Abstract
Human cytomegalovirus modulates macroautophagy in two opposite directions. First, HCMV stimulates autophagy during the early stages of infection, as evident by an increase in the number of autophagosomes and a rise in the autophagic flux. This stimulation occurs independently of de novo viral protein synthesis since UV-inactivated HCMV recapitulates the stimulatory effect on macroautophagy. At later time points of infection, HCMV blocks autophagy (M. Chaumorcel, S. Souquere, G. Pierron, P. Codogno, and A. Esclatine, Autophagy 4:1–8, 2008) by a mechanism that requires de novo viral protein expression. Exploration of the mechanisms used by HCMV to block autophagy unveiled a robust increase of the cellular form of Bcl-2 expression. Although this protein has an anti-autophagy effect via its interaction with Beclin 1, it is not responsible for the inhibition induced by HCMV, probably because of its phosphorylation by c-Jun N-terminal kinase. Here we showed that the HCMV TRS1 protein blocks autophagosome biogenesis and that a TRS1 deletion mutant is defective in autophagy inhibition. TRS1 has previously been shown to neutralize the PKR antiviral effector molecule. Although phosphorylation of eIF2α by PKR has been described as a stimulatory signal to induce autophagy, the PKR-binding domain of TRS1 is dispensable to its inhibitory effect. Our results show that TRS1 interacts with Beclin 1 to inhibit autophagy. We mapped the interaction with Beclin 1 to the N-terminal region of TRS1, and we demonstrated that the Beclin 1-binding domain of TRS1 is essential to inhibit autophagy.
INTRODUCTION
Human cytomegalovirus (HCMV) a member of the Herpesviridae family, is widespread in human populations, with up to 90% of some populations seropositive for the virus (42). Although HCMV infection is usually asymptomatic in healthy individuals, it is a major cause of morbidity and mortality in immunocompromised individuals, such as AIDS patients or bone marrow and solid organ transplant recipients, and it can cause life-threatening infections that compromise long-term graft function. Moreover, HCMV congenital infection is the most common cause of virus-induced birth defects, particularly disorders of the central nervous system.
Macroautophagy (here referred to as autophagy) is a vacuolar homeostatic “self-eating” process that involves the digestion of cytoplasmic components via the lysosomal pathway. During autophagy, part of the cytoplasm is surrounded by a cisternal membrane, known as the phagophore. The phagophore then closes to form a double-membraned vesicle, known as the autophagosome. The autophagosome finally fuses with the lysosome, forming the autolysosome, which digests the sequestered material. The formation of the autophagosome requires the activity of several autophagy-related (Atg) proteins, initially identified in Saccharomyces cerevisiae. Beclin 1, the mammalian counterpart of Atg6 in yeast, is a keystone of this process, because it forms three different complexes involved in both autophagosome formation and maturation. The assembly of the phagophore requires the Beclin 1-phosphatidylinositol 3-kinase (PI3K) complex, consisting of Beclin 1, the class III PI3K hVps34, its regulatory protein kinase hVps15, and Atg14L. This Beclin 1 complex is tightly modulated by the recruitment of positive and negative regulators of autophagy (such as Bcl-2 and Bcl-xL) (48, 51, 52). Regulation is also achieved through posttranslational modifications of Beclin 1 or Bcl-2, through interactions with BH3-only proteins and BH3 mimetics, or by a recently discovered Beclin 1-targeted microRNA (65). This complex and other Atg proteins recruit the Atg5-Atg12-Atg16L multimeric complex and the lipidated form of LC3 (microtubule-associated protein light chain 3) to the phagophore. These last two constituents, which are essential for the forming autophagosome to expand, act sequentially and result from two ubiquitin-like conjugation systems. Two other Beclin 1 complexes regulate the maturation of autophagosomes, via interaction of Beclin 1 with UVRAG (UV radiation resistance-associated gene) and Rubicon (39, 64).
Under normal conditions, autophagy allows cells to degrade long-lived proteins and aged organelles and has two main physiological functions. The first involves recycling macromolecules and long-lived proteins to provide cells exposed to nutrient stress with amino acids and energy. The second function is to help the cell eliminate damaged organelles, such as mitochondria, and toxic aggregate-prone proteins. Autophagy has also recently been shown to be modulated by intracellular pathogens. Whereas some microorganisms, such as poliovirus, use the autophagic machinery for their own benefit such as enhanced replication, autophagy can function in cellular defenses against intracellular pathogens, including viruses, by stimulating both innate and adaptive immunity (reviewed in references 16, 17, and 58). However, viruses including herpes simplex virus 1 (HSV-1) and human herpesvirus 8 (HHV-8) have developed various strategies to counteract the host's antiviral autophagic mechanism (reviewed in references 7 and 17). We and others previously reported that HCMV also modulates autophagy in infected cells (9, 40, 54).
Several viruses are able to counteract autophagy by an interaction between a viral protein and a cellular protein of the autophagy machinery. For example, two members of the Gammaherpesvirinae subfamily, HHV-8 and herpesvirus saimiri, express the viral protein vFLIP, a viral counterpart of cellular FLIP, which is known to regulate apoptosis from death receptors. vFLIP interacts with Atg3 and represses autophagy by preventing the binding of Atg3 to LC3 and consequently the processing of LC3, which is essential for autophagic vesicle expansion (35). Most importantly, Beclin 1 has emerged as the major target for the modulation of autophagy by viruses (24). Indeed, it has been reported that several viral proteins bind and inhibit Beclin 1. HHV-8 and mouse herpesvirus strain 68 (HV-68) express viral homologs of Bcl-2, vBcl-2 and M11, respectively, which achieve inhibition of autophagy by their interaction with Beclin 1 (48, 53). The infected cell protein 34.5 (ICP34.5) of HSV-1 also interacts directly with Beclin 1 and inhibits autophagosome biogenesis (44). Other viral proteins seem to block autophagosome maturation rather than formation by interaction with Beclin 1. The Nef protein of human immunodeficiency virus (HIV) and the matrix protein M2 of influenza A virus interact with Beclin 1 but are responsible for autophagosome accumulation during viral infection, by blocking fusion of the autophagosome with the lysosome (23, 33). These two viral proteins might interact with the other Beclin 1-hVps34 complex containing UVRAG, which is involved in autophagosome maturation.
In this paper, we have explored the modulation of autophagy during HCMV infection and the mechanisms used by HCMV to block autophagy. HCMV induces autophagy independently of viral protein synthesis during the early stages of infection. Later, HCMV actively blocks the autophagosome biogenesis by expression of viral protein(s). We observed viral-induced modifications of Bcl-2, a negative regulator of autophagosome biogenesis. In addition, we identified TRS1 as a new anti-autophagic protein. It has been previously reported that TRS1 blocks the activity of the interferon-induced double-stranded RNA-dependent protein kinase PKR (10, 26). However, we found that the anti-autophagic activity of TRS1 is independent of its interaction with PKR but is related to its binding to Beclin 1.
MATERIALS AND METHODS
Cells and virus.
Primary human embryonic lung fibroblasts MRC5 were purchased from RD-Biotech and used between passages 23 and 28 postisolation. These cells were maintained in minimum essential medium (MEM) (Gibco) supplemented with 10% fetal calf serum (FCS), penicillin G (100 U/ml), streptomycin sulfate (100 μg/ml), l-glutamine (1%), and nonessential amino acids (1%). Cells were cultured at 37°C under 5% CO2. HeLa cells were maintained at 37°C under 5% CO2 in RPMI with 10% FCS. PKR−/− and PKR+/+ mouse embryo fibroblasts kindly provided by B. R. G. Williams (Monash University, Victoria, Australia) were propagated in Dulbecco MEM supplemented with 10% FCS. The AD169 strain of HCMV was obtained from ATCC and was propagated in MRC5 cells as previously described (19). The AD169ΔTRS1 mutant was constructed on the basis of the AD169 bacterial artificial chromosome (BAC), which contains the full-length HCMV AD169 genome (27). Mutagenesis by homologous recombination was performed in Escherichia coli strain DY380 essentially as described previously (5, 38). Briefly, a kanamycin resistance gene (kan) flanked by FLP recombination target sites was PCR amplified using oligonucleotide primers containing 50-nucleotide homologies immediately up- and downstream of TRS1. The PCR product was used to replace the TRS1 gene by homologous recombination. The kan gene was subsequently removed with FLP recombinase as described previously (5). BAC DNA was purified from E. coli using the NucleoBond Midi kit (Clontech). To reconstitute the ΔTRS1 mutant virus, BAC DNA was introduced by electroporation into human foreskin fibroblast (HFF) cells essentially as described previously (38). For gradient purification of HCMV virions, infectious supernatants from infected MRC5 cells with 90 to 100% late-stage cytopathic effects were made cell free by centrifugation for 10 min at 4,000 rpm. Supernatants were then ultracentrifuged for 70 min (23,000 rpm, 4°C, Beckman SW28 rotor). Pellets containing virions and other particles were resuspended in 1 ml of MEM and transferred onto a preformed linear glycerol-tartrate gradient (15 to 35% Na-tartrate and 30 to 0% glycerol in 0.04% Na-phosphate), which was then ultracentrifuged for 45 min (23,000 rpm, 4°C, Beckman SW41 rotor). The virion-containing band was harvested with a syringe, and the virions were washed and pelleted by an additional ultracentrifugation for 70 min at 23,000 rpm. The pellet was resuspended in MEM and stored at −80°C until used for infection experiments. For UV inactivation, HCMV stocks were diluted in MEM, placed in a thin layer in plastic dishes, and irradiated with UV light (254 nm, 6 W, 8 min) at ∼4 cm away from a (VL-6MC) UV lamp (Bioblock Scientific). The efficacy of inactivation was tested by immunofluorescence staining of MRC5 cells infected with UV-HCMV using anti-immediate-early antigens (IEAs) monoclonal antibody (MAb) and pp65 MAb (see below), followed by a secondary fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (data not shown). Successful inactivation was ensured through abrogation of IE antigen expression, whereas pp65 tegument protein detection was comparable to cell infection with nonirradiated virus by using methods previously described (13, 34).
