Abstract
Varicella zoster virus (VZV) is usually associated with mild to moderate illness in immunocompetent patients. However, older age and immune deficiency are the most important risk factors linked with virus reactivation and severe complications. Treatment of VZV infections is based on nucleoside analogues, such as acyclovir (ACV) and its valyl prodrug valacyclovir, penciclovir (PCV) as its prodrug famciclovir, and bromovinyldeoxyuridine (BVDU; brivudin) in some areas. The use of the pyrophosphate analogue foscarnet (PFA) is restricted to ACV-resistant (ACVr) VZV infections. Since antiviral drug resistance is an emerging problem, we attempt to describe the contributions of specific mutations in the viral thymidine kinase (TK) gene identified following selection with ACV, BVDU and its derivative BVaraU (sorivudine), and the bicyclic pyrimidine nucleoside analogues (BCNAs), a new class of potent and specific anti-VZV agents. The string of 6 Cs at nucleotides 493 to 498 of the VZV TK gene appeared to function as a hot spot for nucleotide insertions or deletions. Novel amino acid substitutions (G24R and T86A) in VZV TK were also linked to drug resistance. Six mutations were identified in the “palm domain” of VZV DNA polymerase in viruses selected for resistance to PFA, PCV, and the 2-phophonylmethoxyethyl (PME) purine derivatives. The investigation of the contributions of specific mutations in VZV TK or DNA polymerase to antiviral drug resistance and their impacts on the structures of the viral proteins indicated specific patterns of cross-resistance and highlighted important differences, not only between distinct classes of antivirals, but also between ACV and PCV.
INTRODUCTION
Varicella-zoster virus (VZV), one of the eight human herpesviruses, is classified together with herpes simplex virus types 1 (HSV-1) and 2 (HSV-2) in the subfamily Alphaherpesvirinae. Following primary infection, VZV causes varicella (chickenpox), characterized by a generalized vesicular rash. Varicella, one of the most contagious infectious diseases in humans, is mainly a childhood disease in unvaccinated populations. Although varicella is usually a mild to moderate illness in immunocompetent patients, primary or secondary immunodeficiency is associated with serious complications, such as central nervous system involvement, pneumonia, and secondary bacterial infection, that can lead to the death of the infected patient (22). Older age and possibly pregnancy are also important risk factors associated with severe varicella and fatal outcome (59).
Subsequent to primary infection, the virus remains latent in the dorsal root ganglia of the host, and reactivation may occur several years or decades later, resulting in herpes zoster (HZ), also known as shingles. A painful vesicular rash with a dermatomal distribution characterizes HZ. Postherpetic neuralgia (PHN), the persistence of pain after resolution of the HZ rash, is the most troublesome complication associated with HZ. Worldwide, HZ affects millions of individuals, and a decline in VZV-specific cell-mediated immunity (CMI) because of aging or immunosuppression increases the risks of VZV reactivation (54, 55, 64).
A live-attenuated vaccine is available for the prevention of varicella, and its use in the pediatric population since 1995 has resulted in a significant reduction in the incidence of varicella. However, in rare circumstances in immunosuppressed children who were undiagnosed at the time of vaccination, the vaccine has caused severe infections or disseminated disease that required prompt and prolonged treatment with antivirals (10, 17, 30, 35). A zoster vaccine is also approved by the Food and Drug Administration (FDA) for prevention of HZ in adults 60 years of age and older (19). The vaccine has proven to be safe and partially effective in preventing both HZ and PHN (20, 34, 46).
Varicella is generally a self-limiting disease, and in most cases, symptomatic treatment is sufficient (59), but specific antiviral therapy for varicella is required for groups of people at risk for severe disease (22). Therapy for HZ is warranted to accelerate healing, diminish the duration of acute and chronic pain, and reduce the risk of PHN. Additionally, in immunocompromised patients, antiviral therapy for HZ is needed to limit the risk of dissemination of VZV (1, 68). Acyclovir (ACV), valacyclovir (an oral prodrug of ACV), and famciclovir (an oral prodrug of penciclovir [PCV]) are approved for the treatment of VZV infections. Brivudin (BVDU; bromovinyldeoxyuridine) has been licensed for the therapy of HZ in some European countries. Both brivudin and its arabinofuranosyl analogue sorivudine (BVaraU) are contraindicated in patients with cancer because they can be metabolized to the free base (E)-5-(2-bromovinyl)uracil, which blocks the degradation of the anticancer drug 5-fluorouracil (5-FU), resulting in the accumulation of high (toxic) 5-FU levels (9).
Although ACV resistance (ACVr) is virtually unobserved in immunocompetent patients, it can be a major problem among immunosuppressed hosts, mainly those who have received prolonged ACV therapy (25, 36, 42, 58). Approximately 30% of VZV infections clinically resistant to ACV are associated with virological resistance (our unpublished data). A lack of clinical response to ACV has been correlated with the presence of thymidine kinase (TK)-deficient or TK-altered virus, although a few cases were documented with an alteration in DNA polymerase (Pol) (18). Thus, the molecular basis for resistance to ACV is linked to mutations in either the viral TK or the DNA Pol gene. Herpesvirus TKs are able to transfer a γ-phosphoryl group from ATP to the 5′ hydroxyl group of thymidine (13). In addition, VZV and HSV-1 TKs possess thymidylate kinase activity, which converts dTMP to dTDP. Then, the conversion to dTTP is carried out by cellular kinases. Herpesvirus TKs have a broader substrate specificity than mammalian cytosolic TKs and can phosphorylate many nucleoside analogues, such as ACV, PCV, GCV, BVDU, BVaraU, and the bicyclic pyrimidine nucleoside analogues (BCNAs), a new class of extremely potent and specific anti-VZV agents (5, 38). The difference in substrate specificity between cellular and viral TKs is the basis for the selectivity of nucleoside analogues as antiherpetic compounds.
Because most ACVr VZV strains have alterations in the viral TK (31, 33, 41, 47, 49, 53), foscarnet (PFA) (a direct inhibitor of viral DNA Pol) is the drug of choice to treat ACVr VZV infections (1, 11, 21, 32, 48). A few reports have described VZV mutants with alterations at the level of DNA Pol associated with resistance to PFA (26, 66, 67). The acyclic nucleoside analogues (ANPs), such as cidofovir, which do not require activation by the virus-encoded TK, can be considered alternatives for the treatment of ACVr and/or PFAr mutants, as shown in the cases of HSV-1 and HSV-2.
In several studies, laboratory and clinical ACVr VZV mutants were reported (8, 14, 28, 62). However, investigations correlating a detailed phenotype with specific mutations in the VZV TK and DNA Pol genes are lacking. The analysis of drug resistance with VZV is more complex than with HSV because VZV is normally cell associated, with only a very small fraction of cell-free virus, making it difficult to isolate pure populations of viruses. In the present study, we were interested in genotyping previously isolated drug-resistant VZV emerging under selective pressure with different classes of antiherpesvirus agents, including ACV, PCV, BVDU, BVaraU, four different BCNAs, the pyrophosphate analogue PFA, and two ANPs (i.e., 2-phophonylmethoxyethyl [PME] purine derivatives) (2, 3). Furthermore, the contributions of specific mutations in VZV TK or DNA Pol to antiviral drug resistance and their impacts on the structures of viral proteins were investigated. Our results revealed patterns of cross-resistance associated with specific mutations in VZV TK and DNA Pol and highlight important differences, not only between distinct classes of antivirals, but also between members of one defined structural class of drugs (i.e., ACV and PCV).
MATERIALS AND METHODS
Cells and viruses.
Human embryonic lung (HEL) fibroblasts (ATCC CCL-137) were maintained in minimum essential medium (MEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, and 0.3% sodium bicarbonate. The VZV laboratory strain Oka (ATCC VR-795) was used.
Compounds.
The sources of the compounds were as follows: acyclovir [9-(2-hydroxyethoxymethyl)guanine], GlaxoSmithKline, Stevenage, United Kingdom; the pyrophosphate analogues foscarnet (phosphonoformate sodium salt) and phosphonoacetic acid (PAA), Sigma Chemicals, St. Louis, MO; PMEA {adefovir; 9-[2-(phophonylmethoxyethyl)adenine] and (S)-HPMPC [cidofovir; CDV; (S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine]}, Gilead Sciences, Foster City, CA; (S)-HPMPA [(S)-9-(3-hydroxy-2-phosphonyl-methoxypropyl)adenine], PMEDAP [9-(2-phosphonylmethoxyethyl)-2,6-diaminopurine], Marcela Krécmerova, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Prague, Czech Republic; brivudin [(E)-5-(2-bromovinyl)-1(β-d-2′-deoxyribofuranos-1-yl-uracil], Searle, United Kingdom; sorivudine [1-β-d-arabinofuranosyl-(E)-5-(2-bromovinyl)uracil], Yamasa Shoyu, Koshi, Japan; ganciclovir [GCV; 9-(1,3-dihydroxy-2-propoxymethyl)guanine], Roche, Basel, Switzerland; penciclovir [9-(4-hydroxy-3-hydroxymethylbut-1-yl)guanine], Aventis, Frankfurt, Germany; the BCNAs, i.e., Cf1368 {3-](2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-6-octylfuro[2,3-d]pyrimidin-2(3H)-one}, Cf1603 {6-decyl-3-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]thieno[2,3-d]pyrimidin-2(3H)-one}, Cf1742 {6-(4-hexylphenyl)-3-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]furo[2,3-d]pyrimidin-2(3H)-one}, and Cf1743 {3-[(2R,4S,5R)-4-hydroxy-5-(hydroxymethyl)tetrahydrofuran-2-yl]-6-(4-pentylphenyl)furo[2,3-d]pyrimidin-2(3H)-one}, Chris McGuigan, Cardiff University, United Kingdom.
