Abstract
RNA-dependent RNA polymerases (RdRps) are key to the replication of RNA viruses. A common divalent cation binding site, distinct from the positions of catalytic ions, has been identified in many viral RdRps. We have applied biochemical, biophysical, and structural approaches to show how the RdRp from bacteriophage ϕ6 uses the bound noncatalytic Mn2+ to facilitate the displacement of the C-terminal domain during the transition from initiation to elongation. We find that this displacement releases the noncatalytic Mn2+, which must be replaced for elongation to occur. By inserting a dysfunctional Mg2+ at this site, we captured two nucleoside triphosphates within the active site in the absence of Watson-Crick base pairing with template and mapped movements of divalent cations during preinitiation. These structures refine the pathway from preinitiation through initiation to elongation for the RNA-dependent RNA polymerization reaction, explain the role of the noncatalytic divalent cation in ϕ6 RdRp, and pinpoint the previously unresolved Mn2+-dependent step in replication.
INTRODUCTION
Viral RNA-dependent RNA polymerases (RdRps) share a similar structure and mechanism of catalysis and are potential targets for generic antiviral drugs. These enzymes resemble a closed right hand with fingers, palm, thumb, and fingertips domains (see Fig. 1 A and Fig. 2A for domain structure). As in all known DNA and RNA polymerases, the nucleotidyltransferase reaction (35) involves two metal ions (Mg2+) which are coordinated by invariant aspartate residues located in the palm domain (D324, D453, and D454 in ϕ6 RdRp; Fig. 1C). The remarkable structural similarity between RdRps from viruses that are considered unrelated (6) (e.g., hepatitis C virus [HCV], family Flaviviridae, and bacteriophage ϕ6, family Cystoviridae) indicates an unexpected evolutionary link between the corresponding genes in these viruses, which is reflected in the biochemical properties of these enzymes (see references 15 and 18).
Fig 1.
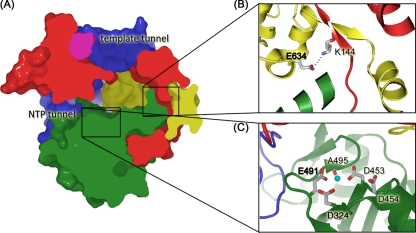
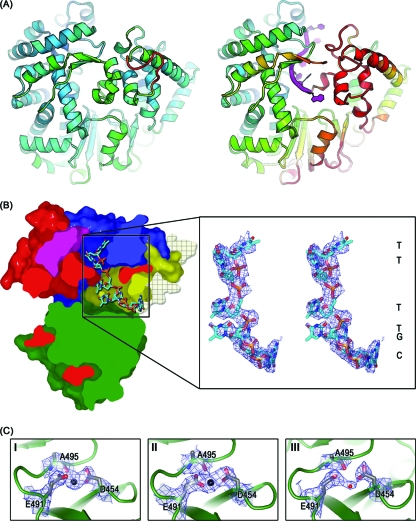
Overview of ϕ6 RdRp structure. (A) A cross section through the surface representation of ϕ6 RdRp structure (PDB accession no. 1HHS) colored according to the generic polymerase domain architecture: red, fingers domain (amino acids 1 to 30, 104 to 276, 333 to 397); green, palm domain (amino acids 277 to 332, 398 to 517); blue, thumb domain (amino acids 37 to 91, 518 to 600); yellow, C-terminal domain (amino acids 604 to 664); pink, connecting chains (amino acids 31 to 36, 92 to 103). The positively charged tunnels for template RNA and substrate NTPs are indicated. In the close-ups (B and C), key amino acids are labeled and are shown in stick representation (white, carbon; blue, nitrogen; red, oxygen; Mn2+ is shown as a cyan sphere). (B) Stabilizing salt bridge. E634 lies within the C-terminal priming domain and forms a salt bridge with K144 within the fingers domain. (C) Active site/noncatalytic ion site. D324 and D453 are the invariant catalytic aspartates involved in the coordination of catalytic Mg2+ ions. The noncatalytic ion site is formed by side chains of D454 and E491 and the backbone carbonyl group of A495. Residues in bold mark the key point mutations (E634 to Q and E491 to Q).
Fig 2.
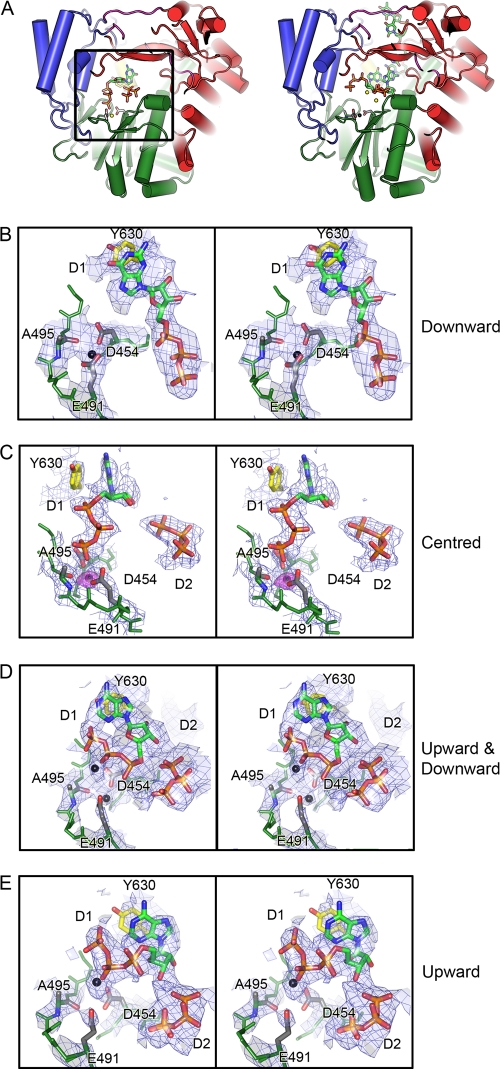
Mg2+ at the noncatalytic ion site and its movements during preinitiation. (A) On the left, WT-5′-AATCT-3′-ATP-GTP-Mg2+ (molecule II) showing secondary structure elements colored as described in the legend to Fig. 1. Part of the fingers domain (red) has been hidden for clarity. Noncatalytic ion site residues D454, E491, and A495 are shown in stick representations (carbon, gray; nitrogen, blue; oxygen, red). Mg2+ coordinated at this site is shown as a yellow sphere. D1 and D2 NTPs (stick representation; carbon, green; nitrogen, blue; oxygen, red; phosphorus, orange) at the active site in the palm domain. D1 NTP base stacks with Y630 (yellow, carbon; red, oxygen) of the CTD (yellow) priming platform. Alongside, on the right, is the initiation complex (1HI0), with oligonucleotide and NTPs drawn for comparison. Coloring is as previously described, with the noncatalytic Mn2+ drawn in black. Stereo image close-ups of WT-5′-AATCT-3′-GTP-Mg2+ (molecule II) (B), WT-5′-AATCT-3′-ATP-GTP-Mg2+ (molecule II) (C), WT-Tri4T-ATP-Mg2+ (molecule I) (D), and WT-Tri4T-ATP-Mg2+ (molecule III) (E) oriented and colored with respect to panel A. BUSTER-derived 2|Fo| − |Fc| SIGMAA electron density maps contoured to 1.2σ are shown as blue wires, and the position of the Mg2+ is indicated on the right. An Fo-Mn − Fo-Mg Phicalc (unbiased) difference electron density map in panel B was calculated at a resolution of 2.9 Å, using amplitudes derived from native WT crystals (with Mn2+ present) and from Mg2+-soaked crystals (WT-5′-AATCT-3′-ATP-GTP-Mg2+). Phases were derived from the high-resolution structure (PDB accession no. 1HHS) that had been refined with the Mn2+ ions removed, to avoid biasing the phases. The difference map (colored purple) is contoured at a level of 6σ (maximum peak height, 8σ). The residual positive density is due to the difference of 13 electrons between Mn2+ and Mg2+. The ligand distances to E491, D454, and the carbonyl of A495 are 2.1, 2.4, and 2.5 Å, respectively.
Previous structural studies identified a high-affinity Mn2+ binding site in the palm domain of bacteriophage ϕ6 RdRp (Fig. 1C) at a position ∼6 Å from the catalytic Mg2+ ions (6). A positionally related noncatalytic ion binding site has been observed in several viral RdRps, including those of poliovirus, foot-and-mouth disease virus, and dengue virus (28). Furthermore, the addition of Mn2+ ions (to the Mg2+-containing reaction buffer) increases the specific activity of ϕ6 RdRp by more than an order of magnitude (21, 28, 42), and similar stimulatory effects have been described for a number of viral RdRps, including those from HCV (and other members of the family Flaviviridae), poliovirus (Picornaviridae) sapovirus (Caliciviridae), reovirus (Reoviridae), brome mosaic virus (Bromoviridae), and bacteriophages ϕ8, ϕ12, and ϕ13 (Cystoviridae) (2, 13, 20, 29, 34, 42, 43, 45). The mechanism of this stimulation is poorly understood, although Mn2+ ions are known to decrease template specificity, lower the Km for nucleotide binding, and reduce the nucleotide specificity in several polymerases (26, 29, 36, 40). Mn2+ ions also support polymerization in the absence of Mg2+ ions (20, 28), suggesting that Mn2+ can replace Mg2+ as the catalytic ion; indeed, initiation complexes with Mn2+ ions coordinating triphosphate moieties of nucleoside triphosphates (NTPs) have been reported (37, 46).
