Abstract
Endogenous retroviruses constitute a significant genomic fraction in all mammalian species. Typically they are evolutionarily old and fixed in the host species population. Here we report on a novel endogenous gammaretrovirus (CrERVγ; for cervid endogenous gammaretrovirus) in the mule deer (Odocoileus hemionus) that is insertionally polymorphic among individuals from the same geographical location, suggesting that it has a more recent evolutionary origin. Using PCR-based methods, we identified seven CrERVγ proviruses and demonstrated that they show various levels of insertional polymorphism in mule deer individuals. One CrERVγ provirus was detected in all mule deer sampled but was absent from white-tailed deer, indicating that this virus originally integrated after the split of the two species, which occurred approximately one million years ago. There are, on average, 100 CrERVγ copies in the mule deer genome based on quantitative PCR analysis. A CrERVγ provirus was sequenced and contained intact open reading frames (ORFs) for three virus genes. Transcripts were identified covering the entire provirus. CrERVγ forms a distinct branch of the gammaretrovirus phylogeny, with the closest relatives of CrERVγ being endogenous gammaretroviruses from sheep and pig. We demonstrated that white-tailed deer (Odocoileus virginianus) and elk (Cervus canadensis) DNA contain proviruses that are closely related to mule deer CrERVγ in a conserved region of pol; more distantly related sequences can be identified in the genome of another member of the Cervidae, the muntjac (Muntiacus muntjak). The discovery of a novel transcriptionally active and insertionally polymorphic retrovirus in mammals could provide a useful model system to study the dynamic interaction between the host genome and an invading retrovirus.
INTRODUCTION
Endogenous retroviruses (ERVs) are initially formed when an exogenous virus is incorporated into the germ line and is subsequently transmitted vertically. ERVs have been found in every vertebrate genome studied; in humans they constitute about 8% of the genome (25), and in mice they represent 10% (51). Because viral germ line invasion events occurred millions of years ago, ERVs are fixed in most host populations. Over time, the ERVs accumulate mutations and deletions at a rate presumed to mirror the neutral mutation rate of the host genome (9, 22). Thus, most ERVs studied to date are littered with mutations that render them replication incompetent. Rare examples of intact and evolutionarily young ERVs that still segregate in the host population have been described in various mammalian species, some of which preserve replication competence (5, 7, 12, 21, 28, 30, 39, 40, 48, 49, 53).
Exogenous retroviruses are important etiological agents of both neoplastic and degenerative diseases involving many cell types (41). The role of ERVs as a direct cause of disease has been more difficult to prove because of their ubiquitous presence in all individuals. Nevertheless, ERVs have been proposed as candidates in human autoimmune conditions and cancer (6, 34, 42). ERVs also can be genomic symbionts. ERV regulatory regions and proteins have been utilized by the host to achieve a new function, e.g., as alternative gene promoters (13) or in the formation of the placenta (32). The presence of ERVs can either positively or negatively influence the outcome of infection by a related exogenous virus (3, 4, 37, 47).
We recently identified a transcribing, novel gammaretrovirus in wild mule deer (52) while conducting a metatranscriptomic study. Although there are no confirmed reports of infectious retroviruses in cervid species, a putative endogenous gammaretrovirus was identified by coculture from black-tailed deer (a subspecies of mule deer), and a wasting disease in moose (Alces alces) has suspected retroviral etiology (1, 31). Because these viruses often cause sporadic acute or chronic progressive disease, their presence in cervid species may have gone undetected. In this work we provide the molecular characterization of a new gammaretrovirus and define its endogenous presence in the genome of mule deer and other cervid species, and we propose the name cervid endogenous gammaretrovirus (CrERVγ). Our data demonstrate that mule deer have experienced continual germ line invasion of CrERVγ since speciation from a last common ancestor, white-tailed deer, and that there is a high degree of CrERVγ insertional polymorphism in contemporary mule deer.
MATERIALS AND METHODS
Animal tissues.
Mule deer, white-tailed deer, and elk retropharyngeal lymph nodes were obtained from animals presented by hunters to check stations several hours after being shot. The geographical origin of all samples is Montana, except for mule deer 556 and 663, which are from Colorado. Because the samples were obtained from legally killed animals, this study is exempt from Pennsylvania State University guidelines governing animal experimentation. Genomic DNAs were prepared from RNAlater (Ambion) preserved tissues using the phenol-chloroform extraction method recommended by the manufacturer.
Southern blotting.
Genomic DNAs (5 μg) were digested with XhoI or EcoRI (NEB) at 37°C overnight, resolved on 1% agarose gels containing ethidium bromide, and denatured by soaking in 1.5 M NaCl-0.5N NaOH for 30 min. The gels were neutralized in 5× SSPE (150 mM NaCl, 10 mM sodium phosphate, 1 mM EDTA) with 10 mM NaOH. The DNA was transferred to a Hybond N+ nylon membrane (GE Healthcare) by capillary force overnight and fixed to the membrane using UV irradiation. DNA probes were generated from PCR products using the [α-32P]dCTP random primer labeling kit (Stratagene), and the hybridizations were performed at 58°C overnight in Church buffer (1% bovine serum albumin [BSA], 1 mM EDTA, 0.5 M sodium phosphate, 7% SDS, pH 7.2). The membranes were washed with 2× SSPE–0.1% SDS and 0.2× SSPE–0.1% SDS at 65°C and exposed for autoradiography at −70°C before development. PCR products used for the generation of probes were amplified from mule deer cDNA using the following primers. The gag-pro probe was amplified with primers 5′-GAAGAACGGATTAGACGGGAGGAG and 5′-GCTGGTTTTCTTAGACATTGGT. The probe is 486 bp long and corresponds to nucleotides 2779 to 3264 of the full-length CrERVγ-in7 provirus sequence. The pol probe was amplified with primers 5′-AGCGGGGACCTCTTACAAAC and 5′-ATCGCTTCGACAGGTATGCT (length, 478 bp; corresponds to nucleotides 4204 to 4681 of CrERVγ-in7 provirus sequence). The env probe was amplified with primers 5′-ATGTGGGGGAGTTGATTCTTTTTA and 5′-AGGTGGCTGATTGATTCTTCTATG (length, 1,108 bp; corresponds to nucleotides 7073 to 8182 of the CrERVγ-in7 provirus sequence).