Selection of drug-resistant virus clones by multiple-step selection.
The drug-resistant virus strains were obtained by serial passage of the reference Oka strain in the presence of increasing concentrations of the compounds, as described previously (2, 3). Briefly, the cell-associated virus was passaged in HEL cells in the presence of the compounds, starting at the 50% inhibitory concentrations (IC50s). The cell cultures were incubated until the virus cytopathic effect (CPE) was about 70%, and the drug concentration was increased 2-fold with every subsequent passage of the virus. Additional passages of the viruses in the absence of compounds were often necessary to amplify virus production before a subsequent passage in the presence of compound. After reaching the highest possible concentration, a final passage was done in drug-free medium in order to obtain the virus stock. The drug-resistant strains were titrated, and their drug susceptibility profiles were determined (2, 3). Subsequently, the different viral strains were plaque purified.
Isolation of plaque-purified viruses.
To ensure the presence of a single virus population, all mutants were plaque purified using cell-free virus. The different drug-resistant strains were plaque purified by limiting dilution using cell-free virus in HEL cells. The cell-free virus stocks were prepared by freezing and thawing, followed by sonication in freezing medium consisting of Recovery Cell Culture Freezing Medium (Invitrogen) supplemented with 5% glycerol and 5% fetal calf serum. Cell-associated virus stocks of the different plaque-purified clones were prepared and used to perform antiviral assays and to isolate viral DNA for sequencing of the viral TK and/or DNA Pol genes.
Antiviral assays.
The drug susceptibilities of the different virus clones were determined by virus plaque reduction assays in HEL cell cultures. Confluent cultures grown in 96-well microtiter plates were inoculated with the various virus clones at an input of 20 PFU/well. These assays were performed as previously described (2). The 50% effective concentration (EC50) was defined as the drug concentration required to reduce viral plaque formation by 50%.
Sequencing of the viral ORF36 (thymidine kinase) and ORF28 (DNA Pol).
VZV drug-resistant plaque-purified strains were cultivated in HEL cells until 70 to 80% CPE was observed. The infected cells were then harvested by trypsinization and rinsed once with phosphate-buffered saline. After centrifugation, the cell pellets were immediately used or stored at −80°C until DNA extraction. DNA was extracted with the QIAamp blood kit (Qiagen) according to the manufacturer's instructions. The entire VZV TK gene was amplified from DNA extracted from infected cells in two overlapping amplicons with the primer sets VZVTKF01 (TCGACTAGCGGACATATTTGAAGTTCC) plus VZVTKR02 (TTGCTTACTCTGGATAAATGACTGG) and VZVTKF02 (AGATACTTAGTGGGAGATATGTCC) plus VZVTKR01 (AACACGTACACGCGAGTATGACAATG) using the FastStart High Fidelity PCR System (Roche). Four sets of primers were used to amplify the entire VZV DNA Pol gene: set A, VZVDPF (TATGTATTAGAAGGGCGTGGGGTTG) and VZVDPR1509 (AGATATGAGACCGTTGATC); set B, VZVDPF1353 (TGATTGGGCGTTTATTATGGAGAAAC) and VZVDPR2219 (ACACCCAGCAGACTTTCTCGAACG); set C, VZVDPF2072 (TTCAGGCCCATAACTTATGTTTTACC) and VZVDPR2946 (TGCATCTGCAATTATGCGTCCAAACC); and set D, VZVDP2793 (TAACGATTATGCCCGCAAACTTG) and VZVDPR (TTACTGTCGGTATGTCGCAACCCAG). The PCR products were purified using a PCR product purification kit (Roche) and sequenced using a cycle-sequencing kit (Dyenamic dye terminator kit; Amersham Biosciences), specific primers targeting both strands of the TK gene or the DNA Pol gene, and a capillary DNA sequencing system (MegaBACE 500; Amersham Biosciences). The data were assembled and compared to the DNA sequences obtained from the wild-type reference Oka strain using VectorNTI (Informax) software.
Modeling of VZV thymidine kinase and DNA Pol.
The model of VZV TK bearing mutations associated with drug resistance was constructed using ArgusLab (Planaria Software LLC, Seattle, WA), based on the crystal structure of the VZV TK complexed to BVdUMP and ADP (Protein Data Bank [PDB] code 1OSN) (7).
A homology model of the VZV DNA Pol active site was built, using the SWISS-MODEL Workspace, based on the structural information available for thermostable B-type DNA Pol from the Thermococcus gorgonarius tridimensional structure (PDB code 1TGO) (23, 57). It shares 22% sequence identity with VZV DNA Pol. The resulting complexes were visualized, and figures were generated with the PyMOL graphic system v.99rc6 (Schrödinger, LLC).
RESULTS
Genotypic characterization of plaque-purified viruses selected under pressure with ACV, BVDU, BVaraU, and BCNAs.
We have previously reported on the phenotypic characterization of VZV mutants selected under pressure with different classes of antiherpesvirus agents (2, 3). Genotyping was performed following plaque purification (using cell-free virus) of each type of virus mutant.
Initially, we genotypically characterized (by sequencing of the viral TK gene ORF36) different clones that were isolated following selective pressure with ACV, PCV, BVDU, and BVaraU (Table 1). A stop codon at position 192 of the viral TK was detected in all six BVDUr clones, leading to a nonfunctional truncated protein. A frameshift reading was observed because of the insertion of a nucleotide at the nucleotide (nt) 493 to 498 stretch of an homopolymer of five cytosines (C) in six out of seven BVaraUr clones or because of a thymidine deletion at position 641 in the 7th BVaraUr clone. All ACVr clones also had a C insertion or a nucleotide deletion in the nt 493 to 498 homopolymer region of the viral TK gene. Four out of 16 ACVr clones contained, in addition to the frameshift reading, a nucleotide substitution that resulted in the T86A amino acid change. Interestingly, none of the PCV clones presented alterations at the level of the TK gene, in agreement with their phenotyping, which suggested no alterations at the level of the gene (2).
Table 1.
Genotypic characterization of drug-resistant VZV mutants obtained in vitro under selective pressure with ACV, BVDU, BVaraU, and the BCNAs
| Resistance selection under pressure with: | No. of clones/total | Changes in viral TK associated with drug resistance |
|
|---|---|---|---|
| Nucleotide | Amino acid | ||
| BVDU | 6/6 | 574-G to T | E192-stop |
| BVaraU | 6/7 | 493–498 C insertion | Frameshift |
| 1/7 | 641-T deletion | Frameshift | |
| ACV-A | 2/4 | 493–498 C deletion | Frameshift |
| 1/4 | 256-A to G | T86A | |
| 493–498 C deletion | Frameshift | ||
| 1/4 | 493–498 C insertion | Frameshift | |
| ACV-B | 4/4 | 493–498 C insertion | Frameshift |
| ACV-C | 4/4 | 493–498 C deletion | Frameshift |
| ACV-E | 1/4 | 493–498 C insertion | Frameshift |
| 3/4 | 256-A to G | T86A | |
| 493–498 C insertion | Frameshift | ||
| PCV | 4/4 | None | None |
| Cf1603 | 6/6 | 493–498 C insertion | Frameshift |
| Cf1368 | 3/5 | 412 insert TA | Frameshift |
| 2/5 | 70-G to A | G24R | |
| Cf1742 | 8/8 | 493–498 C insertion | Frameshift |
| Cf1743 | 17/19 | 493–498 C insertion | Frameshift |
| 2/19 | 256-A to G | T86A | |
| 493–498 C insertion | Frameshift | ||
Due to the high potency and selectivity of the BCNAs as inhibitors of VZV (4, 38) and the recent initiation of clinical trials with one of the BCNA members (37, 38), selection of VZV mutants was also performed for four different BCNAs (i.e., Cf1368, Cf1603, Cf1742, and Cf1743) (Table 1). All Cf1603r, Cf1742r, and Cf1743r clones presented a C insertion within the homopolymeric 493 to 498 C stretch. Two Cf1743r clones had, in addition to the 493 to 498 C insertions, a nucleotide substitution at position 256 that led to the T86A amino acid change. The Cf1368r clones had either an insertion of 2 nt at position 412 of the viral TK gene, leading to a frameshift, or a nucleotide substitution responsible for the G24R amino acid change (two out of five clones).
