consanguineous family of Chinese/Peruvian descent. She was evaluated for immunodeficiency at 3.5 mo of age because of neutropenia (absolute neutrophil count of 0), interstitial pneumonia, and gastroenteritis. The family history was positive for two older brothers and two maternal uncles who died of acute infections between 9 and 18 mo of age (Fig. 1 B). At her initial evaluation, the patient was found to have panhypogammaglobulinemia and <1% CD19+ B cells in the blood and bone marrow (de la Morena et al., 1995). CD56+ NK cells were present but in reduced numbers (11/µl with a normal range of 10–150/µl). The patient was treated with replacement gammaglobulin, and the infections and neutropenia resolved within 3 mo. She did well, with normal growth and development, until 12 yr of age when she developed erythema nodosum. At 15 yr of age, she was treated with TNF inhibitors and methotrexate for juvenile idiopathic arthritis. At 17 yr of age, she was recognized to have recurrent Campylobacter bacteremia and inflammatory bowel disease that has been recalcitrant to therapy. Complete blood counts at 19 yr of age were normal except for anemia of chronic disease. The absolute neutrophil count was 3,528/ml, and the lymphocyte count was 2,700/ml with 99% CD3+ T cells and 1% CD56+/CD16+ NK cells (absolute number 27/µl with a normal range of 50–400/µl). There was a normal distribution of CD4+/CD8+ (64%/29%), normal percentage of naive and memory T cells as evaluated by CD45RA (48% of CD4+ were CD45RA+) and CD45RO (40% of CD4+ were CD45RO+) expression, normal numbers of gamma-delta T cells (7%), normal numbers of regulatory T cells (1.2% CD4+/CD25+), and normal T cell proliferation in response to mitogens and antigens. Fasting glucose (87 mg/dl with normal range 70–125 mg/dl) and insulin levels (7.5 µIU/m with normal range 0–16.9 µIU/m) were normal at 19 yr of age, and there were no signs of hypoglycemia. PCR primers were designed to allow the detection of the wild-type versus mutant sequence by restriction digest with the enzyme BstXI. DNA from 100 controls was analyzed; only the patient and her parents demonstrated the mutant allele (Fig. 1 C).
Flow cytometry
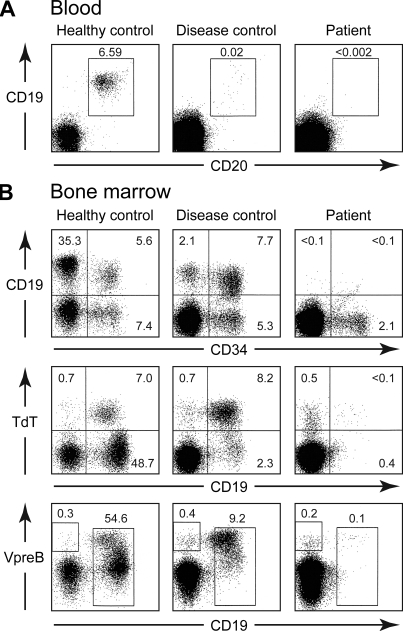
Flow cytometry experiments showed that the patient had <0.002% cells positive for the B cell markers CD19 and CD20 in the peripheral blood (Fig. 2 A). Bone marrow aspirates showed normal cellularity with an almost complete absence of B lineage cells (Fig. 2 B). In healthy controls and in patients with mutations in BTK or μ heavy chain, approximately half the cells that express the stem cell marker CD34 also express CD19; these CD34+CD19+ cells are pro-B cells (Dobbs et al., 2011). Although the patient had a normal percentage of CD34+CD19− cells (normal range 1–10%), she had <0.1% CD34+CD19+ cells (Fig. 2 B). TdT (terminal deoxytransferase) and VpreB, a B lineage–specific protein which forms part of the surrogate light chain, are found in the cytoplasm of B cell precursors before CD19 is expressed on the cell surface (Dobbs et al., 2011). The patient had a normal percentage of these very early B cell precursors. RACE (rapid amplification of cDNA ends) documented a complete absence of rearranged or sterile μ transcripts in the bone marrow sample.