PCR amplifications.
All PCR amplifications used Takara ExTaq DNA polymerase (Takara). To amplify the 3′ ends of the CrERVγ genomic RNA, total RNA was isolated from mule deer lymph nodes using the RNeasy minikit (Qiagen). The 3′ rapid amplification of cDNA ends (RACE) Smart procedure (Clontech) was employed according to the manufacturer's instructions. Total RNA (1.6 μg) was converted to cDNA with AffinityScript multiple-temperature reverse transcriptase (Stratagene) and used as the template for the 3′RACE reaction with virus-specific primer 5′-CCCAATCCTGCTGGCTGTGCT and the 3′RACE UPM primer (Clontech). The PCR products were cloned using the Qiagen PCR cloning kit (Qiagen) and sequenced. An inverse PCR technique was used to obtain the long terminal repeat (LTR) and 5′ flanking region sequences of proviruses CrERVγ-in2, -in3, and -in7. Genomic DNA digested with BamHI was self ligated overnight at 16°C at 15 ng/μl and then amplified with primers 5′-GGTCTTTCATTTGGGGGCTCGTCGGATCC and 5′-GCTGTGTCCAACGCAGTG. The PCR products were cloned using the StrataClone PCR cloning kit (Stratagene) and sequenced. The LTR-BovtA PCR was done using the CrERVγ LTR-specific primer 5′-GAGCAAACATAACGCCATGA and a primer complementary to the BovtA repeats (5′-GCAACCCATTCCAGTATTCTT). Individual PCR products were isolated from the gel and directly sequenced. Based on the sequence obtained by inverse PCR and LTR-BovtA PCR, new primers were designed in the unique flanking host DNA to allow the genotyping of each individual insertion.
For four of the proviruses, the 5′ integration site junctions were obtained and the primer pairs listed below were used for genotyping. The first primer is always complementary to the host 5′ flanking sequence and the second primer to the internal virus untranslated sequence preceding gag: CrERVγ-in2 (5′-GTTGGCTGATGCGTTGAGT and 5′-CGTTCGGATTCTTCCTTCTG), CrERVγ-in3 (5′-GCTGTTCTGACTGGTGCTTG and 5′-CGTTCGGATTCTTCCTTCTG), CrERVγ-in4 (5′-AGCACTTGCATGTGAGGTTG and 5′-CGTTCGGATTCTTCCTTCTG), and CrERVγ-in7 (5′-TCCCTTCCCCTATACCTGCT and 5′-CGTTCGGATTCTTCCTTCTG).
For three of the proviruses, 3′ integration site junctions were obtained and the primer pairs below were used for genotyping. The first primer is always complementary to the host 3′ flanking sequence and the second primer to the internal virus sequence in the env gene: CrERVγ-in1 (5′-GGATGCACAACCAACAAGTG and 5′-CTTACAATTGGGCCTTGCAT), CrERVγ-in5 (5′-AAGGGTTCGTGGAGCCTAAT and 5′-CTTACAATTGGGCCTTGCAT), and CrERVγ-in6 (5′-TCTCCCACAGCCCTTTACTG and 5′-TACAATTGGGCCTTGCATTT).
The missing flanking sequences (5′ or 3′) of the CrERVγ integrations were obtained by designing PCR primers based on the homology of the available mule deer sequence to the bovine genome. This was successful for five of the seven CrERVγ integrations, and the primers were the following: for CrERVγ-in1, primer 5′-CGTGTAAACAAATTGCACATGG; for CrERVγ-in2, primer 5′-ACAGAAGGCGTTCCACAAAG; for CrERVγ-in3, primer 5′-CAGCCTGGGTAGGGATTGTA; for CrERVγ-in5, primer 5′-GACATTGGCAGGTTTGCTTT; and for CrERVγ-in7, primer 5′-CCAACCCTCTCTTTGGGTTT. These primers were used in combination with the primers from the opposite flanking region to amplify the empty preintegration sites. They also were used in combination with the internal virus primers to amplify the missing (5′ or 3′) virus-host junctions.
The complete sequence of the CrERVγ-in7 provirus was amplified in two overlapping PCR products. The 5′ product was approximately 6 kb long and was amplified using the host primer 5′-TCCCTTCCCCTATACCTGCT and the internal virus primer 5′-ACAAAGGCAGGTCCGTTATCAGAG. The 3′ product was approximately 4 kb long and was amplified with the host primer 5′-CCAACCCTCTCTTTGGGTTT and virus primer 5′-AATCCAGCCACGCTCCTAC. The region encompassing the partial pro/pol region was amplified from the white-tailed deer and elk genomic DNA using primers 5′-GCCCTAAGAGGGACTCAAGG and 5′-TCCAACAGTCCCCAGAAACT.
Real-time quantitative PCR.
For the determination of CrERVγ copy numbers in the mule deer genome, four independent quantitative PCR assays were designed employing the B-R SYBR green Supermix (Quanta Biosciences). Multiple alignments of available CrERVγ sequences were used to predict primers in conserved regions of gag (5′-CCAGGTCCCTTATATCGTGGT and 5′-GCAAGAGGCATCCTGAAAGA), pol (5′-CAGCCACGCTCCTACCTAAC and 5′-TTTCTTTGCCCGTCTTTGAC), env (5′-CAAACCAAGGAGCTGTCCTC and 5′-CCCACCTTGCTGAAGAAAAA), and the LTR (5′-CGCTAAATGACCCCTGCTTA and 5′-GACAATGCAAAACGCAAGAA). Primers were designed using the Primer3Plus program (50). Each reaction mixture had a total volume of 20 μl, containing 2 μl of the genomic DNA sample and 300 nM (each) the forward and reverse primers. The samples were run on a Bio-Rad iQ5 iCycler machine with a two-step protocol (1 cycle of 3 min at 95°C and then 40 cycles consisting of 10 s at 95°C and 30 s at 55°C), followed by melting curve analysis to ensure the specificity of the amplification. An absolute standard curve for each assay was obtained by using as templates serial dilutions of a plasmid containing the corresponding amplicon. The results were normalized using the parallel amplification of a single-copy genomic locus derived from flanking sequence of one CrERVγ integration site (primers 5′-GGCCAGGTGCAATAACTGAC and 5′-AGGACCTGGAGTGGGAAACT). To ensure that the genomic locus used is present as a single copy in the mule deer genome, a sequence was chosen that is homologous to single-copy regions in the published cow and sheep genomes. Further, the quantifications were in good agreement with genome copy numbers estimated by spectrophotometer measurements of DNA concentration. A one-way random-effect modeling was performed for normalized results of each of the four assays to assess whether there is a significant presence of variation between individuals.