Antibodies.
Murine MAb E13 was purchased from Argene and recognizes the HCMV nonstructural immediate-early antigens (IEAs) of 52, 72, and 86 kDa. Antibody directed against the C-terminal region of TRS1 protein was a kind gift of T. Shenk (50). In order to detect 6×His-tagged TRS1 constructs, we used a rabbit antibody directed against 6×His (Cell Signaling Technology). For the detection of FLAG-tagged Beclin 1 and FLAG-tagged ICP34.5, we used a rabbit antibody directed against FLAG (Cell Signaling Technology). Antibody against LC3 was produced as previously described (9). Antibodies against p62 (BD Biosciences), Beclin 1 (Santa Cruz), RelB (Santa Cruz), Bcl-2 total (Santa Cruz), and phosphorylated Bcl-2 (Cell Signaling) were used in this study. To explore the JNK pathway, antibody directed against total JNK was obtained from Santa Cruz and antibodies directed against phosphorylated (Thr183/Tyr185) JNK and against total and phosphorylated (Ser73) c-Jun were purchased from Cell Signaling. Anti-actin antibody was supplied by Sigma. Secondary fluorescein isothiocyanate (FITC) and tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse or anti-rabbit IgG were purchased from Jackson. Alexa Fluor 350 donkey anti-rabbit secondary antibody was obtained from Invitrogen.
HCMV infection of fibroblasts.
MRC5 cells were grown in MEM supplemented with 10% FCS. HCMV or UV-inactivated HCMV, diluted in serum-free MEM, or serum-free MEM alone (mock infected) was adsorbed onto cells for 1 h at 37°C at a multiplicity of infection (MOI) of 1 or 3. After the inoculum was removed, the cells were maintained in MEM containing 10% FCS and processed for the different assays at various times postinfection (p.i.). Cell viability was tested by a trypan blue exclusion test, and no significant cell death was observed in HCMV-infected cells at 24 and 48 h p.i. Cells were centrifuged for 45 min at 1,400 × g during the adsorption period, to enhance infectivity of HCMV, in experiments in which MRC5 cells were transfected with green fluorescent protein (GFP)-LC3 (21).
Immunoblot analysis.
MRC5 and HeLa cells were lysed in 65 mM Tris, pH 6.8, 4% SDS, 1.5% β-mercaptoethanol and held at 100°C for 5 min. Proteins were separated on gels containing different percentages of SDS-polyacrylamide: 10% to resolve Beclin 1, His-tagged TRS1, or TRS1, 12% for Bcl-2 and p62, and 15% to resolve LC3 forms I and II. For immunoblot analysis, the proteins were electrotransferred onto a polyvinylidene difluoride membrane (Perkin Elmer, Les Ulis, France). The membrane was incubated for 1 h in Tris-buffered saline (25 mM Tris [pH 7.5], 150 mM NaCl, and 0.05% Tween 20) containing 5% nonfat dry milk and then incubated with primary antibodies at an appropriate dilution. After three washes with Tris-buffered saline, a goat anti-rabbit or anti-mouse horseradish peroxidase-labeled antibody (Amersham, Orsay, France) was used at 1/10,000 dilution as a secondary antibody. Bound immunoglobulins were revealed using the ECL+ detection system under conditions recommended by the manufacturer (Immobilon, Orsay, France). Anti-actin was used to ensure equal loadings. The effect of HCMV on autophagosome maturation is monitored by p62 protein levels and confirmed by comparing untreated samples with those treated with inhibitors of acidic lysosomal proteases E64d/pepstatin A that inhibit degradation of autolysosome content.
Coimmunoprecipitation assays.
To immunoprecipitate Bcl-2 and endogenous Beclin 1 in MRC5 cells, cells were lysed in lysis buffer (20 mM Tris HCl, pH 7.4, 140 mM NaCl, 2 mM EDTA, 0.2% CHAPS, 10% glycerol) for 1 h at 4°C, followed by centrifugation to remove cell debris. Immunoprecipitation was performed overnight at 4°C with a goat polyclonal anti-Beclin 1 antibody (Santa Cruz Biologicals, Santa Cruz, CA) or a control goat serum. Protein G-Sepharose beads (Sigma-Aldrich, St. Louis, MO) were added for 1 h at 4°C and were washed three times with 0.05% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) buffer (20 mM Tris HCl, pH 7.4, 140 mM NaCl, 2 mM EDTA, 0.05% CHAPS, 10% glycerol). Anti-Beclin 1 immunoprecipitates were eluted in Laemmli buffer and subjected to SDS-PAGE, and Bcl-2 was detected by immunoblot analysis. To immunoprecipitate endogenous Beclin 1 and different HCMV TRS1 constructs, HeLa cells were transfected with different TRS1 constructs and then treated as described above. To immunoprecipitate TRS1 and endogenous Beclin 1 in HCMV-infected MRC5 cells, cells were lysed in a different lysis buffer (50 mM Tris HCl, 50 mM NaCl, 0.5% Triton, 0.2% bovine serum albumin [BSA], 25 mM NaPPi, 50 mM Na3VO4) for 1 h at 4°C, followed by ultracentrifugation at 120,000 × g to remove cell debris. Immunoprecipitation was performed overnight at 4°C with a goat polyclonal anti-Beclin 1 antibody or a control goat serum. Protein G-Sepharose beads were added for 1 h at 4°C and were washed three times with buffer (50 mM Tris HCl, 50 mM NaCl, 0.5% Triton, 0.2% BSA, 25 mM NaPPi, 50 mM Na3VO4) and two times with another buffer (20 mM Tris HCl, 50 mM NaCl, 0.2% BSA). Anti-Beclin 1 immunoprecipitates were eluted in Laemmli buffer and subjected to SDS-PAGE, and TRS1 was detected by immunoblot analysis.
Immunofluorescence analysis.
Cell monolayers prepared on glass coverslips were fixed for 10 min at room temperature in 3.5% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) and then washed three times with PBS. Different staining protocols were used for different antibodies. In some experiments, cell permeabilization was performed by incubating the coverslips for 4 min with PBS–0.2% Triton X-100. The cells were stained with a mouse antibody against HCMV IE antigens (1:50; Argene Biosoft) followed by a TRITC-conjugated goat anti-mouse antibody (1:200). To detect 6×His TRS1 construct expression, cells were stained with anti 6×His antibody (1:100; Cell Signaling) followed by TRITC-conjugated goat or Alexa Fluor 350 donkey anti-rabbit secondary antibodies. Cells were also stained overnight with an antibody against phosphorylated Bcl-2 (1:100; Cell Signaling), followed by FITC-conjugated secondary antibody. Coverslips were mounted with Glycergel (Dako) before being analyzed on a Nikon Eclipse 80i epifluorescence microscope. Digitized images were stored and overlaid to evaluate two-color experiments. Photographic images were resized, organized, and labeled using Adobe Photoshop software.
Real-time PCR.
Total RNA was extracted with the aid of TRIzol reagent according to the manufacturer's instructions and then digested with DNase (Life Technologies), extracted with phenol-chloroform, and precipitated with ethanol to remove contaminating DNA. Total RNA (2.5 μg) from mock-infected and HCMV-infected cell samples was reverse transcribed with random hexamers and SuperScript reverse transcriptase (Invitrogen). cDNA quantities were normalized to 18S rRNA quantities. The following primers were used: Bcl-2 forward, 5′-ATGTGTGTGGAGAGCGTCAA-3′, and reverse, 5′-ACAGTTCCACAAAGGCATCC-3′. The primers for Beclin 1 were previously validated and described (18). Reactions were performed in a 10-μl volume that included diluted cDNA sample, primers (0.5 μM), and LightCycler DNA Master SYBR green I mix (Roche) that contained nucleotides, Taq DNA polymerase, and optimized buffer components. Real-time PCRs were performed on a LightCycler 2.0 detection system (Roche). To compare mock-infected and infected cell samples, relative changes in gene expression were determined using the 2−ΔΔCT method.
Transfection.
The GFP-LC3 expression vector and the GFP and red fluorescent protein (RFP) tandemly tagged LC3 construct (mRFP-GFP-LC3) were kindly provided by T. Yoshimori (Research Institute for Microbial Diseases, Osaka University, Osaka, Japan) (30). The pEQ1180 construct, containing a full-length HCMV TRS1 protein cDNA and the pEQ979 (1-738), pEQ1001 (1-679), pEQ1013 (45-795), and pEQ1000 (93-795) constructs, containing the indicated lengths of TRS1, were described previously (25). These constructs were tagged with 6×His. Plasmid FLAG-ICP34.5 was a gift from B. He (University of Illinois, Chicago). Cultures of HeLa cells at 50 to 80% confluence were transiently transfected with empty vector, GFP-LC3, or mRFP-GFP-LC3 plasmids or various TRS1 constructs using FuGENE HD transfection reagent (Roche), according to the manufacturer's instructions. After 4 h, the medium was replaced by MEM-10% FCS, and the cells were incubated for 24 to 48 h. Amino acid starvation by EBSS (starvation medium) was carried out 4 h before fixation. The cells were then fixed in PFA for 10 min at room temperature and washed three times with PBS. Data were statistically compared by using a global Student test (P < 0.01) as indicated in the figure legends.
Proteolysis.
