Abstract
Among different coronavirus genera, the receptor-binding S1 subunits of their spike proteins differ in primary, secondary, and tertiary structures. This study identified shared structural topologies (connectivity of secondary structural elements) in S1 domains of different coronavirus genera. The results suggest that coronavirus S1 subunits share a common evolutionary origin but have attained diverse sequences and structures following extensive divergent evolution. The results also increase understanding of the structures and functions of coronavirus S1 domains whose tertiary structures are currently unknown.
TEXT
Traces of the origins of viruses and viral proteins are often masked or even erased by the high mutation rates, long evolutionary history, and many genetic tricks of the viruses. For viral proteins, evolutionary records are more likely to be conserved in their tertiary structures than in their gene sequences, due to the evolutionary pressure on these proteins to maintain their functions within a certain structural framework. Sometimes, however, evolutionary clues can be lost even in tertiary structures of viral proteins, presenting evolutionary conundrums. This study investigated these conundrums surrounding coronavirus (CoV) spike proteins that have different tertiary structures.
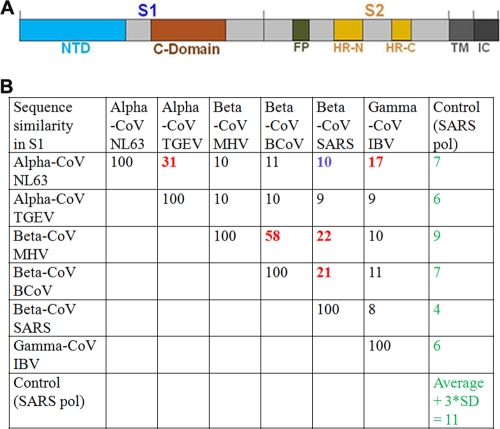
CoVs, a family of enveloped positive-stranded RNA viruses, can be divided into three major genera or groups, the alpha-CoVs (group 1), the beta-CoVs (group 2), and the gamma-CoVs (group 3) (5). The representative members in each genus are listed in Fig. 1. A trimeric spike protein anchored on coronavirus envelopes mediates viral entry into host cells. It contains an ectodomain, a transmembrane anchor, and a short intracellular tail (Fig. 1A). During molecular maturation, the ecotodomain is often cleaved into a receptor-binding S1 subunit and a membrane-fusion S2 subunit. For cell entry, S1 binds to a host receptor for viral attachment, and S2 undergoes dramatic structural changes to fuse the viral and host membranes. The sequences, structures, and membrane-fusion mechanisms of the S2 subunits are conserved among different coronavirus genera (10, 11). However, the S1 subunits from different coronavirus genera share little or no significant sequence similarity (Fig. 1B). Two independent domains have been identified in the S1 subunits from different coronavirus genera, the N-terminal domain (NTD) and C-domain (Fig. 1A), both of which can bind host receptors and hence function as receptor-binding domains (RBDs) (4). Furthermore, coronavirus S1 subunits recognize a variety of host receptors, including proteins and sugars. The diversities in their S1 sequences and receptor usage present evolutionary puzzles surrounding coronavirus spike proteins (5).
Fig 1.
Spike proteins from three coronavirus genera or groups. (A) Schematic representation of coronavirus spike proteins. NTD, N-terminal domain; FP, fusion peptide; HR-N, heptad repeat N; HR-C, heptad repeat C; TM, transmembrane anchor; IC, intracellular tail. (B) Sequence similarities among S1 subunits from representative CoVs (coronaviruses). NL63, human NL63 coronavirus strain Amsterdam I; TGEV, porcine transmissible gastroenteritis virus strain Purdue; MHV, mouse hepatitis coronavirus strain A59; BCoV, bovine coronavirus strain ENT; SARS, SARS coronavirus strain Tor2; IBV, avian infectious bronchitis virus strain M41; SARS pol, SARS polymerase strain Tor02; SD, standard deviation. Alpha-, beta-, and gamma-CoVs can also be referred to as group 1, group 2, and group 3 coronaviruses, respectively. Sequence similarities were calculated using ClusterW (1).
