Protection from replicative stress conferred by Chk1 promotes transformation.
Abstract
Replicative stress (RS) is a type of endogenous DNA damage that cells suffer every time they duplicate their genomes, and which is further boosted by oncogenes. In mammals, the RS response (RSR) is coordinated by ATR and Chk1 kinases. We sought to develop a mammalian organism that is selectively protected from RS. To this end, mice carrying an extra copy of the Chk1 gene were generated. In vitro, Chk1 transgenic cells are protected from RS-inducing agents. Moreover, an extra Chk1 allele prolongs the survival of ATR-Seckel mice, which suffer from high levels of RS, but not that of ATM-deficient mice, which accumulate DNA breaks. Surprisingly, increased Chk1 levels favor transformation, which we show is associated with a reduction in the levels of RS induced by oncogenes. Our study provides the first example where supra-physiological levels of a tumor suppressor can promote malignant transformation, which is a result of the protection from the RS found in cancer cells.
DNA damage is a common source of cancer and aging. Therefore, organisms have evolved a coordinated DNA damage response which detects, signals, and promotes the repair of the lesions that compromise genomic integrity (Jackson and Bartek, 2009). Even though much emphasis has been placed on how organisms respond to DNA double-strand breaks (DSBs), recent works are revealing that replicative stress (RS) might also be a very relevant source of endogenous DNA damage for mammalian disease. For instance, one of the recent models of cancer progression is based on the finding that oncogenes generate RS, placing RS studies at the forefront of cancer research (Halazonetis et al., 2008). At the same time, we and others have previously shown that RS can promote aging in mammals (Ruzankina et al., 2007, 2009; Murga et al., 2009).
Whereas the nature of RS is still poorly defined, it essentially stands for the accumulation of recombinogenic stretches of single-stranded DNA (ssDNA), which can form at processed DSB, but which most frequently derive from stalled replication forks. In mammals, the RS response (RSR) is coordinated by ATR and Chk1 kinases (Cimprich and Cortez, 2008; López-Contreras and Fernandez-Capetillo, 2010).
The essential nature of ATR (Brown and Baltimore, 2000; de Klein et al., 2000) and Chk1 (Liu et al., 2000; Takai et al., 2000) kinases has significantly limited genetic studies addressing the role of the RSR in mammals. To overcome this limitation, we took a complementary genetic approach and decided to generate a mouse model that would be selectively protected from RS. Given that RSR kinases are responsible for the activation of cell cycle checkpoints, we reasoned that an uncontrolled expression of these kinases could be deleterious. Hence, to develop a gain-of-function model of the RSR, we decided to generate a mouse model with just one additional copy of the Chk1 gene. Similar strategies were shown to be successful for other loci where regulation of protein levels is important such as TP53 (García-Cao et al., 2002) or INK4a/ARF (Matheu et al., 2004). The reason for choosing Chk1 was that, in contrast to ATR, which naturally exists in a complex with its binding partner ATRIP (Cortez et al., 2001), Chk1 works in an autonomous manner. Moreover, Chk1 has been shown to be haploinsufficient (Lam et al., 2004), indicating that endogenous Chk1 levels are limiting and that therefore an increased gene dosage could have an effect. We thus decided to develop a mouse strain with one additional copy of the Chk1 gene as a mean to enhance the RSR.
RESULTS AND DISCUSSION
Generation of a mouse model with supra-physiological Chk1 function
Similar approaches in the past relied on the trans-genesis of bacterial artificial chromosomes (BAC) containing the gene of interest (García-Cao et al., 2002; Matheu et al., 2004). However, Chk1 is a small gene and available BACs contained additional genes. Thus, we subcloned a 33.5-kb region from the mouse genome including Chk1 and sequences 5′and 3′ until the next conserved gene was found in either direction (Fig. 1 A). The construct was used for the microinjection of fertilized mouse oocytes and Chk1 transgenic founder lines were established. Southern blot analyses revealed that one of the lines carried a single integration site, for which the intensity of the transgenic band was about half of that caused by endogenous Chk1 (Fig. 1 B). We thus selected this line as a mouse model carrying one additional allele of Chk1 (Chk1Tg).
Figure 1.
