Abstract
Lepidopteran nucleopolyhedroviruses (NPVs) show distinct tissue tropism in host insect larvae. However, the molecular mechanism of this tropism is largely unknown. We quantitatively investigated NPV tissue tropism by measuring mRNA levels of viral genes in 16 tissues from Bombyx mori NPV (BmNPV)-infected B. mori larvae and found clear tissue tropism, i.e., BmNPV replicates poorly in the silk glands, midgut, and Malpighian tubule compared with other larval tissues. We next identified the viral genes determining tissue tropism in NPV infection by investigating the phenotypes of larvae infected with 44 BmNPV mutants in which one gene was functionally disrupted by a LacZ cassette insertion. We found that occlusion body (OB) production was markedly enhanced compared with that of the wild type in the middle silk glands (MSGs) of larvae infected with three mutants in which one of three tandemly arrayed genes (Bm7, Bm8, and Bm9) was disrupted. We generated additional mutants in which one or two genes of this gene cluster were partially deleted and showed that Bm8, also known as BV/ODV-E26, was solely required for the suppression of OB production in the MSGs of BmNPV-infected B. mori larvae. Western blotting showed that a LacZ cassette insertion in Bm7 or Bm9 resulted in aberrant expression of Bm8, presumably leading to abnormal OB production in the MSGs. Larval bioassays also revealed that disruption of Bm8 accelerated the death of B. mori larvae. These results suggest that the group I NPV-specific protein BV/ODV-E26 determines tissue tropism and virulence in host lepidopteran insects.
INTRODUCTION
Baculoviridae is a large family of pathogens that infect insects, particularly the order Lepidoptera. Baculoviruses have a large, circular, supercoiled, and double-stranded DNA genome packaged into rod-shaped virions (58, 59). Baculoviruses are divided phylogenetically into four genera: Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus (23). Alphabaculoviruses can be further subdivided into group I and II nucleopolyhedroviruses (NPVs), based on phylogenetic studies (19). NPVs produce two types of virions during their infection cycle to bring about efficient viral replication within infected larvae and to spread the virus among insects. Occlusion-derived viruses (ODVs), which are occluded in occlusion bodies (OBs), transmit virus from insect to insect via oral infection, whereas budded viruses (BVs) spread infection to neighboring cells (16, 37). At the late stage of infection, a markedly enhanced locomotion behavior of infected larvae is observed (15, 25), which is followed by a dramatic degradation of the host cadaver (11).
orf8 (Bm8) of Bombyx mori NPV (BmNPV), a homolog of Autographa californica NPV (AcMNPV) Ac16 or bv/odv-e26, is one of 17 genes specific to group I NPVs (27). Bm8 is a nucleic acid binding protein that accumulates as distinct foci during the early stage of infection (21). In BmNPV-infected cells, Bm8 colocalizes and interacts with IE1 via the N-terminal region containing the coiled-coil domain, and this interaction is essential for nuclear localization of Bm8 (28). Further microscopic analyses suggest that IE1 and homologous regions (hrs) facilitate the localization of Bm8 to specific nuclear sites (28). Generation of a BmNPV mutant (Bm8D) lacking a functional Bm8 gene revealed that the gene is involved in efficient BV production in B. mori cells (21). Bacmid knockout studies in AcMNPV similarly showed that disruption of Ac16 resulted in a reduction in DNA replication and BV production (46). Recent biochemical approaches have also identified additional Ac16-interacting partners, IE0 and FP25K, and further demonstrated that Ac16 and FP25K form a complex containing cellular actin, which suggests that Bm8/Ac16 is a multifunctional protein (2, 45).
Lepidopteran NPVs demonstrate a unique tissue tropism in host insects. The replication of most NPVs, with the exception of lepidopteran NPVs, is restricted to the midgut, which is the first target tissue in oral infection (11). In contrast, lepidopteran NPVs establish a transient infection in the midgut, without OB production, before the infection spreads to most of the larval tissues. Recently, infection studies using recombinant NPVs expressing marker genes (gfp or LacZ) have allowed us to visually observe the NPV tissue tropism (1, 10, 54). However, the tissue tropism of NPV-infected larvae has not been quantitatively examined yet. Here, we measured the temporal expression levels of four viral genes in 16 tissues from BmNPV-infected B. mori larvae and found clear tissue tropism in the tissues examined. Also, using a series of gene knockout BmNPVs, we identified the gene Bm8 as one of the determinants of BmNPV tissue tropism.
MATERIALS AND METHODS
Insects, cell lines, and viruses.
B. mori larvae were reared as described previously (44). BmN (BmN-4) cells were cultured at 27°C in TC-100 medium supplemented with 10% fetal bovine serum. BmNPV T3 was used as the wild-type virus. The deletion BmNPVs used in this study are listed in Table 1. Virus titers were determined by plaque assay on BmN cells (44).
Table 1.
BmNPV mutants used in this studya
| Knockout mutant | Target gene (locus) | LacZ insertion site(s) | Reference(s) |
|---|---|---|---|
| Bm7D | egt | 7070–7695 | 26 (48) |
| Bm8D | bv/odv-e26 | 8397–8689 | 21 (49) |
| Bm9D | orf9 | 8832 | This study (46) |
| Bm11D | orf11 | 10751 | This study |
| Bm12D | arif-1 | 11776 | This study (56) |
| Bm14D | orf14 | 15049 | This study (42) |
| Bm18D | iap1 | 17760 | (17) 31 |
| Bm21D | orf21 | 20564 | This study |
| Bm24D | vfgf | 23454–23878 | (8) 30 |
| Bm26D | v-ubi | 25199 | 36 (55) |
| Bm27D | 39k | 25555–26071 | 14 (63) |
| Bm29D | nudix | 26601–26953 | This study (13) |
| Bm33D | gta | 30234–30782 | 33 |
| Bm34D | orf34 | 31768 | 34 |
| Bm35D | orf35 | 31931–32137 | This study |
| Bm37D | odv-e66 (chondritinase) | 33019–34082 | This study (62) |
| Bm41D | orf41 | 39246–39510 | 25 |
| Bm44D | orf44 | 41659–41816 | This study |
| Bm47D | ChaB-like | 42851 | This study |
| Bm49D | fp25K | 43922–44023 | (18) 29 |
| Bm51D | orf51 | 46297–46338 | This study |
| Bm52D | gp37 | 46816–47142 | This study (5) |
| Bm56D | orf56 | 54157 | This study (40) |
| Bm60D | orf60 | 56798–57440 | This study |
| Bm70D | vp15 | 66532 | This study |
| Bm71D | cg30 | 67093–67205 | 25 (51) |
| Bm74D | orf74 | 70168 | 25 |
| Bm89D | he65 | 85434–85886 | This study |
| Bm94D | orf94 | 90354–90801 | This study |
| Bm95D | pif-3 | 91697–91814 | 25 (47) |
| Bm96D | orf96 | 92163–92314 | This study |
| Bm106D | p24 | 101749–101999 | This study (57) |
| Bm107D | gp16 | 102387 | 25 |
| Bm108D | pp34 | 102596–103231 | This study (61) |
| Bm111D | orf111 | 105629 | This study (7) |
| Bm112D | p35 | 106000–106672 | (6) 24 |
| Bm115D | p74 | 109478–110200 | 25 (38) |
| Bm121D | orf121 | 116069 | This study (39) |
| Bm124D | odv-e56 (pif-5) | 119158 | (4) 25 |
| Bm127D | ie2 | 120662–121103 | 14 (53) |
| Bm129D | orf129 | 123561 | This study |
| Bm130D | ptp | 124424–124806 | 25 (41) |
| Bm133D | orf133 | 126942–127065 | This study |
| Bm134D | orf134 | 127467 | 25 |
References for the AcMNPV orthologs are in parentheses.