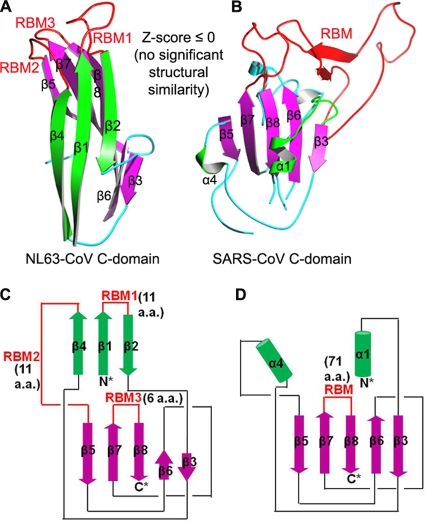
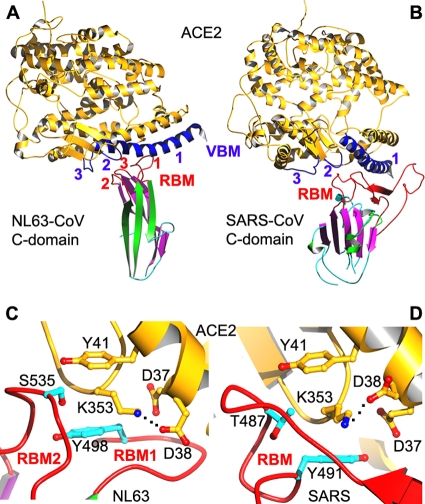
To date, three crystal structures have been available for coronavirus S1 domains: NTD of beta-genus mouse hepatitis coronavirus (MHV) and the C-domains of alpha-genus NL63 coronavirus (NL63-CoV) and beta-genus severe acute respiratory syndrome coronavirus (SARS-CoV) (3, 4, 9). According to the results of searches performed with the DALI protein structure database search server (2), whereas MHV NTD and human galectins share the same fold, the SARS-CoV and NL63-CoV C-domains each represent a novel fold (Fig. 2A and B). Interestingly, despite their different structures, NL63-CoV and SARS-CoV C-domains bind to overlapping regions on their common receptor, human angiotensin-converting enzyme 2 (ACE2) (Fig. 3A and B) (9). We previously hypothesized that SARS-CoV and NL63-CoV C-domains diverged from a common ancestor into different structures and then converged functionally to recognize the same ACE2 receptor (8, 9). In this report, we present evidence to support this hypothesis and expand the evolutionary discussion to include all three coronavirus genera.
Fig 2.
Structures of alpha-coronavirus NL63-CoV and beta-coronavirus SARS-CoV C-domains. (A) Crystal structure of NL63-CoV C-domain (PDB identification no. 3KBH). (B) Crystal structure of SARS-CoV C-domain (PDB identification no. 2AJF). Structural similarity Z-scores were calculated using the DALI server (2). (C) Topology of NL63-CoV C-domain. (D) Topology of SARS-CoV C-domain. RBM, receptor-binding motif.
Fig 3.
Receptor binding by alpha-coronavirus NL63-CoV and beta-coronavirus SARS-CoV C-domains. (A) Crystal structure of NL63-CoV C-domain complexed with human ACE2 (PDB identification no. 3KBH). (B) Crystal structure of SARS-CoV C-domain complexed with human ACE2 (PDB identification no. 2AJF). (C) A hydrophobic tunnel structure at the NL63-CoV/ACE2 interface, comprising residues Tyr41 and Asp37 from ACE2 and Ser535 and Tyr498 from the NL63-CoV C-domain. (D) A hydrophobic tunnel structure at the SARS-CoV/ACE2 interface, comprising residues Tyr41 and Asp37 from ACE2 and Thr487 and Tyr491 from the SARS-CoV C-domain. The hydrophobic tunnels in panels C and D bury a salt bridge between Lys353 and Asp38 from ACE2. ACE2, angiotensin-converting enzyme 2; VBM, virus-binding motif.