Generation of a Chk1Tg strain. (A) Region including Chk1 that was subcloned from the mouse genome. (B) Southern blot with an internal Chk1 probe, illustrating the presence of an integration site (8 kb) on the Chk1Tg strain. The 5-kb band corresponds to the endogenous Chk1. (C) Representative picture of 4-mo-old WT and Chk1Tg littermates. (D) Weight distribution of 1-mo-old WT and Chk1Tg mice. (E) Chk1 Western blot in testis, spleen, and purified B cells after a 2-d stimulation with lipopolysaccharide from WT and Chk1Tg mice. Data are representative of four independent analyses. (F) ATR, Chk1-P, Chk1, RPA-P, and Rad17-P Western blot in WT and Chk1Tg littermate MEF, either untreated (C) or upon treatment with 2 mM HU for 3 h or 10 Gy IR for 1 h. Data are representative of two independent analyses. (G) Chk1-P, RPA-P, and Rad17-P Western blot in WT and Chk1Tg littermate MEF upon treatment with 2 mM HU for 3 h in the presence or absence of 1 µM of ATR inhibitor (ATRi; ETP-46464). Data are representative of two independent analyses. β-Actin was used as a loading control in all Western blots. In D, center lines indicate mean values.
Transgenic mice were viable with no obvious phenotype that would distinguish them from their littermates (Fig. 1, C and D). Nevertheless, tissues from Chk1Tg animals showed increased protein levels of Chk1 (Fig. 1 E). This difference increased in proliferating cells such as B-lymphocytes or mouse embryonic fibroblasts (MEFs; Fig. 1, E and F). Exposure of Chk1Tg MEF to hydroxyurea (HU) or ionizing radiation (IR) led to increased levels of phosphorylation of Chk1 but not other ATR targets such as Rad17 or RPA (Fig. 1, F and G). Hence, an extra allele of Chk1 is compatible with mouse development and leads to increased Chk1 levels, which are susceptible to phosphorylation by ATR.
Protection from RS in Chk1Tg cells
To determine whether increased Chk1 levels provide additional Chk1 function, we first analyzed the response of Chk1Tg MEF to Chk1 inhibition. Transgenic MEFs were resistant to the cytotoxic effects of the Chk1 inhibitor UCN-01 and showed lower amounts of γH2AX in response to the drug (unpublished data), illustrating that the extra Chk1 present on Chk1Tg cells is capable of enhancing Chk1 function beyond WT levels. We next analyzed the response of Chk1Tg MEF to RS-inducing agents other than UCN-01. Colony survival assays revealed that Chk1 transgenic MEFs were more resistant than their WT controls to HU or aphidicolin (APH; Fig. 2 A). Noteworthy, and in agreement with the normal overall size of Chk1Tg mice, replication was not affected by the extra Chk1 allele (WT: 24.6 ± 1.6% vs. Chk1Tg: 24.93 ± 1.5%; P = 0.757; Fig. 2 B). Therefore, the observed survival differences reflected an intrinsic resistance of Chk1Tg MEF to RS. In agreement with these observations, HU- or APH-treated Chk1Tg MEF presented lower amounts of γH2AX (Fig. 2 C). Besides a pan-nuclear γH2AX staining, RS also leads to an accumulation of ssDNA, which can be detected as foci for the ssDNA-binding protein RPA. High-throughput microscopy (HTM) analyses revealed that HU- or APH-treated Chk1Tg MEFs presented significantly lower amounts of chromatin-bound RPA than their WT littermates (Fig. 2 D). Altogether, these results illustrate that an additional allele of Chk1 can provide a supra-physiological protection against RS.
Figure 2.