Construction of gene knockout BmNPVs by inserting a β-galactosidase gene cassette.
We used 44 gene deletion BmNPV mutants, 24 of which (listed in Table 1) we newly generated in this study by homologous recombination, as described previously (29). To construct the plasmids for deletion of BmNPV genes, we first isolated the specific genomic region of each BmNPV gene from the genomic clone of BmNPV T3 (43) and inserted them into plasmid vectors. We then digested the plasmids using convenient restriction enzyme sites within the coding region of each gene and ligated to a β-galactosidase gene (LacZ) cassette containing a Drosophila melanogaster heat shock protein promoter (hsp70-LacZ cassette) (24). The resultant plasmids were cotransfected with T3 DNA into BmN cells using Cellfectin (Invitrogen). BmNPV mutants were isolated by the identification of plaques expressing β-galactosidase. The deletion of each gene was confirmed by PCR as described previously (29). Note that we simply isolated 24 newly isolated viruses, and further detailed characterization has not yet been performed. The BV titers of these mutants propagated in BmN cells are about 5 × 106 to 1 × 108 PFU/ml.
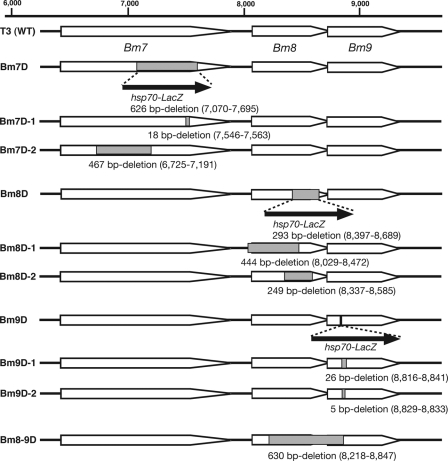
Generation of recombinant BmNPVs with partial deletions in Bm7, Bm8, or Bm9.
We prepared plasmids in which Bm7 (known as the ecdysteroid UDP-glucosyltransferase gene [egt]) was partially deleted to generate Bm7D-1 and Bm7D-2 (see Fig. 4). Deletion plasmids were constructed from the PstI G fragment of BmNPV (43) using a Deletion Kit for Kilo-Sequencing (Takara). The resultant plasmids were cotransfected with Bsu36I-digested Bm7D DNA into BmN cells using Cellfectin (Invitrogen). Five days after transfection, the medium was collected and stored at 4°C until use. Recombinant BmNPVs were isolated by the identification of plaques that did not express β-galactosidase (30). We verified inactivation of EGT by bioassays using day 5 fifth-instar B. mori larvae. Larvae infected with Bm7D-1 and Bm7D-2, as well as Bm7D, started spinning their cocoons by 3 days postinfection (p.i.), whereas T3-infected larvae did not spin cocoons at all. Five more deletion mutants (Bm8D-1, Bm8D-2, Bm9D-1, Bm9D-2, and Bm8-9D) (see Fig. 4) were similarly constructed by homologous recombination in BmN cells. Precise deletion of each gene was confirmed by PCR and DNA sequencing.
Fig 4.
Generation of deletion mutants without an hsp70-LacZ insertion. Shown are schematic representations of all Bm7, Bm8, and Bm9 mutants with or without an hsp70-LacZ insertion. The method for the construction of BmNPV mutants is described in detail in Materials and Methods. WT, wild type.
Western blot analysis of Bm8.
BmN cells were infected with BmNPVs at a multiplicity of infection (MOI) of 5. Cells were collected at 6, 12, 24, and 48 h p.i., and nuclear fractions were subjected to Western blot analysis with anti-Bm8 antibody as described previously (21, 44). Bm8 expression was quantified by densitometry using ImageGauge software (Fujifilm).
qRT-PCR.
Fifth-instar B. mori larvae were starved for several hours and then injected with 50 μl of a viral suspension containing 1 × 105 PFU and returned to an artificial diet at 27°C. Sixteen tissues (brain, corpora allata, central nerve, prothoracic gland, fat body, trachea, hemocytes, testis, ovary, anterior silk gland, middle silk gland [MSG], posterior silk gland [PSG], midgut, Malpighian tubule, integument, and muscle) were dissected from BmNPV-infected B. mori larvae (5 to 30 larvae per tissue) at 1, 2, 3, and 4 days p.i., and total RNA was prepared using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized from 0.2 μg of total RNA, and quantitative reverse transcription (qRT)-PCR was performed using Power SYBR green PCR master mix (Applied Biosystems) with published primers (34). Amplification was detected using the StepOne real-time PCR system (Applied Biosystems) (34).
BmN cells were infected with BmNPVs at an MOI of 5. Cells were collected at 4, 12, 24, and 48 h p.i., and total RNA was prepared. qRT-PCR experiments with Bm7, Bm8, Bm9, and vp39 were performed as described above. The primers used are listed in Table 2.
Table 2.
Primers
| Primer | Sequence | Purpose |
|---|---|---|
| Bm7F | 5′-ACCATATAGTCTACAAAGTG-3′ | qRT-PCR for Bm7 |
| Bm7R | 5′-CTAGTTTCTTGTATTGCTTG-3′ | |
| Bm8F | 5′-AACTCGAAAGTGCACAGAAG-3′ | qRT-PCR for Bm8 |
| Bm8R | 5′-CAATAATTGTGCGAATTGTG-3′ | |
| Bm9F | 5′-GTAGTTTAATGGATACTGAG-3′ | qRT-PCR for Bm9 |
| Bm9R | 5′-ATCGCTAATGCTACAAAAGG-3′ |
Larval bioassays.
The median lethal time (LT50) was determined by intrahemocoelic injection of BVs (50 μl of a viral suspension containing 1 × 105 PFU) into fifth-instar B. mori larvae within 12 h after molting (34). Statistical analyses were performed with Prism 5 (Graphpad). A log rank (Mantel-Cox) test, with Bonferroni correction, was used, comparing each of the mutants with T3 (control virus). At least 20 larvae per dose were used in each experiment.
RESULTS
Tissue tropism of BmNPV in B. mori larvae.