Figure 2.
Analysis of B cell development in the blood and bone marrow by flow cytometry. (A) Ficoll-separated peripheral blood mononuclear cells were stained with PE-labeled anti-CD19 and FITC-labeled anti-CD20. The percentage of cells positive for CD19 and CD20 is indicated. The number of events shown is 20,000 for the healthy control and 250,000 for both the disease control (a patient with a mutation within a transcriptional regulatory element in intron 1 of BTK) and the patient. (B) Bone marrow cells were stained with PE-labeled anti-CD19 and FITC-labeled anti-CD34 or APC-labeled anti-CD19, FITC-labeled anti-TdT, and PE-labeled anti-VpreB. The percentage of cells within the lymphoid gate that fall into each of the sectors is shown. The number of events shown is 20,000 for the healthy control, 125,000 for the disease control (a patient with a mutation at the +1 donor splice site of intron 2 in BTK), and 250,000 for the patient. The peripheral blood analysis was performed on samples obtained from the p85α-deficient patient on three separate occasions. Bone marrow from the patient was obtained once and stained twice.
Expression of PI3K isoforms
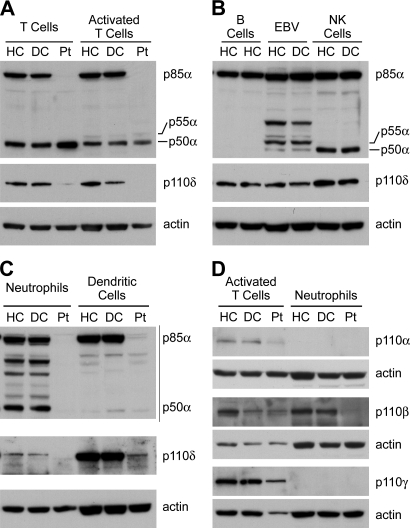
Although the PI3K isoforms have been studied extensively in malignant tissues and cells lines, there is very little information about expression of these proteins in primary hematopoietic cells. Therefore, we used immunoblotting to examine the expression of p85α, p55α, and p50α in purified hematopoietic cells and cell lines from healthy and disease controls (Fig. 3). Normal T cells expressed almost equal amounts of p85α and p50α, and activated T cells contained trace amounts of p55α, seen in long exposures, as well as p85α and p50α. In contrast, normal control B cells contained p85α but no detectable p55α or p50α; EBV-transformed B cell lines expressed some p55α as well as trace amounts of p50α. NK cells contained both p85α and p50α but less p50α than T cells.
Figure 3.
Expression of p85α, p55α, and p50α and the p110 isoforms in hematopoietic cells and cell lines from healthy controls, disease controls, and the patient. (A) Expression of the indicated proteins in peripheral blood T cells isolated by negative selection and in activated T cells that had been stimulated with phytohemagglutinin and then supplemented with 10% IL-2 every 2–3 d for 2 wk. (B) Expression of the same proteins in primary B cells, EBV-transformed B cell lines, and NK cells. Because both the patient (Pt) and the disease controls (DC) lack B cells, expression of PI3K isoforms from two healthy controls (HC) is shown to document that the absence of p50α and p55α was a reproducible finding. The patient had too few NK cells to permit analysis. (C) Expression of PI3K isoforms in neutrophils and DCs. The immunoblots in A–C were sequentially probed with antibodies to p110δ, p85α N terminus, p85α C terminus, and actin. (D) The expression of p110 isoforms in activated T cells and neutrophils is shown. The disease control for NK cell analysis had mutations in λ5. All other disease controls had mutations in BTK.