Resistance to RS in Chk1Tg MEF. (A) Number of colonies per plate in WT and Chk1Tg MEF after treatment with 0.5 mM HU or 1 µM APH for 24 h (normalized to the number of colonies found in the untreated control plates). The experiment was repeated twice with three independent MEF pairs. (B) Representative flow cytometry profiles of WT and Chk1Tg MEF after a 1-h BrdU pulse. Numbers indicate the percentage of BrdU-positive cells. Data are representative of five independent analyses. (C and D) HTM-mediated quantification of γH2AX (C) and RPA (D) intensities in WT and Chk1Tg MEF treated with 0.5 mM HU for 3 h and 5 µM APH for 4 h. Data are representative of four independent analyses. In A, error bars indicate SD; in C and D, center lines indicate mean values. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Chk1Tg alleviates the symptoms of ATR-Seckel but not those of ATM-deficient mice
Given the effects observed on isolated cells, we next sought to evaluate whether an enhanced RSR could have an impact in vivo. One of the most well understood models of an RS-driven disease is ATR-Seckel Syndrome. Patients of this syndrome present reduced levels of the ATR kinase (O’Driscoll et al., 2003). We recently generated a mouse model of ATR-Seckel Syndrome (Murga et al., 2009). ATR mutant animals present an accumulation of RS, which is particularly abundant during embryonic development. Once born, ATR-Seckel (ATRS/S) mice develop a pleiotropic disease, which is manifested by a characteristic cranial appearance and a segmental progeroid disease to which animals succumb early in life. Therefore, and given that Chk1 is phosphorylated and activated by ATR, we investigated the effect of the Chk1 transgene on ATR-Seckel mice.
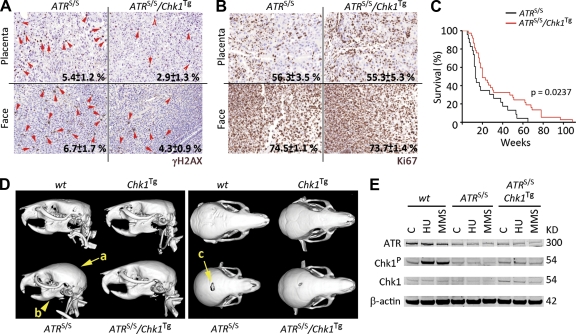
Similar to our observations in MEF, the presence of an extra allele of Chk1 reduced the number of cells presenting a pan-nuclear γH2AX distribution on ATR-Seckel embryos (Fig. 3 A). This effect was not a result of differences in embryonic proliferation rates, as illustrated by Ki67 staining (Fig. 3 B). In addition, there were no significant differences on the weights of Chk1Tg and Chk1+/+ animals at birth (WT: 1.24 ± 0.19 g vs. Chk1Tg: 1.26 ± 0.16 g; P = 0.801), reinforcing that Chk1Tg does not affect fetal proliferation. Importantly, this reduction on embryonic RS correlated with a significant extension of the lifespan on ATRS/S/Chk1Tg animals (Fig. 3 C). Around one fifth of ATRS/S/Chk1Tg animals lived longer than any ATRS/S mouse, with some reaching ages beyond 100 wk. In agreement with this, ATRS/S/Chk1Tg animals showed an alleviation of the craniofacial abnormalities that are present on ATR-Seckel mice. This effect was also present at different degrees, with some of the double mutant mice showing a clear rescue of all measured parameters (Fig. 3 D). Consistent with the in vivo findings, ATRS/S/Chk1Tg MEF showed a partial rescue of the deficient Chk1 phosphorylation that is observed on ATRS/S cells (Fig. 3 E). In summary, all of these results demonstrate that the Chk1 transgene is able to alleviate the symptoms associated to the ATR-Seckel syndrome in mice.
Figure 3.
Alleviation of the ATR-Seckel syndrome by Chk1Tg. (A and B) Representative images of γH2AX (A) and Ki67 (B) on the placenta and face from ATRS/S and ATRS/S/Chk1Tg littermate embryos. Red arrows indicate cells showing a pan-nuclear γH2AX staining. Numbers indicate the mean percentage and SD of positive cells in each case (n = 3). (C) Kaplan-Meyer curves of ATRS/S (n = 23) and ATRS/S/Chk1Tg (n = 36) mice. The p-value was calculated with the Mantel-Cox log-rank test. (D) Computerized Tomography-mediated reconstruction of the heads from WT, Chk1Tg, ATRS/S, and ATRS/S/Chk1Tg mice. Yellow arrows indicate features that show evident rescue such as the shape of the crania (a), the micrognathia (b), and the deficient closure of the fontanelle (c). Data are representative of four independent analyses. (E) ATR, Chk1-P, and Chk1 protein levels in WT, ATRS/S, and ATRS/S/Chk1Tg MEF treated with 0.5 mM HU for 3 h or 10 mM methyl methanesulfonate (MMS) for 4 h. β-Actin was used as a loading control. Data are representative of two independent analyses.