This assay was adapted from that of Bauvy et al. (1). MRC5 cells were incubated for 24 h at 37°C with 0.2 μCi/ml l-[14C] valine. At the end of the radiolabeling period, the cells were washed three times with PBS, pH 7.4. Cells were then incubated in complete medium supplemented with 10 mM cold valine. After incubating for 1 h, by which time short-lived proteins are degraded, the medium was replaced with fresh medium (MEM plus 10% bovine serum albumin and 10 mM cold valine), and the incubation was continued for an additional 4 h. Wortmannin (200 nM) was added to inhibit de novo formation of autophagic vacuoles. Cells and radiolabeled proteins from the 4-h chase medium were precipitated in trichloroacetic acid at a final concentration of 10% (vol/vol) at 4°C. The precipitated proteins were separated from the soluble radioactivity by centrifugation at 600 × g for 10 min and then dissolved in 0.5 ml of 0.2 N NaOH. Radioactivity was determined by liquid scintillation counting. Protein degradation was calculated by dividing the acid-soluble radioactivity recovered from both cells and medium by the radioactivity contained in precipitated proteins from both cells and medium.
Statistics.
Data are expressed as means ± standard errors of the means (SEM) of at least three experiments. The statistical significance was assessed by a Student's t test.
RESULTS
HCMV infection triggers autophagy at early stages of infection.
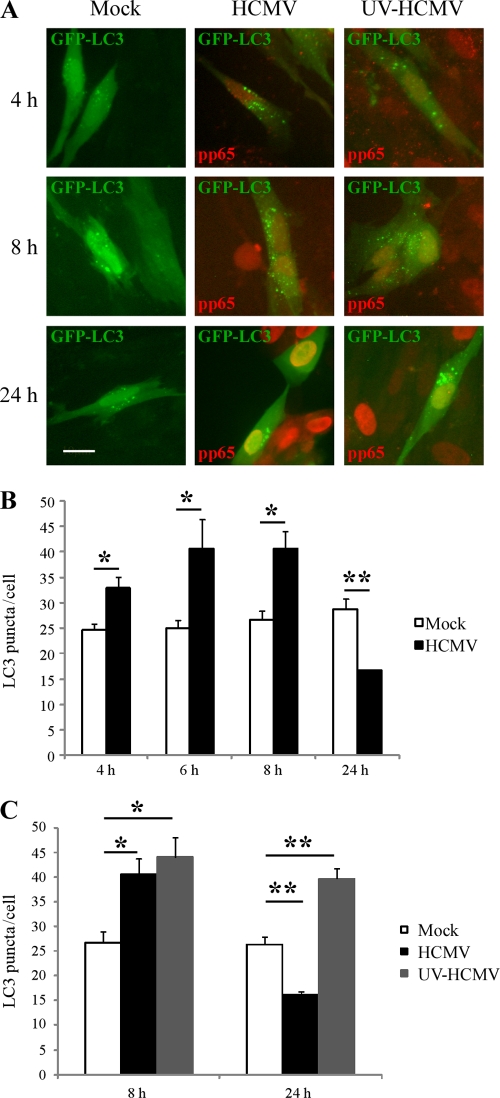
We previously demonstrated that HCMV inhibits autophagy in fibroblasts 24 h postinfection (9). During the first 24 h of infection, HCMV binds to the cell surface, penetrates into the cytoplasm, and targets its genome to the nucleus, and viral immediate early proteins are synthesized. In order to guide our research into the mechanisms of HCMV-mediated inhibition of autophagy, we first investigated at what time this inhibition started. To measure autophagy, we transfected fibroblasts with GFP-LC3, a specific marker of mammalian autophagy. Upon autophagy induction, the lipidated form of LC3 associates with autophagosomal membranes, resulting in the formation of punctuate structures that can be visualized by fluorescence microscopy. MRC5 cells were transfected with GFP-LC3 and then infected with HCMV (MOI = 1) for 4 to 24 h, and the number of puncta of GFP-LC3 per cell was measured only in infected cells, which exhibit pp65 red staining, a viral tegument protein (Fig. 1). We used HCMV particles purified by density gradient centrifugation because we observed a stimulation of autophagy when cells were exposed only to supernatant of HCMV-infected cells, probably via soluble factors (Fig. 2A). In purified HCMV-infected cells, we observed an increase in the number of puncta per cell from 4 to 8 h p.i. but a clear decrease at 24 h p.i. (Fig. 1A and B), confirming our previous results (9). Therefore, these results suggest that autophagy is induced early in response to infection but is then thwarted by the virus by 24 h after infection.
Fig 1.
Autophagosome formation at early and late time points after HCMV infection. MRC5 cells were transfected with GFP-LC3 for 48 h and then mock-infected or infected with HCMV or with UV-inactivated HCMV (UV-HCMV) for 4, 6, 8, and 24 h. Cells were fixed and immunostained for pp65 viral protein (red). (A) Representative images; bar, 10 μm. (B and C) Autophagy was quantified in pp65-positive cells by counting the number of GFP-LC3 puncta per cell. The results are the means of three independent experiments. Between 50 and 100 cells were analyzed per assay. *, P < 0.05; **, P < 0.01 (t test).
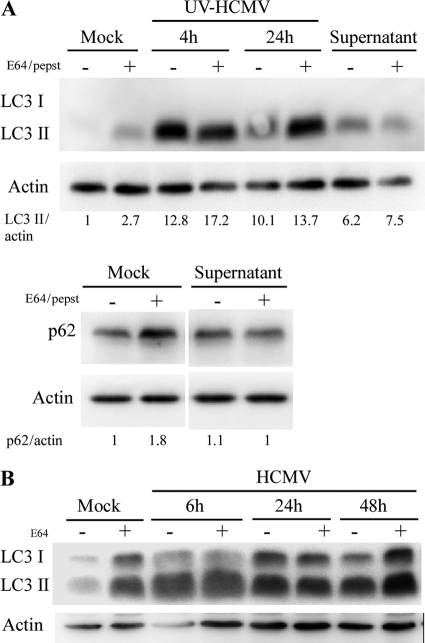
Fig 2.
(A) Supernatant of HCMV-infected cells is able to stimulate autophagy. MRC5 cells were treated during 4 h with supernatant of HCMV-infected cells clarified of viral particles and cellular debris by ultracentrifugation (see Materials and Methods) or infected with UV-inactivated HCMV, followed by immunoblotting with anti-LC3, anti-p62, or anti-actin antibodies. Lysosomal inhibitors E64 and pepstatin A (pepst) were added under all conditions, to block the lysosomal degradation of LC3-II and p62. (B) Accumulation of LC3 during HCMV infection is independent of the autophagic level. MRC5 cells were infected with HCMV at an MOI of 1 during 6, 24, or 48 h and treated or not with E64, followed by immunoblotting with anti-LC3 and anti-actin antibodies. Note that the level of LC3-II in infected cells does not change in the presence of lysosomal inhibitor.
To assess whether the inhibition of autophagy observed in HCMV-infected cells requires expression of viral proteins, we used UV-inactivated HCMV, which is able to interact with cellular receptors and to enter the cells but does not exhibit any viral gene expression. The number of puncta per cell in UV-inactivated infected cells (detected by identification of virion-associated tegument protein pp65 inside the cells) was constantly increased (to around 40 per cell) regardless of the time of infection, compared to that in mock-infected cells (Fig. 1C). Twenty-four hours after infection, a large number of MRC5 cells infected with UV-inactivated HCMV (visualized by pp65 staining) showed a punctuate staining of GFP-LC3 compared to the results in mock- and HCMV-infected cells (Fig. 1A).
However, an increased accumulation of autophagosomes in cells can also correspond to an inhibition of their fusion with lysosomes and to an impaired autophagy (41). Therefore, it is not sufficient to monitor static levels of autophagy, and three additional independent assays were performed to quantify the autophagic flux: the use of a tandem mRFP-GFP LC3 probe, the degradation of p62, and the degradation of [14C]valine-labeled long-lived proteins. We did not use the alternative method of measuring LC3-II by Western blot analysis, because we noticed a constant accumulation of LC3-II, independent of the autophagic level (see Fig. 2B).