LTR aging.
To calculate the time (T) needed to accumulate a given number of differences (N) in the combined LTR length (L), assuming a neutral genomic substitution rate (R) of 2.3 × 10−9 to 5 × 10−9 per site per year, we use the following formula (22, 29): T = N/(R × L).
Illumina sequencing.
Total RNA was extracted from a lymph node sample of mule deer 257 preserved in RNAlater. Lymph node tissue cores were dissected into small pieces and further disrupted, lysed, and homogenized using a TissueLyser with steel beads (Qiagen). Total RNA was isolated using the RNAqueous midi kit (Applied Biosystems) with the inclusion of a DNase I treatment step. The absence of DNA contamination was further confirmed by the absence of intronic hits in several randomly chosen genes. Total RNA then was enriched for bacterial transcripts and viral non-poly(A) transcripts by the depletion of poly(A)-RNA and of host and bacterial rRNA using kits MicroPoly(A) Purist, MICROBEnrich, and MICROBExpress (all from Applied Biosystems). The recovered RNA enriched for microbial transcripts was used for cDNA synthesis (Just cDNA double-stranded cDNA synthesis kit; Stratagene). The resulting cDNA was applied to construct a paired-end DNA library using the DNAseq protocol (Illumina) and run on one lane on the Illumina sequencing platform GA IIx. The paired-end Illumina reads (9.76 million) were aligned to the full-length CrERVγ-in7 sequence using the MAQ alignment software and default settings (27).
Phylogenetic trees.
Sequences were aligned using MEGA version 4 (46) and manually edited. The maximum-likelihood tree was constructed using a heuristic tree search algorithm implemented in PHyML v3.412 (19). The GTR + I + Γ6 model (general time-reversible model with invariant sites and 6 Γ distributed heterogeneous substitution rates) with 500 nonparametric bootstrapping replicates was used. The inferred tree was visualized with MEGA and FigTree version 1.12 (http://tree.bio.ed.ac.uk/software/figtree/).
Splice site determination.
CrERVγ splice donor and acceptor sites first were predicted using the online program available at the Drosophila genome project website (http://www.fruitfly.org). Only the splice donor prediction yielded a strong signal, therefore the splice sites were determined experimentally from mule deer cDNA. The spliced env mRNA was amplified with primers located in the LTR (5′-GAGCAAACATAACGCCATGA) and in the env gene (5′-CCCACCTTGCTGAAGAAAAA). The sequence comparison of the spliced and genomic RNA confirmed the predicted spice donor site and identified the splice acceptor site.
Accession number.
The full-length CrERVγ provirus sequence was submitted to the GenBank database under accession number JN592050.
RESULTS
Presence of endogenous copies of CrERVγ in the mule deer genome.
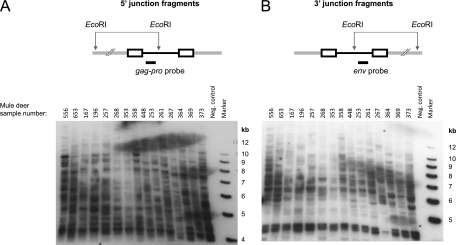
Our metatranscriptomic data from a previous study indicated that there was sequence diversity among the fragments detected in the putative envelope region of a gammaretrovirus (52). We were able to detect additional gammaretrovirus sequences by the low-stringency alignment of Roche-454 sequence reads with several prototype gammaretroviruses. Sequences were empirically confirmed by PCR using genomic DNA from mule deer that were represented in the pooled RNA used for the metatranscriptomic study. Based on these data, probes were developed for both the gag-pro junction and env to assess the retroviral integration patterns by Southern blotting among individual mule deer. The presence of discrete hybridizing bands representing CrERVγ-host junction fragments indicates that virus integrations are clonal and demonstrate that there are multiple copies of CrERVγ present as endogenous viruses in the mule deer genome (Fig. 1). Further, the pattern of bands obtained from individual mule deer genomes was not uniform among the animals tested; some bands were found only in a subset of animals and were absent from the others. These patterns of polymorphic CrERVγ integrations are consistent with a recent origin.
Fig 1.
Southern blot analysis of CrERVγ integrations in the mule deer genome. Mule deer genomic DNAs digested with EcoRI and transferred to a membrane were hybridized with a CrERVγ gag-pro probe to reveal the 5′ virus-host junction fragments (A) or a CrERVγ env probe to reveal the 3′ junction fragments (B). In the full-length CrERVγ genome, the distance of the EcoRI site to the 5′ and 3′ provirus ends is approximately 3.7 and 5.3 kb, respectively. The upper panels show schematic representations of the virus integration sites. The CrERVγ provirus is black, the virus LTR sequences are shown as rectangles, and mule deer flanking genomic DNA is shown as a gray double line. Size markers and mule deer identification numbers are indicated on the blots.
Polymorphic CrERVγ insertions revealed by PCR methods.
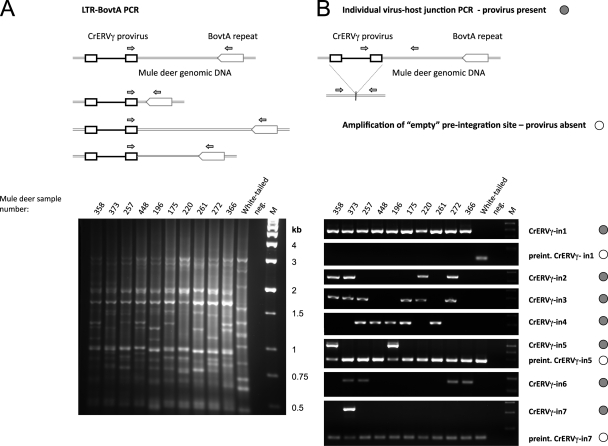
We used PCR-based techniques to confirm the Southern blot results and to characterize the virus integrations at the sequence level. A conserved primer in the CrERVγ 3′ LTR was designed based on Roche-454 sequence data and 3′RACE PCR. A host-specific primer was designed in the BovtA repeat, a ubiquitous short interspersed nuclear element (SINE)-type repeat in ruminants that is present in approximately 1.5 × 106 copies in the bovine genome (2, 44). PCR with these primers yielded bands that each corresponded to an individual virus-host junction fragment. The fragment size corresponds to the distance between the virus 3′ LTR and the BovtA repeat (Fig. 2A). Controls using PCR with only the BovtA primer confirmed that the bands do not represent BovtA repeats in head-to-head orientation (data not shown). The LTR-BovtA PCR supports that CrERVγ integrations are polymorphic among individual mule deer, and all but one (at approximately 3 kb) are clearly unique from those in a single white-tailed deer. Individual bands were obtained from the agarose gel and sequenced, and we confirmed that they correspond to 3′ virus-host junctions. Several additional 5′ virus-host junction fragments were obtained by inverse PCR.