A number of viral RdRps initiate RNA synthesis primer independently (de novo; reviewed in references 17 and 38), and we have described the assembly of such an initiation complex in ϕ6 RdRp (6). A nucleotide complementary to the second nucleotide from the 3′ end of the template (this NTP is defined as D2, and the corresponding nucleotide in the template is defined as T2) is first stabilized by base stacking with a tyrosine residue (Y630) in the C-terminal domain (CTD) of the polymerase (structurally equivalent elaborations are common for RdRps utilizing a de novo initiation mechanism [3, 23, 44, 47]). Subsequently, the template ratchets backwards in the template tunnel so that the D1 NTP, complementary to the 3′-terminal nucleotide of the template (T1), can enter and assemble into the initiation complex for the first nucleotidyl transfer (6).
In the ϕ6 RdRp initiation complex, the CTD blocks the exit path of the newly synthesized double-stranded RNA (dsRNA) and must be displaced for the addition of the third nucleotide of the daughter strand as the polymerase proceeds from initiation to elongation. A similar blocking role is played by the priming loop CTD of flaviviral polymerases. The role of the noncatalytic Mn2+ in this process is the focus of this study. By comparison of the structures of complexes of both wild-type (WT) and mutant (E634 to Q) polymerases with various oligonucleotides and nucleotide substrates, alongside their biophysical and biochemical properties, we are able to tease out the roles of Mn2+ and Mg2+ ions during RNA polymerization. Specifically, the reduced interactions between the finger domain and CTD of the E634Q mutant polymerase allow us to show the displacement of the CTD (Fig. 1B). The noncatalytic Mn2+ lowers the stability of the polymerase so that after the formation of the dinucleotide initiation complex, the CTD can be displaced to allow chain elongation (although Mg2+ can take the position of the noncatalytic Mn2+, it does not destabilize the protein). As the CTD is displaced, the noncatalytic Mn2+ is lost and has to be replaced to allow elongation. It appears that this switch to elongation is the only step in the ϕ6 RdRp-catalyzed polymerization reaction which requires external Mn2+ ions.
MATERIALS AND METHODS
RNA and DNA templates.
Synthetic single-stranded RNAs (ssRNAs) were produced by runoff in vitro transcription with T7 polymerase (12). ϕ6-specific l+ ssRNA (message sense ssRNAs for the large genome segment of ϕ6) was produced from XbaI-cut, mung bean nuclease-treated plasmid pLM687 (22) as previously described (11). Plasmids pEM15 and pEM16 contain the cDNA of ϕ6 S segments with an internal deletion (21) and were used to prepare templates for Δs+ ssRNA with different 3′-end extensions (Δs+, Δs+C, Δs+CCC, and Δs+13) and Δs−, respectively. Templates for Δs+, Δs+C, and Δs+13 ssRNAs were produced as previously described (21, 28). Δs− RNA was transcribed from BpuAI-cut plasmid pEM16. Template for Δs+CCC ssRNA (ϕ6 small genome segment with internal, 2,225-nucleotide [nt]-long deletion and CCC extension at the 3′ end) was PCR amplified from plasmid pEM15 (21) using oligonucleotides T7-1 (42) and 3′end_6 (5′-GGGAGAGAGAGAGCCCCCGA-3′) as upstream and downstream primers, respectively. All produced ssRNAs were successively extracted with TRIzol (Invitrogen)-chloroform (5:1), precipitated with 4 M LiCl and with 0.75 M ammonium acetate, and dissolved in sterile water.
The online Integrated DNA Technologies SciTools OligoAnalyzer (version 3.1) with hairpin function was used to design high-melting-temperature hairpin oligonucleotides. The hairpins, obtained from Operon and used for crystal soaking experiments, were Tetra2T (TTCGCGTAAGCG) and Tetra4T (TTTTCGCGTAAGCG) with hairpin melting temperatures of 72.8°C and Tri2T (TTCGCGTAGCG) and Tri4T (TTTTCGCGTAGCG) with hairpin melting temperatures of 72.1°C (underlined residues base pair). The three 3′-terminal nucleotides of the oligonucleotide 5′-AATCT-3′ correspond to the 3′ end of the positive-sense strands of ϕ6 genomic RNAs (UCUCU-3′).
ϕ6 RdRp purification and polymerase assays.
Escherichia coli BL21(DE3) (Novagen) containing plasmid pEMG2 (27), pSVe4 (28), or pSVe2 (32) was used for expression of WT, E491Q, and E634Q RdRps, respectively. The other mutated ϕ6 RdRps were expressed from plasmids pSVe14 (D453-to-A mutation; SAD) (27), pSVe3 (R30-to-A mutation) (32), pSVe5 (K541-to-L mutation) (32), pMH6 (E165-to-A mutation) (32), and pRT2 (QYKW [629 to 632]-to-SG mutation) (19). Expression conditions and the purification procedure have been described elsewhere (21, 27).
The replication and transcription activity of ϕ6 RdRp was typically assayed in 10-μl reaction mixtures containing 50 mM HEPES-KOH, pH 7.5, 20 mM ammonium acetate, 6% (wt/vol) polyethylene glycol (PEG) 4000, 5 mM MgCl2, 2 mM MnCl2, 0.1 mM EDTA, 0.1% Triton X-100, 0.2 mM UTP and CTP, 1 mM ATP and GTP, 0.8 unit/μl RNasin (Promega), and 0.1 mCi/ml of [α-32P]UTP (3,000 Ci/mmol; Amersham Biosciences). The final concentration of the RNA template was set equimolar to the final ϕ6 RdRp protein concentration (270 nM). Unless otherwise stated, the mixtures were incubated at 30°C for 1 h and stopped with 2× U buffer (25).
For the heparin trap assay, ϕ6 RdRp was preincubated with ssRNA template and selected NTPs as indicated. After 20 min incubation at 30°C, 9.1 mg/ml heparin (final concentration; Sigma) or an equal volume of Milli-Q water (positive control) was added and the incubation was continued for an additional 5 min at 30°C. Subsequently, the reaction mixtures were supplemented with the rest of the NTPs and 0.1 mCi/ml [α-32P]UTP, followed by incubation at 30°C for 1 h (elongation reaction). When required, preassembled elongation complexes were purified through desalt spin columns (Pierce) before heparin addition. Conditions for initiation and elongation reactions were adjusted as indicated.
Reaction products were separated by agarose gel electrophoresis or by denaturing 20% polyacrylamide gel electrophoresis in 50% (wt/vol) urea, Tris-borate-EDTA buffer, and dried on Whatman no. 3 filter paper, followed by autoradiography and/or phosphorimaging (Fuji BAS1500 or FLA 5000) analyses of the product bands. Digital image analysis was performed using TINA (version 2.09c) or AIDA (version 3.44) image analyzer software (Raytest Isotopenmeßgeräte GmbH, Germany). The rate of elongation was estimated as previously described (21). Alternatively, aliquots of the reaction mixture were soaked into Whatman no. 1 filter paper, precipitated with trichloroacetic acid, and washed with acetone to measure the label incorporation by liquid scintillation counting.
Thermal shift assay.
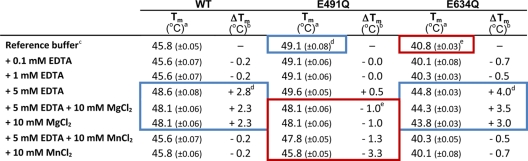
The thermal shift assay (ThermoFluor) was carried out as previously described (28). The melting temperature (Tm) was calculated using GraphPad Prism software and the Boltzmann distribution function. The effect of Mg2+ and Mn2+ was assayed by comparing melting temperatures of protein in reference buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl) with those of protein in reference buffer plus different additives (Table 1).
Table 1.
ThermoFluor statistics
aTm values shown were calculated by applying a Boltzmann distribution equation about the sigmoidal melting curves to obtain the inflection point (the midpoint of the unfolding transition) of the slope (GraphPad Prism software). Values in parentheses indicate the standard error for calculated Tm values.
bAny shift in Tm (ΔTm) from the reference Tm under different conditions indicates a change in protein stability. An increase in Tm (+ΔTm) indicates a stabilization of the protein by an increase in structural order and a reduction in conformational flexibility; a decrease in Tm (−ΔTm) indicates a destabilization (14).
cReference buffer is 50 mM Tris-HCl, pH 8, 50 mM NaCl.
dBlue box highlights significant stabilization compared to WT in reference buffer.
eRed box highlights significant destabilization compared to WT in reference buffer.
Protein crystallization and soaking experiments.
Prior to crystallization by sitting-drop vapor diffusion, protein was concentrated, typically to 10 mg/ml in 10 mM Tris-HCl, pH 8.0, 100 mM NaCl. Optimal crystallization conditions were determined (28) using a Cartesian robot at the Oxford Protein Production Facility (39). A 3-row optimization of initial hits (39) resulted in crystals up to 200 by 110 by 80 μm3 in 20% PEG 4000, 100 mM HEPES (pH 7.5), 8.5% isopropanol, 15% glycerol, 2 mM MnCl2 for WT RdRp and 400 by 200 by 100 μm3 in 12% PEG 20000, 100 mM morpholineethanesulfonic acid (pH 6.5) for E634Q. The conditions used in the soaking experiments are given in Table 2.