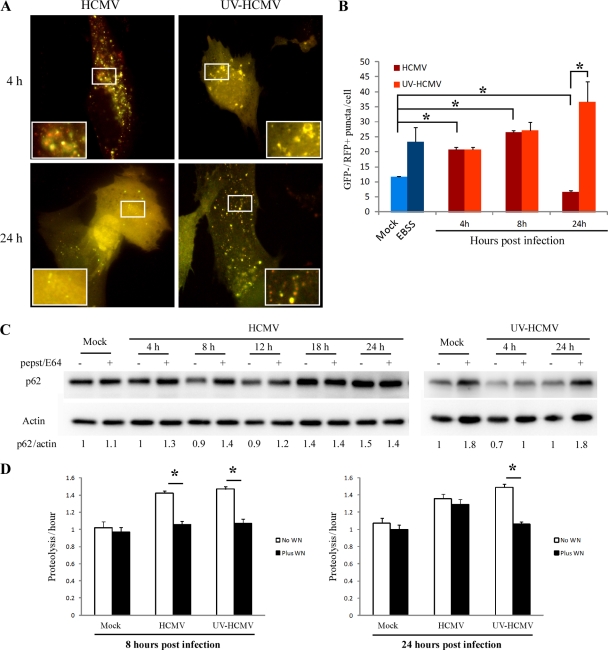
First, we used the tandem mRFP-GFP-LC3 probe developed to differentiate autophagosomes (GFP+ RFP+ or yellow puncta) from autolysosomes (GFP− RFP+ or red puncta), as the GFP signal is quenched in acidic compartments (31). The number of GFP− RFP+ puncta per cell, corresponding to autolysosomes, was measured in MRC5 cells infected with HCMV or UV-inactivated HCMV (Fig. 3A and B). We used amino acid-starved cells as a positive control, as this condition stimulates autophagy. Infection of MRC5 cells by HCMV resulted in an increase in autolysosomes at 4 and 8 h p.i. and in a strong decrease at 24 h p.i. UV-inactivated HCMV-infected cells displayed increased GFP− RFP+ puncta per cell, confirming a constant increase in the number of autolysosomes from 4 to 24 h. We next examined cellular expression levels of the autophagy substrate p62 in UV-inactivated HCMV- and HCMV-infected cells by immunoblotting (Fig. 3C) (2). Whereas p62 protein levels were decreased in HCMV-infected cells compared to control cells at 8 and 12 h p.i., p62 clearly accumulated at 18 and 24 h p.i. Moreover, the presence of lysosomal inhibitors (E64 and pepstatin A) did not further increase p62, suggesting that the autophagy flux is blocked at later stages of infection. In contrast, in UV-inactivated HCMV-treated cells, p62 protein levels were clearly increased in the presence of lysosomal inhibitors at 4 and 24 h p.i., confirming stimulation of the autophagic flux at any time during the treatment (Fig. 3C). Finally, the effects of HCMV on autophagy were confirmed by analyzing the rate of degradation of long-lived proteins. As illustrated in Fig. 3D, UV-inactivated HCMV and HCMV infection increased the rate of long-lived proteins degradation sensitive to wortmannin, an inhibitor of autophagic sequestration (4). However, the stimulation of protein degradation sensitive to wortmannin was no longer observed in cells infected with HCMV at 24 h p.i. Together these results indicate that HCMV stimulates autophagy at early times of infection but then inhibits it by 24 h p.i. The later inhibitory effect requires de novo viral gene expression, since UV-inactivated virus is no longer able to thwart autophagy, consistent with our previous results (9).
Fig 3.
Inhibition of autophagic flux requires expression of viral genes. (A) Representative images of MRC5 cells transfected with mRFP-GFP-LC3 plasmid and infected with HCMV or UV-inactivated HCMV. Red and yellow dots indicate GFP− RFP+ and GFP+ RFP+ puncta, respectively. (B) Autolysosomes were quantified by counting GFP− RFP+ puncta per cell. The results are the means of three independent experiments. Twenty cells were analyzed per assay. *, P < 0.05 (t test). Amino acid-starved cells (EBSS) were used as a positive control (C) Immunoblot analysis of p62 levels in cells infected with HCMV or UV-inactivated HCMV for the indicated times. Lysosomal inhibitors E64/pepstatin A were added to all conditions, to block the lysosomal degradation. Actin immunoblotting was used as a loading control. (D) Effect of HCMV infection on long-lived protein degradation. [14C]valine-radiolabeled MRC5 cells were mock infected or infected with HCMV or UV-inactivated HCMV for 8 or 24 h, and proteolysis was measured as described in Materials and Methods. Cells were treated with 200 nM wortmannin (Plus WN) or not (No WN). The results are the means of three independent experiments. *, P < 0.05.
Expression of Beclin 1 and Bcl-2 during HCMV infection.
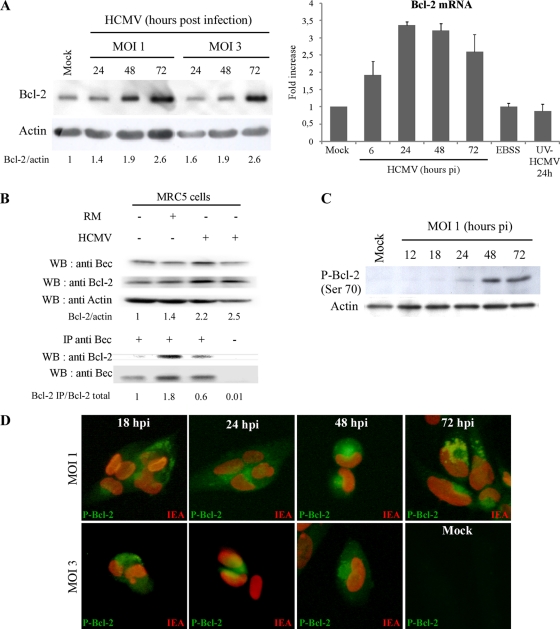
Several viral proteins have been described to interact with Beclin 1 and to modulate its autophagy activity (23, 33, 44). Moreover, it has been reported in the context of HIV infection that the Env-mediated autophagy triggered in T cells correlates with Beclin 1 protein accumulation (22). In a first series of experiments in MRC5 cells, neither Beclin 1 protein expression nor Beclin 1 mRNA level was modified during HCMV infection (data not shown). We then analyzed the impact of HCMV infection on the expression of Bcl-2 protein, a negative regulator of the Beclin 1 complex (48). A significant increase in the amount of Bcl-2 protein was detected in HCMV-infected cells from 1 to 3 days p.i., regardless of the MOI (Fig. 4A). We next analyzed the mRNA level of Bcl-2 in HCMV-infected cells (Fig. 4A). The amount of Bcl-2 mRNA rapidly increased, as early as 6 h p.i. At 24 and 48 h p.i., corresponding to the time when HCMV inhibited autophagy (9), the level of Bcl-2 mRNA was 3-fold higher than in mock-infected cells. We did not observe any modification of Bcl-2 mRNA levels in UV-inactivated HCMV-infected cells compared to mock-infected cells, suggesting that expression of viral proteins is necessary for this induction.
Fig 4.
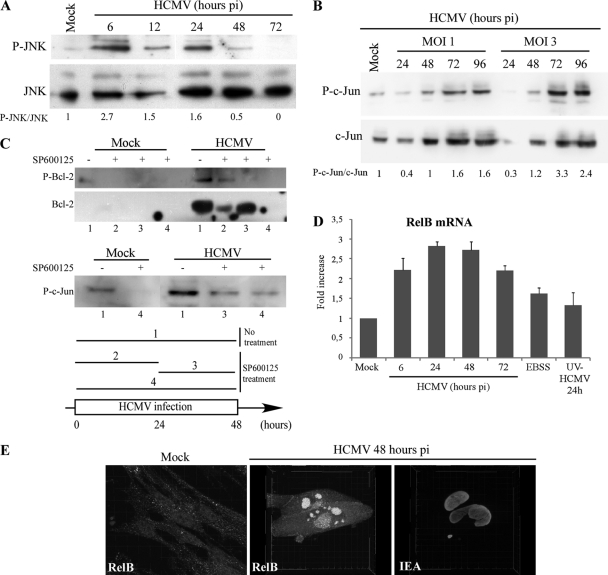
Bcl-2 is not responsible for autophagy inhibition in HCMV-infected cells. (A) Bcl-2 protein and mRNA expression. Immunoblot analysis of Bcl-2 protein in mock-infected cells and in cells infected with HCMV at MOIs of 1 and 3 from 24 to 72 h. Bcl-2 mRNA level was analyzed in control cells, in cells in starvation medium (EBSS), and in cells infected with HCMV at an MOI of 1 from 6 to 72 h or with UV-inactivated HCMV for 24 h. The results are the means of three independent experiments. (B) Coimmunoprecipitation of Beclin 1 and Bcl-2 in MRC5 cells. MRC5 cells were mock infected, infected with HCMV (MOI = 1) for 2 days, or grown in rich medium (RM) for 4 h prior to coimmunoprecipitation. Rich medium is known to inhibit autophagy. Cell lysates were immunoprecipitated with preimmune goat serum or a goat polyclonal anti-Beclin 1 antibody followed by immunoblotting with the indicated antibodies. (C and D) Accumulation of hyperphosphorylated Bcl-2 during HCMV infection in MRC5 fibroblasts. (C) Immunoblot analysis of the phosphorylated Bcl-2 (Ser70). MRC5 cells were mock infected or infected with HCMV at an MOI of 1 from 12 to 72 h. Actin immunoblotting was used as a loading control. (D) MRC5 cells were mock infected or infected with HCMV at an MOI of 1 from 18 to 72 h and at an MOI of 3 from 18 to 48 h. An increased staining of phosphorylated Bcl-2 (juxtanuclear green staining) was observed in the HCMV-infected cells compared to mock-infected cells (nuclear red staining shows HCMV IE antigens).
Upregulation of Bcl-2 is not involved in the HCMV-induced inhibition of autophagy.
It is well known that the overexpression of Bcl-2 inhibits starvation-induced autophagy and that Bcl-2 is a negative regulator of autophagy which interacts with Beclin 1 to avoid stimulation of autophagy (48). We therefore hypothesized that HCMV increases cellular Bcl-2 to potentiate its interaction with Beclin 1 and thereby inhibit autophagy. Fibroblasts were grown 4 h in amino acid excess (rich medium [RM]) to inhibit autophagy (data not shown) and to maximize interaction between Beclin 1 and Bcl-2 (Fig. 4B). Consistent with previous results (48), we observed that Bcl-2 bound to Beclin 1 in normal conditions and that Bcl-2 coimmunoprecipitated more with Beclin 1 during autophagy-inhibiting conditions (RM) (Fig. 4B). However, coimmunoprecipitation of Beclin 1 and Bcl-2 in fibroblasts infected with HCMV showed that, whereas Bcl-2 was overexpressed compared to mock-infected cells, only a small part of the pool of Bcl-2 interacted with Beclin 1 in these cells (Fig. 4B). These results suggest that the inhibition of autophagy by HCMV was not due to the overexpressed Bcl-2 binding to Beclin 1.