Fig 2.
Polymorphic CrERVγ integrations detected by PCR of virus-host junction fragments. (A) Schematic representation and results of the LTR-BovtA PCR are shown. Primers, depicted as small arrows, were designed in a conserved region of the CrERVγ LTR and in a conserved part of the cervid BovtA genomic repeat. The PCR yields a pattern of bands, each representing a CrERVγ integration site with distinct distance to the nearest BovtA repeat. (B) PCR amplification of individual CrERVγ integration sites. The upper panels show schematics of the amplification of individual virus-host junctions, with primers (arrows) designed in internal virus sequences and in flanking mule deer genomic DNA away from the BovtA repeats. The 5′ and 3′ junctions were amplified; the 3′ junction scenario is depicted here. The amplification of the empty preintegration site is depicted schematically with primers in the 5′- and 3′-flanking regions relative to the virus integration point. The results of the PCR amplifications from mule deer and white-tailed deer genomic DNAs are shown below. The junction PCRs that detect the presence of specific virus integration are marked with filled circles, and the PCRs that amplify empty integration sites are marked with empty circles. For CrERVγ-in5 and CrERVγ-in7 the virus-host junctions and the empty integration sites were amplified in duplex PCRs with all primers present. M, molecular size marker; neg., no-template control; mule deer identification numbers are indicated on the gel.
We chose to characterize in detail seven provirus integration sites (CrERVγ-in1 to CrERVγ-in7) with apparent variable distribution among the mule deer evaluated. For each CrERVγ integration, a new primer was designed in the unique mule deer genome flanking sequence that excludes the BovtA repeat region, and it was used with a virus primer placed in conserved regions of the 5′ untranslated region preceding the gag (for 5′ flanks) or env gene (for 3′ flanks) (Fig. 2B). The individual junction PCRs confirm that CrERVγ provirus integrations are polymorphic among the 10 mule deer sampled at all but one of the seven integration sites.
Several patterns of CrERVγ provirus distribution were observed. CrERVγ-in1 is present in all mule deer individuals tested but not in the single white-tailed deer, suggesting that this CrERVγ is fixed in the mule deer population and subsequently integrated into the speciation event that led to these two deer species. CrERVγ-in2, CrERVγ-in3, CrERVγ-in4, CrERVγ-in5, and CrERVγ-in6 each are present in more than one individual, and each is found in a different subset of individuals. CrERVγ-in7 was detected in only a single animal; it was later detected in four additional animals in a screen of more than 200 mule deer across Montana (data not shown). Because the mule deer genome sequence is not available, we tried to obtain the missing flanking sequences (5′ or 3′) of the CrERVγ integrations by designing PCR primers based on homology with the published bovine genome. In five of the seven proviruses this allowed us to obtain both virus-host junctions; for CrERVγ-in4 and CrERVγ-in6 this was not successful. For CrERVγ-in1, -in5, and -in7, empty integration sites were amplified using primers in 5′ and 3′ genomic flanking DNA to determine if CrERVγ was present as a haploid or diploid copy in an individual (Fig. 2B). All CrERVγ-in1 integrations were diploid, which, along with the prevalence in all animals, supports that this provirus has become fixed in this population of mule deer. In contrast, CrERVγ-in5 and CrERVγ-in7 were haploid. White-tailed deer genomic DNA was negative for the presence of all seven CrERVγ integrations, indicating that these integrations are evolutionarily younger than the estimated time of the white-tailed deer/mule deer split at approximately 1.1 MYA (million years ago) (20).
An estimate of the age of an ERV can be determined by comparing the sequence divergence between the 5′ and 3′ LTRs, which were identical when the integration event occurred. Based on a neutral genomic substitution rate of 2.3 × 10−9 to 5 × 10−9 per site per year, the time needed to accumulate a given number of differences in the combined LTR length can be calculated (22, 29). LTR sequences were analyzed for the five proviruses where the sequences of PCR-amplified 5′ and 3′ junctions were available. The CrERVγ-in1 provirus has two differences between 5′ and 3′ LTRs (one of the differences is a 26-bp deletion), resulting in an estimated time of integration of 0.47 to 1 MYA. These data are consistent with this integration appearing after the mule deer/white-tailed deer split. The remaining four integrations (CrERVγ-in2, CrERVγ-in3, CrERVγ-in5, and CrERVγ-in7) have identical LTRs. Therefore, an accurate estimate of integration time cannot be determined based on LTR data alone.
Full-length CrERVγ provirus in the mule deer genome.
