Abstract
The ability to extinguish a viral population of fixed reproductive capacity by causing small changes in the mutation rate is referred to as lethal mutagenesis and is a corollary of population genetics theory. Here we show that coxsackievirus B3 (CVB3) exhibits reduced mutational robustness relative to poliovirus, manifesting in enhanced sensitivity of CVB3 to lethal mutagens that is dependent on the size of the viral population. We suggest that mutational robustness may be a useful measure of the sensitivity of a virus to lethal mutagenesis.
TEXT
RNA virus genomes are replicated at mutation rates orders of magnitude higher than DNA genomes (10, 27) and exist as heterogeneous populations of genetically distinct yet related genomes. Attempts to define these populations have relied upon population genetics theory (5, 6, 34, 35); unfortunately, little empirical evidence exists validating these theories in vivo (30).
Population genetics theory predicts a log-linear relationship between the genomic mutation rate and the fecundity (average number of progeny generated per infectious cycle) needed to avoid extinction and defines a theoretical “extinction threshold” (6). A corollary of this theory is lethal mutagenesis, the ability to drive a viral population to extinction by increasing the error frequency (13).
A high mutation rate can negatively affect viral fitness and may drive a population to extinction. However, the deleterious effects of a high mutation rate may differ between viruses depending on their mutational robustness, i.e., the constancy of a phenotype in the face of deleterious mutations (26). A more robust population is able to minimize the deleterious effects of a high mutation rate by opting not to maximize fitness, a phenomenon termed “survival of the flattest” (35). Previous studies have suggested that the distributions of mutational effects between DNA and RNA virus genomes are similar (25). Therefore, the mutational robustness of a virus might be expected to be similar to that of closely related viruses.
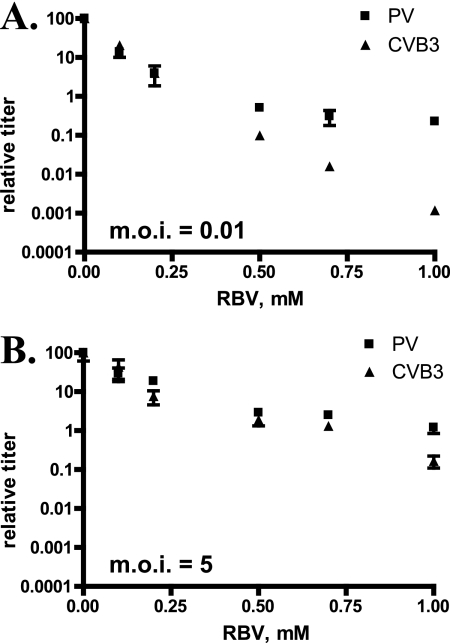
We investigated the mutational robustness of two closely related enteroviruses, poliovirus (PV) and coxsackievirus B3 (CVB3), which employ similar replication strategies and genome organizations and exhibit 59% polyprotein identity (74% similarity) as shown by BLAST analysis (GenBank accession numbers Q5UEA2 and P03300). To compare the characteristics of mutational robustness of these two viruses, we examined the response of PV (Mahoney) and of CVB3 (CVB3/0) to increasing concentrations of the mutagenic nucleoside ribavirin (Fig. 1A). Infection was performed under conditions that included a low multiplicity of infection (MOI) so that the cumulative effect of increased mutation frequency could be observed over two or more replicative cycles. CVB3 was observed to be considerably more sensitive than PV to ribavirin treatment at 0.5 mM or greater. Dramatically increased sensitivity was not observed under conditions of high MOI (Fig. 1B), consistent with the deleterious effect of mutation being reversed by population size (fecundity), as predicted by population genetics theory.
Fig 1.
CVB3 is more sensitive to ribavirin-induced mutagenesis than PV. HeLa S3 cells (1 × 105 in 24-well plates) were pretreated with ribavirin for 1 h prior to infection with either PV or CVB3 at an MOI of 0.01 (A) or 5 (B) as previously described (17). Cells were incubated with ribavirin at the indicated concentration until all cells demonstrated a cytopathic effect or for 6 days, at which time virus was collected and titers were determined using HeLa S3 monolayers. Titers were normalized to controls infected with either virus in the absence of ribavirin. Typical titers for experiments performed at an MOI of 0.01 were 3 × 108 (PV) and 2 × 108 PFU/ml (CVB3) and at an MOI of 5 were 7 × 108 (PV) and 4 × 108 (CVB3). Data are plotted as the means and standard deviations of results determined with 3 independent samples.
