Abstract
Alphavirus budding from the plasma membrane occurs through the specific interaction of the nucleocapsid core with the cytoplasmic domain of the E2 glycoprotein (cdE2). Structural studies of the Sindbis virus capsid protein (CP) have suggested that these critical interactions are mediated by the binding of cdE2 into a hydrophobic pocket in the CP. Several molecular genetic studies have implicated amino acids Y400 and L402 in cdE2 as important for the budding of alphaviruses. In this study, we characterized the role of cdE2 residues in structural polyprotein processing, glycoprotein transport, and capsid interactions. Along with hydrophobic residues, charged residues in the N terminus of cdE2 were critical for the effective interaction of cores with cdE2, a process required for virus budding. Mutations in the C-terminal signal sequence region of cdE2 affected E2 protein transport to the plasma membrane, while nonbudding mutants that were defective in cdE2-CP interaction accumulated E2 on the plasma membrane. The interaction of cdE2 with cytoplasmic cores purified from infected cells and in vitro-assembled core-like particles suggests that cdE2 interacts with assembled cores to mediate budding. We hypothesize that these cdE2 interactions induce a change in the organization of the nucleocapsid core upon binding leading to particle budding and priming of the nucleocapsid cores for disassembly that is required for virus infection.
INTRODUCTION
Alphaviruses, a genus within the Togaviridae family, comprise medically significant pathogens that include western, eastern, and Venezuelan equine encephalitis viruses (VEEV), Chikungunya virus (CHIKV), and Sindbis virus (SINV). These are enveloped arthropod-borne RNA viruses causing diseases ranging from encephalitis to polyarthritis, and they have a wide variety of vertebrate hosts, including humans (31, 38). SINV is the prototype alphavirus transmitted by mosquitoes.
Cryo-electron microscopy (cryo-EM) reconstructions of alphaviruses show the arrangement of the structural proteins in the alphavirus particle (3, 18, 27, 31, 44, 51, 52, 54). SINV has an external diameter of 700 Å and contains 240 copies each of the E2 (423 amino acid residues), E1 (439 amino acid residues), and capsid protein (CP; 64 amino acid residues), all of which are arranged with icosahedral T=4 quasisymmetry (2). A small protein referred to as 6K (55 amino acid residues) is found in substoichiometric amounts in the particle (10, 25). The 11,703-nucleotide (nt) positive-sense viral RNA genome is encapsidated by CPs in the cytoplasm of infected cells to form a nucleocapsid core (NC). The viral envelope is derived from the host plasma membrane (38). The envelope transmembrane glycoproteins E2 and E1 constitute the outer protein shell with spikes formed from a trimer of E2-E1 heterodimers; 80 such spikes are arranged on the icosahedral lattice that overlaps with the NC (3, 34).
Structural proteins are translated from a subgenomic 26S mRNA as a single polyprotein, which is processed cotranslationally into CP, E3, E2, 6K, and E1. E2 is responsible for receptor binding and cell entry and E1 is responsible for cell fusion (37, 52). The newly synthesized CP transiently interacts with ribosomes and finally complexes with genomic RNA resulting in the accumulation of NCs in the cytoplasm. The specific encapsidation of genomic RNA is determined by the interaction of the CP RNA recognition region and specific packaging signal on the RNA. 6K has been suggested to be an ion channel that is involved in virion budding (26). Eventually, budding is initiated by CP-glycoprotein interactions and the nucleocapsid buds through the cell plasma membrane (39, 40).
E2 has a 260-amino-acid-long ectodomain, followed by about 100 amino acids in a stem region and a 30-amino-acid-long transmembrane helix. The 33-amino-acid carboxy-terminal cytoplasmic domain of E2 (cdE2, endodomain) interacts with the NC core (13, 19, 36, 53). There is one-to-one contact between the glycoprotein and the CP across the membrane bilayer through the cdE2 (3, 8, 31). Six carboxy-terminal residues of E1 extend past the inner lipid leaflet into the interior cavity of the virus (27), and mutational analyses ruled out a role of the E1 C-terminal residues in budding (1).
The alphavirus CP has three functional regions: I, II, and III (5). Residues 1 to 80 of region I have been implicated in nonspecific binding, charge neutralization with the viral genomic RNA, and contain a conserved helix, which plays a regulatory role during NC core assembly (15, 32, 33). Amino acids 81 to 113 in region II are involved in specific binding to the encapsidation signal on viral genomic RNA (22, 46, 47). Amino acids 114 to 264 (region III) form the serine protease domain (5). A hydrophobic pocket found within this domain is important for the formation of virus particles (19, 36). In a crystal structure of the SINV CP, amino-terminal arm residues 108 to 111 (L-X-L) were found to bind into the pocket of the neighboring protein composed of residues Y180, W247, and F166 (4, 19). Y400 in cdE2 was found to be important for binding to the CP pocket by hydrophobic interaction (1, 36, 55). This binding involves a conserved Y-X-L tripeptide that is similar to the L-X-L region of the N-terminal arm of the CP. The role of the tripeptide was confirmed by mutational analyses (29). Insertion of the cdE2 residues into the hydrophobic pocket stabilizes the assembled virion and causes a conformational change in the NC such that the core is primed for disassembly (19). The hydrophobic pocket was identified when molecular models of the Semliki Forest virus (SFV) CP and its cdE2 were used to characterize the interaction between these two proteins (36). The essential Tyr residue of the cdE2 could be bound efficiently by aromatic residues in the CP, providing both specificity for spike incorporation and energy for budding by hydrophobic interactions. Three conserved Cys residues and a unique Tyr residue in the cdE2 are modified by palmitoylation and transient phosphorylation, respectively (16, 23). The C-terminal 16 residues of cdE2 have also been implicated in the binding of capsid in an in vitro study using CP constructs and synthetic cdE2 peptides reconstituted into phospholipid vesicles (50).
Reorientation of the C terminus of cdE2 to the cytoplasmic side of the membrane takes place during the transport of E2 to the plasma membrane, and cysteine palmitoylation of cdE2 is believed to be involved in this process (38). The palmitoylated cysteines in cdE2 are essential for the membrane anchoring and correct orientation of cdE2 on the CP (51). The binding of cdE2 into the hydrophobic pocket of the CP is aided by the palmitoylated cysteines 416 and 417. Mutagenesis of these cysteine residues resulted in aberrant particle formation, suggesting that the native structure of cdE2 is essential for proper assembly (16). It has been proposed that cdE2 contains two budding determinants: the first includes Y400 and the second involves the palmitoylated cysteine residues. The function of the latter is to anchor the C terminus of cdE2 against the inner surface of the membrane so that the tyrosine-containing motif is properly presented to the nucleocapsid (55). It was reported earlier that residues from A403 to S420 of cdE2 are important for the specificity of the CP interaction (24). Evidence for the interaction of the C terminus of cdE2 with CP was obtained from genetic studies of revertants, where revertants for a cdE2 mutant were found near the hydrophobic pocket of the capsid (35). The atomic structures of the E1 ectodomain, the capsid protein, and most recently the E2 ectodomain (20, 45) have been fitted into a 7-Å cryo-EM reconstruction of SINV, generating a partial pseudoatomic structure of the virus and identifying the cdE2 density inside the pocket (41).
In alphaviruses, the molecular interactions driving budding are poorly understood, but several lines of evidence suggest that the association between envelope protein cytoplasmic domains and CP plays an important role. In this study, a series of experiments that include molecular genetics, biochemical, and biophysical assays were conducted to investigate the functional roles of cdE2 and its interactions with NC that promote virus assembly and budding. Our findings shed light into the specificity of cdE2 in virus budding. Consistent results obtained from analytical ultracentrifugation (AUC), surface plasmon resonance (SPR), and coimmunoprecipitations (co-IPs) confirmed that in vitro-assembled core-like particles (CLPs) and cytoplasmic cores specifically interact with cdE2. The assays developed in this study could be used to provide greater insight into the budding process and to screen for inhibitors of alphavirus budding.
MATERIALS AND METHODS
Viruses and cells.
BHK-15 cells were propagated in Eagle minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in the presence of 5% CO2. All CP and cdE2 clones were constructed from pToto64, a full-length cDNA clone of SINV that has been previously described (30).
Plasmids and cloning.
The cdE2 mutations were generated in pToto64, using standard overlap PCR mutagenesis procedures. The PCR products were digested with XbaI and BsiWI and inserted into a similarly digested pToto64 vector to generate pToto64–cdE2 mutants. For the construction of Escherichia coli expression plasmids of cdE2, synthetic complementary oligonucleotides with BamHI and XhoI overhangs at the 5′ and 3′ ends, corresponding to the cdE2 amino acids (391 to 423 of E2), were cloned into the BamHI and XhoI sites of the E. coli expression vector pET-GFP (provided by Dinesh Yernool). Table 1 lists the clones used in the present study.
Table 1.
Mutations in SINV cdE2 and GFP-cdE2 fusion proteins
| Construct | cdE2 sequence |
|---|---|
| pToto 64 constructsa | |
| WT cdE2 (391-423) | KARRECLTPYALAPNAVIPTSLALLCCVRSANA |
| 393RRE395/VLA | ..VLA............................ |
| 393RRE395/A3 | ..AAA............................ |
| 397LTPYAL402/A6 | ......AAAAAA..................... |
| 400YAL402/A3 | .........AAA..................... |
| Δ 403APNAVI408 | ............ΔΔΔΔΔΔ............... |
| 409PTSLALL415/A7 | ..................AAAAAAA........ |
| 412LALL415/A4 | .....................AAAA........ |
| 416CC417/A2 | .........................AA...... |
| pET-GFP constructsb | |
| GFP-cdE2 | KARRECLTPYALAPNAVIPTSLALLCCVRSANA |
| GFP-cdE2 391KARREC396/A6 | AAAAAA........................... |
| GFP-cdE2 397LTPYAL402/A6 | ......AAAAAA..................... |
| GFP-cdE2 403APNAVIP409/A7 | ............AAAAAAA.............. |
| GFP-cdE2 410TSLALL415/A6 | ...................AAAAAA........ |
| GFP-cdE2 416CCVRSANA423/A8 | .........................AAAAAAAA |
| GFP-randomized peptide | ANLALPRSLRICVLPEKALSCAVARNATATYCP |
Mutants were constructed in the cdE2 region of full-length SINV cDNA clone pToto64.
Wild-type and mutant cdE2 peptides were purified as C-terminal fusions behind monomeric GFP which has an N-terminal His tag.