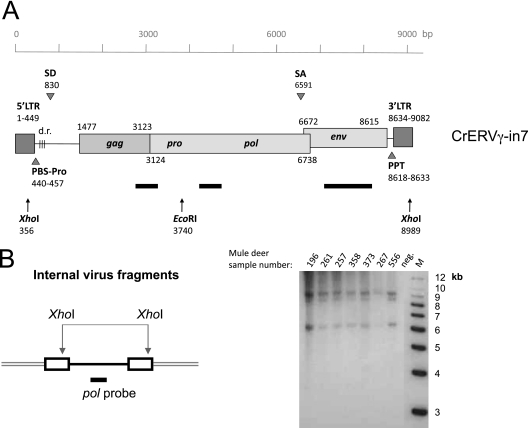
Provirus CrERVγ-in7, which was detected in only 1 of the 10 animals evaluated, was chosen for further sequence analysis because it presumably originated most recently and had the highest probability of containing an intact virus genome. The entire proviral genome was amplified in two overlapping 6- and 4-kb PCR products from the DNA of the single positive animal and was sequenced (GenBank accession number JN592050). The provirus is 9,082 nucleotides long and comprised of complete open reading frames predicted to encode the four gammaretrovirus genes gag, pro, pol, and env (Fig. 3A). The virus sequence is flanked by short 4-bp repeats (GTAA) of the target DNA, a hallmark of retroviral DNA integration (10). The identical upstream and downstream LTRs are 449 bp long, the TATA box is located at position 280, and the poly(A) signal is at position 367. There are three occurrences of the CCAAT box promoter motif. The upstream LTR is followed by a tRNA-proline primer binding site. The 5′ leader region preceding the start of gag is 1,027 bp long and contains three 44-bp direct repeats. A predicted splice donor is located at position 830, and the splice acceptor is at position 6591; the use of these sites was confirmed by sequencing a spliced env transcript (data not shown). The ORF corresponding to gag is predicted to encode a 548-amino-acid protein. It contains a conserved Cys-His box motif in the nucleocapsid region (position 2992) that is important for RNA encapsidation (18). The PPPY late virus budding domain (L-domain) is intact at position 1912, but a PSAP L-domain is not present (14). The pro-pol ORF follows the gag amber stop codon and presumably is translated by the suppression of this leaky stop codon, as is typical in gammaretroviruses (45). It is predicted to encode a 1,205-amino-acid protein. The conserved active-site motif of reverse transcriptase (YXDD) is found at position 4150, and the conserved catalytic motif of integrase (D-D-35X-E) is at positions 5938 to 6166. The env ORF (647 amino acids) partially overlaps with pol in a different reading frame. A conserved CETTG motif that has been described in exogenous gammaretroviruses is located at position 7113 (36). The downstream LTR is preceded by a polypurine tract (PPT).
Fig 3.
Presence of full-length CrERVγ provirus in the mule deer genome. (A) Schematic diagram of full-length CrERVγ-in7 provirus. The scale references nucleotide positions, and the positions of predicted features are indicated (accession number JN592050). Black bars show positions of DNA probes used for Southern blot analysis, and arrows depict restriction enzyme recognition sites. SD, splice donor; SA, splice acceptor; PPT, polypurine tract; PBS, primer binding site; d.r., direct repeats. (B) Mule deer genomic DNAs digested with XhoI and hybridized with a CrERVγ pol probe to reveal the length of complete virus fragments. Three viral species can be detected in animals tested, representing full-length genomes and two with deletions. M, molecular size markers; neg., negative control (no DNA). Mule deer identification numbers are indicated on the gel.
There is an XhoI site found in the R region of the LTR, which was conserved among the evaluated proviruses and in the Roche-454 sequence data. XhoI-digested genomic DNA subjected to Southern blot hybridization revealed three main proviral species. The dominant band corresponded to the expected length of a complete CrERVγ provirus (Fig. 3B). A weaker band, approximately 500 bp shorter, which presumably represents a small deletion in a population of CrERVγ proviruses, was detected in some animals. All animals' genomes showed evidence of deleted CrERVγ proviruses of approximately 6.5 kb in length.
Phylogenetic placement of CrERVγ in gammaretroviruses.
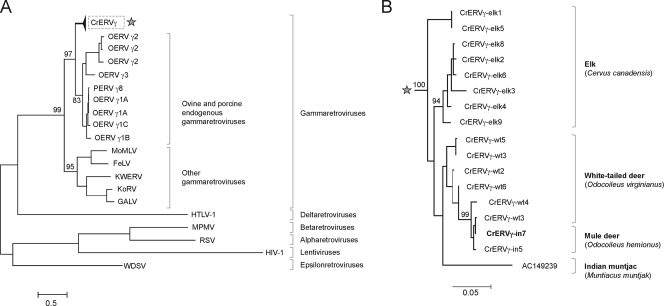
A phylogenetic analysis based on partial pro/pol nucleotide sequences obtained from GenBank grouped the CrERVγ within the gammaretrovirus genus (Fig. 4A), but it demonstrated that CrERVγ is distinct from the best-studied gammaretroviruses. CrERVγ shares a most recent common ancestor with endogenous gammaretroviruses from sheep and pig, specifically the ovine endogenous retroviruses γ1A (OERVγ1A), OERVγ1B, and OERVγ1C and porcine endogenous retrovirus γ8 (PERVγ8). Complete genomes for these porcine and ovine gammaretroviruses are not available; only PCR-amplified sequences encompassing the 0.9-kb partial pro/pol region have been published (23, 24). A search of the current versions of pig and sheep genomes did not find any ERV sequences belonging to these groups or to CrERVγ. However, a BLAST search of GenBank unfinished high-throughput genomic (HTG) sequences with CrERVγ yielded a related sequence (AC149239; positions 12003 to 18957) from another deer species, the Indian muntjac, which was 91% identical to CrERVγ. There are 22 differences between the muntjac 5′ and 3′ LTR sequences, resulting in an estimated time of integration of 5 to 10 MYA.
Fig 4.
Phylogenetic relatedness of CrERVγ. (A) A maximum-likelihood tree based on a 0.9-kb fragment of retroviral pro/pol nucleotide sequence obtained from GenBank and sequences obtained in this study for CrERVγ shows the placement of CrERVγ in the gammaretroviruses. (B) A branch of the tree from panel A (highlighted by an asterisk) representing the CrERVγ sequences from cervid species is shown in expanded form. The same region of pro/pol as that used for panel A was amplified by PCR from elk and white-tailed deer. The Indian muntjac sequence was obtained from GenBank. Bootstrap support is shown at branch nodes. The scale bars refer to the number of substitutions per site. Shown are PERV γ8 (porcine endogenous retrovirus; GenBank accession no. AF511112), OERV (ovine endogenous retrovirus; accession numbers AY193896 through AY193903), MoMLV (Moloney murine leukemia virus; NC_001501), FeLV (feline leukemia virus; NC_001940), GALV (gibbon ape leukemia virus, NC_001885); KoRV (koala retrovirus, AF151794); KWERV (killer whale endogenous retrovirus, GQ222416), HIV-1 (K03455), WDSV (walleye dermal sarcoma virus; AF033822), HTLV-1 (human T-cell lymphotropic virus; D13784), MPMV (Mason-Pfizer monkey virus; NC_001550), and RSV (Rous sarcoma virus; AF033808).