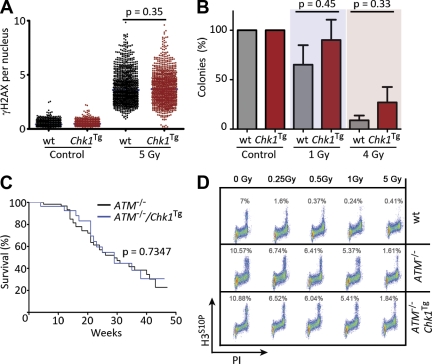
We next evaluated the effect of the Chk1Tg in the context of DNA DSB. In contrast to HU or APH, the presence of the Chk1 transgene did not significantly affect the levels of γH2AX induced by IR (Fig. 4 A). However, Chk1Tg cells presented a hyperactive IR-induced G2/M checkpoint (unpublished data), which is consistent with the role of Chk1 in checkpoint regulation. In spite of this, Chk1Tg MEFs were not radioresistant when compared with their WT littermates (Fig. 4 B). This limited contribution of the G2/M checkpoint on survival to radiation has been previously noticed (Löbrich and Jeggo, 2007) and is consistent with the fact that ATR or Chk1 deficiencies are not particularly radiosensitive.
Figure 4.
Chk1Tg does not rescue ATM deficiency. (A) HTM-mediated quantification of γH2AX in WT and Chk1Tg MEF exposed to 5 Gy of IR for 1 h. Data are representative of three independent experiments. (B) Number of colonies per plate in WT and Chk1Tg MEF after exposure to 1 or 4 Gy of IR (normalized to the number of colonies found in control plates). The experiment was repeated twice with two independent MEF pairs. (C) Kaplan-Meyer curves of ATM−/− (n = 39) and ATM−/−/Chk1Tg (n = 26) mice. The p-value was calculated with the Mantel-Cox log-rank test. (D) Activation of the IR-induced G2/M checkpoint in WT, ATM−/−, and ATM−/−/Chk1Tg cells. Images are representative of two independent experiments. In A, center lines indicate mean values; in B, error bars indicate SD.
To study the impact of the Chk1 transgene on the response to DSB in vivo, Chk1Tg mice were crossed with ATM-deficient mice, which present a deficient response to DSB and die from lymphomas initiated by chromosomal translocations (Barlow et al., 1996). In humans, ATM deficiency leads to another hereditary syndrome known as Ataxia telangiectasia (AT), which is also associated with a high incidence of cancer (Savitsky et al., 1995). In contrast to the effect on ATR-Seckel animals, the Chk1 transgene did not extend the lifespan of ATM−/− mice (Fig. 4 C), the cause of death being lymphoma in both ATM−/− and Chk1Tg/ATM−/− animals. The Chk1 transgene also failed to rescue the IR-induced G2/M checkpoint defect that is observed on ATM−/− cells (Fig. 4 D), which is in agreement with IR-induced Chk1 phosphorylation being ATM dependent (Cuadrado et al., 2006; Jazayeri et al., 2006). In summary, whereas the Chk1Tg allele can alleviate the symptoms of the ATR-Seckel Syndrome, it has no detectable impact on a mouse model of AT.
An extra allele of Chk1 promotes transformation by limiting oncogene-induced RS
Beside external reagents, RS can also occur by endogenous sources that perturb replication. According to the oncogene-induced DNA damage model of cancer progression (Halazonetis et al., 2008), oncogenes would generate substantial amounts of RS which, by activating the DNA damage response, would limit the expansion of the tumor in its initial stages. In this model, ATR and Chk1 would suppress transformation through the activation of p53. We therefore evaluated the response of Chk1Tg MEF to oncogenic transformation. To this end, MEFs were infected with a retroviral plasmid that expressed H-RasV12G and E1A oncogenes (Ras/E1A), a system which is widely used to evaluate transformation in MEF. Moreover, H-RasV12G overexpression promotes Chk1 phosphorylation (Gilad et al., 2010). Surprisingly, transformation with Ras/E1A was consistently and significantly more efficient on Chk1Tg MEF (Fig. 5, A and B) than on WT littermates.