In vitro transcription and transfection.
The wild-type (WT) and mutant cDNA clones were linearized with SacI and in vitro transcribed with SP6 RNA polymerase, transfected into BHK-15 cells by electroporation as previously described (28). Infectious virus produced from the transfected cells was quantified by standard plaque assay using medium over cells collected at 24 h postelectroporation. The presence of cdE2 mutations in each virus was confirmed by sequencing the reverse transcription (RT)-PCR products corresponding to the E2 coding region from RNA purified from cytoplasmic extracts of infected BHK-15 cells.
Analysis of structural polyprotein processing.
Expression and processing of structural proteins were analyzed by performing Western immunoblot analysis on lysates of BHK-15 cells electroporated with Toto64 RNA. Cells were lysed using lysis buffer (TNE [25 mM Tris-HCl (pH 7.4), 100 mM NaCl, and 5 mM EDTA] buffer with 1% Triton X-100) at 12 h postelectroporation, and samples were separated on a 10% or 12% bis-Tris precast SDS-PAGE gel (Bio-Rad). The proteins were subsequently electrophoretically transferred to nitrocellulose membranes and probed either separately or in combination with SINV-specific rabbit polyclonal antibodies, anti-E2 (a kind gift from J. H. Strauss), anti-CP, and anti-E1, as described previously (28, 29, 32). Mouse anti-actin monoclonal antibody (Chemicon) was used as an internal control. Infrared-labeled (IRDye 680 and IRDye 800) goat anti-rabbit and goat anti-mouse secondary antibodies (Li-Cor) were used for the detection of proteins, the blots were scanned with the Odyssey infrared imager (Li-Cor, Lincoln, NE), and protein bands were quantified using Odyssey software version 3. Pulse-chase experiments were carried out as described previously (29) with the following modifications. The medium was replaced at 12 h postelectroporation with 1.5 ml MEM and 5% FBS plus actinomycin D (1.5 μg/ml), and cells were incubated for 2.5 h. After the pulse-chase, the cell lysate was immunoprecipitated using an E2 polyclonal antibody.
IF assay.
Immunofluorescence (IF) was performed on BHK-15 cells electroporated with Toto64 or mutant RNAs as described in “In vitro transcription and transfection” above, and the cells were grown on glass coverslips. Primary antibodies used in the experiments were SINV-specific rabbit polyclonal anti-CP, rabbit polyclonal anti-E1, rabbit polyclonal anti-E2, mouse monoclonal antibodies anti-E2 202, and 127, and rabbit polyclonal anti-giantin (Abcam). IF was performed as previously described (29). The cells were fixed using methanol for permeabilization and cells were fixed with 3.7% paraformaldehyde for the detection of surface expression of proteins. The secondary antibodies used were fluorescein isothiocyanate (FITC)- or tetramethyl rhodamine (TRITC)-conjugated goat anti-rabbit and goat anti-mouse secondary antibodies (Pierce) in phosphate-buffered saline (PBS) (10 mg/ml bovine serum albumin [BSA]). Images were acquired using a Nikon A1R confocal microscope with the 60× oil objective with 1.4 numerical aperture (NA). Images were processed using the NES software, and the brightness and contrast were adjusted using nonlinear lookup tables (LUTs).
FC.
Transport and expression of E2 to the plasma membrane in cdE2 mutants were assayed using flow cytometry (FC) using an anti-E2 antibody (28). Briefly, BHK-15 cells were electroporated with <10 μg of in vitro-transcribed wild-type Toto64 or Toto64 cdE2 mutant RNAs as described in “In vitro transcription and transfection” above. The cells were trypsinized at 12 h posttransfection and resuspended in MEM supplemented with 10% FBS. Cells were washed two times with PBS supplemented with 1% FBS and incubated on ice for 1 h with a 1:100 dilution of a polyclonal antibody against the ectodomain of E2. Subsequently, the cells were washed thrice with PBS (1% FBS) and incubated on ice in the dark for 30 min with a fluorescein-conjugated goat anti-rabbit secondary antibody. The cells were washed thrice with PBS (1% FBS) and suspended in 500 μl of PBS. The cells were analyzed using a FACSCalibur flow cytometer (BD Biosciences, CA). Control staining was performed with mock-transfected cells.
Transmission electron microscopy.
BHK-15 cells were electroporated with in vitro-transcribed RNA from wild-type Toto64 and Toto64 cdE2 mutants as described in “In vitro transcription and transfection” above. Cells were fixed at 12 h posttransfection in a 0.1 M cacodylate buffer (0.1 M Na-cacodylate, 2 mM MgCl2, 2 mM CaCl2, and 0.5% NaCl at pH 7.4) containing 2% glutaraldehyde for 10 min. Cells were then rinsed twice with 0.1 M cacodylate buffer twice for 5 min. Cells were then postfixed with 1% reduced osmium solution (1% osmium tetroxide in 1.5% potassium ferrocyanide) for 5 min and then rinsed in deionized water three times. Cells were then scraped and pelleted in 1.5% low-melting-point agarose gel. Pellets were dehydrated in sequential ethanol concentrations of 10, 30, 50, 70, 90, and 100% for 30 min apiece. The pellets were then washed with 100% acetone twice for 1 h each. Cells were then infiltrated with Durcupan resin in acetone (25% 2 h, 50% overnight, 75% 2 h, and 100% 6 h). Samples were put into blocks to polymerize for 72 h in a 60°C oven. Cell sections were cut to 100 nm on a Leica EM UC7 microtome with a Diatome diamond knife. Sections were collected on 200-mesh carbon-coated copper grids. Sections were post-embed stained with 2% uranyl acetate for 5 min and Sato's lead solution for 1 min. Grids were imaged on an FEI/Philips CM-10 Bio-Twin transmission electron microscope (FEI Company, Hillsboro, OR) at 80 kV.
Purification of CP and nucleocapsid cores.
A truncated CP construct of SINV [CP(19–264)] was expressed in E. coli, purified, and used to assemble core-like particles (CLPs) as previously described (42). For the purification of CLPs, 1 ml of the in vitro assembly reaction mixture was layered onto 10 ml of 0-to-54% Optiprep (60% [wt/vol] solution of iodixanol in water; Sigma) gradient prepared with assembly buffer (25 mM HEPES [pH 7.4], 100 mM potassium acetate, 1.7 mM magnesium acetate). Intracellular NCs (cytoplasmic cores) were isolated from virus-infected cells as previously described (7, 24) and purified using 0-to-54% Optiprep gradient in TNE buffer (25 mM Tris-HCl [pH 7.4], 100 mM NaCl, and 5 mM EDTA) with 0.1% Triton X-100. Both gradients were spun for 2 h at 32,000 rpm at 4°C in an SW41 rotor and fractionated. The presence of cores was assayed by SDS-PAGE. NCs from virus (virus-associated cores) were prepared from purified SINV after NP-40 treatment (7, 11, 24, 34). In an attempt to understand the effects of detergent at different pH conditions, purified virus was treated with β-d-octylglucoside (12) at pH 7.4 (25 mM Tris [pH 7.4], 50 mM NaCl, 5 mM EDTA, and 22 mM β-d-octylglucoside) and at pH 5.5 (100 mM MES [morpholineethanesulfonic acid] [pH 5.5], 50 mM NaCl, 5 mM EDTA, and 22 mM β-d-octylglucoside), and the detergent-treated virus was separated on a density gradient as explained above. Alternatively, different detergents were tested for their ability to solubilize the membrane and completely remove the glycoproteins from the NC. The gradient fractions were analyzed for the presence of E1, E2, and CP on SDS-PAGE followed by Coomassie staining and Western analysis. CLPs, cytoplasmic NCs, and virus-associated cores were concentrated from the fraction using a 100-kDa cutoff concentrator (Amicon Ultra), and their integrity was analyzed by SDS-PAGE, analytical ultracentrifugation, and cryo-electron microscopy.
Purification of cdE2 fusion proteins from E. coli.
For the purification of green fluorescent protein (GFP)-cdE2 fusion proteins, plasmids were transformed into E. coli expression strain Rosetta (DE3) and the cultures were induced with 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the cell density reached 0.5 at A600, and the cultures were grown for 16 h after induction at 19°C. The cells were spun at 5,000 × g at 4°C and then resuspended in binding buffer (20 mM Tris-HCl [pH 7.8], 40 mM imidazole, 300 mM NaCl) and lysed by sonication (pulse, 20 s on and 20 s off; total, 10 min). The cell lysates were centrifuged at 16,000 × g at 4°C for 40 min, and the supernatant was loaded onto a 1-ml HisTrap FF column (GE Healthcare) connected to an Äkta and purified according to the manufacturer's instructions. The integrity of the purified fusion proteins was tested by Western analysis using rabbit polyclonal anti-cdE2 antibody (kindly provided by Milton Schlesinger), penta-His mouse monoclonal antibody (Qiagen), and anti-GFP mouse monoclonal antibody (Molecular Probes).
Co-IP.
For co-IP, purified GFP proteins were mixed with NCs and CLPs in a 1:1 molar ratio (20 mM each CP and GFP) in 100 μl TNE buffer and allowed to interact for 4 h at 4°C. Subsequently 1 μl anti-penta-His mouse monoclonal antibody (Qiagen) was added to the protein mixture, the volume was brought up to 200 μl with TNE buffer, and the mixture was rocked overnight at 4°C. Forty microliters of protein A agarose or Pansorbin (Calbiochem) was added and incubated for 3 h at 4°C. This mixture was spun for 1 min at 14,000 × g, and the pellet was washed with TNE buffer. The pellet was then resuspended in 30 μl of SDS-PAGE buffer (Bio-Rad), boiled for 5 min, and spun at 16,000 × g for 1 min; 25 μl of the supernatant was separated on a 12% bis-Tris precast SDS-PAGE gel (Bio-Rad); and Western analysis was conducted using anti-CP antibody as explained in “Analysis of structural polyprotein processing” above. His tag pulldown was conducted using GFP-cdE2 fusion proteins with the N-terminal His tag and cytoplasmic and virus-associated cores using the Profound pulldown poly-His protein-protein interaction kit (Pierce) according to the manufacturer's instructions. The proteins that were pulled down were analyzed by Western blotting using anti-His antibody (Qiagen) and anti-CP antibody.
AUC.