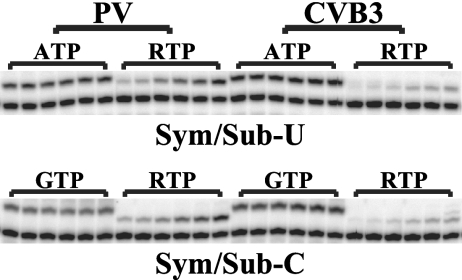
Higher tolerance to mutagens could be explained by a number of mechanisms, including increased replication fidelity (8, 19, 23), increased mutational robustness (7, 26), or specific discrimination against the mutagen (29). It was possible that the low tolerance to mutation of CVB3 was caused by the presence of a lower-fidelity RNA-dependent RNA polymerase (RdRp) in terms of ribavirin incorporation, since at any given concentration of ribavirin, more mutagenesis would occur. To investigate this possibility, we purified the RdRps from both PV and CVB3 (12, 32) and each was incubated with a previously described substrate (sym/sub) and either the “correct” nucleotide or ribavirin triphosphate (RTP) (3, 15). Examination of ribavirin incorporation revealed that CVB3 RdRp incorporated ribavirin much less efficiently than PV RdRp (Fig. 2). Thus, heightened sensitivity of CVB3 to ribavirin treatment was not likely to have been due to a higher rate of genomic incorporation of ribavirin.
Fig 2.
CVB3 encodes an RdRp with higher fidelity than PV. The RdRp from either PV (left) or CVB3 (right) was incubated with sym/sub substrate and either the correct nucleotide substrate or RTP as previously described (3, 15). Panels for each nucleotide correspond to quenching at 30, 60, 120, 300, 600, or 1,800 s.
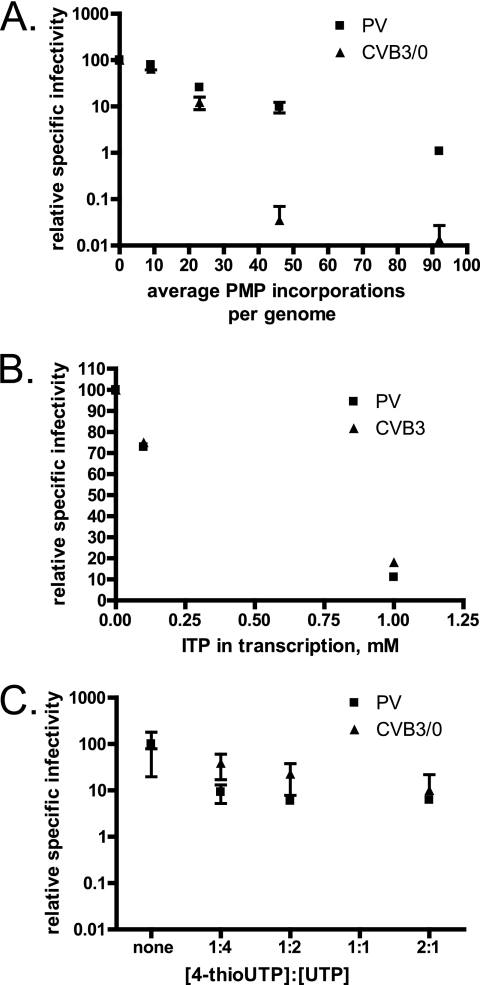
If CVB3 is less genetically robust than PV, this should manifest as a higher proportion of deleterious and/or lethal mutations at a given genomic mutation frequency. To test this hypothesis experimentally, we employed T7 RNA polymerase (T7 RNAP)-directed mutagenesis of viral genomes as previously described using the mutagenic pyrimidine analog P (14, 21, 22). Genomic RNA was in vitro transcribed for either PV or CVB3 under identical conditions to generate a defined number of mutations per genome (14, 16), and the specific infectivity of viral genomes was assessed by an infectious center assay (9). CVB3 showed greatly increased sensitivity when ∼50 or more incorporations of P were made per genome (Fig. 3A). Notably, CVB3 had over 100-fold-lower relative specific infectivity than PV even when less than 1% of the genome was replaced by P. These results are consistent with CVB3 being less genetically robust than PV.