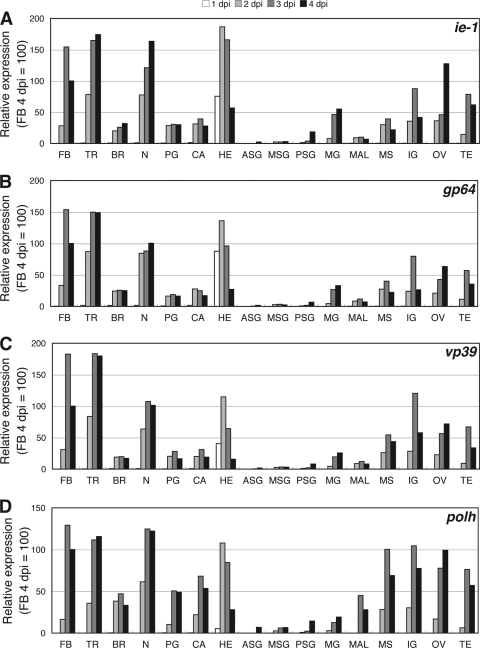
We first determined the tissues where NPVs replicate well in lepidopteran insect larvae. To examine the virus growth quantitatively, we measured the temporal expression levels of four BmNPV genes, i.e., ie1 (immediate early), gp64 (early and late), vp39 (late), and polh (very late), by qRT-PCR of 16 tissues from B. mori larvae intrahemocoelically infected with BmNPV. Of note, this infection is not a normal route under natural conditions. In a previous study using a green fluorescent protein (GFP)-expressing BmNPV, we showed that when a virus was administered through this infection route, virus infection was first observed at 12 h p.i. in hemocytes, and GFP expression in other major tissues, such as fat body and trachea, was clearly detected by 48 h p.i. (32). qRT-PCR analysis showed similar expression patterns for the four genes in BmNPV-infected tissues. At 1 day p.i., viral-gene expression was detected only in hemocytes, confirming that hemocytes are the first target tissue when viruses are inoculated intrahemocoelically (Fig. 1). Viral-gene expression in hemocytes peaked at 2 days p.i. and then gradually declined. In most tissues, expression of virus genes was first detected at 2 days p.i. and peaked at 3 days p.i. This expression pattern is consistent with our previous study using a GFP-expressing BmNPV (32). Interestingly, the three silk glands (anterior silk gland, middle silk gland, and posterior silk gland), midgut, and Malpighian tubule were much less infected than the other 11 tissues (Fig. 1). In addition, we observed that in Malpighian tubules, polh expression was relatively high compared with the expression levels of the other three genes. These results indicate the existence of clear tissue tropism in BmNPV-infected B. mori larvae.
Fig 1.
Expression of viral genes in 16 tissues from BmNPV-infected B. mori larvae. Fifth-instar B. mori larvae were injected with a viral suspension containing 1 × 105 PFU. Sixteen tissues, i.e., brain (BR), corpora allata (CA), central nerve (N), prothoracic gland (PG), fat body (FB), trachea (TR), hemocytes (HE), testis (TE), ovary (OV), anterior silk gland (ASG), middle silk gland (MSG), posterior silk gland (PSG), midgut (MG), Malpighian tubule (MAL), integument (IG), and muscle (MS), were dissected from BmNPV-infected B. mori larvae (5 to 30 larvae per tissue) at 1, 2, 3, and 4 days p.i., and total RNA was prepared. Expression of ie1 (A), gp64 (B), vp39 (C), and polh (D) was examined by qRT-PCR.
Screening of genes involved in tissue tropism of BmNPV in B. mori larvae.
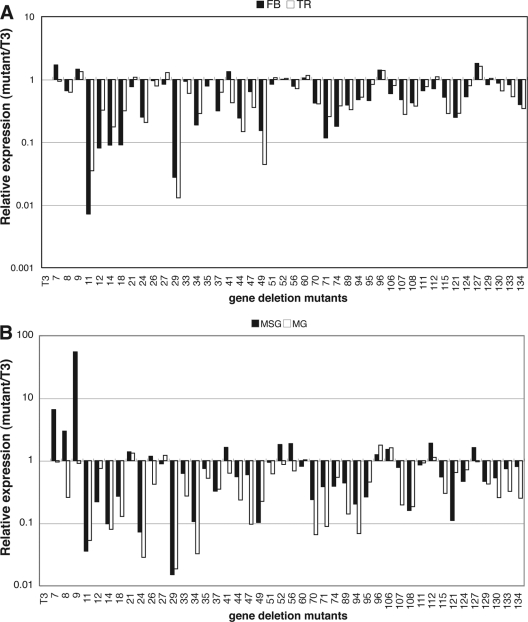
To identify the genes involved in the tissue tropism of BmNPV infection, we screened the phenotypes of 44 BmNPV mutants where one gene was functionally disrupted by the insertion of a LacZ cassette. We measured polh expression in two well-infected (fat body and trachea) and two poorly infected (middle silk gland and midgut) tissues (Fig. 1). Fifth-instar B. mori larvae were injected with BV, and the tissues were dissected from BmNPV-infected B. mori larvae at 4 days p.i. qRT-PCR analysis showed that polh expression was reduced or unchanged in the four tissues infected with most deletion mutants (Fig. 2). However, we found that, compared with the wild-type T3, polh expression was markedly enhanced (3- to 55-fold) in the MSGs of larvae infected with three mutants where one of three tandemly arrayed open reading frames (ORFs) (Bm7, Bm8, and Bm9) was disrupted.
Fig 2.
Expression of polh in the fat body, trachea, middle silk gland, and midgut of B. mori larvae infected with gene deletion BmNPVs. Fifth-instar B. mori larvae were injected with BV of T3 and 44 gene deletion mutants. Four tissues, i.e., fat body (FB), trachea (TR), middle silk gland (MSG), and midgut (MG), were dissected from BmNPV-infected B. mori larvae (five larvae per tissue) at 4 days p.i., and total RNA was prepared. Expression of polh was examined by qRT-PCR. The expression level for T3 was considered 1, and polh expression of the mutants is shown relative to that for T3. The raw data sets are provided as Table S1 in the supplemental material.
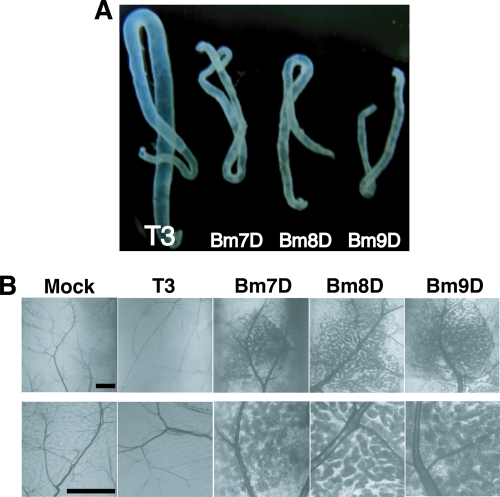
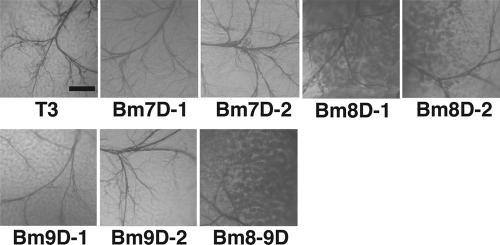
We then examined OB production in the MSGs of B. mori larvae infected with the three mutants. We found that the silk glands of larvae infected with the three mutants were markedly smaller and cloudy compared with T3 (Fig. 3A). Microscopic observations revealed that Bm7D, Bm8D, or Bm9D produced large amounts of OBs around the tracheas of MSGs, whereas OB production was rarely seen in the MSGs of T3-infected larvae (Fig. 3B).