Reaction mixtures of CP and GFP-cdE2 were subjected to sedimentation velocity (SV) studies in a Beckman XLI analytical ultracentrifuge using both Rayleigh interference and absorbance optical systems. Four hundred twenty microliters of reaction mixture was placed in the sample sector of a two-sector carbon-filled centerpiece and 420 μl of reference buffer in the other sector. The 60 Ti four-hole rotor was loaded, placed in the centrifuge, and allowed to equilibrate under vacuum at 20°C for at least 1 h. The rotor was accelerated to 50,000 rpm and data scans were collected at 5-min intervals until the main boundary had reached the bottom of the cell. CLPs and NCs were also studied by AUC. Because these particles sediment with sedimentation coefficients between 60S and 200S, lower speeds of 10,000 to 20,000 rpm were used to get adequate data for analysis. Scans were collected at 1.5- or 3-min intervals. All of the SINV data were analyzed using either c(s) or lsg*(s) methods from SEDFIT or SEDPHAT, which were obtained from Peter Schuck (NIH) (www.analyticalultracentrifugation.com). Loading concentrations were calculated by integrating the peaks of interest and used to normalize the curves. Extinction coefficients were calculated for the Sindbis RNA-core particle from its RNA and CP sequences and for CLPs consisting of 240 copies of both CP(19–264) and a 48-mer of single-stranded DNA (ssDNA-48mer) using an oligonucleotide calculator (http://www.basic.northwestern.edu/biotools/OligoCalc.html) and SEDNTERP (http://www.jphilo.mailway.com/). These values are 156 A260/μM and 146 A260/μM, respectively, for a 1.2-cm path length.
SPR studies.
The effect of GFP-cdE2 mutations on the rate of binding to CP was studied using SPR. The CP was bound transiently to a biotin-labeled 48-mer of single-stranded DNA sequestered on a streptavidin-coated (SA) Biacore chip. SPR studies were conducted using a Biacore 3000 (GE Healthcare). Assembly buffer containing added surfactant (0.005% Tween 20) was filtered and degassed prior to being used in these studies to increase the stability of the CP-oligonucleotide complex. The biotin-labeled 48-mer DNA was purchased from Integrated DNA Technologies (IDT) (42). The CP was preloaded onto the DNA-containing surface at a molar level approaching 5 CP:DNA and then allowed to dissociate until the ratio approached unity. The GFP-cdE2 proteins were injected at this point, and association and dissociation followed for 15 min. The resulting sensorgrams were analyzed using BIA Evaluation software to transform the data into a useful format; Axum was used for graphical purposes.
RESULTS
Mutations in the cytoplasmic domain of E2.
Mutational analysis of cdE2 was conducted to interrogate the regions of cdE2 involved in the multiple roles carried out by this E2 domain. Although numerous mutational studies of the cdE2 region have been reported, a comprehensive interaction map of the entire region is not available. To date, the information obtained by mutation of cdE2 is mainly for the region surrounding the conserved Tyr 400 (SINV numbering) and the palmitoylated cysteine region. Our study was designed to identify the interaction between cdE2 and the nucleocapsid core leading to virus budding and to develop a reliable and quantifiable in vitro assay for cdE2-CP binding. The mutants generated in this study are listed in Table 1. The cdE2 sequence was divided roughly into five regions based on the distribution of conserved cysteines and prolines. The results of mutations in cdE2 are summarized in Table 2, with the majority of the mutants severely affected in terms of virus budding.
Table 2.
Summary of the phenotypic characteristics of cdE2 mutants
| Virus | Plaque sizea | Virus titer (PFU/ml)b | Structural polyprotein processingc | E2 transport to PMd | Virus release measured by qRTe |
|---|---|---|---|---|---|
| WT | LP | 1.2 × 109 | + | + | 100% |
| 393RRE395/A3 | VSP | 2.8 × 102 | + | + | 0.22% |
| 397LTPYAL402/A6 | VSP | 8.0 × 101 | + | + | 0.14% |
| 400YAL402/A3 | VSP | 1.2 × 103 | + | + | 0.70% |
| Δ 403APNAVI408 | SP | 1.2 × 102 | + | + | 0.32% |
| 409PTSLALL415/A7 | NP | − | D | ND | 0.50% |
| 412LALL415/A4 | NP | − | D | ND | 0.15% |
| 416CC417/A2 | VSP | 1.6 × 102 | + | + | 0.24% |
Plaque sizes relative to the wild type are as follows: LP, large plaque (3.5 to 4 mm); SP, small plaque (1 to 2 mm); VSP, very small plaque (≤1 mm); NP, no plaques. Plaque sizes corresponding to the diameter of plaques were measured subsequent to electroporating BHK-15 cells, incubating for 24 h at 37°C, harvesting the medium, and assaying the production of progeny viruses by standard plaque assay after incubation at 37°C for 48 h.
Virus titers were determined by maintaining electroporated BHK cells under liquid medium at 37°C for 24 h, harvesting the medium, and assaying the production of progeny viruses by standard plaque assay. −, no virus production was observed.
Determined by Western analysis using anti-E2 and anti-CP antibodies. D, E2 processing was defective.
Determined by IF and FC. ND, not detected.
Viral RNA molecules released into the medium determined by qRT. Numbers represent percentages relative to WT RNA.
Structural polyprotein processing in cdE2 mutants.
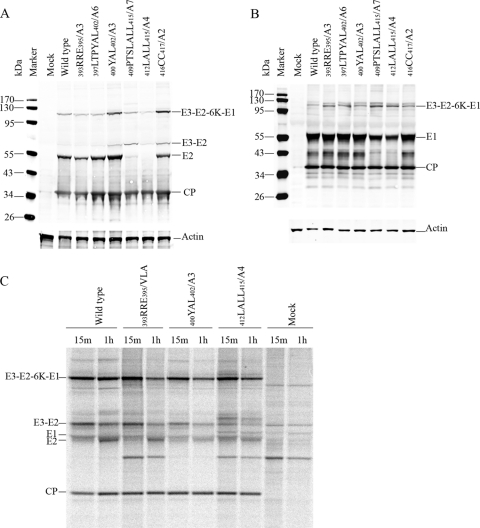
The structural polyprotein processing of cdE2 mutants was analyzed by Western blotting and pulse-chase experiments to assess cleavage intermediates and products (Fig. 1). Mutations were designed such that the signalase cleavage site between E2 and 6K was not altered. Multiple residue substitutions or deletions were generated in order to analyze the collective roles of amino acid side chains in the CP-cdE2 interaction and subsequent budding. The E2 glycoprotein processing of the representative cdE2 mutants is shown in Fig. 1A. The N-terminal cdE2 mutations (393RRE395/A3, 397LTPYAL402/A6, and 400YAL402/A3) were processed like wild-type virus, and the E3-mediated translocation of the polyprotein into the ER was not defective (Fig. 1A). The processed E2 band was reduced in the signal sequence mutants 409PTSLALL415/A7 and 412LALL415/A4, and these mutants showed the accumulation of the unprocessed E3-E2 band. The C-terminal cysteine mutation (416CC417/A2) showed the correct polyprotein processing with some accumulation of the E3-E2 band. The processing of E1 glycoprotein in the wild type and representative cdE2 mutants is shown (Fig. 1B). Although the E1 glycoprotein was processed as for the wild type in all the cdE2 mutants, the amount of E1 was reduced for mutants 409PTSLALL415/A7 and 412LALL415/A4. These experiments were conducted three times with consistent results. Additional protein bands were observed for the mutant 412LALL415/A4 in the pulse-chase analysis, presumably corresponding to the cleavage intermediates E3-E2-6K and E2-6K (Fig. 1C). We hypothesize that this processing defect is due to the failure of the cdE2 signal sequence in the C terminus (409PTSLALLC416) (51) to translocate 6K to the ER lumen, leaving the signalase cleavage site between PE2 and 6K uncleaved, also affecting the orientation of E1 on the ER membrane.
Fig 1.
Analysis of translation and processing of the structural polyprotein. (A) Processing of E2 glycoprotein in BHK-15 cells electroporated with in vitro-transcribed RNA from the wild type (SINV) or mutant clones. At 12 h posttransfection, the cells were harvested and equal amounts of proteins were separated by 10% SDS-PAGE and transferred to nitrocellulose membrane. The Western blots were processed with SINV-specific rabbit polyclonal anti-E2 and anti-CP and mouse monoclonal anti-actin primary antibodies and infrared-labeled (IRDye 680 and IRDye 800) goat anti-rabbit and goat anti-mouse secondary antibodies. (B) Processing of E1 glycoprotein in BHK-15 cells electroporated with in vitro-transcribed RNA from wild-type (SINV) or mutant clones done as for panel A, except that the proteins were separated by 12% SDS-PAGE gel and the Western blots were processed with SINV-specific rabbit polyclonal anti-E1 and anti-CP and mouse monoclonal anti-actin primary antibodies. (C) Pulse-chase analysis of structural polyprotein processing. BHK-15 cells were electroporated with in vitro-transcribed RNA from the wild-type and selected nonbudding mutant clones. At 12 h postelectroporation, the cells were pulsed with [35S]methionine for 15 min and chased for 1 h (pulse, 15 min; chase, 1 h). The cells were lysed and cell lysates were immunoprecipitated using SINV-specific rabbit E2 polyclonal antibody and subjected to 10% Bis-Tris gel SDS-PAGE and autoradiography.
Glycoprotein transport to the plasma membrane is normal in processed cdE2 mutants.