Fig 3.
The CVB3 coding sequence, not the RNA structure, determines the enhanced sensitivity to mutagenic nucleotides relative to PV. Genomic RNA was produced from cDNA in the presence of nucleotide analogs as previously described (14, 16). HeLa S3 cells were transfected with RNA genomes and then serially diluted onto HeLa S3 cell monolayers. An agar overlay was applied to the monolayers prior to incubation for 3 days at 37°C (9, 14). Specific infectivity was defined as the number of plaques resulting from transfection of 1 μg of RNA (9) and normalized so that the number of plaques resulting from RNA transcribed in the absence of mutagen was set to 100. Typical specific infectivities were 2 × 106 (PV) and 2 × 105 (CVB3). The means and standard deviations of the results determined with at least 3 independent samples are reported. PTP (A) or ITP (B) was added at increasing concentrations for T7 RNAP-directed in vitro transcription of PV or CVB3 genomes. 4-thio-UTP (C) was partially substituted for UTP at defined ratios in transcription reactions.
P incorporation into viral RNA may have two unique effects: (i) alteration of codon-anticodon recognition, resulting in amino acid substitutions in viral proteins, and (ii) disruptions in hydrogen-bonding networks essential for structural elements of the RNA. Thus, the deleterious effect of P monophosphate (PMP) substitution may be expressed at the level of either the RNA structure or the encoded proteins. To distinguish between these two possibilities, we employed two other nucleotide analogs, ITP and 4-thiouridine triphosphate (4-thioUTP). Both nucleotides are predicted to cause changes in hydrogen-bonding interactions important in stabilizing tertiary RNA structures while maintaining Watson-Crick pairings and therefore codon integrity (1, 24, 31).
Various concentrations of ITP and 4-thioUTP were employed during transcription of viral genomes, and specific infectivity was monitored as a function of genomic substitution (Fig. 3B and C). While both nucleotides reduced the specific infectivity of viral RNA, the responses observed for PV and CVB3 were identical. PMP substitution sufficient for a 10-fold reduction in PV-specific infectivity resulted in a 1,000-fold reduction in CVB3-specific infectivity (panel A; ∼50 PMP incorporations per genome). However, 10-fold reductions in PV-specific infectivity induced by IMP or 4-thioUMP resulted in reductions of similar magnitude in CVB3-specific infectivity (panel B [1 mM ITP] and panel C [1:4 or greater ratio of 4-thioUTP to UTP]). Thus, our observations suggest that the protein-coding sequence, not the RNA structure, is the determinant of the increased sensitivity of CVB3 to genomic mutation.
Virus populations with lower mutational robustness would be expected to exhibit increased susceptibility to lethal mutagens and be more sensitive to mutation. To investigate this, we generated populations of PV and CVB3 in the presence or absence of 100 μM ribavirin. RNA was isolated from either the total (viable plus nonviable) virus population or from replication-competent virus obtained through plaque purification, and the capsid-coding region of 96 viral genomes was sequenced (4). To permit comparison with previous work, plaque-competent virus was required. The A372V Nancy strain of CVB3 was used, which differs from CVB3/0 at only 12 amino acid positions and carries an identical polymerase (19).
CVB3 indeed showed a lower mutation frequency compared to PV, as predicted by our biochemical data and as would be predicted for a less robust virus (Table 1 [3.8 versus 6.1 mutations per genome in total RNA; 0.7 versus 1.2 in plaque-purified genomes]). We then looked at the general tolerance for mutation with respect to coding changes and silent mutations. Since we expected the total RNA population to contain all genomes (viable and lethally mutagenized) and the polymerase to insert mutations randomly, we were not surprised to find that mutations were evenly distributed among the three codon positions (data not shown) and that most nonsynonymous (NS) mutations occurred in the 1st and 2nd codon positions for both viruses (Table 1). These data show that both PV and CVB3 generate mutations with similar, stochastic distributions in both the presence and absence of mutagen.
Table 1.