Several mechanisms have been described that could explain the disruption of Beclin 1-Bcl-2 interaction during HCMV infection: phosphorylation of either Bcl-2 or Beclin 1 (61, 63), competitive disruption of Beclin 1/Bcl-2 by BH3-only proteins or BH3 mimetics (36), or, potentially, effects of membrane-anchored receptors or their adaptors on Bcl-2/Beclin 1 interactions (57). Interestingly, we observed an increase of Bcl-2 phosphorylation in HCMV-infected cells by immunoblotting as early as 24 h p.i. (Fig. 4C). Moreover, there was no detectable signal of the phosphorylation of Bcl-2 in mock-infected cells but HCMV-infected cells, identified by staining for viral IE antigens, showed strong staining for phosphorylated Bcl-2, visualized by a clear juxtanuclear signal after 24 h of infection (Fig. 4D). Taken together, these results suggest that posttranslational modifications of Bcl-2 might explain why the interaction between Beclin 1 and Bcl-2 is reduced in HCMV-infected cells.
It has been previously reported that the c-Jun N-terminal kinase (JNK) disrupts the Bcl-2/Beclin 1 complex after nutrient deprivation and ceramide-induced autophagy, by phosphorylating Bcl-2 (46, 61). We observed that the JNK pathway was activated during HCMV infection based on accumulation of phosphorylated JNK during the first 24 h p.i. and of its phosphorylated substrate, c-Jun, at later times (Fig. 5A and B). Treatment of infected cells with SP600125, a selective JNK inhibitor, showed that Bcl-2 phosphorylation in infected cells was JNK specific (Fig. 5C). Ectopic expression of HCMV immediate-early protein 1 (IE1) was previously shown to selectively induce RelB NF-κB transcription and activity, via the JNK pathway (59–60). Similarly, we observed in HCMV-infected cells a JNK-mediated upregulation of RelB NF-κB subunit and its nuclear relocalization (Fig. 5D and E), which might be responsible for the HCMV-mediated Bcl-2 upregulation. Finally, our results suggest that HCMV-induced Bcl-2 upregulation and phosphorylation of Bcl-2 are both mediated by JNK activation, either via RelB activity or directly. Taken together, these results could explain the mechanism by which the Beclin 1/Bcl-2 complex is disrupted during HCMV infection. While this dissociation should lead to an increased autophagy, HCMV infection has the opposite effect. Therefore, we still needed to explore the mechanisms for this HCMV-mediated inhibition of autophagy.
Fig 5.
Activation of the JNK pathway during HCMV infection. (A) HCMV activates JNK phosphorylation. MRC5 cells were mock infected or infected with HCMV at MOIs of 1 and 3 from 6 to 72 h. Total cell lysates were prepared, and samples were subjected to immunoblot analysis of JNK and phosphorylated JNK (P-JNK). (B) Immunoblot analysis of the phosphorylated c-Jun during HCMV infection. MRC5 cells were mock infected or infected with HCMV at MOIs of 1 and 3 from 24 to 96 h. (C) Cells were infected with HCMV during 48 h and treated with SP600125, a selective JNK inhibitor, during different time frames as indicated in the diagram (conditions 1 to 4). SP600125 blocks Bcl-2 overexpression when added during the first 24 h of infection (lane 2) and phosphorylation of Bcl-2 when added from 24 until 48 h p.i. (lane 3). SP600125 blocks phosphorylation of c-Jun when added during 24 or 48 h (lanes 3 and 4) (D) RelB mRNA analysis by real-time PCR. MRC5 cells were mock infected or infected with HCMV at an MOI of 1 for 6 to 72 h, infected with UV-inactivated HCMV for 24 h, or grown in starvation medium (EBSS) for 4 h. The results are the means of three independent experiments. (E) Confocal analysis of the expression of RelB in HCMV-infected cells. MRC5 cells were infected with HCMV at an MOI of 1 for 48 h. Whereas RelB presents a marked nuclear staining in HCMV-infected cells, it has a diffuse (nuclear and cytoplasmic) staining in mock-infected cells. The cells were observed under a confocal laser scanning microscope (Zeiss LSM 510) and analyzed with Imaris software for three-dimensional reconstruction.
The viral protein TRS1 inhibits starvation-induced autophagy.
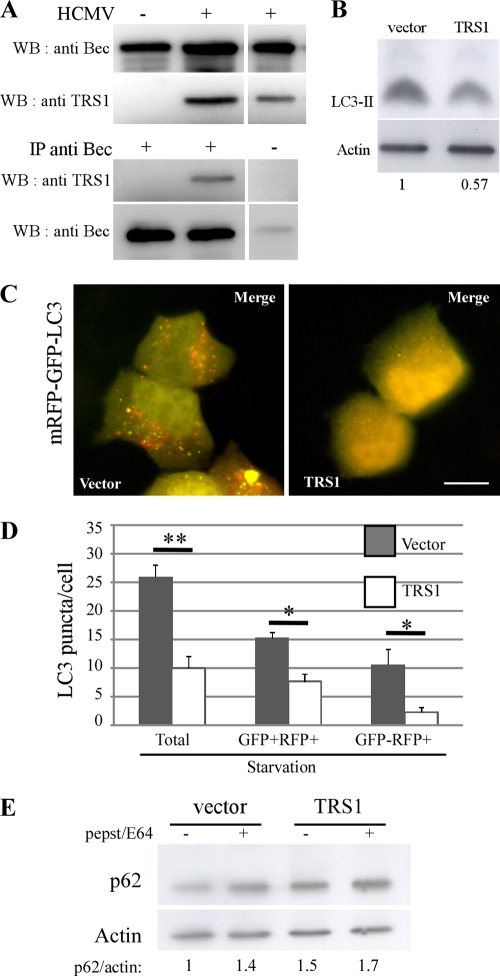
As viral expression during HCMV infection is necessary to inhibit autophagy, we sought to identify the viral protein(s) responsible for autophagy inhibition. It has been previously shown that several viral proteins prevent autophagy induction. The viral proteins ICP34.5 of HSV-1 and vBcl-2 of HHV-8 inhibit autophagy by interaction with Beclin 1, whereas the viral homologue vFLIP of HHV-8 is able to reduce it by interaction with Atg3 (35, 44, 48). Despite the fact that HCMV belongs to the Herpesviridae family, the HCMV genome does not contain any ortholog of the HSV-1 γ134.5 gene or viral Bcl-2 and FLIP homologs, like those known to be present in gammaherpesviruses. However, ICP34.5 was previously described to preclude the host shutoff of protein synthesis via the PKR/eIF2α signaling pathway and HCMV possesses a functional homolog of ICP34.5, called TRS1 (11, 26). TRS1 binds double-stranded RNA and interferon-induced kinase PKR and inhibits the PKR/eIF2α signaling pathway (10, 26). To determine whether TRS1 interacts with Beclin 1 and antagonizes its autophagy function, like ICP34.5, we performed coimmunoprecipitation studies. In MCR5 cells infected with HCMV at an MOI of 1 for 24 h, we found that endogenous Beclin 1 coimmunoprecipitated with TRS1 (Fig. 6A). To assess the impact of TRS1 on the autophagic pathway, different assays to monitor the autophagic flux were performed initially in HeLa cells since they are much more transfectable than MRC5 cells. HeLa cells were transiently transfected with 6×His-tagged TRS1 expression vector or an empty vector, and autophagy was induced by starvation. A decrease of LC3-II was observed by immunoblot analysis in TRS1-transfected cells (Fig. 6B). Comparable results were obtained in HEK cells (data not shown). To determine whether autophagy inhibition caused by TRS1 led to inhibition of the autophagic flux, we used the tandem mRFP-GFP-LC3 probe (Fig. 6C). HeLa cells were transiently cotransfected with TRS1 or empty vector and mRFP-GFP-LC3 plasmids. After starvation for 4 h, TRS1 expressing cells were detected using 6×His antibody by immunofluorescence and GFP+ RFP+ (yellow) or GFP− RFP+ (red) puncta were quantified in those cells (Fig. 6C and D). HeLa cells transfected with TRS1 displayed decreased total LC3 puncta per cell, confirming a decrease in the number of autophagosomes. Similar results were obtained with the GFP-LC3 probe (data not shown). Expression of TRS1 resulted in a decrease of GFP+ RFP+ puncta, corresponding to autophagosome and a strong decrease of GFP− RFP+ puncta, representing autolysosomes. These results indicate that TRS1 inhibits starvation-induced autophagic flux in HeLa cells. To complement these results, we examined cellular expression levels of p62 in TRS1-transfected cells that were cultured in starvation medium. In TRS1-transfected cells, p62 protein levels were approximately increased by 40% compared to control cells, confirming inhibition of the autophagic flux by TRS1 expression (Fig. 6E). Taken together, the decrease of LC3-II, the diminution of autolysosomes per cell detected with mRFP-GFP-LC3 probe, the diffuse staining of GFP-LC3, and the increase of p62 in cells transfected with TRS1 demonstrate that this viral protein can inhibit the formation of autophagosome.
Fig 6.