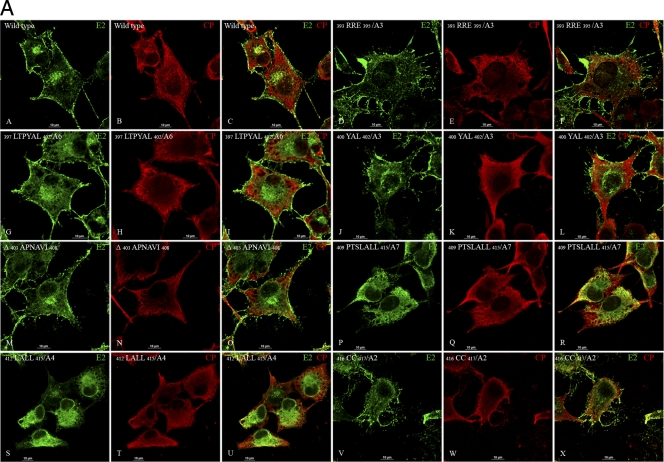
Glycoprotein transport to the plasma membrane and CP-E2 colocalization was examined by immunofluorescence (IF) of the transfected cells (Fig. 2A to C). The colocalization of CP and E2 was studied by confocal microscopy analysis of CP- and E2-stained cells (Fig. 2A). Colocalization of E2 and CP was observed at the plasma membrane in wild-type virus (Fig. 2A, panels A to C). The N-terminal mutant (393RRE395/A3) trafficked E2 normally to the plasma membrane (Fig. 2A, panels D to F). E2 in the hydrophobic pocket-binding mutants (397LTPYAL402/A6 and 400YAL402/A3) was transported to the plasma membrane (Fig. 2A, panels G to L). The deletion mutant Δ 403APNAVI408 showed localization and transport of E2 similar to that of the wild-type virus (Fig. 2A, panels M to O). Interestingly, wild-type-like E2 transport was not observed for the signal sequence mutants 409PTSLALL415/A7 and 412LALL415/A4 (Fig. 2A, panels P to U). E2 was transported to the plasma membrane for the C-terminal 416CC417/A2 mutant (Fig. 2A, panels V to X). The IF experiments confirmed that cdE2 mutations in the N-terminal charged residues (393RRE395), hydrophobic pocket residues (397LTPYAL402), and C-terminal palmitoylated cysteines (416CC417) did not affect the transport of E2 to the plasma membrane and the colocalization of E2 and CP at the plasma membrane. However, the mutations in the C-terminal signal sequence of cdE2 (409PTSLALL415) abrogated the E2 transport to the plasma membrane and its colocalization with CP at the plasma membrane.
Fig 2.
Localization of E2, E1, and CP in SINV RNA transfected BHK-15 cells by immunofluorescence (IF) assay. (A) Localization of CP and E2 by IF analysis. BHK-15 cells were transfected by electroporation with wild-type or mutant in vitro-transcribed RNA. The cells were fixed using methanol at 6 h postelectroporation and subjected to IF using SINV-specific rabbit polyclonal anti-CP and mouse monoclonal anti-E2 antibodies. After the primary antibody washes, the cells were stained with TRITC-conjugated goat anti-rabbit (for CP; red) and FITC-conjugated goat anti-mouse (for E2; green) secondary antibodies. Merged images show the colocalization of E2 and CP. (B) Fluorescence microscopy images showing the distribution of E2 and its colocalization with Golgi marker giantin in BHK-15 cells transfected with in vitro-transcribed RNA from wild-type and cdE2 mutant clones. The cells were fixed with methanol at 6 h postelectroporation and stained with mouse monoclonal anti-E2 antibody and rabbit polyclonal anti-giantin antibody and TRITC-conjugated goat anti-mouse secondary antibody for E2 (red) and FITC-conjugated goat anti-rabbit antibody for giantin (green). The merged images show colocalization of E2 and giantin in yellow. (C) Fluorescence microscopy images showing the cell surface distribution of E2 and E1 in BHK-15 cells transfected with in vitro-transcribed RNA from wild-type and representative cdE2 mutant clones. The cells were fixed at 6 h postelectroporation with 3.7% paraformaldehyde and stained with mouse monoclonal anti-E2 antibody, rabbit polyclonal anti-E1 antibody, and FITC-conjugated goat anti-mouse antibody for E2 (green) and TRITC-conjugated goat anti-rabbit secondary antibody for E1 (red). The merged images show colocalization of E2 and E1 in yellow. Images were acquired using a Nikon A1R confocal microscope with the 60× oil objective with 1.4 NA. Images were processed using NES software, and the brightness and contrast were adjusted using nonlinear LUTs. Bars, 10 μm. Mock-infected cells were not included, as they did not demonstrate any detectable staining.
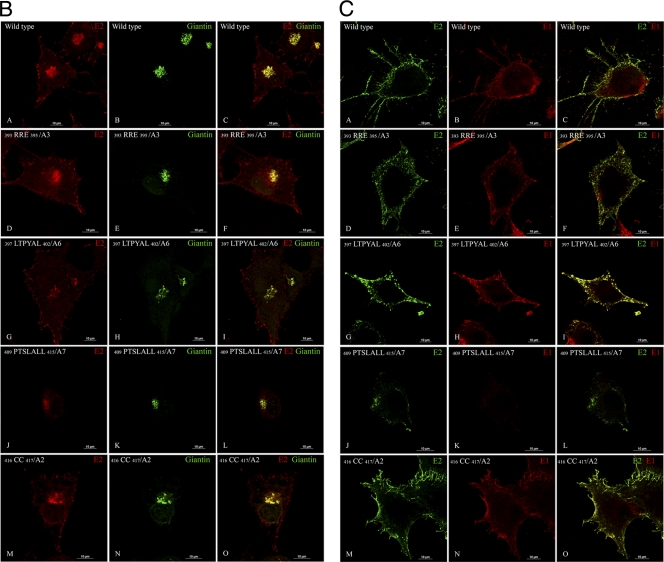
The E2 protein transport defect of some mutants was further confirmed by IF analysis using Golgi marker giantin and E2 antibodies (Fig. 2B). The E2 transport to the plasma membrane was comparable to the wild type (Fig. 2B, panels A to C) for the mutants 393RRE395/A3 (Fig. 2B, panels D to F), 397LTPYAL402/A6 (Fig. 2B, panels G to I), and 416CC417/A2 (Fig. 2B, panels M to O). The signal sequence mutant 409PTSLALL415/A7, which had a protein-processing defect, retained E2 in the ER and Golgi (Fig. 2B, panels J to L). The E2 and E1 surface expression was further analyzed by IF of the paraformaldehyde-fixed cells using anti-E1 and anti-E2 antibodies (Fig. 2C). The surface expression of E2 and E1 was similar to the that of the wild type (Fig. 2C, panels A to C) for the mutants 393RRE395/A3 (Fig. 2C, panels D to F), 397LTPYAL402/A6 (Fig. 2C, panels G to I), and 416CC417/A2 (Fig. 2C, panels M to O). The mutant 409PTSLALL415/A7 did not show the presence of E1 on the cell surface (Fig. 2C, panels J to L). The IF analyses of all the mutants were repeated three times with consistent results; only the representative images are shown.
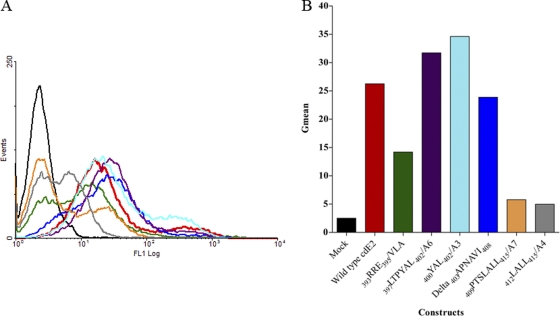
Glycoprotein transport to the plasma membrane was further examined by flow cytometry (FC) analyses (Fig. 3A and B). Although the N-terminal mutant with alanine substitutions (393RRE395/A3) trafficked E2 normally to the plasma membrane in IF, the mutant 393RRE395/VLA was slightly reduced in E2 transport as observed by FC (Fig. 3B). E2 in the mutants with processing defects, 409PTSLALL415/A7 and 412LALL415/A4, was not transported to the plasma membrane (Fig. 3A). In contrast, the nonbudding mutants 397LTPYAL402/A6 and 400YAL402/A3 accumulated E2 on the plasma membrane at levels comparable to those for the wild-type virus (Fig. 3B). The deletion mutant Δ 403APNAVI408 showed slightly reduced transport of E2 to the plasma membrane (Fig. 3B). The FC analysis was conducted only once. The flow cytometry data of the cdE2 mutants was consistent with the results obtained from the IF analysis.
Fig 3.
Analysis of the cell surface expression of E2. (A) BHK-15 cells were electroporated with in vitro-transcribed RNA from wild-type or mutant clones as described. At 12 h posttransfection, the cells were harvested and stained with rabbit polyclonal anti-E2 antibody and a fluorescein-conjugated secondary antibody and analyzed in a FACSCalibur flow cytometer. The black curve represents the signal from mock-transfected cells and red from the wild-type RNA-transfected cells. All the other curves show a shift from the mock and represent the expression of E2 at the cell surface. (B) Graphical representation of the geometric mean of the region of the curve measuring the amount of E2 fluorescence of each cdE2 mutant at the plasma membrane.
Virus budding is defective in the cdE2 mutants.
Virus release was significantly reduced compared to wild-type virus (∼106-fold reduced) in most of the cdE2 mutants (Table 2) as determined by plaque assay. Mutants 409PTSLALL415/A7 and 412LALL415/A4, which showed defective glycoprotein processing and transport, failed to produce plaques. The plaques formed were nonviable after one passage for all the mutants except for Δ 403APNAVI408. All of the other mutants produced very small plaques of less than 1 mm in diameter at 48 h posttransfection. None of these yielded escape mutants, and all retained the original mutations in the RNA sequence obtained from the transfected cells. The mutants were not defective in cytoplasmic core assembly as determined by the core accumulation assay (data not shown). Budding viruses were not observed in the transmission electron microscopy (TEM) analysis of the cdE2 mutants 393RRE395/VLA, 400YAL402/A3, and 412LALL415/A4, although accumulation of cytoplasmic cores was detected in the cell (Fig. 4). TEM analysis was conducted for all the cdE2 mutants listed in Table 1, and only the representative mutants are shown in Fig. 4. The number of viral RNA molecules released into the medium was also quantified for all the mutants using quantitative RT-PCR (qRT-PCR) and found to be reduced (∼103-fold reduction) compared to the wild-type virus (Table 2). The qRT experiments were conducted three times with consistent results. This apparent reduction in virus release was also observed in the number of infectious units released into the medium as determined from plaque assays (Table 2). The mutants that were defective in virus budding were classified as CP-cdE2 interaction-defective mutants are 393RRE395/A3, 397LTPYAL402/A6, and 400YAL402/A3. The deletion mutant also showed a minor interaction deficiency, as observed from the small plaque size and reduced virus titer, and may be caused by shortening the length of cdE2. The defect in the cysteine mutant has been attributed to the lack of palmitoylation (16), which is probably required for the reorientation of the C terminus of cdE2 to the cytoplasmic side of the membrane. The palmitoylated cysteines in cdE2 are located near the lipid head groups of the inner membrane of the lipid bilayer in the high-resolution cryo-EM structure of VEEV (51). The orientation of cysteines in the SINV cryo-EM structure (41) also shows that the cdE2 has indeed flipped back to the internal side of the lipid bilayer after palmitoylation. This suggests that the posttranslational palmitoylation of cysteines triggers the reorientation of the entire E2 C terminus (51). The 416CC417/A2 mutation presumably interferes with the accessibility of cdE2 to the NC core required for virus budding due to the reorientation defect. Consequently, this may also affect the orientation of the 400YAL402 domain to dock into the hydrophobic capsid pocket, leading to severe budding defects.