Figure 5.
Enhanced oncogenic transformation in Chk1Tg MEF. (A) Plates of WT and Chk1Tg MEF 2 wk after infection with a Ras/E1A-expressing retrovirus. Colonies were stained with Methylene blue. Images are representative of six independent experiments. (B) Numbers of transformed colonies per plate in WT and Chk1Tg MEF infected with Ras/E1A. Data derive from three independent experiments. (C) HTM-mediated quantification of γH2AX in WT and Chk1Tg MEF 24 and 48 h after infection with a Ras/E1A-expressing retrovirus. Data are representative of three independent experiments. (D) Percentage of apoptotic cells present in cultures of WT and Chk1Tg MEF 24 and 48 h after infection with a Ras/E1A-expressing retrovirus. The quantification derives from three independent experiments. (E) HTM-mediated quantification of γH2AX in WT and Chk1Tg MEF infected with MycER, treated or untreated with 4-OHT for 8 or 24 h. Data are representative of three independent experiments. (F) MycER-induced apoptosis in WT and Chk1Tg MEF infected with MycER, treated or untreated with 4-OHT for 72 h. Data derive from three independent experiments. (G) HTM-mediated analysis of γH2AX in WT and Chk1Tg MEF infected with MycER and treated with 4-OHT for 24 h, which were previously infected with lentiviruses expressing a p53-specific shRNA (sh p53) or a control shRNA (sh C). Data are representative of three independent experiments. In B, D, and F, error bars indicate SD; in C–G, center lines indicate mean values. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine how Chk1 could facilitate oncogenic transformation, we analyzed the levels of RS present at various times after infection. Interestingly, whereas Ras/E1A expression led to a detectable increase in H2AX phosphorylation in WT MEF, this was significantly reduced on Chk1Tg cells (Fig. 5 C). These data suggest that, whereas Chk1 might be involved in activating p53 in response to oncogenes, it also has a previous function in suppressing oncogene-induced RS. In this context, the enhanced Chk1 levels present on Chk1Tg MEF would be favoring transformation by decreasing the amount of RS generated by the oncogenes, which is intrinsically cytotoxic. In agreement with this, Ras/E1A immortalized Chk1Tg MEFs presented a lower percentage of dead cells than their WT littermates (Fig. 5 D). Moreover, Ras/E1A transformed Chk1Tg colonies were significantly larger in size than those obtained from WT MEF (WT: 1.41 ± 0.68 mm2 vs. Chk1Tg: 2.72 ± 1.48 mm2; P = 0.0003), which would support the idea that Chk1 facilitates the proliferation of cells carrying oncogene-induced RS. In addition to Ras/E1A, Chk1Tg MEFs were also protected from the RS and apoptosis induced by Myc (Fig. 5, E and F). Noteworthy, Chk1Tg-mediated suppression of Myc-induced RS was accentuated upon p53 knockdown (Fig. 5 G), a situation where Myc-induced RS is exacerbated (Murga et al., 2011). Altogether, our data suggest that the RS induced by oncogenes is a toxic by-product of the transformation process, which is not the required to promote transformation, and which can be limited by selectively potentiating the RSR.
Here, we provide proof of concept to show that the mammalian RSR is susceptible of improvement. At the same time, and whereas ATR might have >700 targets (Matsuoka et al., 2007), this study identifies Chk1 as a critical mediator of ATR in vivo, providing the first example of a genetic context which modifies the severity of the Seckel Syndrome. It is possible that the effect of the Chk1Tg might be more marked in the context of other syndromes initiated by replication problems, but which keep intact ATR levels. For instance, recent studies have identified mutations in the pre-replication complex as causative of Meier-Gorlin Syndrome (Klingseisen and Jackson, 2011), which is also characterized by craniofacial abnormalities. To what extent these syndromes are also modulated by the RSR remains to be elucidated.