Mutation frequency and codon substitution distribution in virus populations
| Expt conditions | PV |
CVB3 |
P valueb | ||||
|---|---|---|---|---|---|---|---|
| Mutation frequencya | No. (%) of NS mutations | No. (%) of S mutations | Mutation frequencya | No. (%) of NS mutations | No. (%) of S mutations | ||
| Total RNA no drug | 6.1 | 58 (64) | 32 (36) | 3.8 | 70 (65) | 37 (35) | 1.000 |
| total RNA RBV | 9.6 | 29 (63) | 17 (37) | 6.1 | 28 (67) | 14 (34) | 0.8242 |
| Plaque RNA no drug | 1.2 | 50 (70) | 21 (30) | 0.7 | 22 (48) | 24 (52) | 0.019 |
| Plaque RNA RBV | 4.3 | 24 (33) | 48 (67) | 3.8 | 14 (18) | 62 (82) | 0.041 |
Mutation frequency is represented as the number of mutations per the total 104 nucleotides sequenced.
The statistical significance of the results of comparisons of mutation distributions among codon positions was determined by Fisher's two-tailed test.
We then examined plaque-purified (viable only) virus genomes for mutation profiles. Notably, 70% of the mutations in the untreated PV were nonsynonymous, suggesting a high tolerance for mutation and changes to the protein-coding sequence that did not negatively impact the fitness of these variants with respect to that of the wild type. On the other hand, over 50% of mutations in untreated CVB3 were synonymous (S), suggesting that mostly silent rather than coding mutations are well tolerated in this virus and that it is thus less mutationally robust. This bias for silent mutation was exacerbated upon ribavirin treatment (Table 1). Notably, ribavirin decreased the ratio of NS mutations to S mutations for both viruses, likely due to its property of inducing transition mutations favoring S mutations.
Differences in fecundity or specific infectivity between the viruses might impact their sensitivity to mutagenesis. Thus, to rule out these possibilities, we infected HeLa cells with either PV or CVB3 at an MOI of 0.01 with 0, 200, or 400 μM RBV. After 48 h, virion RNA was extracted, purified, and subjected to quantitative reverse transcription-PCR (qRT-PCR) to determine the total number of genomes produced. Titers of samples were also measured to determine the total number of PFU produced under each set of conditions. We then determined the specific infectivity (PFU/genome) and fecundity (genomes/cell) (Table 2). We observed that the levels of fecundity and specific infectivity of the viruses were similar in the absence of ribavirin. Upon ribavirin treatment, we observed a similar reduction in the fecundity of the viruses but a much more pronounced reduction in specific infectivity for CVB3. This supports the notion that the sensitivity of CVB3 is driven by lethal mutagenesis of packaged genomes and not by a reduction in fecundity.
Table 2.
Specific infectivity of CVB3 is greatly reduced compared to that of PV after ribavirin treatment
| Virus | RBV (μM) | Titer ± SDa (PFU/ml) | No. of virions/ml ± SDb | Fecundityc (no. of virions/cell ± SD) | Specific infectivity ± SDc (× 10−2 PFU/genome) |
|---|---|---|---|---|---|
| PV | 0 | 1.7 × 108 ± 3.1 × 107 | 1.3 × 1010 ± 4.8 × 109 | 4.5 × 104 ± 1.7 × 104 | 1.5 ± 0.84 |
| PV | 200 | 4.0 × 106 ± 9.5 × 105 | 7.8 × 108 ± 6.5 × 107 | 2.8 × 103 ± 2.3 × 102 | 0.51 ± 0.091 |
| PV | 400 | 5.0 × 105 ± 1.4 × 105 | 4.0 × 108 ± 3.2 × 108 | 1.4 × 103 ± 1.1 × 103 | 0.24 ± 0.26 |
| CVB3 | 0 | 4.6 × 107 ± 3.1 × 107 | 4.6 × 109 ± 2.8 × 109 | 1.6 × 104 ± 9.8 × 103 (ns) | 1.2 ± 0.79 (ns) |
| CVB3 | 200 | 1.3 × 105 ± 6.1 × 104 | 2.8 × 108 ± 1.4 × 108 | 9.7 × 102 ± 5.1 × 102 (P < 0.01) | 0.054 ± 0.027 (P < 0.001) |
| CVB3 | 400 | 4.8 × 103 ± 1.7 × 103 | 3.5 × 108 ± 4.1 × 108 | 1.2 × 103 ± 1.4 × 103 (ns) | 0.0035 ± 0.0.0038 (P < 0.05) |
Titers were determined by a standard plaque assay using HeLa cells.