Fig 3.
OB production in MSGs of BmNPV-infected B. mori larvae. (A) MSGs of B. mori larvae infected with T3, Bm7D, Bm8D, and Bm9D at 4 days p.i. (B) Light microscopic observations of MSGs of BmNPV-infected larvae at 4 days p.i. High-magnification images are shown below. The dark blobs are OB-laden nuclei of multinucleated giant cells in MSGs. Bars, 0.2 mm.
Disruption of Bm8 results in abnormal OB production in the MSGs.
Our screening revealed that polh expression and OB production were markedly enhanced in the MSGs of B. mori larvae infected with mutants in which one of three genes (Bm7, Bm8, and Bm9) was disrupted with a LacZ cassette insertion (Fig. 3). We tested whether all three genes are required for proper expression of polh in the MSGs by generating seven additional mutants in which one or two genes were partially deleted without inserting a LacZ cassette (Fig. 4). We examined OB production in the MSGs of B. mori larvae infected with these mutants. As shown in Fig. 5, the functional disruption of Bm7 or Bm9 did not result in increased OB production in the MSGs. However, Bm8D-1 or Bm8D-2, in which only Bm8 is partially deleted, showed a phenotype similar to that observed in B. mori larvae infected with Bm7D, Bm8D, or Bm9D. We also observed that deletion of both Bm8 and Bm9 resulted in enhanced OB production in the MSGs. These results clearly indicate that Bm8 is solely required for proper polh expression in the MSGs of BmNPV-infected B. mori larvae.
Fig 5.
Microscopic observations of OB production in MSGs from larvae infected with BmNPVs with Bm7, Bm8, and Bm9 deleted. Light microscopic observations of MSGs of B. mori larvae infected with Bm7, Bm8, and Bm9 deletion mutants without an hsp70-LacZ insertion at 4 days p.i. are shown. The dark blobs are OB-laden nuclei of multinucleated giant cells in MSGs. Bar, 0.2 mm.
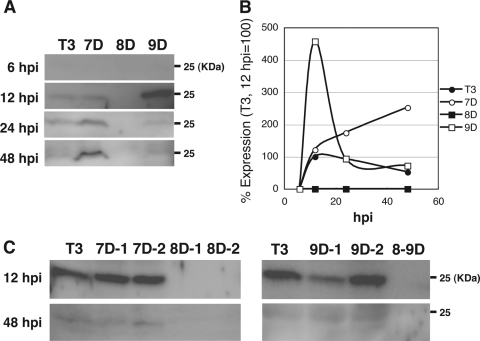
Aberrant expression of Bm8 in cells infected with Bm7D or Bm9D.
To know the reason why Bm7D or Bm9D showed a phenotype similar to that observed in Bm8-mutated BmNPV, we compared the temporal expression patterns of Bm8 in BmN cells infected with T3, Bm7D, Bm8D, or Bm9D. Western blot analysis showed that the level of Bm8 in T3-infected cells peaked at 12 h p.i. and then slightly declined at 48 h p.i. (Fig. 6A and B), which agreed with our previous results (21). In Bm7D- or Bm9D-infected cells, however, the expression pattern was markedly different from that observed in T3-infected cells. In Bm7D-infected cells, Bm8 expression increased gradually and peaked at 48 h p.i. (5-fold expression compared with T3), whereas the expression in Bm9D-infected cells was strikingly enhanced at 12 h p.i. (4.5-fold expression compared with T3) and rapidly declined to the level observed in T3-infected cells by 24 h p.i. (Fig. 6A and B). We then examined Bm8 expression in BmN cells infected with other Bm7, Bm8, and Bm9 mutants. As expected, Bm8 expression was not observed in BmN cells infected with Bm8D-1, Bm8D-2, and Bm8-9D (Fig. 6C). We also found that, unlike LacZ-insertion mutants, Bm7D-1, Bm7D-2, and Bm9D-2 showed expression patterns of Bm8 similar to those observed in T3. On the other hand, we noticed that Bm8 expression was slightly reduced in Bm9D-1-infected cells at 12 h p.i., but its expression was indistinguishable from that of T3 at 48 h p.i. (Fig. 6C). These results suggest that a LacZ cassette insertion, but not a deletion, in Bm7 or Bm9 resulted in enhanced expression of Bm8, presumably leading to abnormal polh expression in the MSGs.
Fig 6.
Aberrant expression of Bm8 during Bm7D and Bm9D infection. (A) BmN cells were infected with T3, Bm7D, Bm8D, and Bm9D, and cells were harvested at the indicated times postinfection. Nuclear fractions were subjected to Western blot analysis with anti-Bm8 antibody. (B) Bm8 expression was quantified by densitometry. (C) Western blot analysis of Bm8 in Bm7D-1-, Bm7D-2-, Bm8D-1-, Bm8D-2-, Bm9D-1-, Bm9D-2-, and Bm8-9D-infected BmN cells. The cells were harvested at 12 and 48 h p.i.