Normal neutrophils expressed approximately equal amounts of p85α and p50α. The p50α band in neutrophils was not seen using an antibody to the N-terminal SH3 domain of p85α, suggesting that the p50α band was not a degradation product. DCs from normal controls had abundant p85α but only trace amounts of p50α. Expression of p85α and p50α in cells from age-matched disease controls, patients with <1% CD19+ cells in the blood, was identical to that seen in healthy adult controls. As expected, the patient had no p85α in T cells, neutrophils, or DCs. The amount of p50α in T cells was normal or slightly increased, whereas the amount in DCs was normal and the amount in neutrophils was decreased. We did not identify a truncated form of p85α, encoded by the first six exons of PIK3R1, when using the antibody to the N-terminal portion of p85α to examine the patient’s T cells. Expression of p110δ, the leukocyte-specific partner of p85α, was decreased in T cells, neutrophils, and DCs from the patient (Fig. 3, A–C). This loss was surprising because the C-terminal p110-binding domain is shared by p85α, p55α, and p50α. The decreased expression of p110δ indicates that the N-terminal end of p85α contributes to the binding and stabilization of p110 or a 50% decrease in the amount of the p110-binding domain results in more significant loss of p110δ.
Expression of other p110 isoforms documented a slight decrease in p110α in the patient’s T cells (Fig. 3 D) and the absence of all p110 isoforms as well as p85α and p50α in the patient’s neutrophils. Although the patient had neutropenia at the time of diagnosis, she had normal neutrophil counts on multiple evaluations after the first year of life, and she had no history of infections typical of neutrophil deficiency. Neutropenia is often seen in patients with severe defects in B cell development who have not yet been started on gammaglobulin replacement (Conley and Howard, 2002). This has been attributed to viral-mediated bone marrow suppression (Conley and Howard, 2002).
Cytokine production by DCs
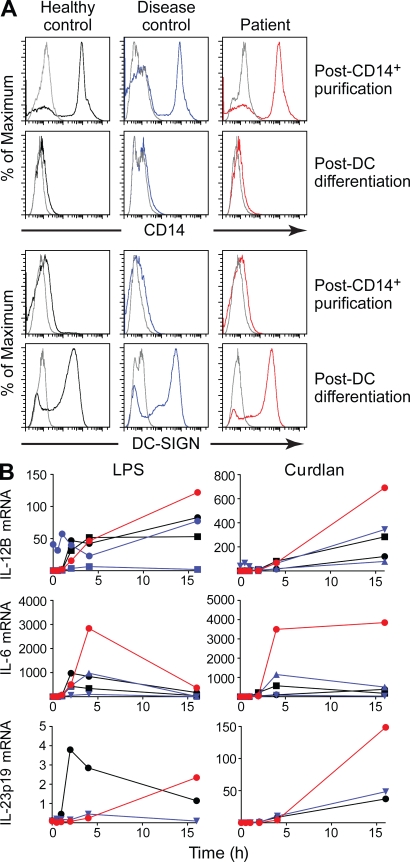
Increased IL-12 production by activated DCs from p85α knockout mice has been reported (Fukao et al., 2002); therefore, DCs from the patient were evaluated. CD14+ monocytes were isolated from the blood of the patient and controls and then cultured for 8 d with GM-CSF and IL-4 with the addition of TNF at day 7. Aliquots taken before and after culture showed that DC maturation was indistinguishable from controls as documented by loss of expression of CD14 and gain of expression of DC-SIGN (Fig. 4 A). DCs from the patient and controls were stimulated with LPS or curdlan for 1–16 h before RNA extraction. Quantitative RT-PCR (qRT-PCR) was used to compare the amount of transcripts for IL-12B, IL-6, and IL-23 in the unstimulated and stimulated samples. The kinetics of cytokine production was similar in the patient and control samples. However, the patient samples generally demonstrated higher peak concentration of IL-12B (encoding IL-p12p40), IL-23p19, and IL-6 transcripts in response to both LPS and curdlan. Production of IL-10, TNF, and IDO1 was similar in the patient and control samples.
Figure 4.
Cytokine production by cultured DCs from healthy controls, disease controls, and the patient. (A) The cell surface expression of monocyte-specific (CD14) and DC-specific (DC-SIGN) surface molecules was analyzed on cells immediately after selection on CD14 beads (post-CD14+ purification) and after a 7–8-d culture in GM-CSF + IL-4 with the addition of TNF for the final 24 h of culture (post-DC differentiation). The isotype control is shown in gray, and the specific staining for the healthy control, disease control, and patient cells are shown in black, blue, and red, respectively. (B) Representative qRT-PCR analysis of inflammatory messenger RNA (mRNA) expression by stimulated DCs. DCs isolated from control (black), disease controls (blue), or the patient (red) were stimulated with 100 ng/ml LPS (left) or 100 µg/ml curdlan (right), and messenger RNAs were analyzed by qRT-PCR with normalization to GAPDH. All the PCRs were performed twice with independent cDNA preps. The disease controls had mutations in BTK. Note the difference in scale in the response to LPS versus curdlan.