Modeling of specific mutations in the VZV TK structure.
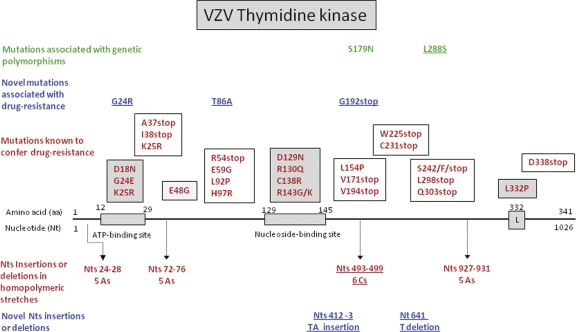
A diagram of VZV TK, including the amino acid changes known to be associated with drug resistance or genetic polymorphisms and mutations identified in the present study, is shown in Fig. 1. The ATP-binding site, the nucleoside-binding site, and the regions that are conserved among Herpesviridae TKs are indicated (Fig. 1 and 2A). Three regions are known to be of particular importance for the function of TK (15). The first critical region is the phosphate-binding loop (P loop), residues 22 to 27 in VZV TK, with a network of hydrogen bonds to the α- and β-phosphates of ATP (HSV-1 TK residues 59 to 64). The second conserved region, containing the DRH/Y/F motif, is present as D129R130H131 in VZV TK (D162R163H164 in HSV TK). Residue Asp162 in HSV-1 TK and the corresponding Asp-129 in VZV are proposed to bind to the ADP/ATP-binding site via a magnesium ion. The arginine in this motif appears to be vital for catalysis. The third region is known as the flap or “lid” in HSV-1 TK (residues 212 to 226). This lysine/arginine-rich loop encloses the ATP site and makes contacts with the phosphates. The lid region has been shown to be flexible in HSV-1 TK to adopt open and closed conformations. However, the equivalent region in VZV TK (residues 184 to 193) is largely disordered, consistent with the presence of the product or an open complex where the release of ADP can be facilitated following catalysis (7).
Fig 1.
Diagram of VZV TK. Mutations that confer drug resistance located in the ATP-binding site, the nucleoside-binding site, and Leu332 are shown in dark shaded boxes. Mutations found in other conserved regions of the viral TK are shown in lightly shaded boxes, while mutations located in nonconserved regions of the viral enzyme are shown in open boxes. Mutations found in the present study are underlined.
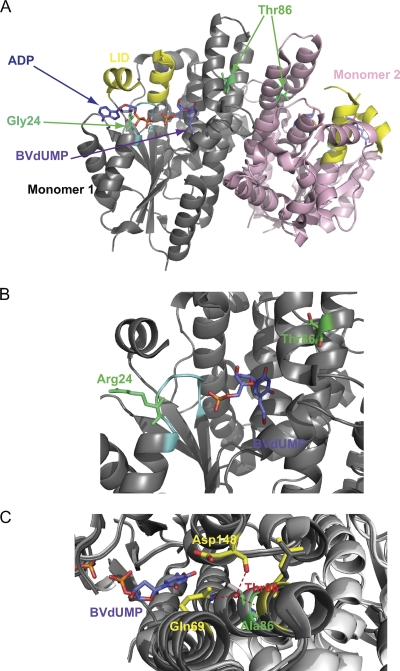
Fig 2.
Models of VZV thymidine kinase bearing point mutations that induce resistance to nucleoside analogues. (A) Dimer of VZV-TK complexed to ADP and BVdUMP (blue). The positions of the mutations are shown in green. (B) Enlarged view of the P loop (cyan) with the G24R mutation (green). BVdUMP (blue) is present in the active site of VZV TK. (C) Wild-type VZV TK superimposed on the protein bearing the G24R mutation. Thr-86 (red) and the T86A mutation (green) are surrounded by four helices at the edge of the dimerization interface. Residues Gln-69 and Asp-148 in interaction with Thr-86 within the monomer are represented in yellow. The hydroxyl moiety of the T86 side chain is involved in the interaction with Q69 and D148, whereas the T86A amino acid change prevents these interactions.
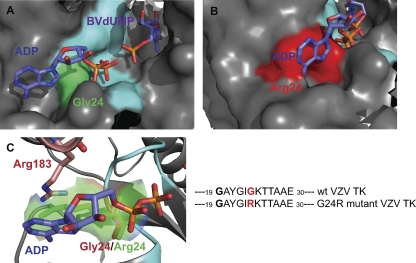
Most of the mutations found in this study were due to insertions or deletions of nucleotides in VZV TK. Only two novel substitutions were found (i.e., G24R and T86A). Two models of VZV TK bearing the mutation G24R (Fig. 2B) or T86A (Fig. 2C) were generated. Gly-24 is located in the P loop of VZV TK, which allows correct positioning of the γ-phosphate (Fig. 3A). The mutation G24R modifies this conserved sequence of the P loop and its structure, and this change is expected to prevent the binding of ATP (ADP in Fig. 3B). Furthermore, the local structure may also be disturbed by electrostatic repulsion with the side chain of Arg-183 from the lid domain (Fig. 3C). The steric hindrance due to the arginine side chain disturbs the base moiety binding. Thus, hydrolysis of the γ-phosphate of the ATP may not be possible, and the protein loses its kinase catalytic activity. Boivin et al. reported a G24E amino acid change in VZV TK that was associated with a TK-deficient phenotype (8). Moreover, Gly-24 in VZV TK is homologous to Gly-61 in HSV-1 TK, and changes at this position in HSV-1 have also been associated with acyclovir resistance (44).
Fig 3.
Enlarged view of the ATP-binding site of VZV TK with Gly-24 (green; wild-type) (A) or Arg-24 (red; mutant) (B). In the wild-type TK, the ATP molecule (ADP in the model) can interact with the P loop, whereas in the G24A mutant, the side chain of the arginine may prevent the binding because of steric hindrance. As a result, the binary complex ATP-VZV TK cannot be formed, and the virus lacks TK activity. (C) Furthermore, Arg-183 might be involved in electrostatic repulsion with Arg-24, disorganizing the local secondary structure. On the right is the TK structure used for modeling studies showing the active site containing a molecule of ADP (one of the products of the enzymatic reaction, while ATP is the substrate (i.e., the phosphate donor).
Thr-86 is located in the α2 helix, which is important for the dimerization of VZV TK. Thr-86 can establish interactions with residues Gln-69 and Asp-148 from the α1 and α5 helices, respectively, and maintains the cohesion of the three helices (α1, α2, and α5) that contribute to the interface of dimerization. The mutation T86A probably destabilizes the tridimensional structure of the monomer by loss of interactions with Gln-69 and Asp-148, leading to a mutant virus lacking TK activity. Gaudreau and collaborators showed in previous studies that the Q104H mutation in HSV-1 TK (homologous to the residue Gln-69 in VZV TK) produced a catalytically deficient TK enzyme (16). Morfin et al. also described the mutation Q104-stop (40).
Phenotypic characterization of plaque-purified viruses bearing mutations in the viral TK gene.
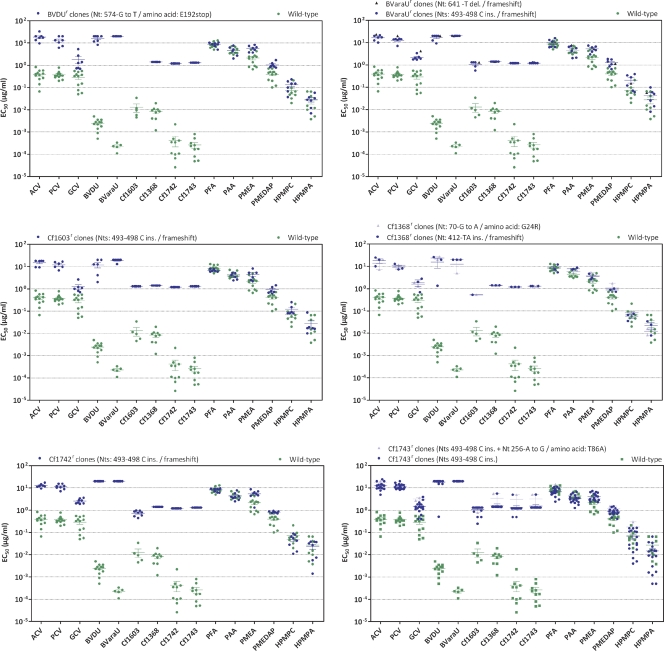
The drug susceptibility/resistance profile of plaque-purified viruses isolated following pressure with the BCNAs was investigated in comparison with mutants selected under pressure with BVDU and BVaraU. Figure 4 displays the phenotypic results of representative clones bearing different types of mutations in the TK gene.