The viral protein TRS1 interacts with Beclin 1 and inhibits autophagic flux. (A) Coimmunoprecipitation of Beclin 1 and TRS1 in HCMV-infected MRC5 cells. MRC5 cells were mock infected or infected with HCMV at an MOI of 1 for 48 h. Cell lysates were immunoprecipitated with preimmune goat serum or a goat polyclonal anti-Beclin 1 antibody followed by immunoblotting with an anti-TRS1 antibody. (B) Immunoblot analysis of LC3 in HeLa cells transfected with TRS1 and cultured in starvation medium. Actin immunoblotting was used as a loading control. (C) Representative images of HeLa cells cotransfected with mRFP-GFP-LC3 and TRS1 plasmids for 24 h and cultured in starvation medium during the last 4 h of infection. Red and yellow dots indicate GFP− RFP+ and GFP+ RFP+ puncta, respectively. Bar, 10 μm. (D) Quantitation of autophagy as measured by the number of LC3 puncta per cell, total puncta per cell (GFP+ RFP+ and GFP− RFP+ puncta), GFP+ RFP+ puncta per cell, and GFP− RFP+ puncta per cell. Only TRS1-expressing cells were quantified as described in Materials and Methods. The results are the means of three independent experiments. Twenty cells were analyzed per assay. *, P < 0.05; **, P < 0.01 (t test). (E) Immunoblot analysis of p62 protein in HeLa cells transfected with TRS1 plasmids and cultured in starvation medium. Actin immunoblotting was used as a loading control.
TRS1 inhibits autophagy independently of its interaction with PKR.
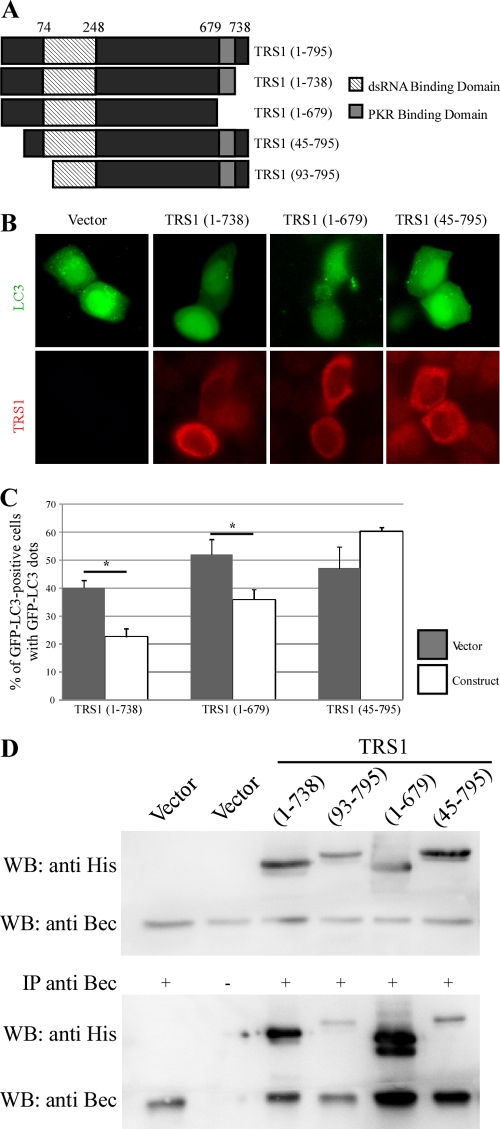
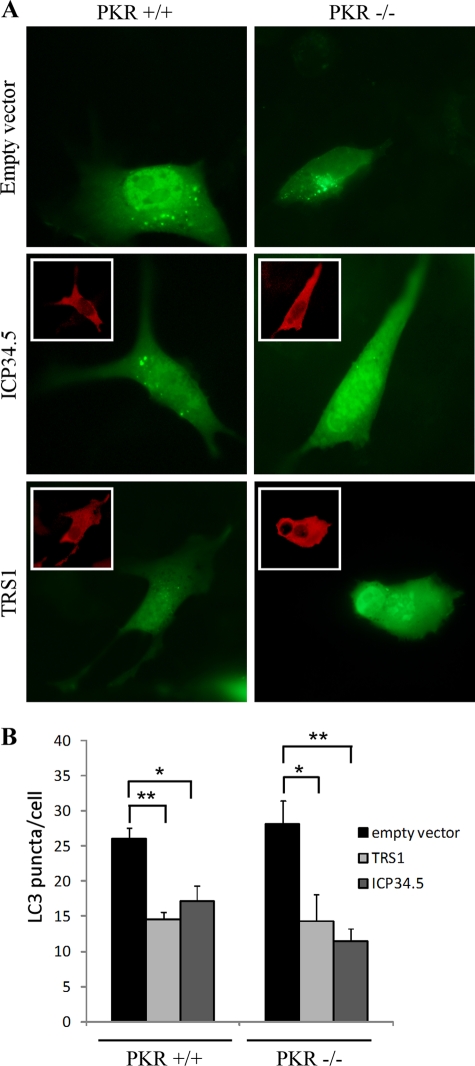
With the finding that TRS1 blocks autophagy, we investigated which domain of TRS1 is responsible for inhibition of autophagy. To accomplish this, we assessed the ability of different 6×His-tagged TRS1 constructs to inhibit starvation-induced autophagy in HeLa cells (Fig. 7). TRS1 contains a domain required for PKR binding located at its carboxy-terminal region and a double-stranded RNA-binding domain (dsRBD) at its amino terminus (Fig. 7A). The percentage of GFP-LC3-positive cells with GFP-LC3 dots was measured in starved cells. We observed that even TRS1 (1-679), which lacks the region required for PKR binding, inhibited starvation-induced autophagy (Fig. 7B and C). This result suggests that PKR binding is not required for autophagy inhibition. This was confirmed by the observation that TRS1 was able to block starvation-induced autophagy in murine fibroblasts lacking PKR (Fig. 8). PKR+/+ and PKR−/− MEFs were cotransfected with plasmids expressing GFP-LC3 and either HSV-1 ICP34.5 or HCMV TRS1. The ability of the viral proteins to block starvation-induced autophagy was compared in wild-type versus PKR-deficient cells. We observed that both TRS1 and ICP34.5 block autophagy, independently of the expression of PKR (Fig. 8). In contrast, an amino-terminal truncation mutant TRS1 (45-795) failed to inhibit starvation-induced autophagy (Fig. 7B and C). The number of GFP-LC3 positive cells was clearly increased after transfection with TRS1 (45-795) and autophagy induction. Thus, the inhibition of autophagy by TRS1 requires the amino terminus of TRS1. Next, we performed a series of coimmunoprecipitation experiments with different His-tagged forms of TRS1 transfected in HeLa cells. We observed that TRS1 (1-738) and (1-679) bound to Beclin 1 whereas TRS1 (45-795) and (93-795) did not bind efficiently to Beclin 1 (Fig. 7D). These results demonstrated that the deletion of the PKR-interacting domain of TRS1 did not impair the interaction of the viral protein with endogenous Beclin 1 but that the N-terminal domain is required for the interaction with Beclin 1. These findings, together with the observation that TRS1 blocks starvation-induced autophagy in PKR−/− cells, support the hypothesis that TRS1 inhibits autophagy as a result of its interaction with Beclin 1.
Fig 7.
The N-terminal region of TRS1 is required for inhibition of autophagy and Beclin 1 binding. (A) Schematic representation of TRS1 showing position of the region required for PKR binding (amino acids 679 to 738) and the dsRNA-binding domain (amino acids 74 to 248). Different TRS1 constructs were used: C-terminal deleted fragment [TRS1 (1-738)], PKR-binding deleted fragment [TRS1 (1-679)], and N-terminal deleted fragments [TRS1 (45-795) and (93-795)]. (B) Representative images of GFP-LC3 staining in HeLa cells cotransfected for 48 h with GFP-LC3 and indicated TRS1 constructs. Four hours before fixation, cells were grown in starvation medium to induce autophagy. TRS1-transfected cells were visualized by an anti-His antibody, and GFP-LC3-positive cells with GFP-LC3 dots were quantified (C). The results are the means of three independent experiments. Between 50 and 100 cells were analyzed per assay. *, P < 0.05 (t test). (D) Coimmunoprecipitation of endogenous Beclin 1 and indicated His-tagged mutant TRS1 in HeLa cells transfected with the indicated plasmids.
Fig 8.
TRS1 inhibits autophagy independently of PKR. Murine PKR−/− and PKR+/+ cells were cotransfected for 48 h with GFP-LC3 and His-TRS1 or FLAG-ICP34.5 constructs. Four hours before fixation, cells were grown in starvation medium to induce autophagy. TRS1-transfected cells were visualized by an anti-His antibody, and ICP34.5-transfected cells were visualized by an anti-FLAG antibody. (A) Representative images of GFP-LC3 in PKR−/− and PKR+/+ cells. Inserts show the viral protein staining. (B) GFP-LC3 positive cells with GFP-LC3 dots were quantified. The results are the means of three independent experiments. Twenty cells were analyzed per assay. *, P < 0.05; **, P < 0.01 (t test).
A TRS1 deletion mutant virus is defective in autophagy inhibition in fibroblasts.