The results of this study indicate that mutations in the N-terminal region of p85α can result in a failure of B cell development. The block in B cell differentiation in the reported patient is earlier and more severe than the block seen in mice that are null for the same protein. The p85α knockout mice demonstrate normal numbers of early B cell precursors, and the block coincides with the expression of the pre-BCR (Suzuki et al., 1999). This suggests that in mice, p85α does not play a critical role before expression of an appropriate rearranged μ heavy chain. In contrast, the block in B cell differentiation in the patient occurs at the earliest stage of commitment to the B cell lineage, which coincides with CD19 expression. This early block is not an acquired defect as the patient had <1% CD19+ cells in the bone marrow and minimal VDJ rearrangement at 6 mo of age (de la Morena et al., 1995).
The signal transduction pathway in humans that requires p85α at the stage of B lineage commitment is not yet clear. One possible candidate is the receptor for SDF-1 (Stromal-derived factor), CXCR4, which is expressed on the cell surface of B lineage precursors at the earliest stage of development and transduces signals through PI3K (Aiuti et al., 1999). Mice that are deficient in CXCR4 have an early block in B cell development that is similar to that seen in the patient (Ma et al., 1998). It may be that signaling through CXCR4 is more dependent on PI3K, or more specifically p85α, for B cell development in humans compared with mice. Impaired signaling through CXCR4 may also be responsible for the decreased number of NK cells in the patient. CXCR4 is expressed on NK cell precursors and is required for mouse NK cell development (Noda et al., 2011). The loss of both p85α and p110δ in the patient’s cells supports the hypothesis that impaired signaling through CXCR4 contributes to her immunodeficiency. Human B cell lines that have been treated with a p110δ inhibitor exhibit reduced signaling through CXCR4 (Hoellenriegel et al., 2011).
Mouse studies have shown that loss of p85α results in increased sensitivity to insulin, defective platelet function, abnormal mast cell development, and increased production of IL-12 by DCs (Fukao et al., 2002; Fruman, 2010). However, we found the clinical consequences of the p85α defect in the patient to be relatively B cell specific. The inflammatory complications seen in the patient, erythema nodosum, arthritis, and colitis, may be caused by the antibody deficiency and resulting chronic Campylobacter infection. Erythema nodosum and arthritis, as well as colitis, have been reported in patients with Campylobacter infection (Ellis et al., 1982; Uotila et al., 2011). This organism, which has been reported in other patients with antibody deficiencies (van den Bruele et al., 2010), can be insidious and difficult to culture. It should also be noted that arthritis and inflammatory bowel disease have been reported in other patients with defects in B cell development (Howard et al., 2006).
Disordered cytokine production related to p85α may also contribute to the patient’s inflammatory complications. Her activated DCs expressed increased amounts of the inflammatory cytokine IL-12. A mouse study showed that decreased PI3K activity in the gut may result in dysregulated inflammation and poor bacterial clearance (Brown et al., 2011). Mice with kinase-impaired p110δ also have decreased regulatory T cell numbers and function (Patton et al., 2006). Normal numbers of regulatory T cells were seen in the peripheral blood of the patient, but it is possible that the number and function of these cells are altered at the site of inflammation. B cells in the gut normally produce the antiinflammatory cytokine IL-10 in response to inflammation (Mizoguchi et al., 2002); it may be that patients who lack B cells are predisposed to colitis.