Fig 4.
Resistance properties of plaque-purified drug-resistant VZV mutants (Oka strain) bearing mutations in the viral TK. HEL cells were grown to confluence in 96-well microplates, infected with 20 PFU of each cloned virus, and incubated for 5 days with serial dilutions of each test compound. At least three independent experiments were performed for each test compound. The data are presented as the EC50 for the drug-resistant clones versus the EC50 for the parent VZV clones. Horizontal lines for each drug and mutant combination indicate the mean values ± standard deviations.
The mutants that had frameshift mutations alone or in combination with the amino acid substitution T86A, the E192-stop mutation, or the G24R amino acid substitution exhibited high levels of resistance to the TK-dependent drugs BVDU, BVaraU, and the BCNAs (>2-log-fold resistance) (Fig. 4). In addition, the acyclic nucleoside analogues ACV, PCV, and GCV proved significantly less active against these mutant viruses, although the increase in EC50s was only 1.5 to 1.8 log units for ACV and PCV and 0.7 to 1.0 log unit for GCV. As expected, the TK mutants remained sensitive to the pyrophosphate analogues PFA and PAA and to the ANPs, including PME and 3-hydroxy-2-phosphonylmethoxypropyl (HPMP) derivatives.
Genotypic characterization of plaque-purified viruses selected under pressure with PCV, PMEA, PMEDAP, and PFA.
Since the PCVr virus had no mutations at the level of the TK (Table 1) and presented a phenotype similar to that observed for mutant viruses isolated under pressure with two PME derivatives (i.e., PMEA and PMEDAP) and the pyrophosphate analogue PFA, the DNA Pol genes (ORF28) of these mutant viruses were analyzed (Fig. 5A). The amino acid substitution V666L appeared to be associated with the resistant phenotype seen for the PCVr virus (Table 2). The plaque-purified viruses selected under pressure with PMEA presented two amino acid substitutions, i.e., E762D and L767S (Table 2). The E762D change can be linked to genetic polymorphism, since an aspartic acid at this position also occurs in the reference VZV strain Dumas. Thus, the L767S change appeared to be responsible for the resistance phenotype of the PMEAr viruses. The mutant viruses isolated under pressure with PFA, PMEA, and PMEDAP also contained the E762D substitution. In addition to this amino acid change, all PMEDAPr clones had the D668Y substitution. Four out of eight clones also contained the M808V substitution, in addition to the D668Y change (Table 2).
Fig 5.
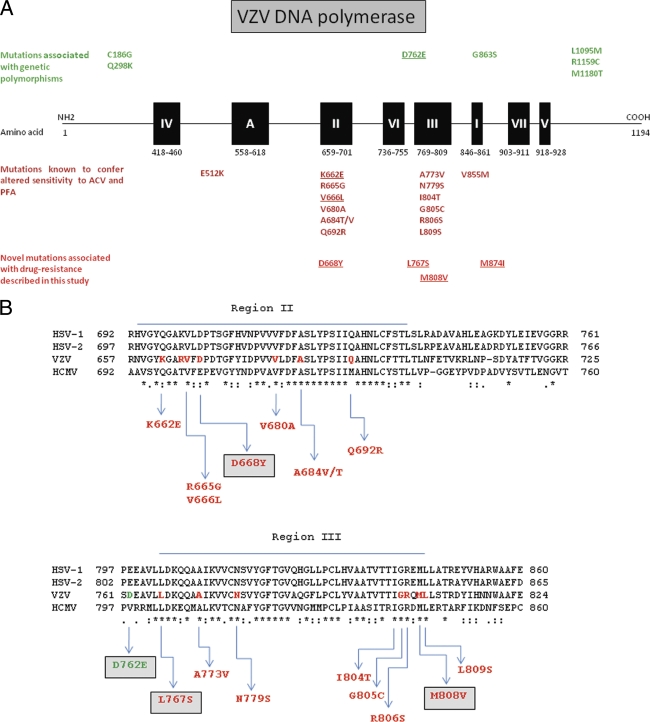
Diagram of VZV DNA Pol. (A) Locations of functional sites on VZV DNA Pol and linear N terminus to C terminus configuration of the polypeptide (not to scale). The solid boxes show regions numbered I to VII based on conservation among DNA Pols and region A, a region showing slight conservation. The locations of point mutations that result in altered drug sensitivity are shown below. Novel mutations associated with drug resistance discovered in the present study are at the bottom of the diagram. (B) Sequence alignment of the conserved regions II and III of HSV-1, HSV-2, human cytomegalovirus (HCMV), and VZV DNA Pols.
Table 2.
Genotypic characterization of drug-resistant VZV mutants obtained in vitro under selective pressure with PMEA, PMEDAP, PFA, and PCV
| Resistance selection under pressure with: | No. of clones/total | Changes in viral DNA Pol associated with drug resistance |
Changes in viral DNA Pol associated with genetic polymorphisms |
||
|---|---|---|---|---|---|
| Nucleotide | Amino acid | Nucleotide | Amino acid | ||
| PMEA | 4/4 | T2300C | L767S | G2286T | E762D |
| PMEDAP | 4/8 | G2002T | D668Y | G2286T | E762D |
| 4/8 | G2002T | D668Y | G2286T | E762D | |
| A2422G | M808V | ||||
| PFA/A | 6/6 | A1984G | K662E | G2286T | E762D |
| PFA/B | 1/2 | A1984G | K662E | G2286T | E762D |
| 1/2 | A1984G | K662E | G2286T | E762D | |
| G2622A | M874I | ||||
| PCV | 4/4 | G1996C | V666L | ||
Two independent selection procedures (A and B) were performed with PFA (Table 2). In all eight clones, the K662E amino acid change was associated with resistance to PFA. One of the clones showed an additional M874I substitution.
Modeling of specific mutations in the VZV DNA Pol structure.
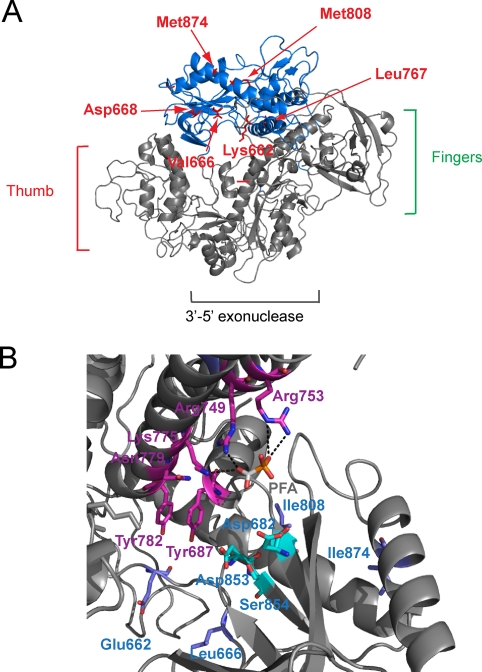
DNA Pols are highly conserved among herpesviruses, and different regions can be distinguished. In VZV DNA Pol, region III (amino acids 769 to 809) plays an important role in nucleotide binding, while region II (amino acids 659 to 701) has a function as the acceptor for hydrolysis of the pyrophosphate (26). In the present study, six mutations were identified in the “palm domain” of the catalytic subunit of VZV DNA Pol: K662E, V666L, and D668Y (subdomain II); L767S (between subdomains VI and III); M808V (subdomain III); and M874I (between subdomains I and VII) (Fig. 5A and 6A). Val-666 is highly conserved at homologous positions among herpesvirus DNA Pols, while Lys-662 and Asp-668 are partially conserved (Fig. 5B). Val-666 and Lys-662 are in direct interaction with the DNA molecule and may affect the binding of the template strand to the active site (Fig. 6A). The modification of a positive charge to a negative one, such as in the K662E substitution, significantly changes the electrostatic environment, causing electrostatic repulsion with the negative charges of the phosphate moiety of the DNA molecule. On the other hand, the V666L change increases steric hindrance, probably affecting the binding of the DNA molecule. The modification of the template strand binding, in both cases, may also affect the positioning of the “incoming” deoxynucleoside triphosphate (dNTP) to have correct base pairing.
Fig 6.
(A) structural model of VZV DNA Pol based on the thermostable B-type DNA Pol from T. gorgonarius tridimensional structure (PDB code 1TGO). It shares 22% sequence identity with VZV DNA Pol. The residues that undergo amino acid changes in the mutant viruses are represented in red. All the mutated positions are present in the palm domain (blue) of VZV DNA Pol (grey). (B) Mutations K662E and V666L (blue) are located in the region of the palm domain in direct interaction with the DNA molecule. The residues that bind the “incoming” dNTP are represented in purple and cyan. In this case, a foscarnet molecule is present in the active site, competing with the pyrophosphate of the dNTP.