Table 2.
Soaking conditions, data collection, and refinement statistics
| Parameter | E634Q | E634Q-Tri2T-AMPPCP-Mg2+ | E634Q-Tri4T-AMPPCP-Mg2+ | E634Q-Tetra2T-AMPPCP-Mg2+ | E634Q-Tetra4T-AMPPCP-Mg2+ | WT-Tri4T-ATP-Mg2+ | WT-5′-AATCT-3′-ATP-GTP-Mg2+ | WT-5′-AATCT-3′-GTP-Mg2+ |
|---|---|---|---|---|---|---|---|---|
| Soaking conditions | ||||||||
| DNA oligonucleotide (all at 0.2 mM) | Tri2T | Tri4T | Tetra2T | Tetra4T | Tri4T | 5′-AATCT-3′ | 5′-AATCT-3′ | |
| NTP(s)a (all at 25 mM) | AMPPCP | AMPPCP | AMPPCP | AMPPCP | ATP | ATP, GTP | GTP | |
| MgCl2 concn (mM) | 10 | 10 | 10 | 10 | 10 | 10 | 10 | |
| Data collection statistics | ||||||||
| X-ray source | ESRF ID14-EH2 | ESRF ID14-EH2 | ESRF ID14-EH2 | ESRF ID14-EH2 | ESRF ID14-EH2 | Diamond I03 | Diamond I03 | Diamond I03 |
| Wavelength (Å) | 0.978 | 0.978 | 0.978 | 0.978 | 0.978 | 0.978 | 0.978 | 0.978 |
| Space group | P21 | P21 | P21 | P21 | P21 | P21 | P21 | P21 |
| Unit cell dimensions | ||||||||
| a, b, c (Å) | 106.2, 92.5, 141.0 | 107.4, 92.9, 141.2 | 107.5, 91.7, 142.1 | 106.9, 91.4, 141.6 | 105.8, 89.3. 141.0 | 105.9, 91.8, 140.5 | 106.3, 92.1, 140.8 | 106.5, 93.0, 140.8 |
| β (o)b | 101.5 | 101.9 | 102.0 | 101.9 | 101.9 | 101.3 | 101.3 | 101.4 |
| Resolution range (Å)c | 69.1–2.7 (2.8–2.7) | 77.2–3.0 (3.1–3.0) | 76.5–2.9 (3.0–2.9) | 104.6–3.0 (3.1–3.0) | 67.6–3.4 (3.5–3.4) | 50.0–3.3 (3.4–3.3) | 58.7–2.9 (3.0–2.9) | 50.0–3.5 (3.6–3.5) |
| No. of observations | 553,323 | 377,954 | 318,362 | 528,644 | 189,936 | 302,147 | 201,019 | 258,004 |
| No. of unique reflections | 74,265 | 50,754 | 53,974 | 51,249 | 30,875 | 39,855 | 55,288 | 34,154 |
| Completeness (%)c | 97.5 (94.0) | 98.4 (96.9) | 98.5 (96.4) | 99.1 (98.1) | 86.8 (61.6) | 100.0 (100.0) | 95.2 (95.5) | 100.0 (100.0) |
| I/σI (%)c | 17.0 (2.3) | 13.3 (2.0) | 12.1 (2.0) | 14.7 (2.4) | 9.7 (2.1) | 6.3 (1.7) | 11.7 (1.6) | 4.9 (2.0) |
| Rmerge (%)d | 9.2 | 18.1 | 19.1 | 17.5 | 17.0 | 26.7 | 9.1 | 32.5 |
| Refinement statistics | ||||||||
| Resolution range (Å) | 69.1–2.7 | 69.6–3.0 | 64.5–2.9 | 104.6–3.0 | 67.6–3.4 | 46.0–3.3 | 46.1–2.9 | 46.5–3.5 |
| No. of reflections (working/test) | 70,513/3,736 | 48,162/2,577 | 51,206/2,742 | 48,616/2,600 | 29,228/1,545 | 37,842/2,000 | 52,462/2,806 | 32,415/1,726 |
| Rwork/Rfreee | 21.5/22.5 | 20.5/22.0 | 21.6/25.0 | 21.0/24.3 | 21.8/25.5 | 24.3/29.0 | 17.9/20.9 | 27.6/30.7 |
| No. of atoms (protein/ligand/water) | 15,795/3/0 | 15,672/253/1 | 15,690/294/1 | 15,631/294/0 | 15,615/274/0 | 15,795/308/0 | 15,795/282/0 | 15,795/235/0 |
| RMSDf | ||||||||
| Bond length (Å) | 0.008 | 0.008 | 0.009 | 0.008 | 0.008 | 0.015 | 0.012 | 0.008 |
| Bond angle (°) | 1.0 | 1.0 | 1.1 | 1.0 | 1.0 | 1.4 | 1.2 | 1.0 |
| Mean B factor (Å2) | 60.6 | 71.8 | 97.3 | 75.4 | 119.2 | 75.2 | 66.9 | 98.6 |
| NCS (% restrained/xyz RMSD [Å]) | 85/0.3 | 75/0.4 | 75/0.5 | 74/0.4 | 68/0.6 | 80/0.5 | 80/0.5 | 81/0.3 |
| RMSD deviation in xyz/B from PDB accession no. 1HHS (Å/Å2) | 0.2/7 | 0.2/8 | 0.2/8 | 0.2/9 | 0.3/11 | 0.4/14 | 0.5/10 | 0.2/11 |
| PDB/SF codes | 4a8o/r4a8osf | 4a8q/r4a8qsf | 4a8s/r4a8ssf | 4a8w/r4a8wsf | 4a8y/r4a8ysf | 4a8f/r4a8fsf | 4a8m/r4a8msf | 4a8k/r4a8ksf |
β,γ-Methyleneadenosine 5′-triphosphate (AMPPCP), ATP, and GTP were all lithium salts from Sigma.
α = γ = 90°.
Values in parentheses refer to the highest-resolution shell.
Rmerge = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣi〈I(h)〉, where Ii(h) is the ith measurement and 〈I(h)〉 is the average of all measurements Ii(h).
Rwork and Rfree are defined by R = Σhkl||Fo| − |Fc||/Σhkl |Fo |, where h, k, and l are the indices of the reflections (used in refinement for Rwork; 5%, not used in refinement, for Rfree), Fo and Fc are the structure factors, deduced from intensities measured and calculated from the model, respectively.
RMSD, root mean square deviation.
Data collection and processing.
X-ray diffraction data for ϕ6 RdRp complex structures were collected at the Diamond Light Source, Oxfordshire, United Kingdom (station I03) and the European Synchrotron Radiation Facility (ESRF), Grenoble, France (station ID14-EH2). All data were collected from crystals cryocooled in a stream of nitrogen gas at 100 K. Prior to cryocooling, the crystals were soaked for 1 to 10 min under the specified condition with 20% (vol/vol) glycerol as cryoprotectant (see Protocol S1 in the supplemental material for details). WT-Tri4T-ATP-Mg2+ and WT-5′-AATCT-3′-GTP-Mg2+ data were integrated and scaled using the HKL-2000 program (24), but for other data, the automated data reduction program XIA2 (41) with the three-dimensional option (Labelit, XDS, and XSCALE algorithms) (16) was used. Data sets were approximately isomorphous to the WT crystals previously described (6, 7, 31), belonging to space group P21, with a ≈ 106 Å, b ≈ 92 Å, c ≈ 142 Å, and β ≈ 102°. Data-processing statistics are given in Table 2.
Structure refinement.
Initial structures were obtained by rigid-body refinement of the three molecules present in the asymmetric unit with the PHENIX program (1), using the WT apo-P21 structure as a starting model. Ligands were built by visual inspection of the SIGMAA 2|Fo| − |Fc| difference-Fourier maps in the COOT program (9) (see Protocol S1 in the supplemental material). Final refinement was done using the BUSTER-TNT program (5), with local structure similarity restraints used not only between the three noncrystallographic symmetry (NCS)-related copies in the asymmetric unit but also to a high-resolution (1.9 Å) target structure (Protein Data Bank [PDB] accession no. 1HHS) (33) (see Table 2 for statistics), and figures were produced with the PyMOL program (8).
RESULTS
In the subsequent sections, we describe the results from eight separate crystal structures, in conjunction with biochemical data, to dissect stages along the catalytic pathway of the ϕ6 RdRp. Despite the modest resolution of several of these structures (the lowest resolution being 3.5 Å), the extensive restraints, both 3-fold NCS and to a high-resolution structure (Table 2), allow us to be confident of the conformational changes and displacement in metal ions described below.
Divalent cations modulate the structural dynamics of ϕ6 RdRp. (i) Effect of Mn2+ and Mg2+ on thermal stability of ϕ6 RdRp.