Because integration sites of a virus related to the one we detected in mule deer were identified in white-tailed deer (Fig. 2), we evaluated white-tailed deer and another member of the Cervidae, elk, to determine if virus sequences could be detected. We obtained a partial fragment of the pro/pol region using primers based on mule deer CrERVγ. All of the CrERVγ sequences from cervids cluster together, and the average pairwise sequence identity in the pro/pol region is 94.9% (standard deviation, 0.03%) (Fig. 4B). Although CrERVγ from elk represents a diverse but distinct grouping, there is insufficient resolution of white-tailed and mule deer CrERVγ using this highly conserved region. However, we cannot exclude that there has been cross-species infection between the two Odocoileus species after they diverged from a common ancestor.
CrERVγ copy numbers in the genome.
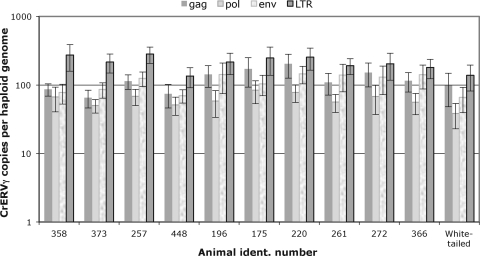
Quantitative real-time PCR assays were devised for gag, pol, env, and LTR sequences and were used on genomic DNA of the panel of 10 mule deer and 1 white-tailed deer (Fig. 5). We employed multiple probes, because the data shown in Fig. 4 revealed that some viruses have a deletion of between 1,500 and 2,000 bp in an unknown region of the genome. Quantification based on gag, pol, and env genes indicated that there are approximately 50 to 150 CrERVγ copies per haploid genome. The quantification based on LTRs shows approximately 2- to 3-fold higher numbers. This is consistent with the presence of two LTRs in each provirus and with the detection of solitary LTRs formed by inter-LTR recombination (8). The assay results for gag and env show a significant variation between individual mule deer animals (P < 0.003 for both gag and env). There is no significant variation in the quantification of pol and LTRs (P = 0.372 for pol and 0.139 for LTR). The differences observed could be due to the polymorphic CrERVγ integrations; we also cannot exclude that somatic integrations, if present, are detected by the quantitative PCR assays.
Fig 5.
CrERVγ genomic copy numbers in the deer genome. Real-time quantitative PCR was used to calculate copy numbers for gag, pol, env, and LTR in the genomic DNA isolated from lymph nodes of 10 mule deer and a single white-tailed deer. Bars indicate average values of four replicates, and standard deviations from PCRs are shown. There are significant differences among numbers of integrations.
Expression of CrERVγ RNA.
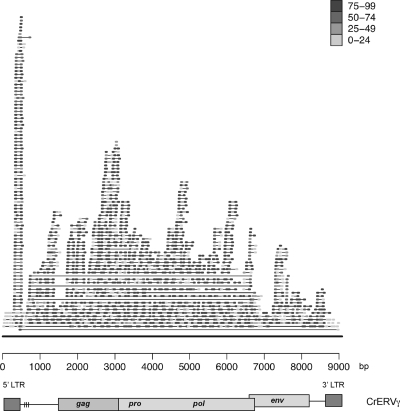
We conducted Illumina sequencing on cDNA isolated from one mule deer (number 257) to determine if CrERVγ RNA is expressed in lymph node tissue. A total of 946 Illumina reads could be aligned to the full-length CrERVγ-in7 sequence, demonstrating the expression of the entire provirus genome (Fig. 6). The depth of read coverage varies across the CrERVγ genome. Various factors could be contributing to the observed variation. It could be due to factors intrinsic to the method used to process the samples for Illumina sequencing, especially by the preferential amplification of different template regions. Alternatively, regions of CrERVγ-in7 that have higher sequence conservation among the expressed CrERVγ proviruses accumulate higher numbers of reads in the alignment.
Fig 6.
CrERVγ transcription in mule deer lymph nodes. Shown is the alignment of paired-end Illumina reads against the full-length CrERVγ sequence. The alignment was done with MAQ alignment software (27). A line connects two reads of a fragment. The scale key represents MAQ mapping qualities. A schematic of the CrERVγ provirus is depicted in the lower part.
DISCUSSION
We have identified a novel endogenous retrovirus in mule deer which forms a unique clade within the Gammaretroviridae. Phylogenetic analysis demonstrates that related viruses can be detected in white-tailed deer, elk, and muntjac, hence we named this a cervid endogenous gammaretrovirus (CrERVγ). The closest relatives of CrERVγ are endogenous gammaretroviruses from sheep and pig. CrERVγ exhibits genomic structure and composition typical for gammaretroviruses. However, all CrERVγ sequenced to date have a long (approximately 1,027-bp) leader sequence, which has been described in a fish retrovirus, salmon swim bladder sarcoma virus (38), but otherwise is unusual for gammaretroviruses.
Consistently with the endogenous nature of CrERVγ, virus-host junction fragment bands were clonal on Southern blot analysis, and all seven integration sites presented here were detected in at least two animals. Retroviruses can target any region of a host genome, and the probability of two independent retrovirus insertions at an identical genomic position is negligible (15, 26, 33). Without having the complete mule deer genome sequence available, the absolute number and diversity of CrERVγ is difficult to determine. Our quantitative PCR estimates suggest that there are, on average, around 100 proviruses closely related to CrERVγ that are integrated into the mule deer genome. The number of LTRs, including the solitary LTRs, is 2- to 3-fold higher. We were able to detect seven copies of CrERVγ LTR in a 52-Mb portion of the white-tailed deer genome that was recently reported (43). Considering an average mammalian haploid genome size of 3,000 Mb and uniform distribution of LTR across the genome, this would yield approximately 400 LTR copies per white-tailed deer genome, a number that is consistent with our quantitative PCR estimates.