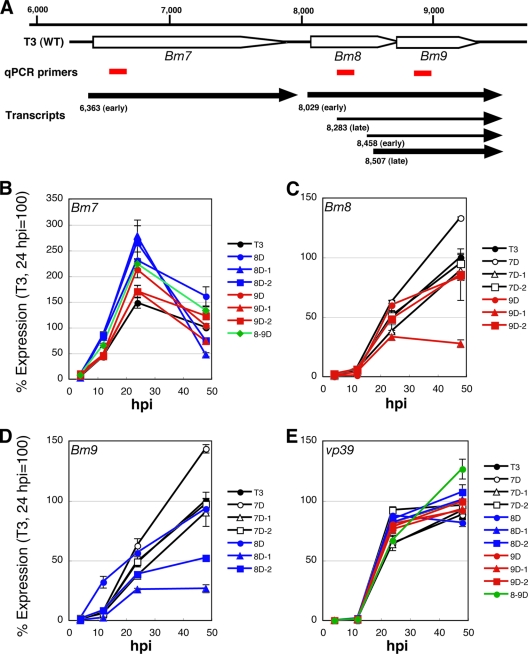
Next, we performed qRT-PCR experiments with Bm7, Bm8, and Bm9 using total RNA prepared from BmN cells infected with each mutant. Bm7 and Bm8 are expressed mainly from early promoters, whereas Bm9 is an early and late transcribed gene (Fig. 7A to D) (35, 64). qRT-PCR analyses showed that the expression level of Bm8 in Bm7D-infected cells was higher than that of T3 from 24 h p.i. In contrast, Bm8 expression in Bm7D-1- and Bm7D-2-infected cells was indistinguishable from that of T3 (Fig. 7C). These results suggest that the enhanced expression of Bm8 protein observed in Bm7D-infected cells is due mainly to the transcriptional upregulation of Bm8. We also noticed Bm9 upregulation in Bm7D-infected cells, and its expression pattern was quite similar to that of Bm8 in Bm7D-infected cells (Fig. 7C and D). As illustrated in Fig. 7A, we previously showed that the Bm8 transcript terminates downstream of the Bm9 coding region (35), indicating that a primer set amplifying Bm9 also detects the Bm8 transcript. This suggests that the observed increase of Bm9 expression in Bm7D-infected cells may result from detection of Bm8 upregulation. Furthermore, we found that mutations in Bm8 had some effects on Bm7 and/or Bm9 expression: all of the Bm8 mutants showed slightly enhanced expression of Bm7, whereas Bm8D-1 and Bm8D-2 expressed low levels of Bm9 compared to T3. As shown in Fig. 7C, Bm9D-1, but not Bm9D-2, showed reduced expression of Bm8, which agreed with the observation of Bm8 protein expression in Bm9D-1-infected cells (Fig. 6C). The expression patterns of vp39 (one of the BmNPV late genes) were similar in all mutants (Fig. 7E).
Fig 7.
Transcriptional analysis of Bm7, Bm8, and Bm9 in BmN cells infected with mutant BmNPVs. (A) Schematic representation of the Bm7-Bm9 region. The positions of primer sets used in the qRT-PCR experiments are indicated by red bars. The arrows show each transcriptional unit revealed by our previous study (35). (B to E) Transcriptional analysis of Bm7, Bm8, and Bm9. BmN cells were infected with each mutant, and cells were harvested at 4, 12, 24, and 48 h p.i. First-strand cDNA was synthesized from total RNA prepared from BmNPV-infected cells, and qRT-PCR experiments with Bm7 (B), Bm8 (C), and Bm9 (D) were performed. vp39 (E) was used as a control. The error bars indicate standard deviations.
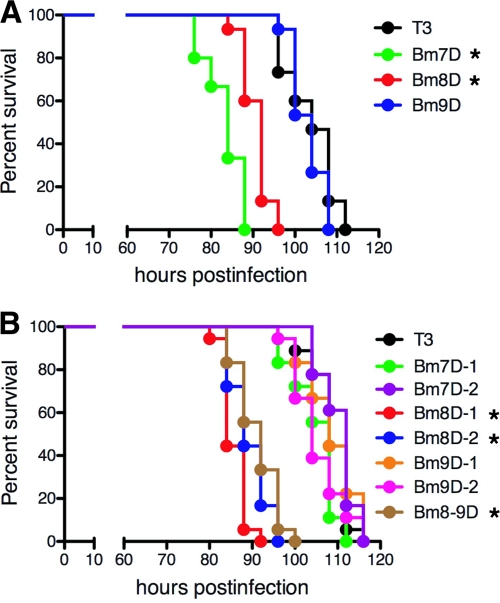
Disruption of Bm8 accelerates the death of B. mori larvae.
Previous studies showed that the deletion of Bm7 (known as the ecdysteroid UDP-glucosyltransferase gene [egt]) from the AcMNPV or BmNPV genome accelerated the death of insect larvae (9, 26, 50). As expected, when we injected BVs into day 0 fifth-instar B. mori larvae (within 12 h after molting), larvae infected with Bm7D died 20 h earlier than those infected with T3 (Fig. 8A). Interestingly, we observed that Bm8D, but not Bm9D, also accelerated larval death, suggesting that enhanced OB production in the MSGs is not directly related to the killing speed in host insects (Fig. 8A). To examine which gene is involved in viral virulence, we performed infection studies using 7 partial-deletion mutants of Bm7, Bm8, and/or Bm9 (Fig. 4) and compared their survival curves using day 0 fifth-instar larvae. As shown in Fig. 8B, mutants with Bm8 deletions showed the virulent phenotype, whereas two Bm7 mutants (Bm7D-1 and Bm7D-2) did not. These results indicate that disruption of Bm8, but not Bm7 (egt), reduced the survival time of BmNPV-infected fifth-instar B. mori larvae and that the earlier death observed in Bm7D-infected larvae may be due mainly to the aberrant expression of Bm8 after a LacZ cassette insertion in Bm7.
Fig 8.
Survival curves for B. mori larvae infected with Bm7-, Bm8-, and Bm9-mutated BmNPVs. (A) The LT50s of T3, Bm7D, Bm8D, and Bm9D were 104 h, 84 h, 92 h, and 104 h, respectively. *, P < 0.05 compared with T3. (B) The LT50s of T3, Bm7D-1, Bm7D-2, Bm8D-1, Bm8D-2, Bm9D-1, Bm9D-2, and Bm8-9D were 108 h, 108 h, 112 h, 84 h, 88 h, 108 h, 104 h, and 92 h, respectively. *, P < 0.05 compared with T3.
DISCUSSION
It is well known that lepidopteran NPVs demonstrate distinct tissue tropism in host insect larvae. For example, lepidopteran NPVs do not produce OBs in the midguts of infected larvae, while OBs are produced in most other larval tissues. To examine the NPV's tissue tropism quantitatively, we first measured mRNA levels of viral genes in 16 tissues from B. mori larvae infected with BmNPV. We found that NPV replicated poorly in the silk glands, midgut, and Malpighian tubule compared with other tissues examined. We then used LacZ insertional mutants of BmNPV to identify the viral genes involved in the tissue tropism of BmNPV. Screening and further experiments identified a gene, Bm8 (also known as bv/odv-e26), required for the suppression of virus growth in the MSGs. The inactivation of Bm8 resulted in abnormally enhanced virus replication and OB production in the MSGs of infected larvae, which probably led to inhibition of silk gland development (Fig. 3A). During the fifth larval stage of B. mori, fibroin and sericin proteins are synthesized and secreted in very high quantities in the PSG and MSG, respectively. The heavy chains of fibroin and sericin are known to be composed of specific amino acid-enriched sequences: glycine (>45%) and alanine (>30%) are enriched in fibroin, whereas serine is enriched in sericin (>30%). Hence, silk gland-specific tRNAs with cognate anticodons for these two proteins accumulate heavily in the PSG and MSG due to massive production of fibroin and sericin (22, 60, 65). Such an abnormal tRNA composition may not be optimal for BmNPV replication and OB production. This seems to be a reason why the virus is not able to propagate well in silk gland cells. It is possible to postulate that loss of Bm8 alters the tRNA composition in infected cells, thereby leading to efficient virus replication and poor development of the silk gland.