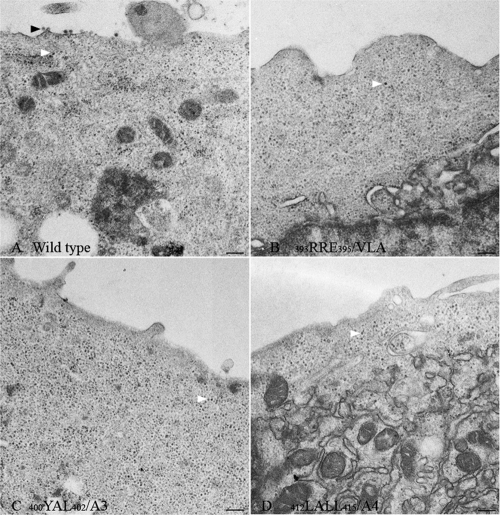
Fig 4.
Electron microscopy analysis of infected cells. Thin-section electron micrographs of BHK-15 cells transfected with wild-type or cdE2 mutant in vitro-transcribed RNA. Representative images of the wild type (A) and selected mutants 393RRE395/VLA (B), 400YAL402/A3 (C), and 412LALL415/A4 (D) are shown. Virus budding (black arrowheads) from the plasma membrane is observed in wild-type SINV-infected cell surface (A) and not from the nonbudding cdE2 mutants (B to D). Cytoplasmic cores are marked by white arrowheads. Bar, 200 nm.
Purification of in vitro-assembled CLPs, cytoplasmic cores, and virus-associated cores.
In order to study the specific interaction of CP with cdE2 in vitro, bacterially expressed CP was used. A truncated version of SINV CP [CP(19–264)] was purified, and CLPs were assembled in vitro using CP and 48-mer DNA according to standard protocols (42, 43). The CLPs were purified through Optiprep density gradient centrifugation to remove the free CP remaining from the assembly reaction. In infected cells, SINV CP assembles into cores with genomic RNA and accumulates in the cytoplasm, where they can be easily isolated as cytoplasmic cores. These cores interact with cdE2 during virus budding to form the assembled virions. Virus-associated cores can also be purified from the released virions by detergent treatment that removes the glycoproteins and lipid bilayer. For the purification of cores from virus, the commonly used detergent is NP-40. However, the absorbance of the detergent interfered with the analytical ultracentrifugation and SPR assays. To overcome this problem, several detergents with low absorbance at A280 were evaluated for their ability to solubilize the viral membrane and remove the glycoproteins while retaining the intact core structure. From the detergents used in the screening, LDAO (lauryldimethylamine-N-oxide; Anatrace) had these optimal properties and was selected for the purification of SINV cores (Fig. 5A). The authenticity of the cores purified using LDAO is shown in Fig. 5B and C. Purified SINV at a concentration of 4 mg/ml was treated with 2% LDAO for 1 h at room temperature, and the glycoproteins were separated from cores by density gradient centrifugation. This detergent was found to be able to remove glycoproteins effectively from cores at an optimal concentration of 87 mM (Fig. 5A). The glycoproteins sedimented in the first three fractions and the cores were confirmed by Western blotting using anti-cdE2 and anti-CP antibodies. The fractions where CP is found (7, 8, and 9) suggest that the cores were not disassembled. Figure 5B shows the Coomassie-stained SDS-PAGE of purified cores. Figure 5C is a cryo-electron microscopy image and confirms the size and shape of the virus-associated cores. While treatment of virus with the detergent β-d-octylglucoside at physiological pH did not remove E2 from the virus (Fig. 5D), using pH 5.5 under the same conditions successfully separated E2 from the cores (Fig. 5E), suggesting that charged interactions are involved in retaining E2 bound to the core in intact virions.
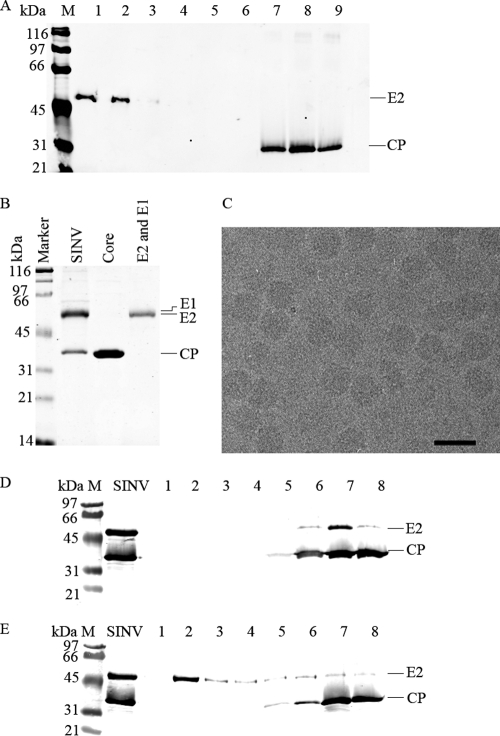
Fig 5.
Purification of NC cores from virus using the detergent LDAO. (A) Western analysis of density gradient fractions from detergent (LDAO)-treated SINV. The fractions are numbered from 1 to 9 from the top to the bottom of the density gradient. The bands were separated on a 10% SDS-PAGE gel, and the Western blot was probed with SINV-specific rabbit polyclonal anti-cdE2 and anti-CP antibodies. Fractions 1, 2, and 3 show the presence of E2 in the top of the gradient and its complete absence in fractions 7, 8, and 9. Cores are present in fractions 7, 8, and 9. (B) Purification of cores and glycoproteins from density gradient fractions of detergent-treated SINV. Coomassie-stained 12% SDS-PAGE showing purified virus (SINV lane 1); NCs purified from fractions 7, 8, and 9 from panel A (core lane); and glycoprotein fractions corresponding to lanes 1 and 2 of panel A purified from SINV using the detergent LDAO (E2 and E1 lane). (C) Cryo-electron microscopy of purified NCs. Bar, 50 nm. (D) Western analysis showing detergent treatment of purified SINV at pH 7.4 (25 mM Tris [pH 7.4], 50 mM NaCl, 5 mM EDTA, and 22 mM β-d-octylglucoside). Detergent-treated virus was separated on a density gradient and fractionated as explained in Materials and Methods. One-milliliter fractions collected from the top of the gradient are marked in lanes 1 to 8. The blot was probed with anti-cdE2 and anti-CP antibodies. (E) Same as panel D with the detergent treatment of purified Sindbis virus at pH 5.5 (100 mM MES [pH 5.5], 50 mM NaCl, 5 mM EDTA, and 22 mM β-d-octylglucoside).
Capsid protein and core-cdE2 interactions.
The interaction of cdE2 with CP, CLPs, and cytoplasmic cores was studied using pulldown assays. Coimmunoprecipitation experiments were conducted to detect the interaction of cytoplasmic cores with GFP-cdE2 and mutants. The GFP-cdE2 and NC complexes were immunoprecipitated using anti-His antibody against the His tag present on the N terminus of GFP. Western blot detection of the immunoprecipitated CP was performed with a polyclonal anti-CP antibody (Fig. 6A). The coimmunoprecipitation confirmed the interaction of cytoplasmic cores with GFP-cdE2. Wild-type cdE2 and all mutants except mutant 391KARREC396/A6 showed significant interaction with cytoplasmic cores. The negative control lane (GFP-randomized peptide) did not show any CP pulldown in Western analysis.
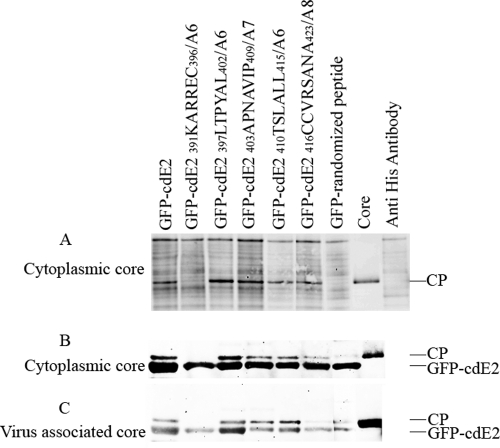
Fig 6.
Coimmunoprecipitation analysis of cdE2 fusion proteins with CP, core-like particles, and cytoplasmic cores. (A) Coimmunoprecipitation (co-IP) of CP and GFP-cdE2. The cytoplasmic cores purified from SINV-infected cells were mixed with GFP-cdE2 or mutant proteins, and the GFP-cdE2 and NC complexes were immunoprecipitated using anti-penta-His mouse monoclonal antibody directed against the N-terminal His tag of GFP. The proteins were detected by Western blot analysis using anti-SINV-specific rabbit polyclonal anti-CP antibody and infrared-labeled goat anti-rabbit secondary antibody. The negative control lane, labeled “core,” represents the core used for the experiment with no antibody, and the anti-His antibody lane represents the antibody used in the pulldown experiment with no cores. (B) His tag pulldown assay of cytoplasmic cores using GFP-cdE2 fusion proteins. The proteins were pulled down using Profound pulldown poly-His protein-protein interaction kit (Pierce) against the N-terminal His tag of GFP and were analyzed by Western blotting using SINV-specific rabbit polyclonal anti-CP and mouse monoclonal anti-penta-His primary antibodies and infrared-labeled goat anti-rabbit and goat anti-mouse secondary antibodies. (C) His tag pulldown assay of virus-associated cores purified from detergent-treated SINV using GFP-cdE2 fusion proteins; experiments were conducted the same way as for B.