Asp-668 is located in the active site of DNA Pol, in proximity to the DNA molecule. A mutation of Asp to Tyr confers resistance to PFA, PMEA, and PMEDAP and, in combination with the M808V change, the drug resistance is extended to ACV. These two amino acid alterations in DNA Pol may cause fewer modifications in the local environment than the other mutations, as shown by the drug resistance profile. Aspartate and tyrosine are polar amino acids and bear a negative charge despite the aromatic side chain of the tyrosine. The change of Met-808 to Val-808 is anticipated to decrease the steric hindrance of the side chain. The residue Met-808 is located in the conserved region III of the viral DNA Pol, where different mutations associated with resistance to PFA and ACV have been described (Fig. 5).
The highly conserved leucine at position 767 establishes hydrophobic interactions with Ile-756, stabilizing the two α-helices (which bear the residues Arg-749, Arg-753, and Lys-775) involved in binding of the “incoming” nucleoside triphosphate. The replacement of a leucine with a serine at position 767 introduces a polar side chain in a nonpolar environment that may disturb the stability of the secondary structure and the orientation of the helices. This mutation confers resistance to the acyclic nucleo-sides (ACV and PCV) and nucleotide analogues (PMEA and PMEDAP), as well as the pyrophosphate analogues (PFA and PAA).
M874I has been found in combination with the K662E mutation (Fig. 6B). The amino acid change at position 874 may locally disturb the secondary structure and prevent DNA Pol inhibition by foscarnet. M874 establishes hydrophobic interactions with Ile-816 that is not as bulky as the methionine. Consequently, the interactions might be weaker, and this might destabilize the local structure, preventing the binding of foscarnet, but also of ACV, PCV, PMEA, and PMEDAP. The mutational combination of K662E and M874I confers cross-resistance to nucleoside (ACV and PCV) and nucleotide (PMEA and PMEDAP) analogues and foscarnet, just as the single point mutation V666L did.
Phenotypic characterization of plaque-purified viruses bearing mutations in the viral DNA Pol gene.
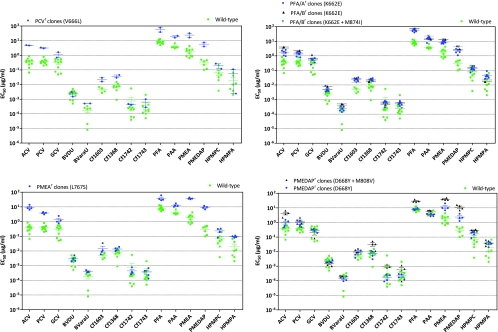
The drug susceptibility/resistance profiles of plaque-purified viruses isolated following pressure with PCV, PFA, and the PME derivatives are shown in Fig. 7. The clones selected under pressure with PCV bearing the V666L substitution showed cross-resistance to ACV and the PME class of ANPs. In addition, this mutation appeared to confer resistance to the pyrophosphate analogues (0.6- to 0.8-log increases in the EC50s) (Fig. 7) but weakly affected sensitivity to GCV and the HPMP derivatives (0.6-log increase in the EC50s).
Fig 7.
Resistance properties of plaque-purified drug-resistant VZV mutants (Oka strain) bearing mutations in the viral DNA Pol. HEL cells were grown to confluence in 96-well microplates, infected with 20 PFU of each cloned virus, and incubated for 5 days with serial dilutions of each test compound. At least three independent experiments were performed for each test compound. The data are presented as the EC50s for the drug-resistant clones versus the EC50s for the parent VZV clones.
The mutant viruses isolated under PFA pressure contained the K662E mutation, and one out of eight clones had a second substitution (i.e., M874I) (Fig. 7). The K662E mutation conferred a 1- to 1.3-log degree of resistance to the pyrophosphate analogues, while the levels of resistance to the PME derivatives, ACV and PCV, were in the range of 0.65 to 1.0 log unit. The extents of resistance associated with the K662E mutation for GCV and the HPMP derivatives were lower (in the range of 0.3 to 0.5 log unit) than with the V666L mutation.
Clones bearing the L767S mutation showed pronounced levels of resistance to the PME derivatives (1.1 to 1.5 log units for PMEA, PMEDAP, ACV, and PCV). The L767S mutation conferred 0.5- to 0.7-log-unit resistance to the pyrophosphate analogues. An increase in the EC50s of 0.7 to 0.9 log unit was recorded for GCV and the HPMP derivatives in the clones bearing the L767S mutation.
Clones selected under pressure with PMEDAP contained two resistance-related mutations (i.e., D668Y and M808V). The D668Y mutation alone conferred lower levels of resistance to the PME derivatives and ACV (0.4 to 0.9 log unit) than to the double mutants (D668Y and M808V) (1.0 to 1.6 log units). While the D668Y mutation alone did not affect sensitivity to PFA, this mutation, in combination with the M808V mutation, afforded a 0.5-log-fold level of resistance to PFA. Neither the D668Y mutation alone nor in combination with the M808V mutation had an impact on susceptibility to GCV, while they affected sensitivity to the HPMP derivatives and PCV by approximately 0.5 log unit.
Sensitivity to BVDU and BVaraU was not significantly affected by any of the mutations in viral DNA Pol. The mutant viruses showed a relatively small increase in EC50s for the BCNAs (0.4 to 0.8 log unit) compared to the mutants selected under pressure with the BCNA-bearing mutations at the level of the TK gene.
DISCUSSION
In the first part of this study, we correlated specific mutations in VZV TK with detailed phenotyping and analyzed the impacts of amino acid substitutions associated with drug resistance on the structure of the viral enzyme. We showed here that the genetic basis for resistance to the BCNAs, as well as to ACV, BVDU, and BVaraU, was alterations in the viral TK gene. Mutants arising under selective pressure with four different BCNAs were investigated, including Cf1743, whose 5′ valyl ester (i.e., FV-100) has been evaluated recently in a clinical phase 2 trial (4, 38, 43). The data show a therapeutic benefit of FV-100 given at 200 or 400 mg/day in reducing the severity and duration of shingles-related pain, the incidence of postherpetic neuralgia, and the time to lesion healing. A phase 2b trial is envisioned to further explore its antiviral efficacy. In cell culture, BCNAs display highly potent anti-VZV activity and have an absolute requirement for VZV TK-mediated activation for antiviral activity. They were shown to be efficiently phosphorylated by VZV TK, but not by the TKs of other herpesviruses, such as HSV-1 and HSV-2 (6). Obligate intracellular 5′ monophosphorylation of Cf1743 is required for biological activity (37).
Among 67 plaque-purified clones analyzed for alterations at the level of VZV TK (emerging under ACV, BVDU, BVaraU, and the four different BCNAs), 55 showed an insertion or a deletion of a C in a string of 6 Cs at nucleotides 493 to 498 of the viral TK gene. An insertion or a deletion in this run of Cs was found in 8 out of 10 independent selection procedures. Hence, this homopolymer region appears to function as a hot spot within the VZV TK gene, producing nonfunctional, truncated TK proteins. This run of 6 Cs at nucleotides 493 to 498 in the VZV TK gene is not found at homologous positions in the HSV-1 or HSV-2 TK gene. Two other frameshift mutations in VZV TK were due to an insertion of 2 nucleotides at position 412 or to a T deletion at nucleotide 641. These changes did not occur in homopolymeric repeats of VZV TK. It is intriguing to observe that the major predominant resistance mechanism against VZV TK for the VZV TK-specific BCNAs is the 493 to 498 C insertion that is also observed for BVaraU and ACV. Given the high selectivity of BCNAs for VZV TK, but not HSV TK, one might have expected unique amino acid mutations that do not occur with other antiherpesvirus drugs in VZV TK or in the HSV-1 TK.
In contrast to VZV, sequencing of the TK genes of HSV from in vitro-isolated and clinical ACV-resistant mutants revealed the presence of several homopolymeric nucleotide (G or C) stretches that function as hot spots (16, 27, 40, 51, 52, 56). Approximately 50% of the cases of ACV resistance in HSV are due to nucleotide insertions or deletions in homopolymers of Gs or Cs in the viral TK gene, mainly in the two longest homopolymers, one of 7 Gs and one of 6 Cs. It appears that the TK genes of high-GC-content herpesviruses, such as HSV (GC content, 68%) and bovine herpesvirus type 1 (BHV-1) (GC content, 72%), contain several stretches of Gs or Cs where insertions or deletions of nucleotides lead to frameshift mutations (39, 51). VZV, which has a markedly lower GC content (46%) than HSV or BHV, has only a few homopolymer stretches (three of 5 As, one of 5 Ts, and one of 6 Cs). According to our results, the stretch of 6 Cs at nucleotides 493 to 498 emerged as the most important hot spot in the VZV TK gene for insertions or deletions. Hot spots for mutations in canine herpesvirus, another herpesvirus with a low GC content (32%), has also been shown to occur in only two stretches of 8 As in the viral TK gene (69).