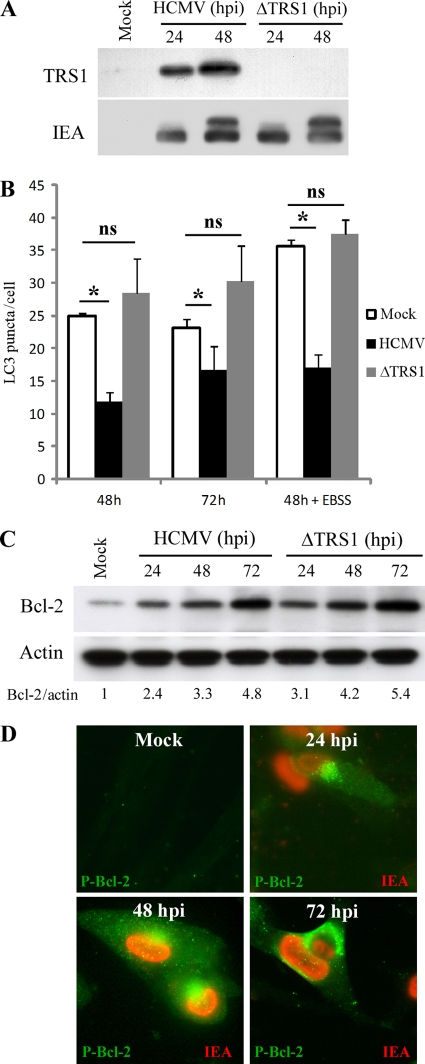
The observations that TRS1 interacts with Beclin 1 and inhibits starvation-induced autophagy to the same extent as ICP34.5 support the role of TRS1 in the inhibition of autophagy in HCMV-infected cells. To test whether the virally expressed TRS1 is necessary for the inhibition of autophagy in HCMV-infected cells, we analyzed autophagy in cells infected with an HCMV mutant lacking the TRS1 gene (ΔTRS1) (Fig. 9A). We observed that ΔTRS1 infected cells exhibit levels of autophagy indistinguishable from those of mock-infected fibroblasts (Fig. 9B). In contrast, the number of autophagosomes was significantly decreased in HCMV-infected cells. Moreover, the number of GFP-LC3 dots increased after nutrient deprivation (EBSS) and the ΔTRS1 mutant virus was not able to block starvation-induced autophagy. We found that Bcl-2 is overexpressed and hyperphosphorylated in ΔTRS1 mutant virus-infected cells similarly to results in HCMV-infected cells (Fig. 9C and D). These results confirmed that increased expression of Bcl-2 plays no role in inhibition of autophagy by HCMV.
Fig 9.
TRS1 is required for inhibition of autophagy in fibroblasts. (A) Expression of TRS1 and IEA in MRC5 cells infected with HCMV wild-type and ΔTRS1 mutant virus. (B) MRC5 cells were infected with HCMV wild-type or ΔTRS1 mutant virus at an MOI of 1 during 48 or 72 h and transfected for 48 h with GFP-LC3. When indicated, cells were grown in starvation medium (EBSS) 4 h before fixation to induce autophagy. Autophagy was quantified in IEA-positive cells by counting the number of GFP-LC3 puncta per cell. The results are the mean of three independent experiments. Twenty cells were analyzed per assay. *, P < 0.05; ns, not significant (t test). (C) Immunoblot analysis of Bcl-2 protein in cells infected with wild-type HCMV or ΔTRS1 mutant virus at an MOI of 1, from 24 to 72 h. (D) Accumulation of hyperphosphorylated Bcl-2 during ΔTRS1 mutant virus infection in MRC5 fibroblasts at 24, 48, and 72 h p.i. Nuclear red staining corresponds to viral IE antigens.
DISCUSSION
In this report, we report that HCMV stimulates autophagy in human fibroblasts during the early stages of infection independently of viral protein synthesis and is able to inhibit autophagosome formation and maturation by 24 h postinfection, as a result of expression of one or more viral proteins. We identify the viral protein TRS1 of HCMV as a negative modulator of autophagy, independently of its previously described function on the kinase PKR but likely via its interaction with the autophagy protein Beclin 1 (26).
The observation that autophagy is stimulated at early time points after infection independently of the viral replication is in a good agreement with a recent study by McFarlane et al. (40). The significance of the early induction of autophagy by HCMV remains to be determined. It could represent a cell stress response to infection and may aid in an adaptive immune response by promoting viral antigen presentation by major histocompatibility complex (MHC) class II molecules. It might also contribute to an early innate immune response by degradation of viruses into autophagolysosomes, referred to as xenophagy (45, 56). In fact, other viruses, such as HIV or measles virus, also trigger autophagy by interacting with cell surface receptors and/or soon after their entry in the cell cytoplasm (22, 28). For example, the binding of measles virus (Edmonston strain) to CD46 at the surface of human cells induces de novo formation of autophagosome (28). The mechanism by which HCMV triggers autophagy remains to be better defined. McFarlane et al. proposed that autophagy is stimulated as a response to the HCMV DNA genome (40). However, it may be that contact of the viral particle with the cell surface or interaction between a viral structural component and a cellular sensor triggers autophagy. In line with this possibility, we observed that the binding of HCMV to cell surface heparan sulfate proteoglycan is required to stimulate autophagy (data not shown). HCMV interacts with Toll-like receptor 2 (TLR2) via two viral envelope glycoproteins and initiates inflammatory cytokine secretion independently of viral entry (29). Since TLR activation by different ligands induces autophagy, it will be interesting to test whether activation of TLR2 by HCMV triggers autophagy (15).
Whatever the mechanism responsible for the induction of autophagy by HCMV, we observed that the virus is able to inhibit autophagy at later time points (reference 9 and the present study). The inhibition of autophagy has been shown by the use of different readouts for autophagy: accumulation of GFP-LC3 and GFP-RFP-LC3 puncta, degradation of the autophagy cargo p62, and rate of long-lived protein degradation in the presence and absence of wortmannin. On the basis of these criteria, we conclude that HCMV blocks autophagy at the stage of autophagosome formation. This inhibition is dependent on the synthesis of viral proteins because we observed that autophagy is stimulated in cells infected with UV-inactivated HCMV, as recently reported by McFarlane et al. (40). Contrary to our results, these authors suggest that HCMV is still able to stimulate autophagy after 24 h of infection. This conclusion relies on the analysis of the accumulation of LC3-II by Western blotting, a usually reliable method to detect the form of the protein associated with autophagosomes. However, we previously noticed that LC3-I and -II accumulated in HCMV-infected cells independently of the accumulation of autophagosomes (9) (Fig. 2). Reggiori and colleagues reported that LC3-I is hijacked by coronaviruses to provide the membranous support for viral replication complex, in an autophagy-independent way (49). Moreover, the final envelopment of HCMV in the viral assembly complex requires a remodeling of membranes from different organelles, such as the trans-Golgi network (TGN) and endosomes (8, 14), that are also reservoirs for autophagy proteins such as Atg9 (37, 62). Therefore, it would be interesting to investigate whether LC3 and/or some Atg proteins could have a role during HCMV infection, independently of their function in autophagy.
To understand the mechanisms by which HCMV inhibits autophagy, we decided to focus on the HCMV protein TRS1, which appears to be a functional homolog of the HSV-1 ICP34.5, regarding the PKR/eiF2α pathway. Activation of this signaling pathway triggers autophagy in the context of infection with a Δ34.5-HSV-1 recombinant virus, and the viral protein ICP34.5 is capable of inhibiting autophagy (55). In this report, we identified for the first time TRS1 as a HCMV protein able to block autophagy and to interact with Beclin 1. Consistent with the timing of HCMV-mediated autophagy inhibition, we observed that TRS1 accumulates predominantly in the cytoplasm of infected cells as early as 24 h p.i. (data not shown). Two functional domains of TRS1 have been identified: the carboxy-terminal part, which is required for interaction with PKR, and an unconventional dsRNA-binding domain located in a 175-amino-acid stretch close to the N terminus (between amino acids 74 and 248) (25). We thus explored which domain of TRS1 is involved in the inhibition of the autophagy process. As the domain of TRS1 required for PKR binding is necessary for the prevention of PKR activation and thereby preventing phosphorylation of eIF2α, we were surprised to discover that this domain is not necessary for inhibiting autophagy in our model. However, a truncated form of TRS1 in the N-terminal part is no longer able to block autophagy. This observation, together with the fact that the N-terminal part of TRS1 is required for strong binding to Beclin 1, suggests that TRS1 blocks autophagy by interaction with Beclin 1. It is interesting that both ICP34.5 and TRS1 inhibit autophagy not via their ability to interfere with the activation of PKR but via their interaction with Beclin 1 (44). Since numerous viral proteins block the PKR-eIF2α signaling pathway to preclude the shutoff of protein synthesis, one important question that arises is whether autophagy can be inhibited by the unique blockade of PKR-dependent signaling by a viral protein.
We observed that the form of virally expressed TRS1 is sufficient for the inhibition of autophagy in HCMV-infected cells (Fig. 9). It is interesting to note that even though ΔTRS1 mutant virus did not inhibit autophagy, we did not observe a stimulation of autophagy such as the one induced by UV-HCMV. HCMV expresses a second gene product, IRS1, that is highly homologous to TRS1 (10). Thus, it will be important to explore whether IRS1 can also antagonize autophagy. It is likely that IRS1, which has an amino terminus identical to that of TRS1, duplicates this TRS1 function. Indeed, the N-terminal 549-amino-acid regions of the two proteins are identical and although their C-terminal regions diverge, they nevertheless remain 55% identical in the divergent region (3). Construction of ΔTRS1/ΔIRS1 recombinant mutant virus revealed that IRS1 can compensate for TRS1 regarding PKR inactivation (38). Unfortunately, the mutant virus lacking both genes is totally unable to replicate in human fibroblasts (38). An alternative approach could be to use a chimeric Δ34.5-HSV-1 recombinant virus which expresses the HCMV gene TRS1. Indeed, Cassady showed that TRS1 can complement Δ34.5-HSV-1 recombinant virus regarding wild-type viral protein synthesis in infected cells (6). Whereas infection with a Δ34.5-HSV-1 recombinant virus stimulates autophagy, it is not known whether TRS1 can also restore the inhibitory effect of ICP34.5 on autophagy in infected cells. Monitoring the autophagy level after infection with the HCMV/HSV-1 chimeric virus would address this possibility.