The narrow phenotype of p85α deficiency in the reported patient is striking, considering the broad expression of this protein and the many signal transduction pathways that use it as a regulatory component. The earlier and more severe block in B cell differentiation in the patient, compared with mouse models, is also noteworthy. Both findings may reflect evolutionary requirements permitting survival of the relatively long-lived human species. Metabolic pathways may be more redundant in man to protect against hypoglycemia, and there may be more stringent control of B cell development to avoid complications of old age, including autoimmune disease and cancer. The phenotypic differences between p85α deficiency in humans and mice provide valuable information about the potential effects of PI3K inhibitors, which are currently being considered for the treatment of cancer and autoimmune disease (Engelman et al., 2006; Vanhaesebroeck et al., 2010).
MATERIALS AND METHODS
Patients.
All participants were enrolled in a research study to characterize genetic immunodeficiencies approved by the St. Jude Children’s Research Hospital Institutional Review Board. The disease controls were patients with reduced or absent B cells and mutations in BTK or IGLL1(λ5). Written informed consent was obtained from all participants and/or parents including the healthy controls.
Whole exome sequencing.
Exome enrichment of 3 µg of genomic DNA was performed using the SureSelect Human All Exon kit capture library (G3362) according to the manufacturer’s protocol (SureSelect Human All Exon Illumina Paired-End protocol version 1.0.1; Agilent Technologies). The capture library, containing regions totaling ∼38 Mb, is designed to target all human exons corresponding to exons in the NCBI consensus coding sequence (CCDS) database.
Captured DNAs were sequenced on a GAIIx sequencer (Illumina) with 76-bp pair-end reads. A single lane on a flow cell was used for the sample to generate sufficient reads for sequence alignment. Image analyses and base calling were performed using the Genome Analyzer Pipeline software (GAPipeline version 1.5 or greater; Illumina) with default parameters. Reads were aligned to a human reference sequence (UCSC assembly hg19, NCBI build 37), and genotypes were called at all positions where there were high quality sequence bases (Phred-like Q20 or greater) at minimum coverage of five, using Genomics Workbench version 4.0.2 (CLC Bio). Approximately 55% of 57 million reads were uniquely mapped to the targeted human exon regions to give a mean coverage of 52 (median of 39). Under such coverage, >95% of targeted regions were covered by 5 reads or more, and ∼80% were covered by >20 reads.
To identify the pathogenic mutation, we first discarded the variants that did not change the amino acid sequence. We then used an exclusion of alleles present in dbSNP131, reasoning that the causative variants were uncommon and unlikely to be registered there; and the variants were further filtered using our in-house database (collected exome data from >500 individuals) with similar methodology. The filtering flow is shown in Scheme 1.
 |
SCHEME 1 |
Because the patient was the product of a first cousin marriage, we placed a priority on homozygous alterations. Of 248 mutations and variants, the patient initially appeared to be homozygous for 6 variants. The single nucleotide variants (SNVs) in PPARGC1B and PLXNA4 demonstrated low coverage and on further analysis were found to be heterozygous. The single nucleotide variant in RBBP8 (Q414N) was tolerated by SIFT analysis and categorized as benign by PlyPhen-2 analysis. One of the insertion/deletion variants was in a pseudogene, LOC126987; the other, in FAM90A12, occurred in a copy number variant. Thus, only PIK3R1 was a plausible candidate.
Mutation screening.
PCR was used to screen DNA from controls for the mutation found in the patient. The primers 5′-TCATAAAAGTTATAGAAATTTTAATCCCAA-3′ and 5′-GAAGCTGTGTTACTTCAAAGG-3′ were used to amplify a 156-bp fragment that was digested with BstXI restriction enzyme (New England Biolabs, Inc.). The primers match the wild-type sequence except for the underlined C in the first primer. The recognition site for BstXI is CCANNNNNNTGG. The underlined C in the recognition sequence corresponds to the underlined C in the first primer. The underlined TGG in the recognition site is present in the wild-type sequence but not in the patient. Digestion of the PCR product from controls resulted in 35- and 121-bp fragments, which were separated by electrophoresis using a 4% agarose gel.
Flow cytometry and RACE analysis.
The techniques and reagents used for flow cytometry and RACE have been previously reported (Dobbs et al., 2011).
Cell separations and Western blots.