In a recent study, the proportions of short sequence repeats (SSRs), which include minisatellites, microsatellites, and homopolymers, were compared among different herpesviruses and mimivirus (61). Although larger SSRs are more obvious to the eye, homopolymers are the most abundant class of SSRs in all viral genomes analyzed thus far. SSRs are likely to be a common property of large DNA viruses, and the authors suggest that homopolymers across the viral genome are mutational hot spots for evolutionary diversity in herpesviruses. VZV and HSV homopolymer regions are likely sites of mutation, both within the TK gene and in the other genes. This study also points to a higher average protein-coding variation for HSV-1 than for VZV: 1.3% versus 0.2%, respectively. The higher variation in HSV-1 correlated with a higher G/C percentage overall (68% for HSV-1 versus 46% for VZV) and in SSRs (84% for HSV-1 and 47% for VZV). Furthermore, a comparison of interstrain variation in homologous proteins of HSV-1, VZV, and pseudorabies virus (PRV) across 3 HSV-1, 3 PRV, and 18 VZV strains highlighted several proteins that appear to vary more substantially in one virus than another. Interestingly, UL23 (HSV-1 thymidine kinase) showed a higher percentage of amino acid variation (1.33%) than the homologs in VZV (ORF36) and PRV (UL23) with, respectively, 0.39% and 0.31% amino acid variation across strains.
Another remarkable difference between the HSV and VZV TKs is the smaller number of genetic polymorphisms in VZV TK than in HSV-1 and HSV-2 TKs (44). It appears that VZV TK is relatively conserved, and only a very low number of genetic polymorphisms have been described (i.e., L288S and S179N). In our study, the T86A amino acid change in VZV TK always occurred in combination with a deletion or an insertion of a C in the homopolymeric stretch of 6 Cs (nt 493 to 498). Considering the high degree of conservation of VZV TK, the location of Thr-86 in a region where mutations have been associated with drug resistance, and its effect on the structure of the viral protein, it can be assumed that the T86A change may affect the activity of the enzyme. Two other novel mutations were found in VZV TK that could be linked to drug resistance: E192-stop (selected under pressure with BVDU) and the G24R substitution in the ATP-binding site (upon selection with one of the BCNAs). The importance of the changes at position G24 in VZV TK is highlighted by its impact on the structure of the viral enzyme. Furthermore, in a previous work, Boivin et al. characterized the G24E mutation in the ATP-binding site of VZV TK, and the phenotype of the clinical strain bearing this mutation was shown to be TK deficient (8). Mutations at homologous residues in HSV-1 and HSV-2 (i.e., G61) have also been linked to drug resistance (44).
Although ACV resistance in HSV and VZV may involve different mechanisms (i.e., reduction or loss of viral TK activity, alteration of substrate specificity for viral TK, and mutations affecting DNA Pol), most of the ACVr HSV and VZV strains are TK deficient (18). Our studies are in agreement with these findings (Table 1). Such frameshift mutations, as also observed to occur in the VZV TK gene under BCNA drug pressure, resulted in poor, if any, phosphorylating activity of the mutant TK enzyme, as examined in the extracts of HEL cell cultures infected with mutant VZV strains that were selected in the presence of Cf1603 or Cf1743 (data not shown). In contrast, the PCVr viruses had a mutation at the level of the DNA Pol gene. Furthermore, Ida and collaborators found that the emergence frequency of resistant VZV mutants was significantly higher following ACV exposure than following PCV treatment (24). No data are available regarding the isolation of PCV-resistant mutants in the clinic. This can be explained by the longer time of exposure to PCV required for selection of resistance compared to ACV and by the type of mutants isolated, since TK is not vital for the virus while only a limited number of mutations that are nonlethal for the virus can occur in viral DNA Pol.
The differences between HSV and VZV regarding selection of ACVr and PCVr mutants can be explained either by the different natures of the viruses or by subtle structural differences between PCV and ACV. First, there is a higher number of homopolymer stretches in HSV TK than in VZV TK, and no long homopolymers of Gs are found in VZV TK. VZV TK contains only one run of 6 Cs that is a hot spot for insertion or deletion of G. Second, the triphosphate form of ACV (ACV-TP) acts as an obligate DNA chain terminator when incorporated into the growing DNA strain, while the triphosphate form of PCV (PCV-TP) can be incorporated into the growing DNA chain, resulting in limited chain elongation (65). Third, the affinities and therefore the molecular interactions of ACV, PCV, and their triphosphates with the viral TK and DNA Pol differ. In herpesvirus-infected cells, the efficiency of phosphorylation to the triphosphate derivatives is higher for PCV than for ACV. Instead, ACV-TP is a more potent inhibitor of HSV-1 and VZV DNA polymerases than PCV-TP (12). Fourth, the triphosphate forms of ACV and PCV differ in their intracellular half-lives in infected cells (much longer for PCV-triphosphate than for ACV-triphosphate). Fifth, it has been proposed that ACV-TP may be capable of modifying the enzymatic or proofreading properties of HSV DNA Pol to selectively slip or stutter in regions containing G-C homopolymers and induce frameshift mutations (50, 51). Although the Ki of PCV-TP with DNA Pol is significantly higher than that of ACV-TP, PCV-TP is actually a more efficient and complete DNA chain terminator under physiological conditions (with competing natural nucleotides present) than is ACV-TP (12, 45). In conclusion, the more complete DNA chain termination activity of PCV-TP, the weaker interaction with DNA Pol than ACV-TP, and the lack of homopolymeric stretches of Gs in the VZV TK and Pol genes may explain the differences observed between ACV and PCV in the present study. It is worth mentioning that different mutational patterns in TK genes of HSV were reported with viruses selected in cell culture for resistance to ACV or PCV (50, 60). Additionally, PCV, similar to GCV, may be phosphorylated, not only by VZV TK, but also by the protein kinase ORF47, which is homologous to the HCMV protein kinase UL97.
VZV encodes a protein kinase (ORF47) that is homologous to the HCMV protein kinase UL97. As HCMV does not encode a TK, the UL97 protein kinase is responsible for GCV activation in infected cells. Koyano et al. (29) analyzed the phosphorylation pathways of antiherpesvirus nucleoside analogues by VZV-specific enzymes. Cells expressing VZV TK proved to be susceptible to ACV and GCV, as well as oxetanocin analogues. In contrast, VZV PK-expressing cells were susceptible only to GCV and oxetanocin analogues, indicating that VZV PK phosphorylates certain nucleoside analogues, for example, GCV and oxetanocin. Further evidence for the activation of GCV and oxetanocin analogues was provided by the fact that these compounds were able to inhibit the replication of TK-negative VZV strains at concentrations 10 times higher than those at which they inhibited wild-type VZV. These results are in agreement with our phenotyping data for mutant VZVs bearing different types of changes in the viral TK gene, which showed an increase in EC50s of only 0.7 to 1.0 log unit compared to 1.5 to 1.8 log units for ACV and PCV and >2 log units for BVDU and the BCNAs (Fig. 4).
In the present study, we also investigated the DNA Pol changes in drug-resistant virus mutants. Since the PCVr plaque-purified isolates did not show alterations at the level of the TK gene and showed cross-resistance to the pyrophosphate analogues and the PME derivatives, we performed genetic analysis of the DNA Pol gene. PCV seemed to select for virus strains that contain mutations in VZV DNA Pol (i.e., V666L) that were also reported to occur under PFA pressure (66, 67). Among the four novel mutations (i.e., D668Y, L767S, M808V, and M874I) in the viral DNA Pol that could be linked to drug resistance, two positions (i.e., Met-808 and Leu-767) are conserved among HSV, VZV, and HCMV, while two are partially conserved (i.e., Asp-668 and Met-874). Their effects on the structure of VZV DNA Pol (Fig. 6 and 7) highlight the impact of mutations at these positions.
A recent study has highlighted the limitations of employing the crystal structures of related polymerases, such as that of the bacteriophage RB69 or Thermococcus gorgonariusas, as model systems to explain mechanisms of inhibition of DNA synthesis and drug resistance (63). It was demonstrated that the RB69 DNA polymerase (gp43) is ∼400-fold less sensitive to the pyrophosphate analogue PFA than the HCMV DNA polymerase (UL54). These data point to profound differences in sensitivity and selectivity to antiviral drugs when the viral enzyme is compared with gp43. Critical regions of the nucleotide-binding site of gp43 were replaced by equivalent regions of the HCMV enzyme. Interestingly, the chimeric gp43-UL54 enzymes containing residues of the helix N and helix P of UL54 were resensitized to PFA and ACV. Hence, chimeric enzymes can provide a valuable tool to study the mechanisms of action and resistance to PFA and to nucleotide analogue inhibitors.