SARS-CoV and NL63-CoV C-domains differ in primary, secondary, and tertiary structures. The sequence similarity between their S1 subunits is 10%, not significantly higher than the similarity between two random protein sequences (Fig. 1B). The core structure of the NL63-CoV C-domain is a β-sandwich consisting of two β-sheet layers that stack against each other. The two β-sheet layers consist of five strands (β5-β7-β8-β6-β3) and three strands (β4-β1-β2), respectively (Fig. 2A and B). On the other hand, the core structure of the SARS-CoV C-domain is a single-layer five-strand β-sheet (β5-β7-β8-β6-β3) with two α-helices (α4-α1) stacked against it (Fig. 2A and B). The DALI Z-score determined in comparisons of the SARS-CoV and NL63-CoV C-domains suggests no significant similarity in their tertiary structures.
Surprisingly, despite their different primary, secondary, and tertiary structures, SARS-CoV and NL63-CoV C-domains share related structural topologies (i.e., connectivity of secondary structural elements) (Fig. 2C and D). Close inspections of NL63-CoV and SARS-CoV C-domains show that the following structural differences exist between the two proteins. First, strands β-4 and β-1 in NL63-CoV become helices α4 and α1 in SARS-CoV, respectively. Second, strand β-2 in NL63-CoV is missing in SARS-CoV. Third, strands β-6 and β-3 are located at the center of the β-sheet in SARS-CoV but have been moved to one side of the β-sheet in NL63-CoV. Taking these structural differences into account, virtually all of the secondary structural elements in the two C-domains are connected in the same order from the N terminus to C terminus. The shared structural topology in their C-domains strongly suggests that the S1 subunits of NL63-CoV and SARS-CoV have the same evolutionary origin and that the current structural differences between them result from extensive divergent evolution.
Despite their related structural topologies, the NL63-CoV and SARS-CoV C-domains bind to their common ACE2 receptor by the use of different molecular mechanisms. The SARS-CoV C-domain contains a single long continuous subdomain that binds ACE2 (Fig. 2B and D). The subdomain has been termed the receptor-binding motif, or RBM. The NL63-CoV C-domain contains three short and discontinuous ACE2-binding loops that are termed RBM1, RBM2, and RBM3 (Fig. 2A and C). The SARS-CoV RBM is topologically equivalent to NL63-CoV RBM3, because they both connect strands β-7 and β-8. However, compared with NL63-CoV RBM3, the SARS-CoV RBM is much longer and makes many more contacts with ACE2. On the other hand, although the NL63-CoV and SARS-CoV C-domains both bind to the same virus-binding motifs (VBMs) on ACE2, ACE2 is bound in different orientations when the two C-domains are structurally aligned (Fig. 3A and B). Furthermore, although the NL63-CoV and SARS-CoV C-domains both form an energetically stabilizing hydrophobic tunnel structure with ACE2, RBM1 and RBM2 from the NL63-CoV C-domain and RBM from the SARS-CoV C-domain are involved in these interactions but not the NL63-CoV RBM3 that is topologically equivalent to the SARS-CoV RBM (Fig. 3C and D). Overall, because of the distinct molecular mechanisms that the NL63-CoV and SARS-CoV C-domains use to recognize ACE2, ACE2 binding is likely the outcome of convergent evolution of the two C-domains recognizing a common virus-binding hot spot on ACE2.