The EasySep Magnet and antibodies (STEMCELL Technologies) were used to purify primary T, B, and NK cells by negative selection. Neutrophils were purified by dextran sedimentation using 3% dextran T-500 (Sigma-Aldrich) with the addition of Mini protease inhibitor (Roche) at each step of separation. 1.5–5 × 106 lymphocytes, neutrophils, or DCs were resuspended in lysis buffer and incubated on ice for 10 min. Cell lysates equivalent to 1–5 × 105 cells were heated at 95°C for 5 min and subjected to SDS-PAGE on 9% acrylamide minigels. After protein transfer onto polyvinylidene fluoride membranes by electroblotting, membranes were blocked with 5% nonfat dry milk in TBS containing 0.1% Tween 20 overnight at 4°C, washed with TBS/Tween, and incubated in TBS/Tween containing 0.5% nonfat dry milk and primary antibodies against p85α, p85α N-SH3, p110δ, p110α, p110β, p110γ (all from Millipore), or β-actin (Sigma-Aldrich). For detection of p110δ and p85α N-SH3, we used horseradish peroxidase–conjugated goat anti–rabbit and goat anti–mouse (Thermo Fisher Scientific), respectively, followed by SuperSignal West Dura ECL (Thermo Fisher Scientific). To detect all other isoforms, we used horseradish peroxidase–conjugated goat anti–rabbit or goat anti–mouse (Promega), followed by Amersham ECL (GE Healthcare).
Purification and stimulation of DCs.
Monocytes from healthy control subjects and the patient were isolated from peripheral blood using Ficoll gradients followed by purification with CD14 magnetic beads according to the manufacturer’s instructions (Miltenyi Biotech). Monocytes were counted and plated in 80 ng/ml GM-CSF (Invitrogen) and 50 ng/ml IL-4 (Invitrogen) in RPMI containing 10% fetal bovine serum for 7–8 d with feeding by demi-depletion every other day using methods optimized from published protocols (Sallusto and Lanzavecchia, 1994). 10 ng/ml TNF was added during the last 24 h of culture as described by Sallusto and Lanzavecchia (1994). Cell samples before and after magnetic bead isolation and after culture were analyzed by flow cytometry using antibodies to CD11B, DC-SIGN, HLA-DR, and CD14 (BD). After differentiation, cells were counted and replated in complete media followed by stimulation with 100 ng/ml Escherichia coli LPS or 100 µg/ml curdlan (LeibundGut-Landmann et al., 2007). Cells were centrifuged, and total RNA was isolated by TRIZOL and subjected to qRT-PCR analysis using normalization with GAPDH-specific primers (Applied Biosystems). We used full-length human cDNAs encoding each inflammatory mediator (OriGene) as standards for each PCR assay. Patient samples were prepared in duplicate and analyzed in two separate experiments along with control samples.
Acknowledgments
We thank the patient and her family as well as our healthy and disease controls for their participation in this study. We also thank Melissa Mann and Linda Hendershot for technical advice and Betsy Williford for help in preparation of the figures.
This study was supported in part by grants from the National Institutes of Health (AI25129), National Cancer Institute grant P30 CA21765, American Lebanese Syrian Associated Charities, and by funds from the Federal Express Chair of Excellence and The Hartwell Foundation.
None of the authors report financial conflicts of interest.