It is possible that HCMV possesses more than one mechanism for inhibiting the autophagic pathway. Cellular Bcl-2 (cBcl-2) is an anti-autophagy protein which acts via its inhibitory interaction with Beclin 1 (48). Despite the fact that other herpesviruses, such as HHV8 and MHV-68, express a viral homolog of Bcl-2 able to inhibit autophagy, HCMV does not possess one. Although HCMV stimulates the expression of cBcl-2, it does not use this strategy to block autophagy. Indeed, Bcl-2 is overexpressed in ΔTRS1 mutant virus-infected cells whereas autophagy is not inhibited. We also observed an accumulation of the phosphorylated form of Bcl-2 that does not interact with Beclin 1. This phosphorylation, as well as the induction of the expression of Bcl-2, is dependent on the activity of JNK. Previous studies have shown that the JNK-dependent phosphorylation of Bcl-2 triggers the dissociation of its complex with Beclin 1 (47, 61). It is interesting to note that the gammaherpesvirus Bcl-2 homologue constitutively blocks autophagy via its interaction with Beclin 1, because vBcl-2 does not have the N-terminal loop that contains amino acid substrates for JNK and thus cannot be phosphorylated (61). Another possibility concerns the autophagy regulatory complex mTORC1 (mammalian target of rapamycin complex 1), which is stimulated by HCMV during infection (32). Activation of mTORC1 not only turns on the protein synthesis involved in cell growth but also suppresses autophagy (reviewed in reference 20). HCMV influences multiple cellular pathways that communicate with mTORC1, such as inhibition of the tuberous sclerosis complex TSC1/2 by the HCMV UL38 protein (43). Moreover, the activation and the perinuclear localization of mTORC1 are maintained in HCMV-infected cells, even during nutrient starvation, which normally inhibits mTOR and activates autophagy (12).
In conclusion, HCMV uses a strategy related to the one used by HSV-1 to block autophagy by interacting with Beclin 1. Herpesviruses have developed redundant strategies to block autophagy by interfering with the function of Beclin 1 or other Atg proteins involved in the biogenesis of autophagosomes. Our results demonstrate that two viruses of the herpes family use a similar strategy to block autophagy independently of their ability to produce a viral form of Bcl-2.
ACKNOWLEDGMENTS
We thank T. Yoshimori for providing us with the GFP-LC3 and mRFP-GFP-LC3 constructs. We are very grateful to Chantal Bauvy for immunoprecipitation and proteolysis experiments. We thank Claudine Deloménie (IFR141 innovation thérapeutique, Châtenay-Malabry) for providing assistance with quantitative reverse transcription-PCR studies and Caroline Tran Van (UPS, Châtenay-Malabry).
This work was supported by institutional funding from The Institut National de la Santé et de la Recherche Médicale (INSERM) and from Paris Sud University, by grants from the Agence Nationale de la Recherche (ANR MIME 2007) to A.E., from the National Institutes of Health (AI26672) to A.P.G., and the Deutsche Forschungsgemeinschaft (BR1730/3-1) to W.B.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Bauvy C, Meijer AJ, Codogno P. 2009. Assaying of autophagic protein degradation. Methods Enzymol. 452:47–61 [DOI] [PubMed] [Google Scholar]
- 2. Bjorkoy G, et al. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blankenship CA, Shenk T. 2002. Mutant human cytomegalovirus lacking the immediate-early TRS1 coding region exhibits a late defect. J. Virol. 76:12290–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 243:240–246 [DOI] [PubMed] [Google Scholar]
- 5. Brune W, Nevels M, Shenk T. 2003. Murine cytomegalovirus m41 open reading frame encodes a Golgi-localized antiapoptotic protein. J. Virol. 77:11633–11643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cassady KA. 2005. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J. Virol. 79:8707–8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cavignac Y, Esclatine A. 2010. Herpesviruses and autophagy: catch me if you can! Viruses 2:314–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cepeda V, Esteban M, Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol. 12:386–404 [DOI] [PubMed] [Google Scholar]
- 9. Chaumorcel M, Souquere S, Pierron G, Codogno P, Esclatine A. 2008. Human cytomegalovirus controls a new autophagy-dependent cellular antiviral defense mechanism. Autophagy 4:1–8 [DOI] [PubMed] [Google Scholar]
- 10. Child SJ, Hakki M, De Niro KL, Geballe AP. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chou J, Kern ER, Whitley RJ, Roizman B. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262–1266 [DOI] [PubMed] [Google Scholar]
- 12. Clippinger AJ, Maguire TG, Alwine JC. 2011. Human cytomegalovirus infection maintains mTOR activity and its perinuclear localization during amino acid deprivation. J. Virol. 85:9369–9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Compton T, et al. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Das S, Pellett PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J. Virol. 85:5864–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. 2008. Toll-like receptors control autophagy. EMBO J. 27:1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deretic V. 2011. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240:92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deretic V, Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5:527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Djavaheri-Mergny M, et al. 2006. NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J. Biol. Chem. 281:30373–30382 [DOI] [PubMed] [Google Scholar]
- 19. Esclatine A, et al. 2001. Differentiation-dependent redistribution of heparan sulfate in epithelial intestinal Caco-2 cells leads to basolateral entry of cytomegalovirus. Virology 289:23–33 [DOI] [PubMed] [Google Scholar]
- 20. Esclatine A, Chaumorcel M, Codogno P. 2009. Macroautophagy signaling and regulation. Curr. Top. Microbiol. Immunol. 335:33–70 [DOI] [PubMed] [Google Scholar]
- 21. Esclatine A, Lemullois M, Servin AL, Quero AM, Geniteau-Legendre M. 2000. Human cytomegalovirus infects Caco-2 intestinal epithelial cells basolaterally regardless of the differentiation state. J. Virol. 74:513–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Espert L, et al. 2006. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J. Clin. Invest. 116:2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gannage M, et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gannage M, Ramer PC, Munz C. 2010. Targeting Beclin 1 for viral subversion of macroautophagy. Autophagy 6:166–167 [DOI] [PubMed] [Google Scholar]
- 25. Hakki M, Geballe AP. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. J. Virol. 79:7311–7318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hakki M, Marshall EE, De Niro KL, Geballe AP. 2006. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J. Virol. 80:11817–11826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joubert PE, et al. 2009. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 6:354–366 [DOI] [PubMed] [Google Scholar]
- 29. Juckem LK, Boehme KW, Feire AL, Compton T. 2008. Differential initiation of innate immune responses induced by human cytomegalovirus entry into fibroblast cells. J. Immunol. 180:4965–4977 [DOI] [PubMed] [Google Scholar]
- 30. Kimura S, Fujita N, Noda T, Yoshimori T. 2009. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 452:1–12 [DOI] [PubMed] [Google Scholar]
- 31. Kimura S, Noda T, Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3:452–460 [DOI] [PubMed] [Google Scholar]
- 32. Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78:11030–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kyei GB, et al. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186:255–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee AW, et al. 2006. Human cytomegalovirus alters localization of MHC class II and dendrite morphology in mature Langerhans cells. J. Immunol. 177:3960–3971 [DOI] [PubMed] [Google Scholar]
- 35. Lee JS, et al. 2009. FLIP-mediated autophagy regulation in cell death control. Nat. Cell Biol. 11:1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maiuri MC, et al. 2007. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy 3:374–376 [DOI] [PubMed] [Google Scholar]
- 37. Mari M, et al. 2010. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190:1005–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marshall EE, Bierle CJ, Brune W, Geballe AP. 2009. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J. Virol. 83:4112–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsunaga K, et al. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11:385–396 [DOI] [PubMed] [Google Scholar]
- 40. McFarlane S, et al. 2011. Early induction of autophagy in human fibroblasts after infection with human cytomegalovirus or herpes simplex virus 1. J. Virol. 85:4212–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mocarski ES, Jr, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2772 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 43. Moorman NJ, et al. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Orvedahl A, et al. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1:23–36 [DOI] [PubMed] [Google Scholar]
- 45. Paludan C, et al. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307:593–596 [DOI] [PubMed] [Google Scholar]
- 46. Pattingre S, et al. 2009. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J. Biol. Chem. 284:2719–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pattingre S, Bauvy C, Levade T, Levine B, Codogno P. 2009. Ceramide-induced autophagy: to junk or to protect cells? Autophagy 5:558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pattingre S, et al. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939 [DOI] [PubMed] [Google Scholar]
- 49. Reggiori F, et al. 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7:500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Romanowski MJ, Garrido-Guerrero E, Shenk T. 1997. pIRS1 and pTRS1 are present in human cytomegalovirus virions. J. Virol. 71:5703–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ropolo A, et al. 2007. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J. Biol. Chem. 282:37124–37133 [DOI] [PubMed] [Google Scholar]
- 52. Shi CS, Kehrl JH. 2008. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 283:33175–33182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sinha S, Colbert CL, Becker N, Wei Y, Levine B. 2008. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy 4:989–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Smith JD, de Harven E. 1978. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells. III. Cytochemical localization of lysosomal enzymes in infected cells. J. Virol. 26:102–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Talloczy Z, et al. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 99:190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Talloczy Z, Virgin HW, Levine B. 2006. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2:24–29 [DOI] [PubMed] [Google Scholar]
- 57. Vicencio JM, et al. 2009. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1. Cell Death Differ. 16:1006–1017 [DOI] [PubMed] [Google Scholar]
- 58. Virgin HW, Levine B. 2009. Autophagy genes in immunity. Nat. Immunol. 10:461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X, et al. 2007. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat. Cell Biol. 9:470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang X, Sonenshein GE. 2005. Induction of the RelB NF-kappaB subunit by the cytomegalovirus IE1 protein is mediated via Jun kinase and c-Jun/Fra-2 AP-1 complexes. J. Virol. 79:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. 2008. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 30:678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Young AR, et al. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119:3888–3900 [DOI] [PubMed] [Google Scholar]
- 63. Zalckvar E, et al. 2009. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 10:285–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhong Y, et al. 2009. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11:468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu H, et al. 2009. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 5:816–823 [DOI] [PMC free article] [PubMed] [Google Scholar]