This study provides evidence, for the first time, that the S1 subunits of different coronavirus genera share the same evolutionary origin but have undergone extensive divergent evolution. It suggests that the two S1 domains, the NTD and C-domain, from different coronavirus genera have related structural topologies (Fig. 4). To date, the tertiary structures of the alpha-CoV and gamma-CoV NTDs and gamma-CoV C-domains have remained unknown. Here we can infer that the alpha-CoV and gamma-CoV NTDs likely share similar structural topologies with the beta-CoV MHV NTD, which is believed to have originated from a host galectin but to have later evolved a carcinoembryonic antigen-related cell adhesion molecule (CEACAM1)-binding function (4). Indeed, sugar-binding functions have been preserved in all three major coronavirus genera (6, 7), suggesting that the galectin fold of the beta-CoV MHV NTD also exists in the alpha-CoV and gamma-CoV NTDs. In addition, we can infer that gamma-CoV C-domains likely share similar structural topologies with alpha-coronavirus NL63-CoV and beta-coronavirus SARS-CoV C-domains. In fact, the sequence similarity between the NL63-CoV S1 and gamma-CoV IBV S1 is higher than that between the NL63-CoV S1 and SARS-CoV S1, whose C-domains have related structural topologies (Fig. 1B). The alpha-CoV C-domains may have undergone divergent evolution to acquire APN- or ACE2-binding functions, whereas alpha-coronavirus NL63-CoV and beta-coronavirus SARS-CoV C-domains may have undergone first divergent evolution and then convergent evolution to both acquire ACE2-binding functions. During the long and complicated evolutionary history of coronaviruses, it is likely that sugars have been serving as the primordial and fallback receptors for coronaviruses, allowing coronaviruses to search for additional and high-affinity protein receptors. Overall, this study has enhanced our understanding of the origin, evolution, structures, and functions of coronavirus spike proteins. Future structural determinations of coronavirus S1 domains whose atomic structures are currently unknown will further clarify the curious evolutionary relationships among coronavirus spike proteins.
Fig 4.
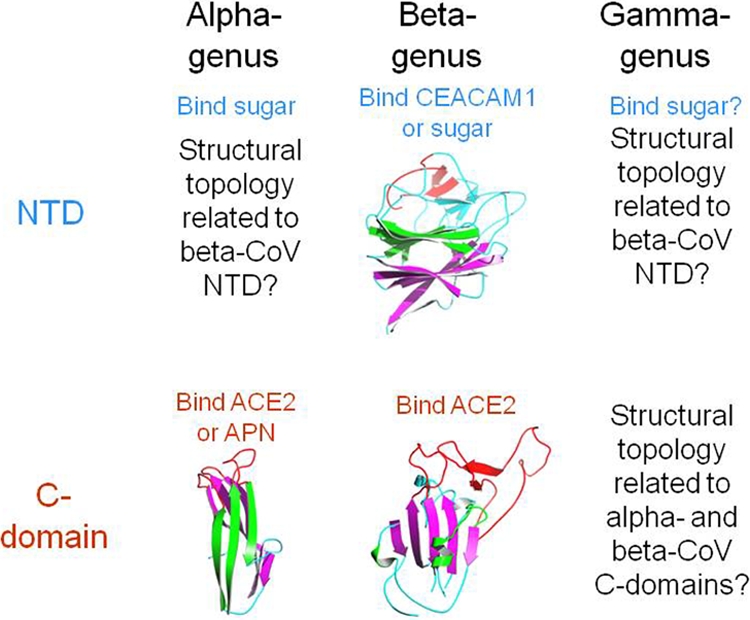
Summary of structure, function, and evolution of coronavirus S1 domains. APN, aminopeptidase N; CEACAM1, carcinoembryonic antigen-related cell adhesion molecule.
ACKNOWLEDGMENTS
I thank Kathryn Holmes and Zhaohui Qian for discussions and comments.
This work was supported by NIH grant R01AI089728 and by a University of Minnesota AHC Faculty Research Development grant. Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Goujon M, et al. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Holm L, Sander C. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li F, Li WH, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868 [DOI] [PubMed] [Google Scholar]
- 4. Peng GQ, et al. 2011. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. U. S. A. 108:10696–10701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwegmann-Wessels C, Herrler G. 2006. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 23:51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wickramasinghe INA, de Vries RP, Grone A, de Haan CAM, Verheije MH. 2011. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J. Virol. 85:8903–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu K, et al. 2011. A virus-binding hot spot on human angiotensin-converting enzyme 2 is critical for binding of two different coronaviruses. J. Virol. 85:5331–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu KL, Li WK, Peng GQ, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. U. S. A. 106:19970–19974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Y, et al. 2004. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 279:49414–49419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng Q, et al. 2006. Core structure of S2 from the human coronavirus NL63 spike glycoprotein. Biochemistry 45:15205–15215 [DOI] [PubMed] [Google Scholar]