In what relates to cancer, our work provides a more complex view of the role of Chk1 and RS during carcinogenesis. Recent data have shown that an extra supply of nucleotides decreases the RS induced by E6/E7 oncogenes, which correlated with reduced transformation (Bester et al., 2011). In contrast, several studies have now shown that, over a threshold, the RS induced by Myc, Ras, or cyclin E oncogenes can be cytotoxic (Gilad et al., 2010; Murga et al., 2011; Toledo et al., 2011; Schoppy et al., 2012), which would rather argue that oncogene-induced RS limits transformation. The data reported here favor the latter view, supporting that oncogene-induced RS is a toxic byproduct of transformation, which is not needed by the oncogenes to transform, and which decreases the fitness of transformed cells. This does not exclude that RS is the cause of genomic rearrangements that are selected during cancer evolution, but rather helps to understand how cancer cells might cope with high levels of RS.
A recent review classified tumor suppressors in different categories (Berger et al., 2011). Tumor suppressors were originally classified according to the “two-hit” model, whereby deletion of both alleles was necessary for cancer development (i.e., Rb). However, several exceptions to this class emerged such as haploinsufficiency whereby loss of one allele is sufficient to promote malignancy (i.e., p53). More intricate relationships like obligatory haploinsufficiency have also been defined. This is the case of PTEN, in which heterozygosity favors transformation but full deletion can lead to senescence. In this context, our data provide the first example of a tumor suppressor (Chk1) for which a supra-physiological dose facilitates transformation through diminishing oncogene-induced stress. Interestingly, Chk1 expression is under the control of Myc and E2F oncogenes, and increased Chk1 levels have been observed in lymphomas and breast carcinomas (Verlinden et al., 2007; Höglund et al., 2011). Moreover, public microarray repositories show that up-regulation of Chk1, rather than down-regulation, is a very frequent event in cancer (http://www.oncomine.org). Even though deletions of RSR genes have not been found in cancer as a result of their essential nature, it would be interesting to see whether amplification or overexpression of these factors is a frequent event during tumor evolution that favors the growth of cells with high loads of RS.
MATERIALS AND METHODS
Mouse work.
ATR-Seckel (Murga et al., 2009) and ATM−/− (Barlow et al., 1996) mice have been described before. For the generation of Chk1Tg mice, a 33.5-kb region from the mouse genome which encompassed the Chk1 gene was first cloned into a minimal vector by recombineering (Gene Bridges). The linearized vector was used for the microinjection of fertilized oocytes. Transgenic animals were first identified by Southern Blot through standard procedures, and subsequently followed by PCR with primers amplifying a 600-bp sequence from the vector (available upon request). Mice were kept under standard conditions at serum-pathogen free facility of the Spanish National Cancer Centre in a mixed C57BL/6-129/Sv background. All mouse work was performed in accordance with the Guidelines for Humane Endpoints for Animals Used in Biomedical Research, and under the supervision of the Ethics Committee for Animal Research of the Instituto de Salud Carlos III.
MEF work.
MEFs from embryonic day 13.5 post coitum embryos were obtained by standard methods and grown in DMEM (Invitrogen) supplemented with 10% FBS (Hyclone). UCN-01, HU, and APH (Sigma-Aldrich) were added at the indicated concentrations. For clonogenic survival analyses, 103 cells were plated per 10-cm plate and the number of colonies was counted after 10 d. For the work on oncogenes, pBabe-RASV12/E1A-puro or pBabe-puro (gift from M. Barbacid, Spanish National Cancer Research Centre, Madrid, Spain) were retrovirally transduced according to standard procedures. Infected cells were selected with 2 µg/ml puromycin for 2 d, and then 2,000 cells were seeded in 10-cm diameter plates. After 7 d, colony formation was assessed by Methylene blue staining. Apoptosis was quantified by flow-cytometry as the fraction of cells with a DNA content lower than G1. Lentiviruses expressing p53-specific shRNAs and their controls (gift from J.M. Silva, Institute for Cancer Genetics, Columbia University, New York, NY) were infected according to standard procedures. For all experiments, cells were grown in 5% oxygen to minimize the exposure to reactive oxygen species, and low (less than three) passage MEFs were used.
Immunoblotting.