Our previous studies suggested that Mn2+ ions modulate the flexibility of ϕ6 RdRp and that mutation of E491 to Q, which reduces the affinity for Mn2+ (28), stabilizes the enzyme. Here, we use comprehensive thermal denaturation assays to probe the stability of the WT RdRp, E491Q, and a new mutation, E634Q, under different divalent cation conditions. These assays show that saturating WT or E634Q RdRps with 10 mM MgCl2 is sufficient to outcompete the destabilizing effect of the noncatalytic Mn2+ and results in an increase in thermal stability similar to that observed by introducing 5 mM EDTA (sufficient to remove the bound ion from the RdRp) (Table 1; see Fig. S1A and C in the supplemental material). Furthermore, the melting curves obtained for the E634Q mutant, in which the mutation prevents the formation of the E634-K144 salt bridge, pinning the CTD to the main body of the protein (32) (Fig. 1B and Table 1; see Fig. S1C in the supplemental material), revealed a significantly lower thermal stability than that for the WT polymerase (melting temperature, 41°C, compared to 46°C for WT). This decrease in thermal stability is presumably due to increased flexibility of the CTD in the absence of the stabilizing salt bridge.
(ii) Stability of ϕ6 RdRp · Mn2+complex.
ThermoFluor analysis (Table 1; Fig. S1 in the supplemental material) indicates that a 3,850 molar excess of the divalent cation-chelating agent EDTA over ϕ6 RdRp (in 5 mM and 1.3 μM concentrations, respectively) is sufficient to extract the noncatalytic Mn2+ ion from the RdRp, while in the presence of a 770 molar excess of EDTA (1 mM EDTA; Table 1), the RdRp · Mn2+ complex is stable for the time period of the measurement (see Fig. S1 in the supplemental material). Based on this information and the known dissociation constant, Kd, of the Na2EDTA · Mn2+ complex (1.6 × 10−14; and a corresponding stability constant, log K, of 13.8 [30]), we can estimate that the Kd for the ϕ6 RdRp · Mn2+ complex is in the range of 10−17 to 10−18. These data are supported by crystallographic analysis of a crystal soaked in 50 mM EDTA, which shows the quantitative elimination of Mn2+ from the noncatalytic site (see Fig. S2 in the supplemental material).
(iii) Structure reveals the basis for Mg2+-induced thermal stabilization.
To establish if the Mg2+-induced increase in thermal stability arises from replacement of the noncatalytic Mn2+ by Mg2+, WT ϕ6 RdRp crystals were soaked with 10 mM MgCl2 together with an oligonucleotide template and an NTP(s) (WT crystals; Table 2). Replacement of the noncatalytic Mn2+ by Mg2+ was confirmed by differences in electron density: Mn2+ and Mg2+ possess 23 and 10 electrons, respectively, and a simple isomorphous difference map, calculated using the observed structure factor amplitudes from Mn2+- and Mg2+-soaked crystals, revealed strong positive density at the noncatalytic divalent cation site (Fig. 2). The disposition of the surrounding ligands confirmed that the residual density was attributable to Mg2+ rather than a water molecule (for distances, see Fig. 2). A final piece of evidence is that no peaks are seen in anomalous difference maps, although 8σ peaks are seen for a data set collected at the same wavelength but containing Mn2+ at the noncatalytic site (data not shown). This Mg2+ was found in place of the noncatalytic Mn2+ in all three molecules of the crystallographic asymmetric unit (molecules denoted I, II, and III) of the P21 WT crystals (Fig. 2). Thus, Mg2+ can occupy this binding site (Fig. 2), but ThermoFluor data demonstrate that it does not act as a destabilizing factor (Table 1; see Fig. S1 in the supplemental material).
Stepwise assembly of initiation complex.
The replacement of Mn2+ by Mg2+ has allowed us to capture crystallographically preinitiation stages not observed when Mn2+ is bound, presumably reflecting changes in the kinetics of complex formation due to stabilization of the enzyme.
The three RdRp molecules in the crystal asymmetric unit exhibit different features (Table 2). Molecules I and II are stabilized by a substantial number of crystallographic contacts and tend to behave similarly, in terms of ligand binding, whereas molecule III has fewer contacts and exhibits much greater flexibility. Furthermore, the differences in crystal packing trapped an incoming oligonucleotide between molecules II and III, but not at other positions (see below and Fig. 3).
Fig 3.
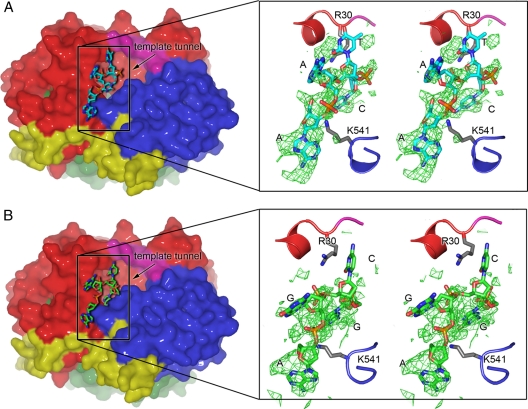
Template binding on the surface of the ϕ6 RdRp. WT-5′-AATCT-3′-ATP-GTP-Mg2+ (molecule II) (A) and WT-Tri4T-ATP-Mg2+ (molecule II) (B) structures. Molecular surfaces (left) and fragments of polypeptide chains in the close-up stereo images (right) colored as described in the legend to Fig. 1. Oligonucleotides are depicted using stick presentation (carbon, cyan or green; nitrogen, blue; oxygen, red; phosphorus, orange). The template tunnel entrance is indicated by an arrow (black). (Right) |Fo| − |Fc| SIGMAA weighted electron density maps contoured to 3σ for the oligonucleotide (green wire); phases were calculated from models with the oligonucleotide removed. Residues R30 and K541 (stick presentation; carbon, gray; nitrogen, blue; oxygen, red), which sit on either side of the template tunnel, stabilize the incoming template. Due to local disorder in the bases, nucleotides were assigned by best fit of sequence into the available electron density; assigned bases and positions of R30 and K541 are indicated.
(i) Template entry.
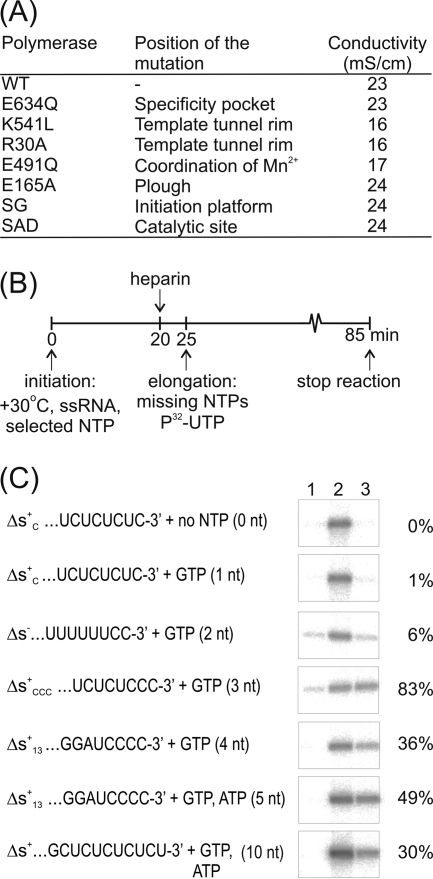
Replacement of the noncatalytic Mn2+ by Mg2+ clearly affected the ability of the WT polymerase to bind template. Neither of two tested oligonucleotides (5′-AATCT-3′ or Tri4T) could enter the template tunnel (Fig. 3), although strong density for the template in the tunnel was previously detected in Mn2+-bound WT structures (6, 31). This is in line with the weakened template binding of the E491Q mutant (28). Although template could not be detected inside the binding tunnel, 4 nucleotides of DNA could be fitted into electron density at the outermost region of the tunnel of molecule II for all the three WT complexes (Fig. 3; Table 2). The template enters the tunnel over a positively charged groove on the outer surface of the RdRp (Fig. 3). Residues R30 and K541, which lie in the center of two positively charged patches on either side of the template tunnel, make direct contacts with the phosphate backbone of the incoming template (Fig. 3), potentially facilitating the movement of the template toward the active site. This explains mutagenesis results demonstrating a key role for R30 and K541 in template RNA binding and preinitiation events (32) as well as in heparin binding (see below and Fig. 4A). Interestingly, the structure showed that the predicted hairpin of the Tri4T was opened on the surface of the RdRp (Fig. 3B), suggesting that interactions with the molecular surface of the RdRp may allow denaturation of RNA secondary structures before the template enters into the template tunnel.
Fig 4.
Heparin sensitivity of the ϕ6 RdRp-catalyzed RNA polymerization reaction. (A) Affinity difference of heparin binding by WT RdRp and different ϕ6 mutant polymerases. Purified RdRps were eluted from a heparin agarose column with an increasing concentration of NaCl, and the electrical conductivity of the RdRp peak fraction was measured. (B) Diagram of the heparin trap assay and experimental setup. (C) Heparin-sensitive phases of the ϕ6 RdRp-catalyzed RNA polymerization reaction. The nucleotide substrates in the initiation mixtures and the templates with their 3′-terminal sequences are indicated (left). The radioactively labeled full-length dsRNA products were separated in a standard agarose gel and autoradiographed (right). Heparin was added to the initiation mixture before nucleotides were added (negative control; lane 1), no heparin was added (positive control; lane 2), or heparin was added after 20 min incubation of the initiation mixture (lane 3). The maximal length of the nascent RNA strand (in nucleotides) synthesized prior to the addition of the elongation mixture is in parentheses. The RNA polymerization products were quantitated by phosphorimager analysis. Lane 1 values were set to 0%, and lane 2 values were set to 100%. Percentages on the right depict the polymerization activity measured in heparin-trapped reactions (lane 3) (mean of two independent experiments).