We detected abnormal OB production in the MSGs of B. mori larvae infected with three LacZ cassette insertion mutants, i.e., Bm7D, Bm8D, and Bm9D. Further experiments using mutants with partial deletions in Bm7, Bm8, and Bm9 clearly showed that this phenotypic abnormality was solely caused by disruption of Bm8. Western blot experiments also showed that the expression pattern of Bm8 was deregulated in Bm7D- or Bm9D-infected cells (Fig. 6A and B). Overall, we concluded that the disruption or abnormal expression of Bm8 may result in abnormal polh expression in the MSGs, thereby leading to enhanced OB production in the MSGs. We also observed that Bm8 expression was reduced in Bm9D-1-infected cells at 12 h p.i. (Fig. 6C), suggesting that a slight decrease in Bm8 at an early stage of infection has little effect on polh expression in the MSG. We then asked why the insertion of a LacZ cassette into Bm7 or Bm9 resulted in the aberrant expression of Bm8. Transcriptome analysis of the Bm8 region of BmNPV (35) and AcMNPV (46) showed that the Bm8 transcript terminates downstream of the Bm9 coding region (Fig. 7A). This indicates that a LacZ insertion in Bm9 disrupts the 3′ untranslated region of Bm8, which may affect the transcriptional regulation of Bm8. However, we did not observe a difference between Bm8 expression in T3- and Bm9D-infected cells (Fig. 7C). In Bm9D-infected cells, Bm8 expression was transiently enhanced at 12 h p.i. compared with T3 (Fig. 6A), suggesting that a LacZ insertion into the untranslated region of Bm8, which corresponds to the Bm9 coding region, enhances the early-phase expression of the Bm8 protein without altering Bm8 transcription. Unlike Bm8 expression in T3-infected cells, its expression in Bm7D-infected cells increased gradually, and it did not decline at the late stage of infection (48 h p.i.). Although the mechanism of Bm8 deregulation in Bm7D-infected cells remains unknown, qRT-PCR experiments revealed that a LacZ cassette insertion into the Bm7 coding region abnormally activates the Bm8 promoter throughout the infection (Fig. 7C). Taken together, these results suggest that insertions and/or deletions in the Bm7-Bm9 cluster sometimes result in aberrant expression of each gene. Deep RNA sequencing and fine mapping of transcripts expressed from this region are required to clarify the complex regulation of these three genes.
Previous studies showed that removal of a functional egt gene (Bm7) from the genome of AcMNPV accelerated the death of host insects, Spodoptera frugiperda and Trichoplusia ni (approximately 20 h earlier than the wild-type virus infection) (9, 50). The dose-mortality responses were not significantly different between wild-type and egt deletion viruses. Early degeneration of the Malpighian tubules in Spodoptera exigua larvae infected with an egt-mutated AcMNPV was observed (12); however, the mechanism of the fast-killing phenotype remains elusive. In this study, we confirmed a previous study showing that Bm7D, an egt deletion mutant of BmNPV with a LacZ cassette insertion, also resulted in accelerated death of fifth-instar B. mori larvae (26) (Fig. 8A). However, it was surprising that the LT50s of two other egt deletion mutants, Bm7D-1 and Bm7D-2, did not show the same phenotype. These results indicate that EGT activity is not related to the fast-killing phenotype observed in Bm7D-infected fifth-instar B. mori larvae and that inactivation of BmNPV EGT does not affect the survival time of fifth-instar larvae. The larval instar and infection period are likely critical factors for killing speed, because the length of each instar is different, and passing through the molting stage is extremely stressful for the infected larvae. We examined the effects of egt deletion on survival time using day 0 and day 3 fourth-instar larvae. When we used day 0 fourth-instar larvae, we obtained almost the same results as with day 0 fifth-instar larvae. However, we found that three mutants with Bm7 deletion (Bm7D, Bm7D-1, and Bm7D-2) showed the fast-killing phenotype using day 3 fourth-instar larvae (data not shown). In addition, for some insects, there are reports that killing speed is not affected by the deletion of egt (3, 52). Therefore, we conclude at present that the effects of egt deletion from the NPV genome on the survival time are different depending on the larval instar, infection period, and insect species. We also found that Bm8D, but not Bm9D, accelerated the speed of death in fifth-instar larvae, showing that abnormal production of OBs in the MSGs does not solely lead to the fast killing. Western blot analysis of Bm8 in Bm7D-infected cells (Fig. 6A) also suggested that the fast-killing phenotype found in Bm7D-infected fifth-instar B. mori larvae may be due mainly to the aberrant expression of Bm8 by marker cassette insertion into the Bm7 locus.
NPVs induce abnormal wandering behavior in host larvae at the late stage of infection. It is believed that this behavior contributes to OB contamination on a larger surface area of the host plant, which contributes to the efficient dispersal of the virus. Recently, BmNPV protein tyrosine phosphatase (25)- and Lymantria dispar NPV (LdMNPV)-encoded egt genes (20) have been shown to be required for enhanced locomotory activity (ELA), but the molecular mechanism is still unknown. In the course of infection experiments, we noticed that larvae infected with Bm8-mutated viruses and Bm7D did not exhibit distinct wandering behavior throughout the infection, whereas the typical virus-induced behavior was observed in larvae infected with T3, Bm7D-1, Bm7D-2, and Bm9-mutated viruses (data not shown). These results suggest that Bm8 is a third gene required for NPV-induced enhanced behavior. Furthermore, deletion of egt alone did not affect the host behavior in BmNPV, unlike a group II LdMNPV. Survival curves clearly showed that these wandering-defective mutants exhibited the fast-killing phenotype. The LT50 of T3 was 104 to 108 h, whereas those of Bm8D, Bm8D-1, Bm8D-2, Bm8-9D, and Bm7D were 84 to 92 h (Fig. 8). Locomotion enhancement in T3-infected fifth-instar B. mori larvae was observed from approximately 92 to 96 h p.i., at which time over 50% of the larvae infected with Bm8-mutated viruses and Bm7D had died. We suggest that collectively, Bm8-mutated viruses kill larvae before hijacking the host neuronal circuit and/or other organs required for wandering, which would lead to viruses failing to manipulate host behavior.
In conclusion, we discovered that the group I NPV-specific protein BV/ODV-E26 is crucial for the suppression of virus growth in the MSGs of host lepidopteran insects. To our knowledge, this is the first report to identify a gene(s) determining the sites where NPVs replicate in host larvae. Elucidating how this protein functions in the silk glands of infected larvae will allow us to understand some of the mechanisms whereby baculoviruses select tissues for optimal replication in infected hosts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Munetaka Kawamoto and Minako Suzuki for clerical assistance.
This work was supported by grants from MEXT (no. 19688004 and 22380033 to S.K. and no. 17018007 to T.S.) and NIAS/MAFF (Agrigenome Research Program), Japan.