Footnotes
Abbreviations used:
- qRT-PCR
- quantitative RT-PCR
References
- Aiuti A., Tavian M., Cipponi A., Ficara F., Zappone E., Hoxie J., Peault B., Bordignon C. 1999. Expression of CXCR4, the receptor for stromal cell-derived factor-1 on fetal and adult human lympho-hematopoietic progenitors. Eur. J. Immunol. 29:1823–1831 [DOI] [PubMed] [Google Scholar]
- Brown J.B., Cheresh P., Goretsky T., Managlia E., Grimm G.R., Ryu H., Zadeh M., Dirisina R., Barrett T.A. 2011. Epithelial phosphatidylinositol-3-kinase signaling is required for β-catenin activation and host defense against Citrobacter rodentium infection. Infect. Immun. 79:1863–1872 10.1128/IAI.01025-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley M.E., Howard V. 2002. Clinical findings leading to the diagnosis of X-linked agammaglobulinemia. J. Pediatr. 141:566–571 10.1067/mpd.2002.127711 [DOI] [PubMed] [Google Scholar]
- Conley M.E., Mathias D., Treadaway J., Minegishi Y., Rohrer J. 1998. Mutations in Btk in patients with presumed X-linked agammaglobulinemia. Am. J. Hum. Genet. 62:1034–1043 10.1086/301828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley M.E., Dobbs A.K., Farmer D.M., Kilic S., Paris K., Grigoriadou S., Coustan-Smith E., Howard V., Campana D. 2009. Primary B cell immunodeficiencies: comparisons and contrasts. Annu. Rev. Immunol. 27:199–227 10.1146/annurev.immunol.021908.132649 [DOI] [PubMed] [Google Scholar]
- de la Morena M., Haire R.N., Ohta Y., Nelson R.P., Litman R.T., Day N.K., Good R.A., Litman G.W. 1995. Predominance of sterile immunoglobulin transcripts in a female phenotypically resembling Bruton’s agammaglobulinemia. Eur. J. Immunol. 25:809–815 10.1002/eji.1830250327 [DOI] [PubMed] [Google Scholar]
- de Weers M., Brouns G.S., Hinshelwood S., Kinnon C., Schuurman R.K.B., Hendriks R.W., Borst J. 1994. B-cell antigen receptor stimulation activates the human Bruton’s tyrosine kinase, which is deficient in X-linked agammaglobulinemia. J. Biol. Chem. 269:23857–23860 [PubMed] [Google Scholar]
- Dobbs A.K., Bosompem A., Coustan-Smith E., Tyerman G., Saulsbury F.T., Conley M.E. 2011. Agammaglobulinemia associated with BCR− B cells and enhanced expression of CD19. Blood. 118:1828–1837 10.1182/blood-2011-01-330472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M.E., Pope J., Mokashi A., Dunbar E. 1982. Campylobacter colitis associated with erythema nodosum. Br. Med. J. (Clin. Res. Ed.). 285:937 10.1136/bmj.285.6346.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman J.A., Luo J., Cantley L.C. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7:606–619 10.1038/nrg1879 [DOI] [PubMed] [Google Scholar]
- Fruman D.A. 2010. Regulatory subunits of class IA PI3K. Curr. Top. Microbiol. Immunol. 346:225–244 10.1007/82_2010_39 [DOI] [PubMed] [Google Scholar]
- Fruman D.A., Snapper S.B., Yballe C.M., Davidson L., Yu J.Y., Alt F.W., Cantley L.C. 1999. Impaired B cell development and proliferation in absence of phosphoinositide 3-kinase p85alpha. Science. 283:393–397 10.1126/science.283.5400.393 [DOI] [PubMed] [Google Scholar]
- Fukao T., Tanabe M., Terauchi Y., Ota T., Matsuda S., Asano T., Kadowaki T., Takeuchi T., Koyasu S. 2002. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat. Immunol. 3:875–881 10.1038/ni825 [DOI] [PubMed] [Google Scholar]
- Guo B., Kato R.M., Garcia-Lloret M., Wahl M.I., Rawlings D.J. 2000. Engagement of the human pre-B cell receptor generates a lipid raft-dependent calcium signaling complex. Immunity. 13:243–253 10.1016/S1074-7613(00)00024-8 [DOI] [PubMed] [Google Scholar]