(ii) Divalent cation movements direct NTPs toward the initiation platform.
Although replacement of the noncatalytic Mn2+ by Mg2+ prevented the entry of the template, a GTP molecule (Fig. 2B) or two NTPs (Fig. 2C to E) were observed at the active site. In the GTP-bound structure, the Mg2+ at the noncatalytic site is coordinated by the side chains of D454 and E491, as well as the carbonyl backbone of A495, as seen in the Mn2+-bound state (Fig. 2B, downward position). In this complex, the triphosphate group of the GTP is stabilized by the basic residues K223, R225, R268, and R270 that form interrogation site I (6). However, in the structures with two bound NTPs, two clearly distinct states were captured for a bound Mg2+ (Fig. 2C to E, centered and upward). In the centered position (Fig. 2C), an Mg2+ sits directly between the side chains of D454 and E491, coordinated additionally by A495 and one of the phosphate groups of the D1 NTP. In the upward position (Fig. 2E), the Mg2+ sits above the side chains of D454 and E491 and is also coordinated by the β- and γ-phosphates of the D1 NTP (no interaction with A495). We predict that Mg2+ continually flips between a downward and upward position about the D454 and E491 axis of the noncatalytic ion site, stabilizing the nearby negative charge of the D1 NTP triphosphate backbone when both NTPs are bound during preinitiation. In agreement with this, the electron density in WT-Tri4T-ATP-Mg2+ (molecule I) suggests partial occupancy of Mg2+ in both upward and downward states (Fig. 2D). It is important to bear in mind that some of the positions of NTPs and Mg2+ in these preinitiation structures (Fig. 2) may not be on the reaction pathway, and these results are probably best interpreted as snapshots of NTPs interrogating the active site at sufficiently high NTP concentrations (25 mM) to be captured by crystallography.
Biochemical dissection of the steps leading to elongation. (i) Heparin and template RNA binding sites overlap.
The initial phase of the ϕ6 RdRp-catalyzed polymerization reaction is sensitive to heparin, but elongation is not (43). To understand the mechanism of heparin inhibition and to localize the heparin binding site in ϕ6 RdRp, we analyzed the relative strength of the interaction between heparin and a battery of mutant enzymes (Fig. 4A). Mutations that reduce the binding affinity for template RNA (E491Q, R30A, and K541L) (28, 32) also significantly reduce the affinity for heparin (Fig. 4A), suggesting that the binding sites for RNA and heparin in the ϕ6 RdRp molecule overlap. This implies that the heparin inhibition assay is based on competition between heparin and the template and that the efficiency of inhibition reflects the stability of the initiation/replication complex.
(ii) Heparin blocks steps preceding the transition to elongation.
To determine the precise stage in the ϕ6 RdRp-catalyzed polymerization reaction that accounts for the heparin sensitivity, standard polymerization reactions were carried out using a set of template-NTP combinations designed to stall the reactions at different stages and thus restrict the maximal length of the nascent strand (Fig. 4B and C). The complexes formed were analyzed for sensitivity to heparin (see Fig. 4B and C for the experimental setup). As expected, the overall efficiency of the polymerization reaction in the presence of heparin (Fig. 4C, lane 3) was always less than that for the positive control (with no heparin, lane 2), and pretreatment with heparin efficiently abolished RNA synthesis (negative control, lane 1). Apparently, if the initiation (or elongation) complexes dissociate, heparin will bind to the released apo-RdRp, decreasing the number of successful elongation events and consequently biasing the results somewhat toward reduced polymerization activity. Heparin, as a divalent metal ion chelate, also reduces the concentration of free divalent metal ions and consequently shifts the conditions slightly from the optima. There was substantial template-dependent variability in the heparin-treated reactions (Fig. 4C, lane 3). However, it was possible to detect a clear transition from heparin sensitivity to heparin resistance when the third nucleotide is added to the dinucleotide initiation complex. This agrees with existing structural data (6) and suggests that the heparin trap assay provides a way to dissect the initiation and elongation phases.
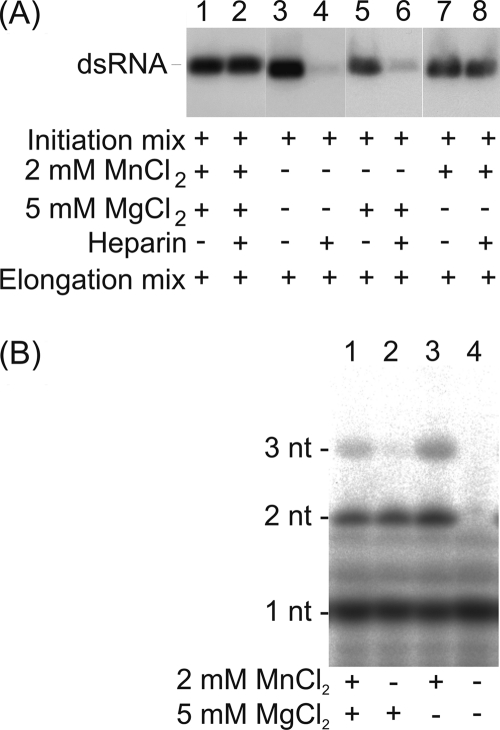
Initiation to elongation switch is significantly stimulated in the presence of external Mn2+. (i) Manganese ions are required to reach the heparin-resistant stage in the ϕ6 RdRp-catalyzed polymerization reaction.
In the absence of external MnCl2, the reaction was unable to reach the heparin-resistant elongation state (Fig. 5A, lanes 4 and 6). This implies that Mn2+ is essential for the initial phase of polymerization, prior to the formation of the nascent trinucleotide product. This Mn2+ dependence is unlikely to arise from requirement of the ion for template association, as RNA binding by ϕ6 RdRp is independent of the divalent cation conditions applied (no ions, 2 mM MnCl2, or 5 mM MgCl2 [28]).
Fig 5.
Mn2+ dependence of the initial phase of the ϕ6 RdRp-catalyzed RNA polymerization reaction. ϕ6 RdRp was incubated with ssRNA template sΔ+CCC and GTP substrate (synthesis of 3 nucleotides) under various divalent cation conditions for 20 min. (A) Heparin trap assay adding divalent cations at up to optimal concentrations into the elongation reaction mixture. The reaction products were analyzed by standard agarose gel electrophoresis and autoradiography. (B) [γ-32P]GTP substrate was applied, and the reaction products of the initiation mixture were purified using microspin columns (Pierce) before analysis by urea-containing 20% polyacrylamide gel and phosphorimaging. The mobility of dsRNA (A) or mono-, di-, and trinucleotides (B) is indicated on the left, and the divalent cation concentrations in the initiation mixtures and the heparin additions are shown below.
(ii) External Mn2+ ions are not required for the formation of the initiation complex.
The role of Mn2+ ions was analyzed in more detail by separating the products of the initiation reaction (sΔ+CCC template and [γ-32P]GTP substrate) in a high-resolution urea-containing polyacrylamide gel (Fig. 5B). Under optimal conditions (2 mM MnCl2, 5 mM MgCl2), a trinucleotide product (pppGpGpG) was synthesized, as expected (Fig. 5B, lane 1). However, if Mn2+ ions were omitted from the mixture, mainly the dinucleotide initiation complex was produced (Fig. 5B, lane 2). Due to abortive initiation, this was a predominant product also in the presence of Mn2+. In the absence of both Mn2+ and Mg2+, no catalysis occurred (Fig. 5B, lane 4). Furthermore, Mn2+ ions could support the formation of trinucleotides in the absence of Mg2+ ions (Fig. 5B, lane 3). This indicates that the addition of Mn2+ ions is not required for the formation of the initiation complex but is essential for the subsequent nucleotide addition.
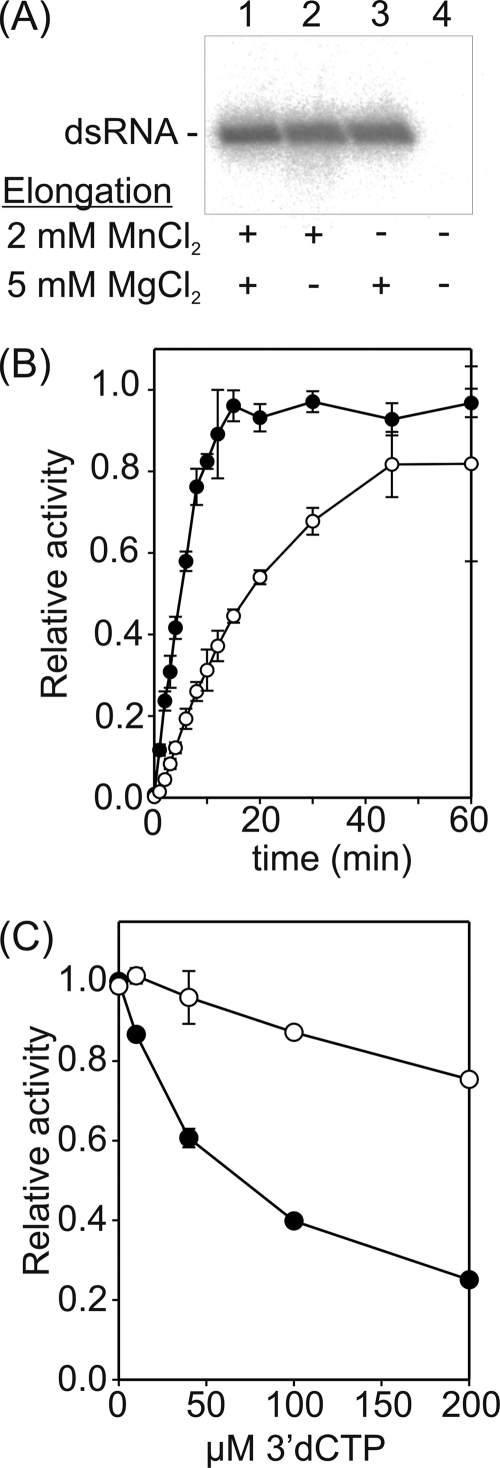
(iii) Mn2+ is not an essential component of the elongation reaction.