Footnotes
Published ahead of print 21 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Barrett JW, Brownwright AJ, Primavera MJ, Palli SR. 1998. Studies of the nucleopolyhedrovirus infection process in insects by using the green fluorescence protein as a reporter. J. Virol. 72:3377–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beniya H, Braunagel SC, Summers MD. 1998. Autographa californica nuclear polyhedrosis virus: subcellular localization and protein trafficking of BV/ODV-E26 to intranuclear membranes and viral envelopes. Virology 240:64–75 [DOI] [PubMed] [Google Scholar]
- 3. Bianchi FJ, et al. 2000. Biological activity of SeMNPV, AcMNPV, and three AcMNPV deletion mutants against Spodoptera exigua larvae (Lepidoptera: noctuidae). J. Invertebr. Pathol. 75:28–35 [DOI] [PubMed] [Google Scholar]
- 4. Braunagel SC, Elton DM, Ma H, Summers MD. 1996. Identification and analysis of an Autographa californica nuclear polyhedrosis virus structural protein of the occlusion-derived virus envelope: ODV-E56. Virology 217:97–110 [DOI] [PubMed] [Google Scholar]
- 5. Cheng X, Krell P, Arif B. 2001. P34.8 (GP37) is not essential for baculovirus replication. J. Gen. Virol. 82:299–305 [DOI] [PubMed] [Google Scholar]
- 6. Clem RJ, Fechheimer M, Miller LK. 1991. Prevention of apoptosis by a baculovirus gene during infection of insect cells. Science 254:1388–1390 [DOI] [PubMed] [Google Scholar]
- 7. Clem RJ, Robson M, Miller LK. 1994. Influence of infection route on the infectivity of baculovirus mutants lacking the apoptosis-inhibiting gene p35 and the adjacent gene p94. J. Virol. 68:6759–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Detvisitsakun C, Hutfless EL, Berretta MF, Passarelli AL. 2006. Analysis of a baculovirus lacking a functional viral fibroblast growth factor homolog. Virology 346:258–265 [DOI] [PubMed] [Google Scholar]
- 9. Eldridge R, O'Reilly DR, Hammock BD, Miller LK. 1992. Insecticidal properties of genetically engineered baculoviruses expressing an insect juvenile hormone esterase gene. Appl. Environ. Microbiol. 58:1583–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engelhard EK, Kam-Morgan LN, Washburn JO, Volkman LE. 1994. The insect tracheal system: a conduit for the systemic spread of Autographa californica M nuclear polyhedrosis virus. Proc. Natl. Acad. Sci. U. S. A. 91:3224–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Federici BA. 1997. Baculovirus pathogenesis, p 33–59 In Miller LK. (ed), The baculoviruses. Plenum Press, New York, NY [Google Scholar]
- 12. Flipsen JT, Mans RM, Kleefsman AW, Knebel-Mörsdorf D, Vlak JM. 1995. Deletion of the baculovirus ecdysteroid UDP-glucosyltransferase gene induces early degeneration of Malpighian tubules in infected insects. J. Virol. 69:4529–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ge J, et al. 2007. AcMNPV ORF38 protein has the activity of ADP-ribose pyrophosphatase and is important for virus replication. Virology 361:204–211 [DOI] [PubMed] [Google Scholar]
- 14. Gomi S, Zhou CE, Yih W, Majima K, Maeda S. 1997. Deletion analysis of four of eighteen late gene expression factor gene homologues of the baculovirus, BmNPV. Virology 230:35–47 [DOI] [PubMed] [Google Scholar]
- 15. Goulson D. 1997. Wipfelkrankheit: manipulation of host behavior by a baculovirus. Oecologia 109:219–228 [DOI] [PubMed] [Google Scholar]
- 16. Granados RR, Lawler KA. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108:297–308 [DOI] [PubMed] [Google Scholar]
- 17. Griffiths CM, et al. 1999. In vitro host range of Autographa californica nucleopolyhedrovirus recombinants lacking functional p35, iap1 or iap2. J. Gen. Virol. 80:1055–1066 [DOI] [PubMed] [Google Scholar]
- 18. Harrison RL, Summers MD. 1995. Mutations in the Autographa californica multinucleocapsid nuclear polyhedrosis virus 25 kDa protein gene result in reduced virion occlusion, altered intranuclear envelopment and enhanced virus production. J. Gen. Virol. 76:1451–1459 [DOI] [PubMed] [Google Scholar]
- 19. Herniou EA, et al. 2001. Use of whole genome sequence data to infer baculovirus phylogeny. J. Virol. 75:8117–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hoover K, et al. 2011. A gene for an extended phenotype. Science 333:1401. [DOI] [PubMed] [Google Scholar]
- 21. Imai N, Kurihara M, Matsumoto S, Kang W. 2004. Bombyx mori nucleopolyhedrovirus orf8 encodes a nucleic acid binding protein that colocalizes with IE1 during infection. Arch. Virol. 149:1581–1594 [DOI] [PubMed] [Google Scholar]
- 22. International Silkworm Genome Consortium 2008. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38:1036–1045 [DOI] [PubMed] [Google Scholar]
- 23. Jehle JA, et al. 2006. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 151:1257–1266 [DOI] [PubMed] [Google Scholar]
- 24. Kamita SG, Majima K, Maeda S. 1993. Identification and characterization of the p35 gene of Bombyx mori nuclear polyhedrosis virus that prevents virus-induced apoptosis. J. Virol. 67:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamita SG, et al. 2005. A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc. Natl. Acad. Sci. U. S. A. 102:2584–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang KD, Lee EJ, Kamita SG, Seong SI. 1998. Effect of the ecdysteroid UDP-glucosyltransferase gene of the Bombyx mori nucleopolyhedrovirus on the development of the silkworm, Bombyx mori. Korean J. Seric. Sci. 40:105–110 [Google Scholar]
- 27. Kang W. 2009. Molecular dissection of Bombyx mori nucleopolyhedrovirus orf8 gene. Virol. Sin. 24:315–322 [Google Scholar]
- 28. Kang W, Imai N, Kawasaki Y, Nagamine T, Matsumoto S. 2005. IE1 and hr facilitate the localization of Bombyx mori nucleopolyhedrovirus ORF8 to specific nuclear sites. J. Gen. Virol. 86:3031–3038 [DOI] [PubMed] [Google Scholar]
- 29. Katsuma S, Noguchi Y, Zhou CL, Kobayashi M, Maeda S. 1999. Characterization of the 25K FP gene of the baculovirus Bombyx mori nucleopolyhedrovirus: implications for post-mortem host degradation. J. Gen. Virol. 80:783–791 [DOI] [PubMed] [Google Scholar]
- 30. Katsuma S, Horie S, Daimon T, Iwanaga M, Shimada T. 2006. In vivo and in vitro analyses of a Bombyx mori nucleopolyhedrovirus mutant lacking functional vfgf. Virology 355:62–70 [DOI] [PubMed] [Google Scholar]
- 31. Katsuma S, Kawaoka S, Mita K, Shimada T. 2008. Genome-wide survey for baculoviral host homologs using the Bombyx genome sequence. Insect Biochem. Mol. Biol. 38:1080–1086 [DOI] [PubMed] [Google Scholar]
- 32. Katsuma S, Horie S, Shimada T. 2008. The fibroblast growth factor homolog of Bombyx mori nucleopolyhedrovirus enhances systemic virus propagation in B. mori larvae. Virus Res. 137:80–85 [DOI] [PubMed] [Google Scholar]