For total protein extracts, cells were washed once with PBS, collected by directly adding 2× NuPAGE LDS Sample buffer, and incubated for 5 min at 95°C. For soluble protein extracts, cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, and 1 mM EDTA) containing protease and phosphatase inhibitors (Sigma-Aldrich). Samples were resolved by SDS-PAGE and analyzed by standard Western blotting techniques. Antibodies against p53, RPA, Rad17-S645P, and Chk1-S345P (Cell Signaling Technology), γH2AX (Millipore), ATR (Serotec), β-actin (Sigma-Aldrich), RPA-S4P/S8P (Bethyl Laboratories), and Chk1 (Novocastra) were used. Protein blot analyses were performed on the LICOR platform (eBioscience).
HTM analyses.
MEFs were grown on μCLEAR bottom 96-well plates (Greiner Bio-One) and γH2AX and RPA immunofluorescence were performed using standard procedures. In the case of RPA, a detergent extraction step was done previous to fixation which eliminates the nucleosoluble pool of RPA and leaves the chromatin-bound fraction. Images were automatically acquired from each well by an Opera High-Content Screening System (Perkin Elmer). A 40× magnification lens was used and pictures were taken at nonsaturating conditions. Images were segmented using the DAPI staining to generate masks matching cell nuclei from which the mean γH2AX and RPA signals were calculated.
Cell cycle analysis.
Cells were resuspended in a PBS solution containing 1% (wt/vol) BSA, 10 µg/ml propidium iodide, and 0.5 mg/ml RNase A and were analyzed by flow cytometry in a FACSCalibur machine (BD). To monitor replication, cells were incubated with BrdU for 1 h at 37°C. After fixation with 4% (wt/vol) paraformaldehyde for 30 min, cells were processed with a FITC BrdU Flow Cytometry kit as recommended by the manufacturer (BD). For G2/M checkpoint analysis, proliferating B-lymphocytes were exposed to IR at the indicated doses, and the percentage of mitotic cells was calculated 1 h after the treatment by a dual staining with propidium iodide (DNA content) and the mitotic marker H3S10P (Millipore).
Whole body imaging.
Whole-body imaging was performed on anesthetized mice using the eXplore Vista PET-CT (GE Healthcare) and a 7-tesla Pharmascan (Bruker). MMWKS software (GE Healthcare) was used for the quantifications.
Acknowledgments
We thank Dr. M. Serrano and A. Nussenzweig for critical comments on the manuscript.
A.J. López-Contreras is recipient of a Juan de la Cierva fellowship (JCI-2009-05099) from the Spanish Ministry of Science. Work in the O. Fernandez-Capetillo laboratory is supported by grants from the Spanish Ministry of Science (CSD2007-00017 and SAF2011-23753), the Association for International Cancer Research (12-0229), and the European Research Council (ERC-210520).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- APH
- aphidicolin
- DSB
- double-strand break
- HTM
- high-throughput microscopy
- HU
- hydroxyurea
- IR
- ionizing radiation
- MEF
- mouse embryonic fibroblast
- RS
- replicative stress
- RSR
- RS response
- ssDNA
- single-stranded DNA
References
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D., Wynshaw-Boris A. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 86:159–171 10.1016/S0092-8674(00)80086-0 [DOI] [PubMed] [Google Scholar]
- Berger A.H., Knudson A.G., Pandolfi P.P. 2011. A continuum model for tumour suppression. Nature. 476:163–169 10.1038/nature10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester A.C., Roniger M., Oren Y.S., Im M.M., Sarni D., Chaoat M., Bensimon A., Zamir G., Shewach D.S., Kerem B. 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 145:435–446 10.1016/j.cell.2011.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D. 2000. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14:397–402 [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A., Cortez D. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616–627 10.1038/nrm2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S.J. 2001. ATR and ATRIP: partners in checkpoint signaling. Science. 294:1713–1716 10.1126/science.1065521 [DOI] [PubMed] [Google Scholar]
- Cuadrado M., Martinez-Pastor B., Murga M., Toledo L.I., Gutierrez-Martinez P., López E., Fernandez-Capetillo O. 2006. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 203:297–303 10.1084/jem.20051923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klein A., Muijtjens M., van Os R., Verhoeven Y., Smit B., Carr A.M., Lehmann A.R., Hoeijmakers J.H. 2000. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr. Biol. 10:479–482 10.1016/S0960-9822(00)00447-4 [DOI] [PubMed] [Google Scholar]