Several lines of evidence suggest that at least some of the CrERVγ proviruses integrated relatively recently into the mule deer genome. CrERVγ-in1 is present in all mule deer evaluated to date (approximately 250 animals; data not shown) but not in any white-tailed deer. This suggests that CrERVγ-in1 integrated after the estimated split of white-tailed deer and mule deer approximately 1.1 MYA. A coarse estimate based on the divergence of the 5′ and 3′ LTRs is consistent with this time frame. A caveat of this method is that both homologous and heterologous recombinations between different proviruses can introduce large errors in the estimated age (22, 29). All six of the other CrERVγ proviruses are present as haploid copies and are still segregating in the mule deer population; of the four we have evaluated, all have identical LTRs. Further, the CrERVγ-in7 genome is intact and has conserved functional motifs. Interestingly, the env gene of CrERVγ-in7 contains a CETTG motif. The presence of this motif was found to be invariant in exogenous gammaretroviruses and absent from ERVs, including the very recent endogenous koala retrovirus (KoRV) insertions in koalas (36).
Insertional polymorphism is unusual for an endogenous retrovirus; most cases of insertional polymorphism documented in humans involved only a few provirus copies (7, 21, 28, 49). In mice, the most active ERV families are the retrovirus-like intracisternal A particles (IAP) and the MusD/early transposons (ETn) (30, 53). Insertional polymorphism has been described among breeds of domestic animals. In cats, a group of 29 endogenous feline leukemia viruses (enFELVs) was found to exhibit significant insertional polymorphism among various cat breeds (39, 40). The sheep genome contains at least 27 copies of endogenous Jaagsiekte sheep retrovirus (enJSRV), a betaretrovirus. Most of them are fixed in domestic sheep, but at least seven are insertionally polymorphic both between and within sheep breeds (5, 12). The most recent endogenization process has been documented for KoRV, which is associated with hematopoietic neoplasia in its host. KoRV has probably entered the koala genome during the last 200 years, and there are still geographical regions where the koala populations are virus free (48). We have tested mule deer from Montana and Colorado, which is a small portion of their North American range, and detected significant insertional polymorphisms of CrERVγ proviruses. This represents a unique example of an ERV in a nondomestic animal that exhibits recent activity of a virus that has been present in the host species and closely related species for an extended evolutionary period of time.
The source of the polymorphic CrERVγ integrations could be an active endogenous provirus, which would be consistent with the transcriptional activity that we detected in the lymph node. However, we cannot exclude the existence of a currently circulating virus closely related to CrERVγ, an alternative that we are actively pursuing. An additional process that leads to the recent amplification of ERVs has been described in marsupials. The kangaroo endogenous retrovirus (KERV) was massively amplified in Macropus rufogriseus through genome duplications limited to centromeric regions (16). We can only estimate the chromosomal distribution of CrERVγ integrations by comparing the flanking sequences to the published ungulate genomes (cow, pig, and sheep). The matched genomic positions are distributed across various chromosomes without any obvious bias. Moreover, all integrations are flanked by the unique short target site duplications, which supports that at least the proviruses evaluated here were formed by independent unique virus integrations and not by genome duplication events.
We could detect CrERVγ sequences in the three cervid species that we tested, and sequences related to CrERVγ can be identified in the muntjac genome. This raises the possibility that an ancestral CrERVγ entered the cervid genome before the cervid family radiation around 10 MYA (17). This would be consistent with the estimated age of the provirus present in the muntjac (5 to 10 MYA). If this were the case, all cervid species should share a subset of CrERVγ integration sites, as has been described for JSRV (5, 12). None of the seven CrERVγ proviruses we have identified to date is found in white-tailed deer, the closest relative to mule deer. The CrERVγ sequences that we obtained by PCR from elk are phylogenetically distinct, but those from white-tailed and mule deer do not cluster strictly by species of origin (Fig. 4B). Although this could be due to insufficient diversity in the highly conserved pro/pol fragment utilized, it is possible that cross-species infections have occurred. In addition, mule deer and white-tailed deer are unique among large mammals because they can form viable interspecies hybrids (11). Interestingly, KERV amplification presumably was caused by the epigenetic deregulation of the chromosomal locus in interspecific marsupial hybrids (35). Thus, another possibility is that mule deer/white-tailed deer hybridization is a driving force in CrERVγ insertional polymorphism.
The lymph node samples for this study were collected from healthy deer at hunter check stations. Therefore, we cannot associate CrERVγ with any disease state or other phenotype. A C-type retrovirus in moose has been proposed to have causal association with wasting syndrome (31). Also, a replication-competent gammaretrovirus was isolated from black-tailed deer, a subspecies of mule deer (1). There is no sequence data for either isolate available, which precludes comparison with CrERVγ. However, the discovery of a novel transcriptionally active and insertionally polymorphic retrovirus in cervids provides a unique model to study the dynamic interaction between a mammalian genome and an invading retrovirus.
ACKNOWLEDGMENTS
We thank Paul Cross for supplying samples for this study, Josh Marceau and Xiaoyan Gong for excellent technical assistance, and Yee Ling Chong for help with phylogenetic analysis. The comments from three anonymous reviewers substantially improved the manuscript.