- 33. Katsuma S, Fujii T, Kawaoka S, Shimada T. 2008. Bombyx mori nucleopolyhedrovirus SNF2 global transactivator homologue (Bm33) enhances viral pathogenicity in B. mori larvae. J. Gen. Virol. 89:3039–3046 [DOI] [PubMed] [Google Scholar]
- 34. Katsuma S, Shimada T. 2009. Bombyx mori nucleopolyhedrovirus ORF34 is required for efficient transcription of late and very late genes. Virology 392:230–237 [DOI] [PubMed] [Google Scholar]
- 35. Katsuma S, et al. 2011. Mass identification of transcriptional units expressed from the Bombyx mori nucleopolyhedrovirus genome. J. Gen. Virol. 92:200–203 [DOI] [PubMed] [Google Scholar]
- 36. Katsuma S, Tsuchida A, Matsuda-Imai N, Kang W, Shimada T. 2011. Role of the ubiquitin-proteasome system in Bombyx mori nucleopolyhedrovirus infection. J. Gen. Virol. 92:699–705 [DOI] [PubMed] [Google Scholar]
- 37. Keddie BA, Aponte GW, Volkman LE. 1989. The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science 243:1728–1730 [DOI] [PubMed] [Google Scholar]
- 38. Kuzio J, Jaques R, Faulkner P. 1989. Identification of p74, a gene essential for virulence of baculovirus occlusion bodies. Virology 173:759–763 [DOI] [PubMed] [Google Scholar]
- 39. Lapointe R, et al. 2004. Characterization of two Autographa californica nucleopolyhedrovirus proteins, Ac145 and Ac150, which affect oral infectivity in a host-dependent manner. J. Virol. 78:6439–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li G, Wang J, Deng R, Wang X. 2008. Characterization of AcMNPV with a deletion of ac68 gene. Virus Genes 37:119–127 [DOI] [PubMed] [Google Scholar]
- 41. Li Y, Miller LK. 1995. Properties of a baculovirus mutant defective in the protein phosphatase gene. J. Virol. 69:4533–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lung OY, Cruz-Alvarez M, Blissard GW. 2003. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J. Virol. 77:328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maeda S, Majima K. 1990. Molecular cloning and physical mapping of the genome of Bombyx mori nuclear polyhedrosis virus. J. Gen. Virol. 71:1851–1855 [DOI] [PubMed] [Google Scholar]
- 44. Nakanishi T, et al. 2010. Comparative studies of lepidopteran baculovirus-specific protein FP25K: development of a novel Bombyx mori nucleopolyhedrovirus-based vector with a modified fp25K gene. J. Virol. 84:5191–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nie Y, Fang M, Theilmann DA. 2009. AcMNPV AC16 (DA26, BV/ODV-E26) regulates the levels of IE0 and IE1 and binds to both proteins via a domain located within the acidic transcriptional activation domain. Virology 385:484–495 [DOI] [PubMed] [Google Scholar]
- 46. Nie Y, Theilmann DA. 2010. Deletion of AcMNPV AC16 and AC17 results in delayed viral gene expression in budded virus infected cells but not transfected cells. Virology 404:168–179 [DOI] [PubMed] [Google Scholar]
- 47. Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. 2005. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J. Virol. 79:15258–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. O'Reilly DR, Miller LK. 1989. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyl transferase. Science 245:1110–1112 [DOI] [PubMed] [Google Scholar]
- 49. O'Reilly DR, Passarelli AL, Goldman IF, Miller LK. 1990. Characterization of the DA26 gene in a hypervariable region of the Autographa californica nuclear polyhedrosis virus genome. J. Gen. Virol. 71:1029–1037 [DOI] [PubMed] [Google Scholar]
- 50. O'Reilly DR, Miller LK. 1992. Improvement of a baculovirus pesticide by deletion of the egt gene. Biotechnology 9:1086–1089 [Google Scholar]
- 51. Passarelli AL, Miller LK. 1994. In vivo and in vitro analyses of recombinant baculoviruses lacking a functional cg30 gene. J. Virol. 68:1186–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Popham HJR, Li Y, Miller LK. 1997. Genetic improvement of Helicoverpa zea nuclear polyhedrosis virus as a biopesticide. Biol. Control 10:83–91 [Google Scholar]
- 53. Prikhod'ko EA, Lu A, Wilson JA, Miller LK. 1999. In vivo and in vitro analysis of baculovirus ie-2 mutants. J. Virol. 73:2460–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rahman MM, Gopinathan KP. 2004. Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res. 101:109–118 [DOI] [PubMed] [Google Scholar]
- 55. Reilly LM, Guarino LA. 1996. The viral ubiquitin gene of Autographa californica nuclear polyhedrosis virus is not essential for viral replication. Virology 218:243–247 [DOI] [PubMed] [Google Scholar]
- 56. Roncarati R, Knebel-Mörsdorf D. 1997. Identification of the early actin-rearrangement-inducing factor gene, arif-1, from Autographa californica multicapsid nuclear polyhedrosis virus. J. Virol. 71:7933–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schetter C, Oellig C, Doerfler W. 1990. An insertion of insect cell DNA in the 81-map-unit segment of Autographa californica nuclear polyhedrosis virus DNA. J. Virol. 64:1844–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Summers MD, Anderson DL. 1972. Granulosis virus deoxyribonucleic acid: a closed, double-stranded molecule. J. Virol. 9:710–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Summers MD, Anderson DL. 1973. Characterization of nuclear polyhedrosis virus DNAs. J. Virol. 12:1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takasu Y, et al. 2007. Identification and characterization of a novel sericin gene expressed in the anterior middle silk gland of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 37:1234–1240 [DOI] [PubMed] [Google Scholar]
- 61. van Lent JW, et al. 1990. Localization of the 34 kDa polyhedron envelope protein in Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. Arch. Virol. 111:103–114 [DOI] [PubMed] [Google Scholar]
- 62. Xiang X, et al. 2011. Autographa californica multiple nucleopolyhedrovirus odv-e66 is an essential gene required for oral infectivity. Virus Res. 158:72–78 [DOI] [PubMed] [Google Scholar]
- 63. Yamagishi J, Burnett ED, Harwood SH, Blissard GW. 2007. The AcMNPV pp31 gene is not essential for productive AcMNPV replication or late gene transcription but appears to increase levels of most viral transcripts. Virology 365:34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang ZN, et al. 2009. Bombyx mori nucleopolyhedrovirus ORF9 is a gene involved in the budded virus production and infectivity. J. Gen. Virol. 90:162–169 [DOI] [PubMed] [Google Scholar]
- 65. Zhou CZ, et al. 2000. Fine organization of Bombyx mori fibroin heavy chain gene. Nucleic Acids Res. 28:2413–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.