- Hoellenriegel J., Meadows S.A., Sivina M., Wierda W.G., Kantarjian H., Keating M.J., Giese N., O’Brien S., Yu A., Miller L.L., et al. 2011. The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood. 118:3603–3612 10.1182/blood-2011-05-352492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard V., Greene J.M., Pahwa S., Winkelstein J.A., Boyle J.M., Kocak M., Conley M.E. 2006. The health status and quality of life of adults with X-linked agammaglobulinemia. Clin. Immunol. 118:201–208 10.1016/j.clim.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Jou S.T., Carpino N., Takahashi Y., Piekorz R., Chao J.R., Carpino N., Wang D., Ihle J.N. 2002. Essential, nonredundant role for the phosphoinositide 3-kinase p110delta in signaling by the B-cell receptor complex. Mol. Cell. Biol. 22:8580–8591 10.1128/MCB.22.24.8580-8591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D., Kudo A., Schaal S., Müller W., Melchers F., Rajewsky K. 1992. A critical role of lambda 5 protein in B cell development. Cell. 69:823–831 10.1016/0092-8674(92)90293-L [DOI] [PubMed] [Google Scholar]
- LeibundGut-Landmann S., Gross O., Robinson M.J., Osorio F., Slack E.C., Tsoni S.V., Schweighoffer E., Tybulewicz V., Brown G.D., Ruland J., Reis e Sousa C. 2007. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat. Immunol. 8:630–638 10.1038/ni1460 [DOI] [PubMed] [Google Scholar]
- Ma Q., Jones D., Borghesani P.R., Segal R.A., Nagasawa T., Kishimoto T., Bronson R.T., Springer T.A. 1998. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc. Natl. Acad. Sci. USA. 95:9448–9453 10.1073/pnas.95.16.9448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi A., Mizoguchi E., Takedatsu H., Blumberg R.S., Bhan A.K. 2002. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity. 16:219–230 10.1016/S1074-7613(02)00274-1 [DOI] [PubMed] [Google Scholar]
- Noda M., Omatsu Y., Sugiyama T., Oishi S., Fujii N., Nagasawa T. 2011. CXCL12-CXCR4 chemokine signaling is essential for NK-cell development in adult mice. Blood. 117:451–458 10.1182/blood-2010-04-277897 [DOI] [PubMed] [Google Scholar]
- Pappu R., Cheng A.M., Li B., Gong Q., Chiu C., Griffin N., White M., Sleckman B.P., Chan A.C. 1999. Requirement for B cell linker protein (BLNK) in B cell development. Science. 286:1949–1954 10.1126/science.286.5446.1949 [DOI] [PubMed] [Google Scholar]
- Patton D.T., Garden O.A., Pearce W.P., Clough L.E., Monk C.R., Leung E., Rowan W.C., Sancho S., Walker L.S., Vanhaesebroeck B., Okkenhaug K. 2006. Cutting edge: the phosphoinositide 3-kinase p110 delta is critical for the function of CD4+CD25+Foxp3+ regulatory T cells. J. Immunol. 177:6598–6602 [DOI] [PubMed] [Google Scholar]
- Rawlings D.J., Saffran D.C., Tsukada S., Largaespada D.A., Grimaldi J.C., Cohen L., Mohr R.N., Bazan J.F., Howard M., Copeland N.G., et al. 1993. Mutation of unique region of Bruton’s tyrosine kinase in immunodeficient XID mice. Science. 261:358–361 10.1126/science.8332901 [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179:1109–1118 10.1084/jem.179.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Terauchi Y., Fujiwara M., Aizawa S., Yazaki Y., Kadowaki T., Koyasu S. 1999. Xid-like immunodeficiency in mice with disruption of the p85alpha subunit of phosphoinositide 3-kinase. Science. 283:390–392 10.1126/science.283.5400.390 [DOI] [PubMed] [Google Scholar]
- Uotila T., Antonen J., Laine J., Kujansuu E., Haapala A.M., Lumio J., Vuento R., Oksa H., Herrala J., Kuusi M., et al. ; Pirkanmaa Waterborne Outbreak Study Group. 2011. Reactive arthritis in a population exposed to an extensive waterborne gastroenteritis outbreak after sewage contamination in Pirkanmaa, Finland. Scand. J. Rheumatol. 40:358–362 10.3109/03009742.2011.562533 [DOI] [PubMed] [Google Scholar]
- van den Bruele T., Mourad-Baars P.E., Claas E.C., van der Plas R.N., Kuijper E.J., Bredius R.G. 2010. Campylobacter jejuni bacteremia and Helicobacter pylori in a patient with X-linked agammaglobulinemia. Eur. J. Clin. Microbiol. Infect. Dis. 29:1315–1319 10.1007/s10096-010-0999-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. 2010. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11:329–341 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]