We analyzed the effect of Mg2+ and Mn2+ on RNA chain elongation after the formation of a 3-nucleotide product (Fig. 6). Assembled elongation complexes were purified and elongation reactions were carried out under a range of divalent cation conditions (Fig. 6A). dsRNA was produced regardless of the presence of Mn2+ ions in the elongation reaction mixture if Mg2+ was present (Fig. 6A, lanes 1 to 3). However, when all divalent cations were omitted, no dsRNA was detected (Fig. 6A, lane 4), confirming that there was no carryover of divalent cations to the elongation reaction.
Fig 6.
Mn2+ dependence of the ϕ6 RdRp-catalyzed polymerization reaction after transition to elongation. ϕ6 RdRp was incubated with ssRNA template sΔ+CCC and GTP (A and B; synthesis of 3 nucleotides) or l+ ssRNA, GTP, and ATP (C; synthesis of 10 nucleotides) under optimal divalent cation conditions for 20 min. Subsequently, reaction complexes were purified through desalting spin columns (Pierce), and the formation of new initiations was suppressed by heparin. (A) Effect of Mn2+ on ϕ6 RdRp-catalyzed elongation reaction; (B) time-dependent accumulation of replication elongation products in the presence (filled circles) and absence (open circles) of Mn2+; (C) polymerization activity in the presence of increasing concentrations of 3′-dCTP under different divalent cation conditions (open circles, 5 mM MgCl2; filled circles, 2 mM MnCl2 and 5 mM MgCl2). The reaction products were analyzed by standard agarose gel electrophoresis and phosphorimaging analysis (A and B), or label incorporation was measured by liquid scintillation counting (C). The divalent cations in the elongation mixtures (A) are indicated below the autoradiogram. The graphs are normalized so that the highest observed value within each plot is set to 1 (B and C). The error bars represent the standard deviation of the mean from three (B) or two (C) replicates.
More detailed characterization revealed that the rate of elongation was reduced to approximately one-fourth (6 nt versus 23 nt) if the elongation mixture did not contain Mn2+ (Fig. 6B). As an increase in elongation rate could lead to reduced fidelity, the effect of Mn2+ on the incorporation of 3′-dCTP by ϕ6 RdRp was evaluated (Fig. 6C). 3′-dCTP, lacking the 3′ hydroxyl group, acts as chain terminator. Accordingly, addition of increasing concentrations of 3′-dCTP to the reaction mixture decreased the overall polymerization activity regardless of the conditions applied (Fig. 6C). However, the effect was milder if Mn2+ was omitted from the reaction buffer. This implies that Mn2+ ions reduce discrimination against 3′-dCTP, possibly because the catalytic Mg2+ ions are at least partially replaced by Mn2+ ions.
C-terminal domain movement opens a path for template exit. (i) Movement of the C-terminal domain demonstrated using a mutant RdRp with increased flexibility.
Since we have been unable to obtain crystallographic evidence for CTD movement with WT enzyme, we investigated the structure of the E634Q mutant where the association of the CTD with the main body of the protein is likely to be weaker (Fig. 1B and Table 1; see Fig. S1C in the supplemental material).
The crystal structure of the E634Q mutant revealed that Mn2+ bound at the noncatalytic sites of all three molecules in the crystal asymmetric unit (Table 2). Furthermore, consistent with the WT structures (Fig. 2), the Mn2+ could be replaced by Mg2+ by soaking with MgCl2 (Table 2). The structure of Mn2+-bound E634Q was essentially indistinguishable from that of the WT enzyme (6), apart from the lack of the salt bridge between K144 and E634 (Fig. 1B), conferring increased flexibility on residue Q634 (data not shown).
Unlike the WT enzyme (Fig. 3), the E634Q mutant retained its relatively high intrinsic flexibility (Table 1; see Fig. S1C in the supplemental material) and ability to bind DNA in the template binding tunnel even in the presence of 10 mM MgCl2 (Fig. 7 and other E634Q-hairpin-AMPPCP-Mg2+ structures, where AMPPCP is β,γ-methyleneadenosine 5′-triphosphate [Table 2]). Under these conditions, the DNA hairpin was unwound and threaded through the template tunnel (Fig. 7) so that 3, 4, and 6 bases of DNA could be modeled into the central portion of the template tunnel of molecules I, II, and III, respectively, with the best-ordered DNA density being seen in the least constrained molecule, molecule III. In the presence of template, molecule III also showed evidence for CTD movement: in all complexes with DNA oligonucleotides (Table 2), residues 606 to 614 and 631 to 647 are highly disordered, with little or no evidence of electron density (Fig. 7 and data not shown). The disordering is apparently induced by the deeper penetration of the oligonucleotide into the template tunnel, passing the active site and mimicking the switch to elongation.
Fig 7.
CTD disordering and loss of the noncatalytic ion. (A) Cartoon of ϕ6 RdRp colored by B factor. (Left) The high-resolution WT structure (PDB accession no. 1HHS) is drawn looking into the C-terminal domain, color ramped between 10 and 110 Å2 as blue via green to red; (right) the model for E634Q-Tri4T-AMPPCP-Mg2+ (molecule III) is drawn in the same view, with the RNA oligonucleotide in purple, highlighting the disordered sections of the C-terminal domain. To maintain the same baseline, the color ramp varies from 50 to 150 Å2. (B) Cross-section of molecular surfaces of E634Q-Tri4T-AMPPCP-Mg2+ (molecule III) with secondary structure elements colored as described in the legend to Fig. 1. CTD residues 606 to 614 and 631 to 647 are highly disordered and are missing from the structure, allowing exit of unwound Tri4T hairpin (carbon, cyan; nitrogen, blue; oxygen, red; phosphorus, orange). Molecular surfaces of disordered CTD residues are modeled onto the structure (crosshatched semitransparent wheat) and depict the entire CTD blocking the template exit path. The right panel is a stereo image close-up of unwound Tri4T hairpin colored and oriented with respect to the left panel. 2|Fo| − |Fc| SIGMAA electron density maps (blue wire) are shown and contoured to 1.2σ. Due to local disorder in the bases, nucleotides were assigned by best fit of the unwound sequence into the available electron density; one-letter codes mark the assigned bases (right). (C) Close-up of the noncatalytic ion site for molecules I to III of E634Q-Tri4T-AMPPCP-Mg2+. Palm domain (green) with stick representations of amino acids D454, E491, and A495 (carbon, gray; nitrogen, blue; oxygen, red). Mg2+ (a large black sphere) in molecules I and II and a water molecule (a small red sphere) in molecule III are shown within the 2|Fo| − |Fc| SIGMAA electron density maps contoured to 1.2σ (blue wire).
(ii) Loss of the noncatalytic ion upon C-terminal domain displacement.
In molecules I and II, which possess an ordered CTD, an Mg2+ ion is seen at the noncatalytic ion site coordinated by D454 and E491 in the standard downward position (Fig. 7B); however, there is little electron density at this site in molecule III (Fig. 7B), suggesting that disordering of the CTD has released the bound divalent cation.
(iii) NTPs carry Mg2+ ions to the catalytic site.
The electron density maps for the different E634Q-hairpin-AMPPCP-Mg2+ complexes (Table 2) showed evidence for an AMPPCP molecule bound at the interrogation site (6). As expected, the backbone triphosphates were coordinated by positively charged residues K223, R225, R268, and R270, and the sugar and base moieties were disordered. An Mg2+ ion was found in molecule I of two of the E634Q complexes (E634Q-Tri4T-AMPPCP-Mg2+ and E634Q-Tetra2T-AMPPCP-Mg2+), coordinated by the triphosphates of AMPPCP, the side chain of D324, and the backbone carbonyl group of V325 (data not shown). This is 5 Å away from the closest of the catalytic Mg2+ sites previously identified in the ϕ6 RdRp initiation complex structure (6) and suggests that NTP · Mg2+ enters the catalytic site as a preformed complex.