It should be noted that mutations in viral TK are usually associated with high levels of resistance (≥1 log10-fold increase in EC50s). In contrast, mutations in viral DNA Pol result in smaller differences in EC50s between the wild-type and mutant viruses.
Our results illustrate the complexity of the mechanisms of herpesvirus resistance to antiviral drugs. Although there is conservation among viral TKs and DNA Pols, marked differences between VZV and HSV could be found. In addition, differences between HSV and VZV regarding the types of mutants selected under ACV and PCV pressure were observed. The relevance of these findings to the in vivo situation warrants further investigation.
ACKNOWLEDGMENTS
This work was supported by the Belgium Federal Public Service “Public Health, Food Chain Safety and Environment, action 29 of the National Cancer Plan”; by the Fonds voor Wetenschappelijk Onderzoek (FWO) Krediet no. G-0608-08; and by the Program Financing (PF-10/18) of the K. U. Leuven.
We are grateful to Anita Camps, Lies Van Den Heurck, Steven Carmans, and Lizette van Berckelaer for excellent technical assistance.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Ahmed AM, Brantley JS, Madkan V, Mendoza N, Tyring SK. 2007. Managing herpes zoster in immunocompromised patients. Herpes 14:32–36 [PubMed] [Google Scholar]
- 2. Andrei G, De Clercq E, Snoeck R. 2004. In vitro selection of drug-resistant varicella-zoster virus (VZV) mutants (OKA strain): differences between acyclovir and penciclovir? Antiviral Res. 61:181–187 [DOI] [PubMed] [Google Scholar]
- 3. Andrei G, et al. 2005. Susceptibilities of several clinical varicella-zoster virus (VZV) isolates and drug-resistant VZV strains to bicyclic furano pyrimidine nucleosides. Antimicrob. Agents Chemother. 49:1081–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andrei G, Snoeck R. 2011. Emerging drugs for varicella-zoster virus infections. Expert Opin. Emerg. Drugs 16:507–535 [DOI] [PubMed] [Google Scholar]
- 5. Balzarini J, McGuigan C. 2002. Chemotherapy of varicella-zoster virus by a novel class of highly specific anti-VZV bicyclic pyrimidine nucleosides. Biochim. Biophys. Acta 1587:287–295 [DOI] [PubMed] [Google Scholar]
- 6. Balzarini J, et al. 2002. Lack of susceptibility of bicyclic nucleoside analogs, highly potent inhibitors of varicella-zoster virus, to the catabolic action of thymidine phosphorylase and dihydropyrimidine dehydrogenase. Mol. Pharmacol. 61:1140–1145 [DOI] [PubMed] [Google Scholar]
- 7. Bird LE, et al. 2003. Crystal structure of varicella zoster virus thymidine kinase. J. Biol. Chem. 278:24680–24687 [DOI] [PubMed] [Google Scholar]
- 8. Boivin G, et al. 1994. Phenotypic and genotypic characterization of acyclovir-resistant varicella-zoster viruses isolated from persons with AIDS. J. Infect. Dis. 170:68–75 [DOI] [PubMed] [Google Scholar]
- 9. Breuer J, Whitley R. 2007. Varicella zoster virus: natural history and current therapies of varicella and herpes zoster. Herpes 14(Suppl. 2):25–29 [PubMed] [Google Scholar]
- 10. Chouliaras G, Spoulou V, Quinlivan M, Breuer J, Theodoridou M. 2010. Vaccine-associated herpes zoster ophthalmicus [correction of opthalmicus] and encephalitis in an immunocompetent child. Pediatrics 125:e969–e972 [DOI] [PubMed] [Google Scholar]
- 11. Crassard N, et al. 2000. Acyclovir-resistant varicella infection with atypical lesions in a non-HIV leukemic infant. Acta Paediatr. 89:1497–1499 [DOI] [PubMed] [Google Scholar]
- 12. Earnshaw DL, et al. 1992. Mode of antiviral action of penciclovir in MRC-5 cells infected with herpes simplex virus type 1 (HSV-1), HSV-2, and varicella-zoster virus. Antimicrob. Agents Chemother. 36:2747–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. El Omari K, Liekens S, Bird LE, Balzarini J, Stammers DK. 2006. Mutations distal to the substrate site can affect varicella zoster virus thymidine kinase activity: implications for drug design. Mol. Pharmacol. 69:1891–1896 [DOI] [PubMed] [Google Scholar]
- 14. Fillet AM, et al. 1998. Acyclovir-resistant varicella-zoster virus: phenotypic and genetic characterization. J. Med. Virol. 55:250–254 [PubMed] [Google Scholar]
- 15. Gardberg A, Shuvalova L, Monnerjahn C, Konrad M, Lavie A. 2003. Structural basis for the dual thymidine and thymidylate kinase activity of herpes thymidine kinases. Structure 11:1265–1277 [DOI] [PubMed] [Google Scholar]
- 16. Gaudreau A, Hill E, Balfour HH, Jr, Erice A, Boivin G. 1998. Phenotypic and genotypic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297–303 [DOI] [PubMed] [Google Scholar]
- 17. Gershon AA. 2003. Varicella vaccine: rare serious problems—but the benefits still outweigh the risks. J. Infect. Dis. 188:945–947 [DOI] [PubMed] [Google Scholar]
- 18. Gilbert C, Bestman-Smith J, Boivin G. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updat. 5:88–114 [DOI] [PubMed] [Google Scholar]
- 19. Gnann JW., Jr 2008. Vaccination to prevent herpes zoster in older adults. J. Pain 9:S31–S36 [DOI] [PubMed] [Google Scholar]
- 20. Hambleton S, Steinberg SP, Larussa PS, Shapiro ED, Gershon AA. 2008. Risk of herpes zoster in adults immunized with varicella vaccine. J. Infect. Dis. 197(Suppl. 2):S196–S199 [DOI] [PubMed] [Google Scholar]
- 21. Hatchette T, et al. 2008. Foscarnet salvage therapy for acyclovir-resistant varicella zoster: report of a novel thymidine kinase mutation and review of the literature. Pediatr. Infect. Dis. J. 27:75–77 [DOI] [PubMed] [Google Scholar]
- 22. Heininger U, Seward JF. 2006. Varicella. Lancet 368:1365–1376 [DOI] [PubMed] [Google Scholar]
- 23. Hopfner KP, et al. 1999. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl. Acad. Sci. U. S. A. 96:3600–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ida M, et al. 1999. Emergence of resistance to acyclovir and penciclovir in varicella-zoster virus and genetic analysis of acyclovir-resistant variants. Antiviral Res. 40:155–166 [DOI] [PubMed] [Google Scholar]
- 25. Jacobson MA, et al. 1990. Acyclovir-resistant varicella zoster virus infection after chronic oral acyclovir therapy in patients with the acquired immunodeficiency syndrome (AIDS). Ann. Intern. Med. 112:187–191 [DOI] [PubMed] [Google Scholar]
- 26. Kamiyama T, Kurokawa M, Shiraki K. 2001. Characterization of the DNA polymerase gene of varicella-zoster viruses resistant to acyclovir. J. Gen. Virol. 82:2761–2765 [DOI] [PubMed] [Google Scholar]
- 27. Kit S, et al. 1987. Nucleotide sequence changes in thymidine kinase gene of herpes simplex virus type 2 clones from an isolate of a patient treated with acyclovir. Antimicrob. Agents Chemother. 31:1483–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kodama E, Mori S, Shigeta S. 1995. Analysis of mutations in the thymidine kinase gene of varicella zoster virus associated with resistance to 5-iodo-2′-deoxyuridine and 5-bromo-2′-deoxyuridine. Antiviral Res. 27:165–170 [DOI] [PubMed] [Google Scholar]
- 29. Koyano S, Suzutani T, Yoshida I, Azuma M. 1996. Analysis of phosphorylation pathways of antiherpesvirus nucleosides by varicella-zoster virus-specific enzymes. Antimicrob. Agents Chemother. 40:920–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kramer JM, et al. 2001. Disseminated vaccine strain varicella as the acquired immunodeficiency syndrome-defining illness in a previously undiagnosed child. Pediatrics 108:E39. [DOI] [PubMed] [Google Scholar]
- 31. Lacey SF, Suzutani T, Powell KL, Purifoy DJ, Honess RW. 1991. Analysis of mutations in the thymidine kinase genes of drug-resistant varicella-zoster virus populations using the polymerase chain reaction. J. Gen. Virol. 72:623–630 [DOI] [PubMed] [Google Scholar]
- 32. Lee MY, Kim KS, Lee WK. 2011. Intravitreal foscarnet for the treatment of acyclovir-resistant acute retinal necrosis caused by varicella zoster virus. Ocul. Immunol. Inflamm. 19:212–213 [DOI] [PubMed] [Google Scholar]