Numbers of virions per milliliter were determined by qRT-PCR using virion RNA and the following primers and probes: for CVB3, the forward primer was GATCGCATATGGTGATGATGTGA, the reverse primer was AGCTTCAGCGAGTAAAGATGCA, and the 6-carboxyfluorescein (FAM) probe was CGCATCGTACCCATGG; and for PV, the forward primer was CCCGTCCAAAACCAAGCTT, the reverse primer was CCTTCACCCCTTCAAACACATAG, and the FAM probe was ACCCAGTGCTTTCC.
Statistical significance was determined by a two-tailed unpaired t test; n = 3.
We have shown that CVB3 has higher polymerase fidelity and a lower basal mutation rate than PV, is more adversely affected by genomic mutation at a given frequency, and is less tolerant of protein-coding changes. In sum, these results support the notion of lower mutational robustness for CVB3 populations compared to PV. At this time, we cannot rule out the possibility that PV and CVB3 exhibit the same deleterious mutation rate, with CVB3 exhibiting a higher lethal mutation rate. The tempo for extinction is dictated by the rate of accumulation of lethal changes. One consequence of faster extinction is a reduction in opportunities for evolution of resistance.
The biological significance of the lower robustness of CVB3 is still unknown. Maintenance of host tropism may constrain robustness by imposing selective restraints. Nonsynonymous mutations that are fitness neutral in one host or cell type may nonetheless reduce fitness in other cell types. This may explain the lower robustness of CVB3, as this virus has been implicated in the infection of numerous organs, including the heart, pancreas, and lymphocytes, as well as the central nervous system, despite its primary route of infection and transmission being the enteric system (2, 11, 18, 20, 28). Restriction of PV genetic diversity was found to greatly impact pathogenesis and tissue distribution (33), suggesting a fundamental link between mutation rate and pathology. The possibility also exists that PV has evolved higher robustness based on environmental fluctuations and/or its higher basal mutation frequency.
The suggestion that RNA viruses obey the strictures of lethal mutagenesis theory has significant implications for antiviral therapy. Subtle (<2 log) decreases in virus titer may be sufficient to reduce the population size below the limit of viability. The stringent criteria applied to the selection of antiretroviral therapeutics for clinical studies may be overkill for selection of therapeutics against classical RNA viruses. The observation that differences between closely related viruses in drug sensitivity can be attributed to differences in mutational robustness may provide an explanation for the loss or absence of efficacy of drugs in the absence of a drug-resistant target.
ACKNOWLEDGMENTS
Financial support was provided by the National Institutes of Health (grant AI053531 to C.E.C.), the American Heart Association (established investigator award 0340028N to C.E.C.), and ERC Starting grant 242719 (to M.V.).
We thank Nora Chapman and Steven Tracy (University of Nebraska Medical Center) and Kirk Knowlton (University of California, San Diego) for providing CVB3 cDNA. We thank David Loakes and Daniel Brown (MRC Laboratory of Molecular Biology) for a gift of PTP and Claus Wilke (University of Texas at Austin) for critical review of the manuscript.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Aboul-ela F, Koh D, Tinoco I, Jr, Martin FH. 1985. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 13:4811–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson DR, et al. 1996. Direct interactions of coxsackievirus B3 with immune cells in the splenic compartment of mice susceptible or resistant to myocarditis. J. Virol. 70:4632–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold JJ, Cameron CE. 2000. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275:5329–5336 [DOI] [PubMed] [Google Scholar]
- 4. Beaucourt SP, et al. 16 June 2011, posting date Isolation of fidelity variants of RNA viruses and characterization of virus mutation frequency. J. Vis. Exp. 2011:2953 doi:10.3791/2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bull JJ, Meyers LA, Lachmann M. 2005. Quasispecies made simple. PLoS Comput. Biol. 1:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bull JJ, Sanjuan R, Wilke CO. 2007. Theory of lethal mutagenesis for viruses. J. Virol. 81:2930–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Codoñer FM, Daros JA, Sole RV, Elena SF. 2006. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS Pathog. 2:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Coffey LL, Beeharry Y, Borderia AV, Blanc H, Vignuzzi M. 2011. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. U. S. A. 108:16038–16043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crotty S, Cameron CE, Andino R. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. U. S. A. 98:6895–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 96:13910–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Feuer R, et al. 2009. Viral persistence and chronic immunopathology in the adult CNS following coxsackievirus infection during the neonatal period. J. Virol. 83:9356–9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gohara DW, et al. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17:128–138 [DOI] [PubMed] [Google Scholar]