A His tag pulldown assay was conducted using GFP-cdE2 and mutants with various cores to reconfirm the results obtained for the interaction of GFP-cdE2 with cytoplasmic and virus-associated cores without the use of antibodies. Cores were allowed to mix with GFP-cdE2, and components bound to His-tagged beads were coeluted after washing. The interaction of GFP-cdE2 and mutants with cytoplasmic cores (Fig. 6B) and with cores purified from virus (Fig. 6C) is shown. The interaction of GFP-cdE2 is observed with both cytoplasmic and virus-associated cores. Along with the negative control, mutant 391KARREC396/A6 showed no interaction with cytoplasmic cores or virus-associated cores. Unexpectedly, mutant 397LTPYAL402/A6 showed positive interaction with both cytoplasmic and virus-associated cores. Mutants 403APNAVIP409/A7, 410TSLALL415/A6, and 416CCVRSANA423/A8 showed reduced binding to NCs. The mutant 416CCVRSANA423/A8 did not show a significant interaction with virus-associated cores. Although the GFP-randomized peptide did not demonstrate an interaction with cytoplasmic cores in the coimmunoprecipitation assay, this mutant exhibited a reduced binding to virus-associated cores in the His tag pulldown assay.
SV studies of CLPs and cdE2.
Sedimentation velocity (SV) experiments were conducted using free CP, CLPs, or virus-associated cores together with a monomeric form of GFP-cdE2. The use of a monomeric GFP tag also facilitates the simultaneous detection of core (absorbance at 260 nm) and GFP-cdE2 (absorbance at 393 nm) in AUC. Normalized distribution plots are shown in each panel. The SV analyses of GFP-cdE2 (Fig. 7A, blue) showed that the molecule is a monomer with an s20,w of 2.1S and a mass of 32 kDa, which is slightly lower than the sequence mass, 36.3 kDa. The first c(s) distribution peak in Fig. 7A (red) is for CP(19-264) with a sequence mass of 27.2 kDa. The observed values for s20,w and mass are 1.7S and 28 kDa, respectively. Thus, in the absence of nucleic acid, CP(19-264) is also a monomer. A mixture of these two species, containing an excess of GFP-cdE2 (see below), produced a faster-sedimenting peak as shown by the normalized c(s) distribution for absorbance data collected at 393 nm (green), which is one of the absorbance maxima for GFP-cdE2. This species sediments with an s20,w of 2.6S with an apparent mass of 43 kDa. Absorbance data at 270 nm and interference data were also collected during this experiment. In neither the A270 nor the interference c(s) distributions are there a peak at 1.7S for the CP(19-264) species. However, an analysis of the loading concentrations of the 2.6S peak from the interference data and the A393 data indicates that the actual concentration of CP is approximately one-third that of the GFP-cdE2 present in the peak, suggesting that this peak is composed of mixture of GFP-cdE2 and a complex between GFP-cdE2 and CP(19-264). The presence of nonresolved free GFP-cdE2 explains the less-than-expected mass for the complex, since a weight-average mass is calculated by Sedfit for the complex. GFP alone does not bind detectibly to CP(19-264) by analytical ultracentrifugation (data not shown).
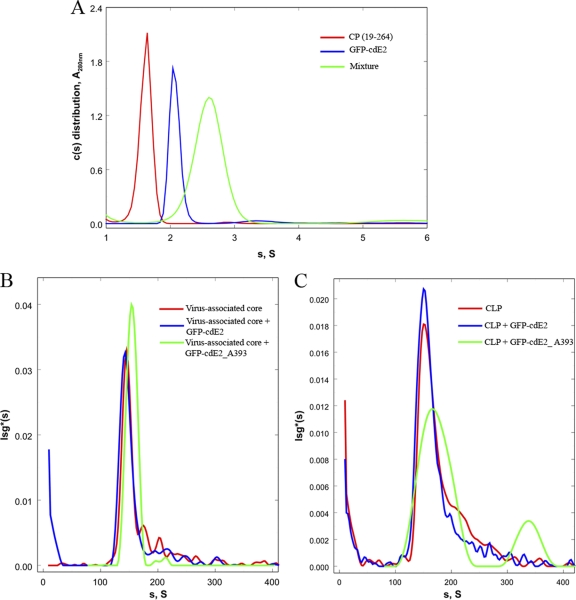
Fig 7.
Sedimentation velocity studies of CP, virus-associated core, and CLPs with GFP-cdE2. (A) Sedimentation velocity studies on the interaction between CP(19–264) and GFP-cdE2 are shown. Normalized sedimentation velocity c(s) distributions for CP(19–264) (red line), GFP-cdE2 (blue line) and a mixture (green) of the two containing an excess of GFP-cdE2 are shown. Scans were collected at 280 nm for all the runs and 393 nm for the samples containing the GFP-cdE2. Loading concentrations were calculated and used for calculating the normalized curves (CP, 0.95 A280; GFP-cdE2, 0.394 A393; and mixture, 0.051 A393). (B) Sedimentation velocity studies on the interaction of virus-associated core and GFP-cdE2 are shown. Normalized lsg*(s) distributions are shown for velocity scans collected at 260 nm for sedimentation of the virus-associated core (red) and for the mixture of this core plus GFP-cdE2 (blue). Also shown is the distribution for the scans collected at 393 nm (green). Loading concentrations were calculated and used for calculating the normalized curves (core, 0.13 A260; core plus GFP-cdE2, 0.093 A393; and mixture, 0.0002 A393). (C) Sedimentation velocity studies on the interaction of CLP and GFP-cdE2 are shown. Normalized lsg*(s) distributions are shown for velocity scans collected at 260 nm for sedimentation of both CLP (red) and a mixture of CLP and GFP-cdE2 (blue), and for the mixture of this core and GFP-cdE2 (green). Loading concentrations were calculated and used for calculating the normalized curves (CLP, 0.12 A260; core plus GFP-cdE2, 0.24 A393; and mixture, 0.003 A393).
The interaction between GFP-cdE2 and the virus-associated core, as well as the CLP from the assembly reaction between a DNA 48-mer and CP(19-264), were also studied by sedimentation velocity. In Fig. 7B, normalized lsg*(s) distributions collected in the absence of GFP-cdE2 and at 260 nm are shown for the virus-associated core (red line) and core in the presence of GFP-cdE2 (blue and green lines). The concentration of the 156S core alone was calculated from its loading concentration and an estimate of the extinction coefficient of this core based on the protein sequence and the RNA composition at about 0.8 nM virus-associated core. The peak (blue) in this panel was calculated from data where the sample also contained, in addition to RNA core, 6 μM GFP-cdE2. We estimate that the concentration of core in the mixture is about 0.6 nM. (Note that complete saturation of the core with GFP-cdE2 will contribute less than 10% to the total A260 of this peak). The normalized distribution A393 also shows a peak at 156S with a loading concentration of 0.0002 A393, which corresponds to a concentration of 1.7 nM bound GFP-cdE2 or about 3 GFP-cdE2/core. Since particles of >105 gm/mole are known to scatter UV light significantly, and both the virus-associated core and the CLP are considerably larger, ∼4 × 106 g/mole, we suspect most of the virus-associated core absorbance but not that for CLP (see below) results from light scattering by the larger core particles rather than bound GFP-cdE2. Hence, the virus-associated core likely does not interact detectably with GFP-cdE2. In Fig. 7C, normalized lsg*(s) distributions collected in the absence GFP-cdE2 and at 260 nm are shown for CLP (red line) and CLP in the presence of GFP-cdE2 (blue and green lines). The calculated loading concentration of the CLP for the 160S peak in this panel in the absence of GFP-cdE2 (red) is about 0.9 nM. For the study with GFP-cdE2, the calculated molar concentration of CLP in the peak (blue) was 1.5 nM. In addition, a normalized distribution peak (green) with a loading concentration of 30 nM (0.003 A393) is observed at 164S. We suggest that this peak represents the CLP-GFP-cdE2 complex, with about 20 GFP-cdE2 bound per core, which would increase the mass of the core by about 10%. Based on the judgment that the ratio of S values for two spheres increases by the two-thirds power of the increase in the ratio of their masses, we estimate that a core with 20 GFP-cdE2 bound should increase by no more than 10S. Hence, we conclude that GFP-cdE2 binds to both CP(19-264) and CLPs but not to virus-associated cores. The detergent Triton X-100 used for the isolation of cytoplasmic cores interfered with AUC and the cytoplasmic cores were not examined.
SPR of CLPs and cdE2.
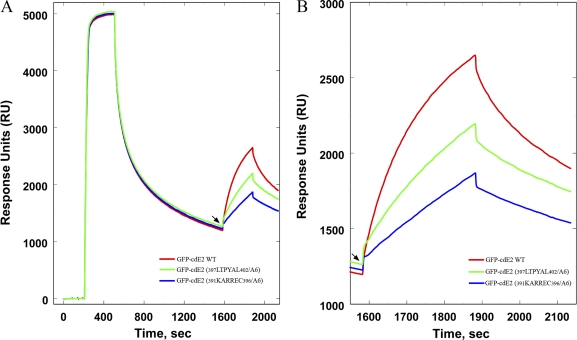
To quantitatively study the extent of interaction of various cdE2 residues in CP interaction, surface plasmon resonance (SPR) experiments were conducted using CP with GFP-cdE2 and mutants. This experiment was conducted using a streptavidin (SA)-containing chip to bind a biotin-labeled, single-stranded DNA 48-mer essentially irreversibly. Using the SA-DNA surface, CP was reacted with the surface at a concentration range of 1 to 10 μg/ml until on average ∼5 mol of DNA-CP was bound. Prior to adding the GFP-cdE2 and cdE2 mutants, excess CP was allowed to dissociate until a ratio of ∼1 CP per DNA molecule was attained (Fig. 8A). Injection of wild-type GFP-cdE2 and mutants was carried out at this point (Fig. 8A, arrowhead). Fig. 8B shows an amplification of the reaction profile that is observed after the addition of wild-type GFP-cdE2 and mutants. The differences in the reaction profiles suggest that these molecules bind to the DNA-CP surface and that the extent of binding of the mutants 391KARREC396/A6 and 397LTPYAL402/A6 are bound to the chip less tightly than the wild-type GFP-cdE2. The GFP-cdE2 mutant proteins 403APNAVIP409/A7, 410TSLALL415/A6, and 416CCVRSANA423/A8 were also tested in the SPR experiments, and they bound to the DNA-CP surface with levels between the wild-type GFP-cdE2 and the 397LTPYAL402/A6 levels (data not shown). Combining these results with those obtained from sedimentation velocity studies suggests that differential binding of the various GFP-cdE2 proteins occurs and that it depends on the sequence of the cdE2.
Fig 8.