This work was supported by USGS grant 06HQAG0131.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Aaronson SA, Tronick SR, Stephenson JR. 1976. Endogenous type C RNA virus of Odocoileus hemionus, a mammalian species of New World origin. Cell 9:489–494 [DOI] [PubMed] [Google Scholar]
- 2. Adelson DL, Raison JM, Edgar RC. 2009. Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc. Natl. Acad. Sci. U. S. A. 106:12855–12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson MM, Lauring AS, Burns CC, Overbaugh J. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828–1830 [DOI] [PubMed] [Google Scholar]
- 4. Armezzani A, et al. 2011. The signal peptide of a recently integrated endogenous sheep betaretrovirus envelope plays a major role in eluding gag-mediated late restriction. J. Virol. 85:7118–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arnaud F, et al. 2007. A paradigm for virus-host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog. 3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balada E, Vilardell-Tarres M, Ordi-Ros J. 2010. Implication of human endogenous retroviruses in the development of autoimmune diseases. Int. Rev. Immunol. 29:351–370 [DOI] [PubMed] [Google Scholar]
- 7. Belshaw R, et al. 2005. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J. Virol. 79:12507–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belshaw R, et al. 2007. Rate of recombinational deletion among human endogenous retroviruses. J. Virol. 81:9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blikstad V, Benachenhou F, Sperber GO, Blomberg J. 2008. Evolution of human endogenous retroviral sequences: a conceptual account. Cell. Mol. Life Sci. 65:3348–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown PO. 1997. Integration. In Coffin JM, Hughes SH, Varmus HE. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY: [PubMed] [Google Scholar]
- 11. Carr SM, Ballinger SW, Derr JN, Blankenship LH, Bickham JW. 1986. Mitochondrial DNA analysis of hybridization between sympatric white-tailed deer and mule deer in west Texas. Proc. Natl. Acad. Sci. U. S. A. 83:9576–9580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chessa B, et al. 2009. Revealing the history of sheep domestication using retrovirus integrations. Science 324:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448:105–114 [DOI] [PubMed] [Google Scholar]
- 14. Demirov DG, Freed EO. 2004. Retrovirus budding. Virus Res. 106:87–102 [DOI] [PubMed] [Google Scholar]
- 15. Elleder D, Pavlíček A, Pačes J, Hejnar J. 2002. Preferential integration of human immunodeficiency virus type 1 into genes, cytogenetic R bands and GC-rich DNA regions: insight from the human genome sequence. FEBS Lett. 517:285–286 [DOI] [PubMed] [Google Scholar]
- 16. Ferreri GC, et al. 2011. Recent amplification of the kangaroo endogenous retrovirus, KERV, limited to the centromere. J. Virol. 85:4761–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilbert C, Ropiquet A, Hassanin A. 2006. Mitochondrial and nuclear phylogenies of Cervidae (Mammalia, Ruminantia): systematics, morphology, and biogeography. Mol. Phylogenet. Evol. 40:101–117 [DOI] [PubMed] [Google Scholar]
- 18. Gorelick RJ, Henderson LE, Hanser JP, Rein A. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. U. S. A. 85:8420–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 20. Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22:2971–2972 [DOI] [PubMed] [Google Scholar]
- 21. Jha AR, et al. 2011. Human endogenous retrovirus K106 (HERV-K106) was infectious after the emergence of anatomically modern humans. PLoS One 6:e20234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson WE, Coffin JM. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 96:10254–10260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klymiuk N, Muller M, Brem G, Aigner B. 2003. Characterization of endogenous retroviruses in sheep. J. Virol. 77:11268–11273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klymiuk N, Muller M, Brem G, Aigner B. 2002. Characterization of porcine endogenous retrovirus gamma pro-pol nucleotide sequences. J. Virol. 76:11738–11743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- 26. Lewinski MK, Bushman FD. 2005. Retroviral DNA integration–mechanism and consequences. Adv. Genet. 55:147–181 [DOI] [PubMed] [Google Scholar]
- 27. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Macfarlane C, Simmonds P. 2004. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J. Mol. Evol. 59:642–656 [DOI] [PubMed] [Google Scholar]
- 29. Mager DL, Freeman JD. 1995. HERV-H endogenous retroviruses: presence in the New World branch but amplification in the Old World primate lineage. Virology 213:395–404 [DOI] [PubMed] [Google Scholar]
- 30. Maksakova IA, et al. 2006. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merza M, Larsson E, Steen M, Morein B. 1994. Association of a retrovirus with a wasting condition in the Swedish moose. Virology 202:956–961 [DOI] [PubMed] [Google Scholar]
- 32. Mi S, et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789 [DOI] [PubMed] [Google Scholar]
- 33. Mitchell RS, et al. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moyes D, Griffiths DJ, Venables PJ. 2007. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet. 23:326–333 [DOI] [PubMed] [Google Scholar]
- 35. O'Neill RJ, O'Neill MJ, Graves JA. 1998. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393:68–72 [DOI] [PubMed] [Google Scholar]
- 36. Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. 2007. Changes in viral protein function that accompany retroviral endogenization. Proc. Natl. Acad. Sci. U. S. A. 104:17506–17511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Overbaugh J, Riedel N, Hoover EA, Mullins JI. 1988. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature 332:731–734 [DOI] [PubMed] [Google Scholar]
- 38. Paul TA, et al. 2006. Identification and characterization of an exogenous retrovirus from Atlantic salmon swim bladder sarcomas. J. Virol. 80:2941–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roca AL, Nash WG, Menninger JC, Murphy WJ, O'Brien SJ. 2005. Insertional polymorphisms of endogenous feline leukemia viruses. J. Virol. 79:3979–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roca AL, Pecon-Slattery J, O'Brien SJ. 2004. Genomically intact endogenous feline leukemia viruses of recent origin. J. Virol. 78:4370–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosenberg N, Jolicoeur P. 1997. Retroviral pathogenesis. In Coffin JM, Hughes SH, Varmus HE. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY: [PubMed] [Google Scholar]
- 42. Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. 2008. Endogenous retroviruses and cancer. Cell. Mol. Life Sci. 65:3366–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Seabury CM, et al. 2011. Genome-wide polymorphism and comparative analyses in the white-tailed deer (Odocoileus virginianus): a model for conservation genomics. PLoS One 6:e15811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shimamura M, Abe H, Nikaido M, Ohshima K, Okada N. 1999. Genealogy of families of SINEs in cetaceans and artiodactyls: the presence of a huge superfamily of tRNA(Glu)-derived families of SINEs. Mol. Biol. Evol. 16:1046–1060 [DOI] [PubMed] [Google Scholar]
- 45. Swanstrom R, Wills JW. 1997. Synthesis, assembly, and processing of viral proteins. In Coffin JM, Hughes SH, Varmus HE. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, New York, NY: [PubMed] [Google Scholar]
- 46. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 47. Tandon R, et al. 2011. Identification of human endogenous retrovirus-specific T cell responses in vertically HIV-1-infected subjects. J. Virol. 85:11526–11531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tarlinton RE, Meers J, Young PR. 2006. Retroviral invasion of the koala genome. Nature 442:79–81 [DOI] [PubMed] [Google Scholar]
- 49. Turner G, et al. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11:1531–1535 [DOI] [PubMed] [Google Scholar]
- 50. Untergasser A, et al. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Waterston RH, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562 [DOI] [PubMed] [Google Scholar]
- 52. Wittekindt NE, et al. 2010. Nodeomics: pathogen detection in vertebrate lymph nodes using meta-transcriptomics. PLoS One 5:e13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. 2008. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 4:e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]