- García-Cao I., García-Cao M., Martín-Caballero J., Criado L.M., Klatt P., Flores J.M., Weill J.C., Blasco M.A., Serrano M. 2002. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21:6225–6235 10.1093/emboj/cdf595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad O., Nabet B.Y., Ragland R.L., Schoppy D.W., Smith K.D., Durham A.C., Brown E.J. 2010. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 70:9693–9702 10.1158/0008-5472.CAN-10-2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T.D., Gorgoulis V.G., Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science. 319:1352–1355 10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- Höglund A., Nilsson L.M., Muralidharan S.V., Hasvold L.A., Merta P., Rudelius M., Nikolova V., Keller U., Nilsson J.A. 2011. Therapeutic implications for the induced levels of Chk1 in Myc-expressing cancer cells. Clin. Cancer Res. 17:7067–7079 10.1158/1078-0432.CCR-11-1198 [DOI] [PubMed] [Google Scholar]
- Jackson S.P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C., Lukas J., Jackson S.P. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8:37–45 10.1038/ncb1337 [DOI] [PubMed] [Google Scholar]
- Klingseisen A., Jackson A.P. 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 25:2011–2024 10.1101/gad.169037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M.H., Liu Q., Elledge S.J., Rosen J.M. 2004. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 6:45–59 10.1016/j.ccr.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Liu Q., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., et al. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14:1448–1459 10.1101/gad.14.12.1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löbrich M., Jeggo P.A. 2007. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer. 7:861–869 10.1038/nrc2248 [DOI] [PubMed] [Google Scholar]
- López-Contreras A.J., Fernandez-Capetillo O. 2010. The ATR barrier to replication-born DNA damage. DNA Repair (Amst.) 9:1249–1255 10.1016/j.dnarep.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A., Pantoja C., Efeyan A., Criado L.M., Martín-Caballero J., Flores J.M., Klatt P., Serrano M. 2004. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 18:2736–2746 10.1101/gad.310304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., III, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316:1160–1166 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- Murga M., Bunting S., Montaña M.F., Soria R., Mulero F., Cañamero M., Lee Y., McKinnon P.J., Nussenzweig A., Fernandez-Capetillo O. 2009. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 41:891–898 10.1038/ng.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M., Campaner S., López-Contreras A.J., Toledo L.I., Soria R., Montaña M.F., D’Artista L., Schleker T., Guerra C., Garcia E., et al. 2011. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 18:1331–1335 10.1038/nsmb.2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., Goodship J.A. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 33:497–501 10.1038/ng1129 [DOI] [PubMed] [Google Scholar]
- Ruzankina Y., Pinzon-Guzman C., Asare A., Ong T., Pontano L., Cotsarelis G., Zediak V.P., Velez M., Bhandoola A., Brown E.J. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 1:113–126 10.1016/j.stem.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y., Schoppy D.W., Asare A., Clark C.E., Vonderheide R.H., Brown E.J. 2009. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat. Genet. 41:1144–1149 10.1038/ng.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S., et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 268:1749–1753 10.1126/science.7792600 [DOI] [PubMed] [Google Scholar]
- Schoppy D.W., Ragland R.L., Gilad O., Shastri N., Peters A.A., Murga M., Fernandez-Capetillo O., Diehl J.A., Brown E.J. 2012. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J. Clin. Invest. 122:241–252 10.1172/JCI58928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Tominaga K., Motoyama N., Minamishima Y.A., Nagahama H., Tsukiyama T., Ikeda K., Nakayama K., Nakanishi M., Nakayama K. 2000. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 14:1439–1447 [PMC free article] [PubMed] [Google Scholar]
- Toledo L.I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J.R., Fernandez-Capetillo O. 2011. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 18:721–727 10.1038/nsmb.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinden L., Vanden Bempt I., Eelen G., Drijkoningen M., Verlinden I., Marchal K., De Wolf-Peeters C., Christiaens M.R., Michiels L., Bouillon R., Verstuyf A. 2007. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 67:6574–6581 10.1158/0008-5472.CAN-06-3545 [DOI] [PubMed] [Google Scholar]