- 13. Graci JD, Cameron CE. 2002. Quasispecies, error catastrophe, and the antiviral activity of ribavirin. Virology 298:175–180 [DOI] [PubMed] [Google Scholar]
- 14. Graci JD, et al. 2007. Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue. J. Virol. 81:11256–11266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Graci JD, et al. 2008. Lethal mutagenesis of picornaviruses with N-6-modified purine nucleoside analogues. Antimicrob. Agents Chemother. 52:971–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harki DA, et al. 2006. Synthesis and antiviral activity of 5-substituted cytidine analogues: identification of a potent inhibitor of viral RNA-dependent RNA polymerases. J. Med. Chem. 49:6166–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harki DA, et al. 2002. Synthesis and antiviral evaluation of a mutagenic and non-hydrogen bonding ribonucleoside analogue: 1-beta-D-ribofuranosyl-3-nitropyrrole. Biochemistry 41:9026–9033 [DOI] [PubMed] [Google Scholar]
- 18. Kim KS, Hufnagel G, Chapman NM, Tracy S. 2001. The group B coxsackieviruses and myocarditis. Rev. Med. Virol. 11:355–368 [DOI] [PubMed] [Google Scholar]
- 19. Levi LI, et al. 2010. Fidelity variants of RNA dependent RNA polymerases uncover an indirect, mutagenic activity of amiloride compounds. PLoS 6:e1001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mena I, et al. 2000. Coxsackievirus infection of the pancreas: evaluation of receptor expression, pathogenesis, and immunopathology. Virology 271:276–288 [DOI] [PubMed] [Google Scholar]
- 21. Moriyama K, et al. 1998. Synthesis and RNA polymerase incorporation of the degenerate ribonucleotide analogue rPTP. Nucleic Acids Res. 26:2105–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moriyama K, Otsuka C, Loakes D, Negishi K. 2001. Highly efficient random mutagenesis in transcription-reverse-transcription cycles by a hydrogen bond ambivalent nucleoside 5′-triphosphate analogue: potential candidates for a selective anti-retroviral therapy. Nucleosides Nucleotides Nucleic Acids 20:1473–1483 [DOI] [PubMed] [Google Scholar]
- 23. Pfeiffer JK, Kirkegaard K. 2003. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U. S. A. 100:7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ryder SP, Ortoleva-Donnelly L, Kosek AB, Strobel SA. 2000. Chemical probing of RNA by nucleotide analog interference mapping. Methods Enzymol. 317:92–109 [DOI] [PubMed] [Google Scholar]
- 25. Sanjuán R. 2010. Mutational fitness effects in RNA and single-stranded DNA viruses: common patterns revealed by site-directed mutagenesis studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:1975–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sanjuán R, Cuevas JM, Furio V, Holmes EC, Moya A. 2007. Selection for robustness in mutagenized RNA viruses. PLoS Genet. 3:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J. Virol. 84:9733–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Selinka HC, et al. 1998. Coxsackie B virus and its interaction with permissive host cells. Clin. Diagn. Virol. 9:115–123 [DOI] [PubMed] [Google Scholar]
- 29. Sierra M, et al. 2007. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. J. Virol. 81:2012–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Springman R, Keller T, Molineux IJ, Bull JJ. 2010. Evolution at a high imposed mutation rate: adaptation obscures the load in phage T7. Genetics 184:221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Testa SM, Disney MD, Turner DH, Kierzek R. 1999. Thermodynamics of RNA-RNA duplexes with 2- or 4-thiouridines: implications for antisense design and targeting a group I intron. Biochemistry 38:16655–16662 [DOI] [PubMed] [Google Scholar]
- 32. van Ooij MJ, Glaudemans DH, Heus HA, van Kuppeveld FJ, Melchers WJ. 2006. Structural and functional integrity of the coxsackievirus B3 oriR: spacing between coaxial RNA helices. J. Gen. Virol. 87:689–695 [DOI] [PubMed] [Google Scholar]
- 33. Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wilke CO. 2005. Quasispecies theory in the context of population genetics. BMC Evol. Biol. 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. 2001. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 412:331–333 [DOI] [PubMed] [Google Scholar]