DISCUSSION
Noncatalytic ion sites corresponding to the site seen in the ϕ6 RdRp have been observed in a number of other viral RNA polymerases (28) (see Table S1 in the supplemental material). We have demonstrated that at a high concentration (10 mM), Mg2+ can replace Mn2+ at this position in the ϕ6 enzyme (Table 1; Fig. 2). Since several of the crystallization conditions for other RdRps had high concentrations of Mg2+ salts (up to 300 mM; see Table S1 in the supplemental material), it is conceivable that Mn2+ could be the natural ion at this site in all of these polymerases. Where no divalent ions have been seen, this might reflect their loss during purification due to chelating agents being included (e.g., West Nile virus, foot-and-mouth disease virus, Norwalk virus, and reovirus; see Table S1 in the supplemental material). Thus, the important role of Mn2+ in controlling the switch to elongation which we have identified for ϕ6 RdRp (Fig. 5) might also apply to other viral RdRps, explaining their Mn2+ dependence.
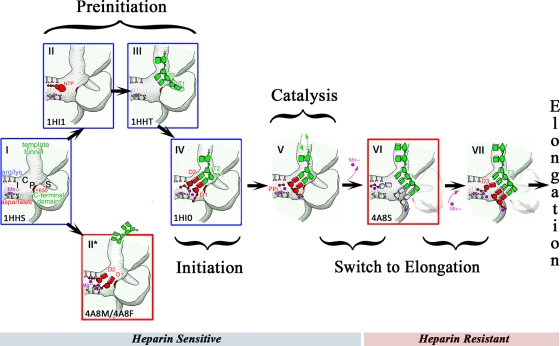
We have put together the results from this study in Fig. 8, which rationalizes the sequence of events covering the early stages through initiation to elongation for the ϕ6 polymerase model system. Each stage of the model is outlined below, focusing on the role of Mn2+ at the noncatalytic ion site in de novo RNA synthesis.
Fig 8.
Cartoon model for ϕ6 RdRp-catalyzed polymerization. Key steps have been characterized crystallographically (red box, determined as part of this study; blue box, determined previously; codes in the bottom left-hand corner of each box, the relevant PDB accession numbers). Step I, polymerase with Mn2+ (magenta sphere) bound at noncatalytic ion site. The positions of the interrogation site (Arg/Lys, site I), catalytically relevant NTP binding site (site C), priming platform (Y630, site P), and specificity pocket (site S) are indicated; nomenclature is according to reference 6. The template tunnel, C-terminal domain (CTD), and the catalytic aspartates are also indicated. Steps II and III (preinitiation), continuous entry and exit of template and/or D1 and D2 NTPs with Mg2+ ions (purple spheres). NTPs in the absence of Watson-Crick base pairing with template can be stabilized about the noncatalytic ion site if occupied by Mg2+ (step II*). The 3′ end of the template can enter the tunnel with Mn2+ at the noncatalytic ion site and binds at site S (step III) (6) so that the penultimate nucleotide (T2) is held at the active site (site P); arrows (black) indicate different preinitiation routes (NTPs or template binding first) that are sensitive to divalent cation conditions. Step IV (assembly of initiation complex), after binding of template at site S, D2 NTP shifts to base pair with the T2 nucleotide, at which point the template ratchets back to allow base pairing of the D1 NTP with the T1 nucleotide of template (see reference 6 for more detail); Mg2+ ions shift into catalytic position, and Mn2+ occupies the noncatalytic ion site, establishing the correct geometry for catalysis. Step V (catalysis), the released pyrophosphate (PPi) diffuses out from the polymerase with one bound Mg2+; the other catalytic Mg2+ ion stays associated with the 5′-triphosphate group of the daughter strand; template can still be lost from the tunnel at this stage (green arrows), allowing competition with heparin for entry (see reference 31 for more detail and PDB accession no. 1UVK). Heparin-sensitive and heparin-resistant steps are indicated by a bar below the cartoon. Steps VI and VII (switch to elongation), two-step mechanism involving CTD displacement (loss of Mn2+) and elongation complex assembly (recapture of Mn2+). Step VI, first, CTD displacement is facilitated by bound destabilizing Mn2+. The dinucleotide initiation complex shifts down, placing the template T3 nucleotide in the active site, allowing it to base pair with an incoming D3 NTP. Upon displacement of the CTD, there is transient weakening in the high-affinity noncatalytic ion site and concomitant loss of the bound Mn2+; the reaction stalls at this point in the absence of external Mn2+ ions (in the cartoon, catalytic Mg2+ ions [purple spheres] and the positions of the daughter strand and incoming D3 NTP [light gray nucleotides] are shown for completeness but were not captured in the E634Q-hairpin-AMPPCP-Mg2+ structures). Step VII, the CTD is at its new position, and the high-affinity Mn2+ binding site is re-formed. The second step in the transition mechanism involves the D3 NTP moving from site I to base pair with the T3 nucleotide, now positioned at the active site. External Mn2+ ions promote efficient rebinding of Mn2+ at the noncatalytic ion site and establish the correct geometry for elongation complex assembly and catalysis of the third nucleotide; if external Mn2+ ions are not present, the reaction does not proceed. Synthesis of the trinucleotide daughter strand marks the beginning of elongation. The CTD is locked into its displaced position, Mn2+ is stably bound, and there is no further requirement for external Mn2+ (however, the presence of external Mn2+ increases the rate of elongation).
Apoenzyme and Mn2+-bound state (step I).
Our data reveal that the noncatalytic divalent cation binding site has sufficiently high affinity for Mn2+ that it copurifies with the polymerase (Fig. 8, step I). The estimated Kd for the ϕ6 RdRp · Mn2+ complex is extremely high but comparable, for instance, with that reported for the extrinsic 23-kDa protein of photosystem II (4). The high affinity for Mn2+ is essential, given its low physiological concentration (10 μM) (10). The stable binding of Mn2+ increases the structural fluidity of the RdRp (Table 1), which is critical for its activity (see below), whereas the replacement of Mn2+ by Mg2+ (Table 1) results in stabilization of the enzyme.
Preinitiation (steps II and III).
Our crystallographic data reveal that Mg2+ ions enter into the polymerase as stable NTP · Mg2+ complexes and can have a number of geometries about the noncatalytic ion site (Fig. 2 and Fig. 8, steps II and II*). Additionally, we have demonstrated that the template enters over a positively charged groove on the outside of the polymerase (Fig. 8, step II*) and is postulated to be guided toward the catalytic site by two positively charged patches on either side of the template tunnel (Fig. 3; for genetic evidence, see reference 32). Entry of template into the tunnel with Mn2+ at the noncatalytic site has been observed previously (6, 31) (Fig. 8, step III) and is essential for initiation complex assembly. Replacement of the noncatalytic Mn2+ by Mg2+ prevented template entry (Fig. 3); also, mutation of E491 to Q reduces template binding (28) and consequently the assembly of the initiation complex. Nevertheless, our structural data suggest that Mg2+ can be drawn from the canonical noncatalytic Mn2+ site to either a central or upward position by the D1 and D2 NTPs present at the active site in the absence of Watson-Crick base pairing with the template (Fig. 2C to E and Fig. 8, step II*).
Initiation (step IV).
At the point of initiation, the D1 and D2 NTPs (coordinated at the noncatalytic site) shift 6 Å from the preinitiation position to base pair with the ultimate (T1) and penultimate (T2) nucleotides of the template, respectively (Fig. 8, step IV). As the NTPs move, the bound Mg2+ ions lock into their catalytic position. The strong affinity of the noncatalytic site for Mn2+ is critical for the formation of the functional initiation complex. In line with this, the E491Q mutant lacking Mn2+ formed an inactive initiation complex (28). The positions of divalent cations and NTP in a productive initiation complex are depicted in Fig. 8, step IV (6).
Catalysis (steps V and VI).
After linkage of the first two NTPs (which is dependent on the noncatalytic Mn2+ site being occupied to correctly structure the active site), the released pyrophosphate and one bound Mg2+ ion diffuse out of the active site, while the second catalytic Mg2+ ion stays associated with the 5′-triphosphate group of the initiated daughter strand. We previously captured a dead-end structure (PDB accession no. 1UVK) (31) of the products of this reaction where the oligonucleotide template has been lost from the binding tunnel, demonstrating that at this stage the template is not locked into the polymerase (31) (Fig. 8, step V). In line with this, our heparin trap assay demonstrates how RNA template and heparin compete for entry at the template binding tunnel during initiation, until a locked-in heparin-resistant elongation state (trinucleotide product) is reached (Fig. 4A and C).
Template movement and C-terminal domain displacement (step VI).
The E634Q mutant (which binds Mn2+ strongly, like WT) has a significantly lower thermal stability than WT even in the presence of 10 mM MgCl2 (Table 1). In this mutant the applied DNA template can shift past the active site, displacing part of the CTD (Fig. 7). This transition, which in the WT RdRp is facilitated by the bound Mn2+ but in the E634Q mutant also occurs in the Mg2+-bound stage, transiently weakens the high-affinity Mn2+ binding site so that the ion occupying this position is released (this is observed only in those molecules where the transition to elongation has occurred) (Fig. 7C, panel III; Fig. 8, step VI). In the absence of external Mn2+, the reaction stalls at this point (Fig. 5).
Mn2+-dependent assembly of the elongation complex (step VII).