Surface plasmon resonance studies on the interaction of cdE2 and mutants with immobilized capsid protein. CP(19–264) was immobilized by injecting it over a Biacore streptavidin chip containing single-stranded DNA 48-mer bound via a biotin group. The CP was initially allowed to dissociate until the molar ratio of its bound concentration to that of DNA was about 1 as indicated by the arrow at 1600 s. At this point, either GFP-cdE2 or the mutants indicated below were injected into the chip. (B) Amplification of the sensorgram from the arrow in A for binding of GFP-cdE2 (red) plus the mutants 391KARREC396/A6, (blue) and 397LTPYAL402/A6 (green) to the CP(19–264)-DNA complex (the same scheme is used in A).
DISCUSSION
We have characterized polyprotein processing of SINV cdE2 mutants using Western and pulse-chase analyses and found that the mutations in the C-terminal signal sequence of cdE2 reduced glycoprotein processing and thereby transport to the plasma membrane. An earlier report on the Semliki Forest virus (SFV) signal sequences by Liljestrom and Garoff (21) has suggested that the C-terminal region of E2 protein which preceded the 6K protein on the polyprotein sequence functions as a signal sequence for the 6K protein. Recently, Zhang et al. (51) described the residues between 409 and 416 of VEEV E2 (409PFCLAVLC416) as the C-terminal signal sequence. The SINV amino acids corresponding to the VEEV E2 C-terminal signal sequence are 409PTSLALLC416, and we describe the mutations in this region as the signal sequence mutations (409PTSLALL415/A7 and 412LALL415/A4). Mutants with changes near the signalase cleavage sites between E2 and 6K have been previously shown to be defective in proteolytic processing (9). Except for the two C-terminal signal sequence mutations, all other cdE2 mutations were processed correctly. Deletions in the C-terminal region of cdE2 (Δ406–407, Δ409–411, and Δ414–417) have previously been shown to affect virus assembly in which the nucleocapsids failed to attach to membranes (49). Our experiments suggest that mutations in the signal sequence region of cdE2 (409PTSLALL415/A7 and 412LALL415/A4) affect the glycoprotein processing (Fig. 1A and C) and transport to the plasma membrane. These signal sequence mutations involving multiple alanine substitutions (409PTSLALL415/A7 and 412LALL415/A4) affect polyprotein translation and probably disrupt the incorporation of 6K into the endoplasmic reticulum (ER) membrane in the correct orientation. We have constructed a double alanine substitution mutant, 416CC417/A2, as a control for our experiment. This mutant was previously characterized by Ivanova and Schlesinger (16), and we have used this mutant as a control for our experiments. Also, only some representative mutants are shown in each experiment. This mutant was not affected in polyprotein processing (Fig. 1A) or transport of E2 and E1 to the plasma membrane (Fig. 2A to C), but the virus production was severely affected (Table 2) in agreement with earlier findings implicating the cysteines in palmitoylation (16). Unlike for the signal sequence mutants, the colocalization of CP and cdE2 was observed on the plasma membrane in this mutant. We hypothesize that the flipping of the C terminus of cdE2 to the cytoplasmic side of the membrane that is believed to occur in the Golgi is affected (51). This mutant was found to have wild-type-like virus colocalization with the Golgi marker giantin (Fig. 2B). Our finding supports earlier hypotheses about the involvement of palmitoylated cysteines in virus budding.
Several studies have indicated that the cdE2 residues 397LTPYAL402 bind into the hydrophobic pocket of CP (19, 27, 29). The 397LTPYAL402/A6 and 400YAL402/A3 mutants yielded very little budding virus, although not affected in nucleocapsid formation (data not shown) or glycoprotein trafficking (Fig. 2A to C and 3B). As shown by FC analysis and IF, the glycoproteins in these mutants accumulate on the plasma membrane, presumably due to the lack of interaction with CP. The accumulation of E2 on the plasma membrane for these mutants was significantly increased at 12 h and 18 h posttransfection (data not shown). IF experiments using wild-type virus indicate that E2 and CP colocalize before reaching the plasma membrane (Fig. 2A, panels A to C), although the majority of CP and E2 colocalization was observed at the plasma membrane. It was previously suggested that aromatic residues in the CP serve to interact with the side chain of the essential Y400, providing both specificity for spike incorporation and energy for budding (36). The critical role of cdE2 residue L402 in the nucleocapsid core interaction was confirmed by molecular genetic studies (29). It is believed that there is a critical minimal spanning distance from the K391 and the conserved Y400, and mutations that affect positioning of the 398TPY400 domain into the putative CP binding cleft were found to produce very low levels of virus (13, 14). Deletion of residues 403APNAVI408 was not lethal for the virus, although virus release was severely affected. The glycoprotein processing and transport were unaffected in this mutant (Fig. 2A, panels M to O), suggesting that the defect probably is in virus assembly. We hypothesize that the topology of cdE2 inside the pocket is affected by this mutation, leading to slower assembly and virus release. The residues in this region are probably involved in providing cdE2 with the appropriate spacing for the C terminus to reach the lipid bilayer.
We have observed that similar to the hydrophobic cdE2 residues, mutation of the N-terminal charged residues (393RRE395) also affects virus budding. It is likely that the transport of E2 to the plasma membrane is slightly reduced in the 393RRE395/VLA mutant when compared to the wild type, as observed from the FC analysis. The polyprotein processing was unaffected in this mutant as observed by Western analysis, and the glycoproteins were expressing on the plasma membrane, as evidenced from the IF analysis. The 393RRE395/A3 mutant E2 protein gets transported to the plasma membrane but fails to interact with cores and does not produce particles, suggesting that there is a charged interaction between the N-terminal region of cdE2 and CP required for virus budding. Lopez and coworkers (24) have shown that the E395 is important for the growth of SINV. Recently from the cryo-EM structure of SINV, it was identified that the N-terminal cdE2 residues form critical interactions with the surface-exposed residues of CP that are important for virus assembly and budding (41). The proximity of cdE2 and CP residues was also observed in the high-resolution cryo-EM structure of VEEV (51). It was identified earlier that the cluster of charged residues marks the hydrophobic transmembrane helix of E2 and cytoplasmic boundaries in which the cytoplasmic charge cluster is positive with respect to the luminal boundary. Although the mutagenesis of the charge cluster did not change the topology of PE2 in the membrane or inhibit the subsequent transport of the protein to the plasma membrane, these changes did lower the stability of the anchoring of the protein in cellular membranes (6, 38). No quantitative information on E2 transport was available from this earlier study. Our FC analysis suggests that even though reduced in E2 transport, when compared to the WT and the hydrophobic residue mutation 397LTPYAL402/A6, the charged-residues mutant 393RRE395/VLA did show E2 expression on the plasma membrane as seen in IF analysis. However, the reduction in virus budding was not directly proportional to the reduction in E2 transport of the charged-residues mutant. While the decreased stability of E2 on the membrane may be one reason that possibly could affect the virus budding for the charged-residues mutants, based on the latest information obtained from the cryo-EM structure of SINV (41) we believe that the charged residues are equally involved in the CP interaction that is absolutely required for alphavirus budding. Budding viruses were not observed for any of the cdE2 mutants in the TEM analysis of infected cells (Fig. 4).
Further evidence for this putative charged interaction was obtained from detergent treatment of purified virus at a different pH. In an attempt to examine the involvement of charged interactions between cdE2 and NC in SINV, we used low-pH buffer and low ionic strength on purified virions to test the nature of the cdE2 interaction with cores. It was observed that E2 was released from the core at low pH and low ionic strength in the presence of detergent β-d-octylglucoside. SINV treated with β-d-octylglucoside at physiological pH retained E2 on the virus (Fig. 5D), while pH treatment at 5.5 removed E2 from the core (Fig. 5E). Thus, interactions of cdE2 and core are sensitive to low pH, suggesting that ionic interactions play critical roles in virus budding and possibly entry of alphaviruses. It was reported earlier that the spike glycoproteins remain attached to the core at neutral pH and low ionic strength but not at elevated pH and ionic strength (12). The sensitivity to elevated pH and salt concentration suggested that the interactions attaching the spike proteins to the nucleocapsids depend on charged groups (12). The cryo-EM structure of SINV has shown that the region of cdE2 from K391 to T398, which contains four charged residues, is the part that interacts with the NC (27). The amino acids involved in these charged interactions are the surface-exposed CP residues Y162 and K252. These CP residues form charged interactions with R393 and E395 of cdE2 (41), and these interactions with the surface-exposed CP residues that cover the hydrophobic pocket of CP presumably occur prior to the virus budding.
Coimmunoprecipitation and His tag pulldown experiments were performed (Fig. 6A to C) to study the in vitro interaction of cores with GFP-cdE2 and its mutants (Table 1). The efficiency of nucleocapsid cores to bind various GFP-cdE2 mutants was determined by precipitation of CP by the anti-His antibody (Fig. 6A). The lowest binding was observed for the mutant 391KARREC396/A6. This observation supports our hypothesis regarding the involvement of the charged residues during the interaction of cdE2 with cores. As observed in the coimmunoprecipitation experiments, the interaction was abrogated with mutation of the N-terminal six residues. Another mutant that showed reduced interaction with cores in the His tag pulldown experiments is 416CCVRSANA423. Surprisingly, in both coimmunoprecipitation experiments and His tag pulldown experiments, the mutation of residues 397LTPYAL402 did not abolish the interaction of cdE2 with cores. Therefore, the in vitro binding with 400YAL402 does not recapitulate the in vivo results.
We pursued analyzing the cdE2-core interaction using in vitro biophysical methods as well. A complex between CP(19-264) and GFP-cdE2 in the absence of oligonucleotides was also detected by sedimentation velocity centrifugation (Fig. 7A). Based on the AUC studies, we conclude that cdE2 will bind to both CP and CLP (Fig. 7C). Specific interactions of GFP-cdE2 with CP were further confirmed by SPR. Complexes between CP and GFP-cdE2 could be detected by immobilizing CP on an SPR active streptavidin surface that contained the DNA 48-mer used in the CLP assembly studies sequestered to the surface using a biotin-containing linkage. Consistent and stable interactions were observed in vitro with wild-type GFP-cdE2 and CP bound to DNA. This is the first clear demonstration of this interaction in an in vitro assay. In general, most of the cdE2 mutants bound less tightly than the wild type, and the mutant that had the weakest binding was 391KARREC396/A6 (Fig. 8A and B), suggesting the involvement of charged interactions with CP. It has been demonstrated earlier that a peptide consisting of the N-terminal 14 residues of SFV cdE2 when microinjected into cells infected with SFV could inhibit virus assembly (17). It is possible that since the SPR experiments do not contain lipids they may artificially favor hydrophilic charged interactions. The mutant 397LTPYAL402/A6 also binds less efficiently to the CP (Fig. 8A and B), in agreement with the observation that the 398TPY400 motif is important for the capsid-cdE2 interaction (48). Evidence for direct interaction between the C-terminal 16 residues of cdE2 and the SINV CP(19-264) was shown earlier using CP and synthetic cdE2 peptides reconstituted into phospholipid vesicles (50).