- 33. Levin MJ, et al. 2003. Development of resistance to acyclovir during chronic infection with the Oka vaccine strain of varicella-zoster virus, in an immunosuppressed child. J. Infect. Dis. 188:954–959 [DOI] [PubMed] [Google Scholar]
- 34. Levin MJ, Gershon AA, Dworkin RH, Brisson M, Stanberry L. 2010. Prevention strategies for herpes zoster and post-herpetic neuralgia. J. Clin. Virol. 48(Suppl. 1):S14–S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levy O, et al. 2003. Disseminated varicella infection due to the vaccine strain of varicella-zoster virus, in a patient with a novel deficiency in natural killer T cells. J. Infect. Dis. 188:948–953 [DOI] [PubMed] [Google Scholar]
- 36. Linnemann CC, Jr, Biron KK, Hoppenjans WG, Solinger AM. 1990. Emergence of acyclovir-resistant varicella zoster virus in an AIDS patient on prolonged acyclovir therapy. AIDS 4:577–579 [DOI] [PubMed] [Google Scholar]
- 37. McGuigan C, Balzarini J. 2009. FV100 as a new approach for the possible treatment of varicella-zoster virus infection. J. Antimicrob. Chemother. 64:671–673 [DOI] [PubMed] [Google Scholar]
- 38. McGuigan C, et al. 2007. Preclinical development of bicyclic nucleoside analogues as potent and selective inhibitors of varicella zoster virus. J. Antimicrob. Chemother. 60:1316–1330 [DOI] [PubMed] [Google Scholar]
- 39. Mittal SK, Field HJ. 1989. Analysis of the bovine herpesvirus type 1 thymidine kinase (TK) gene from wild-type virus and TK-deficient mutants. J. Gen. Virol. 70:901–918 [DOI] [PubMed] [Google Scholar]
- 40. Morfin F, et al. 2000. Genetic characterization of thymidine kinase from acyclovir-resistant and -susceptible herpes simplex virus type 1 isolated from bone marrow transplant recipients. J. Infect. Dis. 182:290–293 [DOI] [PubMed] [Google Scholar]
- 41. Morfin F, et al. 1999. Phenotypic and genetic characterization of thymidine kinase from clinical strains of varicella-zoster virus resistant to acyclovir. Antimicrob. Agents Chemother. 43:2412–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pahwa S, et al. 1988. Continuous varicella-zoster infection associated with acyclovir resistance in a child with AIDS. JAMA 260:2879–2882 [PubMed] [Google Scholar]
- 43. Pentikis HS, et al. 2011. Pharmacokinetics and safety of FV-100, a novel oral anti-herpes zoster nucleoside analogue, administered in single and multiple doses to healthy young adult and elderly adult volunteers. Antimicrob. Agents Chemother. 55:2847–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Piret J, Boivin G. 2011. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob. Agents Chemother. 55:459–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reardon JE, Spector T. 1989. Herpes simplex virus type 1 DNA polymerase. Mechanism of inhibition by acyclovir triphosphate. J. Biol. Chem. 264:7405–7411 [PubMed] [Google Scholar]
- 46. Reynolds MA, Chaves SS, Harpaz R, Lopez AS, Seward JF. 2008. The impact of the varicella vaccination program on herpes zoster epidemiology in the United States: a review. J. Infect. Dis. 197(Suppl. 2):S224–S227 [DOI] [PubMed] [Google Scholar]
- 47. Roberts GB, Fyfe JA, Gaillard RK, Short SA. 1991. Mutant varicella-zoster virus thymidine kinase: correlation of clinical resistance and enzyme impairment. J. Virol. 65:6407–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Safrin S, et al. 1991. Foscarnet therapy in five patients with AIDS and acyclovir-resistant varicella-zoster virus infection. Ann. Intern. Med. 115:19–21 [DOI] [PubMed] [Google Scholar]
- 49. Saint-Leger E, et al. 2001. Clinical and virologic characterization of acyclovir-resistant varicella-zoster viruses isolated from 11 patients with acquired immunodeficiency syndrome. Clin. Infect. Dis. 33:2061–2067 [DOI] [PubMed] [Google Scholar]
- 50. Sarisky RT, et al. 2001. Characterization of herpes simplex viruses selected in culture for resistance to penciclovir or acyclovir. J. Virol. 75:1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sasadeusz JJ, et al. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872–3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sauerbrei A, Deinhardt S, Zell R, Wutzler P. 2010. Phenotypic and genotypic characterization of acyclovir-resistant clinical isolates of herpes simplex virus. Antiviral Res. 86:246–252 [DOI] [PubMed] [Google Scholar]
- 53. Sauerbrei A, Taut J, Zell R, Wutzler P. 2011. Resistance testing of clinical varicella-zoster virus strains. Antiviral Res. 90:242–247 [DOI] [PubMed] [Google Scholar]
- 54. Schmader K. 2007. Herpes zoster and postherpetic neuralgia in older adults. Clin. Geriatr. Med. 23:615–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmader K, Gnann JW, Jr., Watson CP. 2008. The epidemiological, clinical, and pathological rationale for the herpes zoster vaccine. J. Infect. Dis. 197(Suppl. 2):S207–S215 [DOI] [PubMed] [Google Scholar]
- 56. Schmit I, Boivin G. 1999. Characterization of the DNA polymerase and thymidine kinase genes of herpes simplex virus isolates from AIDS patients in whom acyclovir and foscarnet therapy sequentially failed. J. Infect. Dis. 180:487–490 [DOI] [PubMed] [Google Scholar]
- 57. Shi R, Azzi A, Gilbert C, Boivin G, Lin SX. 2006. Three-dimensional modeling of cytomegalovirus DNA polymerase and preliminary analysis of drug resistance. Proteins 64:301–307 [DOI] [PubMed] [Google Scholar]
- 58. Snoeck R, et al. 1994. Meningoradiculoneuritis due to acyclovir-resistant varicella zoster virus in an acquired immune deficiency syndrome patient. J. Med. Virol. 42:338–347 [DOI] [PubMed] [Google Scholar]
- 59. Steiner I, Kennedy PG, Pachner AR. 2007. The neurotropic herpes viruses: herpes simplex and varicella-zoster. Lancet Neurol. 6:1015–1028 [DOI] [PubMed] [Google Scholar]
- 60. Suzutani T, et al. 2003. Differential mutation patterns in thymidine kinase and DNA polymerase genes of herpes simplex virus type 1 clones passaged in the presence of acyclovir or penciclovir. Antimicrob. Agents Chemother. 47:1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Szpara ML, et al. 2011. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 7:e1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Talarico CL, Phelps WC, Biron KK. 1993. Analysis of the thymidine kinase genes from acyclovir-resistant mutants of varicella-zoster virus isolated from patients with AIDS. J. Virol. 67:1024–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tchesnokov EP, Obikhod A, Schinazi RF, Gotte M. 2009. Engineering of a chimeric RB69 DNA polymerase sensitive to drugs targeting the cytomegalovirus enzyme. J. Biol. Chem. 284:26439–26446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thomas SL, Hall AJ. 2004. What does epidemiology tell us about risk factors for herpes zoster? Lancet Infect. Dis. 4:26–33 [DOI] [PubMed] [Google Scholar]
- 65. Vere Hodge RA, Sutton D, Boyd MR, Harnden MR, Jarvest RL. 1989. Selection of an oral prodrug (BRL 42810; famciclovir) for the antiherpesvirus agent BRL 39123 [9-(4-hydroxy-3-hydroxymethylbut-l-yl)guanine; penciclovir]. Antimicrob. Agents Chemother. 33:1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Visse B, Dumont B, Huraux JM, Fillet AM. 1998. Single amino acid change in DNA polymerase is associated with foscarnet resistance in a varicella-zoster virus strain recovered from a patient with AIDS. J. Infect. Dis. 178(Suppl. 1):S55–S57 [DOI] [PubMed] [Google Scholar]
- 67. Visse B, Huraux JM, Fillet AM. 1999. Point mutations in the varicella-zoster virus DNA polymerase gene confers resistance to foscarnet and slow growth phenotype. J. Med. Virol. 59:84–90 [PubMed] [Google Scholar]
- 68. Whitley RJ, Gnann JW., Jr 2009. Herpes zoster in the age of focused immunosuppressive therapy. JAMA 301:774–775 [DOI] [PubMed] [Google Scholar]
- 69. Yamada S, Matsumoto Y, Takashima Y, Otsuka H. 2005. Mutation hot spots in the canine herpesvirus thymidine kinase gene. Virus Genes 31:107–111 [DOI] [PubMed] [Google Scholar]