The transition from initiation to elongation is the only step in the ϕ6 RdRp-catalyzed polymerization reaction that is entirely dependent on the presence of external Mn2+ (Fig. 5 and 6). Such activation could potentially arise from some undefined effect of Mn2+ displacing the catalytic ions at this specific step of the polymerization reaction. However, since there is no evidence for such an effect, we prefer the following mechanism: following displacement of the CTD, the high-affinity binding site for Mn2+ re-forms to allow exogenous Mn2+ to bind and establish the correct geometry for the attachment of the third nucleotide (D3 NTP base pairing with the T3 template nucleotide). After this two-step transition involving the loss and rebinding of Mn2+, synthesis of the trinucleotide daughter strand proceeds. From this point on Mn2+ remains stably bound, and external ions are not required for elongation (Fig. 6A and B).
Conclusion.
Using a combination of structural and biochemical techniques, we have been able to precisely define all the stages in the polymerization process for a model RNA-dependent RNA polymerase. In particular, we are now able to provide a satisfying explanation for the role of the noncatalytic Mn2+ ion in this process. We suggest that noncatalytic ions are likely to play a similar role in a range of other viruses.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to R. Tarkiainen and S. Vehma for technical assistance and S. Graham for assistance with data collection and processing, which was also supported by staff at station I03 of Diamond Light Source, Oxfordshire, United Kingdom, and station ID14-EH2 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France. M. Koivunen is acknowledged for designing the oligonucleotide 3′end_6.
D.I.S. and S.W. are funded by the Medical Research Council (MRC), M.M.P. is funded by the Academy of Finland (grants 250113 and 256069), and M.M.P. and D.H.B. are funded by the Sigrid Jusélius Foundation. This work was supported by the Finnish Centre of Excellence Programme 2006-2011 (Academy of Finland grant 1129684 to D.H.B.) and by EC project SPINE2-Complexes (LSHG-CT-2006-031220). The Wellcome Trust is acknowledged for providing administrative support (grant 075491/Z/04).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Adams PD, et al. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 2. Arnold JJ, Ghosh SK, Cameron CE. 1999. Poliovirus RNA-dependent RNA polymerase (3Dpol). Divalent cation modulation of primer, template, and nucleotide selection. J. Biol. Chem. 274:37060–37069 [DOI] [PubMed] [Google Scholar]
- 3. Behrens SE, Tomei L, De Francesco R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12–22 [PMC free article] [PubMed] [Google Scholar]
- 4. Bondarava N, Un S, Krieger-Liszkay A. 2007. Manganese binding to the 23 kDa extrinsic protein of photosystem II. Biochim. Biophys. Acta 1767:583–588 [DOI] [PubMed] [Google Scholar]
- 5. Bricogne G, et al. 2010. BUSTER version 2.9. Global Phasing Ltd, Cambridge, United Kingdom [Google Scholar]
- 6. Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI. 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410:235–240 [DOI] [PubMed] [Google Scholar]
- 7. Butcher SJ, Makeyev EV, Grimes JM, Stuart DI, Bamford DH. 2000. Crystallization and preliminary X-ray crystallographic studies on the bacteriophage phi6 RNA-dependent RNA polymerase. Acta Crystallogr. D Biol. Crystallogr. 56:1473–1475 [DOI] [PubMed] [Google Scholar]
- 8. Delano WL. 2002. The PyMOL molecular graphics system. Delano Scientific, Palo Alto, CA [Google Scholar]
- 9. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 10. Finney LA, O'Halloran TV. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931–936 [DOI] [PubMed] [Google Scholar]
- 11. Frilander M, Bamford DH. 1995. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage phi 6: the three segments modulate each other's packaging efficiency. J. Mol. Biol. 246:418–428 [DOI] [PubMed] [Google Scholar]
- 12. Frilander MJ, Turunen JJ. 2005. RNA ligation by T4 DNA ligase, p 36–52 In Handbook of RNA biochemistry. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 13. Fullerton SW, et al. 2007. Structural and functional characterization of sapovirus RNA-dependent RNA polymerase. J. Virol. 81:1858–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geerlof A, et al. 2006. The impact of protein characterization in structural proteomics. Acta Crystallogr. D Biol. Crystallogr. 62:1125–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hong Z, et al. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6–11 [DOI] [PubMed] [Google Scholar]
- 16. Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795–800 [Google Scholar]
- 17. Kao CC, Singh P, Ecker DJ. 2001. De novo initiation of viral RNA-dependent RNA synthesis. Virology 287:251–260 [DOI] [PubMed] [Google Scholar]
- 18. Laurila MR, Makeyev EV, Bamford DH. 2002. Bacteriophage phi 6 RNA-dependent RNA polymerase: molecular details of initiating nucleic acid synthesis without primer. J. Biol. Chem. 277:17117–17124 [DOI] [PubMed] [Google Scholar]
- 19. Laurila MR, Salgado PS, Stuart DI, Grimes JM, Bamford DH. 2005. Back-priming mode of phi6 RNA-dependent RNA polymerase. J. Gen. Virol. 86:521–526 [DOI] [PubMed] [Google Scholar]
- 20. Luo G, et al. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makeyev EV, Bamford DH. 2000. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage phi6. EMBO J. 19:124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. 1994. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage phi 6. Virology 202:258–263 [DOI] [PubMed] [Google Scholar]
- 23. Ng KK, et al. 2004. Crystal structure of Norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J. Biol. Chem. 279:16638–16645 [DOI] [PubMed] [Google Scholar]
- 24. Otwinowski Z. 1997. Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 25. Pagratis N, Revel HR. 1990. Detection of bacteriophage phi 6 minus-strand RNA and novel mRNA isoconformers synthesized in vivo and in vitro, by strand-separating agarose gels. Virology 177:273–280 [DOI] [PubMed] [Google Scholar]
- 26. Palmenberg A, Kaesberg P. 1974. Synthesis of complementary strands of heterologous RNAs with Qbeta replicase. Proc. Natl. Acad. Sci. U. S. A. 71:1371–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poranen MM, Koivunen MR, Bamford DH. 2008. Nontemplated terminal nucleotidyltransferase activity of double-stranded RNA bacteriophage phi6 RNA-dependent RNA polymerase. J. Virol. 82:9254–9264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poranen MM, et al. 2008. Structural explanation for the role of Mn2+ in the activity of phi6 RNA-dependent RNA polymerase. Nucleic Acids Res. 36:6633–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ranjith-Kumar CT, Gutshall L, Kim MJ, Sarisky RT, Kao CC. 2002. Requirements for de novo initiation of RNA synthesis by recombinant flaviviral RNA-dependent RNA polymerases. J. Virol. 76:12526–12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sadek FS, Reilley CN. 1959. A survey on the application of visual indicators for the chelometric calcium determination in serum. J. Lab. Clin. Med. 54:621–629 [PubMed] [Google Scholar]
- 31. Salgado PS, et al. 2004. The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase. Structure 12:307–316 [DOI] [PubMed] [Google Scholar]
- 32. Sarin LP, et al. 2009. Insights into the pre-initiation events of bacteriophage phi 6 RNA-dependent RNA polymerase: towards the assembly of a productive binary complex. Nucleic Acids Res. 37:1182–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smart O, et al. 2008. Refinement with local structure similarity restraints (LSSR) enables exploitation of information from related structures and facilitates use of NCS, Abstr. TP139, p. 117 Annu. Meet. Am. Crystallogr. Assoc., Knoxville, TN [Google Scholar]
- 34. Starnes MC, Joklik WK. 1993. Reovirus protein lambda 3 is a poly(C)-dependent poly(G) polymerase. Virology 193:356–366 [DOI] [PubMed] [Google Scholar]
- 35. Steitz TA. 1998. A mechanism for all polymerases. Nature 391:231–232 [DOI] [PubMed] [Google Scholar]
- 36. Tabor S, Richardson CC. 1989. Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. U. S. A. 86:4076–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tao Y, Farsetta DL, Nibert ML, Harrison SC. 2002. RNA synthesis in a cage—structural studies of reovirus polymerase lambda3. Cell 111:733–745 [DOI] [PubMed] [Google Scholar]
- 38. van Dijk AA, Makeyev EV, Bamford DH. 2004. Initiation of viral RNA-dependent RNA polymerization. J. Gen. Virol. 85:1077–1093 [DOI] [PubMed] [Google Scholar]
- 39. Walter TS, et al. 2005. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 61:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang TS, Korn D. 1982. Specificity of the catalytic interaction of human DNA polymerase beta with nucleic acid substrates. Biochemistry 21:1597–1608 [DOI] [PubMed] [Google Scholar]
- 41. Winter G. 2010. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43:186–190 [Google Scholar]
- 42. Yang H, Makeyev EV, Bamford DH. 2001. Comparison of polymerase subunits from double-stranded RNA bacteriophages. J. Virol. 75:11088–11095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang H, Makeyev EV, Butcher SJ, Gaidelyte A, Bamford DH. 2003. Two distinct mechanisms ensure transcriptional polarity in double-stranded RNA bacteriophages. J. Virol. 77:1195–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yap TL, et al. 2007. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-Å resolution. J. Virol. 81:4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yi GH, Zhang CY, Cao S, Wu HX, Wang Y. 2003. De novo RNA synthesis by a recombinant classical swine fever virus RNA-dependent RNA polymerase. Eur. J. Biochem. 270:4952–4961 [DOI] [PubMed] [Google Scholar]
- 46. Zamyatkin DF, et al. 2008. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283:7705–7712 [DOI] [PubMed] [Google Scholar]
- 47. Zhong W, Gutshall LL, Del Vecchio AM. 1998. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J. Virol. 72:9365–9369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.