Based on the information obtained in this study from molecular genetic and biochemical experiments, we propose that there are three major interactions involving cdE2 that occur at various cellular compartments at three different stages of the virus life cycle. The N-terminal charged interaction of cdE2 and CP takes place after the polyprotein is translocated across the ER membrane and the signalase cleavage of PE2 and 6K has occurred. Reorientation of the C terminus of cdE2 in the Golgi membrane is aided by the palmitoylation of cysteines, and the placement of all three palmitoylated cysteines on the membrane gives cdE2 the “hydrophobic cleft” topology required to fit into the hydrophobic pocket of CP. Simultaneous to the furin cleavage of E3 in the late Golgi, the E1 and E2 heterodimers form spikes composed of trimers of three heterodimers and form an ordered lattice on the plasma membrane. Presumably at this stage, the hydrophobic interactions occur between cdE2 and CP: three cdE2s form a single spike at a quasisymmetric position binding different CP pockets from three distinct capsomeres. This in turn gives the strict T=4 icosahedral symmetry to the core and the glycoprotein layer and virus buds from the cell. This irreversible cdE2-mediated structural rearrangement of the cytoplasmic cores that takes place during budding primes the cores for disassembly.
The reemergence of CHIKV places a new emphasis on control and dissemination of alphavirus disease. There are currently no antivirals or effective vaccines available for alphavirus containment. It has been proposed earlier that the cdE2-mediated budding signal offers a sensitive point in the viral life cycle that may be amenable to therapeutic attack (17). We show here that it is possible to quantitatively determine the interaction of cdE2 with capsid protein using SPR, and this can be developed into a high-throughput assay for screening inhibitory compounds. In the future, in vitro interaction assays developed in the present study may be used for evaluating compounds that inhibit the alphavirus cdE2-capsid interaction and virus budding.
ACKNOWLEDGMENTS
We thank Chanakha Navaratnarajah, Aaron Taylor, and Dinesh Yernool for creative discussions, Paul Chipman for EM work, and Anita Robinson for clerical and administrative support. We thank M. J. Schlesinger for providing the anti-E2 cytoplasmic domain antibodies and J. H. Strauss for supplying polyclonal anti-E2 antibodies. We acknowledge the use of the Bioscience Imaging Facility, the Biophysical Analysis Lab, and the Flow Cytometry and Cell Separation Facility of the Bindley Bioscience Center and Life Science Microscopy Facility, Purdue University.
This work was supported by NIH grant GM56279 to R.J.K.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Barth BU, Suomalainen M, Liljestrom P, Garoff H. 1992. Alphavirus assembly and entry: role of the cytoplasmic tail of the E1 spike subunit. J. Virol. 66:7560–7564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caspar DL, Klug A. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27:1–24 [DOI] [PubMed] [Google Scholar]
- 3. Cheng RH, et al. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choi HK, et al. 1996. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J. Mol. Biol. 262:151–167 [DOI] [PubMed] [Google Scholar]
- 5. Choi HK, et al. 1991. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature 354:37–43 [DOI] [PubMed] [Google Scholar]
- 6. Cutler DF, Garoff H. 1986. Mutants of the membrane-binding region of Semliki Forest virus E2 protein. I. Cell surface transport and fusogenic activity. J. Cell Biol. 102:889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Forsell K, Griffiths G, Garoff H. 1996. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 15:6495–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fuller SD, Berriman JA, Butcher SJ, Gowen BE. 1995. Low pH induces swiveling of the glycoprotein heterodimers in the Semliki Forest virus spike complex. Cell 81:715–725 [DOI] [PubMed] [Google Scholar]
- 9. Gaedigk-Nitschko K, Schlesinger MJ. 1991. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology 183:206–214 [DOI] [PubMed] [Google Scholar]
- 10. Gaedigk-Nitschko K, Schlesinger MJ. 1990. The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology 175:274–281 [DOI] [PubMed] [Google Scholar]
- 11. Garoff H, Hewson R, Opstelten DJ. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Helenius A, Kartenbeck J. 1980. The effects of octylglucoside on the Semliki forest virus membrane. Evidence for a spike-protein–nucleocapsid interaction. Eur. J. Biochem. 106:613–618 [DOI] [PubMed] [Google Scholar]
- 13. Hernandez R, Ferreira D, Sinodis C, Litton K, Brown DT. 2005. Single amino acid insertions at the junction of the Sindbis virus E2 transmembrane domain and endodomain disrupt virus envelopment and alter infectivity. J. Virol. 79:7682–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hernandez R, Lee H, Nelson C, Brown DT. 2000. A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly. J. Virol. 74:4220–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hong EM, Perera R, Kuhn RJ. 2006. Alphavirus capsid protein helix I controls a checkpoint in nucleocapsid core assembly. J. Virol. 80:8848–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ivanova L, Schlesinger MJ. 1993. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol. 67:2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kail M, et al. 1991. The cytoplasmic domain of alphavirus E2 glycoprotein contains a short linear recognition signal required for viral budding. EMBO J. 10:2343–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kostyuchenko VA, et al. 2011. The structure of barmah forest virus as revealed by cryo-electron microscopy at a 6-angstrom resolution has detailed transmembrane protein architecture and interactions. J. Virol. 85:9327–9333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee S, et al. 1996. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4:531–541 [DOI] [PubMed] [Google Scholar]
- 20. Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liljestrom P, Garoff H. 1991. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 65:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Linger BR, Kunovska L, Kuhn RJ, Golden BL. 2004. Sindbis virus nucleocapsid assembly: RNA folding promotes capsid protein dimerization. RNA 10:128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu N, Brown DT. 1993. Transient translocation of the cytoplasmic (endo) domain of a type I membrane glycoprotein into cellular membranes. J. Cell Biol. 120:877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopez S, Yao JS, Kuhn RJ, Strauss EG, Strauss JH. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lusa S, Garoff H, Liljestrom P. 1991. Fate of the 6K membrane protein of Semliki Forest virus during virus assembly. Virology 185:843–846 [DOI] [PubMed] [Google Scholar]
- 26. Melton JV, et al. 2002. Alphavirus 6K proteins form ion channels. J. Biol. Chem. 277:46923–46931 [DOI] [PubMed] [Google Scholar]
- 27. Mukhopadhyay S, et al. 2006. Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses. Structure 14:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Navaratnarajah CK, Kuhn RJ. 2007. Functional characterization of the Sindbis virus E2 glycoprotein by transposon linker-insertion mutagenesis. Virology 363:134–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Owen KE, Kuhn RJ. 1997. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 230:187–196 [DOI] [PubMed] [Google Scholar]
- 30. Owen KE, Kuhn RJ. 1996. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 70:2757–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paredes AM, et al. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. U. S. A. 90:9095–9099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Perera R, Navaratnarajah C, Kuhn RJ. 2003. A heterologous coiled coil can substitute for helix I of the Sindbis virus capsid protein. J. Virol. 77:8345–8353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perera R, Owen KE, Tellinghuisen TL, Gorbalenya AE, Kuhn RJ. 2001. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 75:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pletnev SV, et al. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryan C, Ivanova L, Schlesinger MJ. 1998. Mutations in the Sindbis virus capsid gene can partially suppress mutations in the cytoplasmic domain of the virus E2 glycoprotein spike. Virology 243:380–387 [DOI] [PubMed] [Google Scholar]
- 36. Skoging U, Vihinen M, Nilsson L, Liljestrom P. 1996. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 4:519–529 [DOI] [PubMed] [Google Scholar]
- 37. Smith TJ, et al. 1995. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. U. S. A. 92:10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strauss JH, Strauss EG, Kuhn RJ. 1995. Budding of alphaviruses. Trends Microbiol. 3:346–350 [DOI] [PubMed] [Google Scholar]
- 40. Suomalainen M, Garoff H. 1992. Alphavirus spike-nucleocapsid interaction and network antibodies. J. Virol. 66:5106–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tang J, Jose J, Chipman P, Zhang W, Kuhn RJ, Baker TS. 2011. Molecular links between the envelope glycoprotein and nucleocapsid core in Sindbis virus. J. Mol. Biol. 414:442–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tellinghuisen TL, Hamburger AE, Fisher BR, Ostendorp R, Kuhn RJ. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309–5319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tellinghuisen TL, Perera R, Kuhn RJ. 2001. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 75:2810–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Venien-Bryan C, Fuller SD. 1994. The organization of the spike complex of Semliki Forest virus. J. Mol. Biol. 236:572–583 [DOI] [PubMed] [Google Scholar]
- 45. Voss JE, et al. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712 [DOI] [PubMed] [Google Scholar]
- 46. Warrier R, Linger BR, Golden BL, Kuhn RJ. 2008. Role of Sindbis virus capsid protein region II in nucleocapsid core assembly and encapsidation of genomic RNA. J. Virol. 82:4461–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weiss B, Geigenmuller-Gnirke U, Schlesinger S. 1994. Interactions between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 22:780–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. West J, Brown DT. 2006. Role of a conserved tripeptide in the endodomain of Sindbis virus glycoprotein E2 in virus assembly and function. J. Gen. Virol. 87:657–664 [DOI] [PubMed] [Google Scholar]
- 49. West J, Hernandez R, Ferreira D, Brown DT. 2006. Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly. J. Virol. 80:4458–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wilkinson TA, Tellinghuisen TL, Kuhn RJ, Post CB. 2005. Association of sindbis virus capsid protein with phospholipid membranes and the E2 glycoprotein: implications for alphavirus assembly. Biochemistry 44:2800–2810 [DOI] [PubMed] [Google Scholar]
- 51. Zhang R, et al. 2011. 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30:3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang W, Heil M, Kuhn RJ, Baker TS. 2005. Heparin binding sites on Ross River virus revealed by electron cryo-microscopy. Virology 332:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang W, et al. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang X, Kielian M. 2005. An interaction site of the envelope proteins of Semliki Forest virus that is preserved after proteolytic activation. Virology 337:344–352 [DOI] [PubMed] [Google Scholar]
- 55. Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljestrom P. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]