Abstract
Dicistroviridae and Picornaviridae are two phylogenetically related families of positive-sense single-stranded RNA viruses in the picornavirus-like superfamily with similar gene contents but different genome organizations and hosts. In a surveillance study involving 1,472 samples from 368 dogs over a 22-month period, we identified a novel picornavirus-like virus from 47 fecal and urine samples by the use of reverse transcription-PCR (RT-PCR). Sequencing and phylogenetic analysis of three complete genomes revealed that, although it seemed that the virus was most closely related to other picornaviruses, P1, P2, and P3 of the virus possessed very low amino acid identities of <30% to those of all other known picornaviruses and that the amino acid identities between the 3Dpol and 2C of the virus and the RNA-dependent RNA polymerases and helicases of all other picornaviruses were <35%. Distinct from other picornaviruses, the genomes of the virus contain two putative internal ribosome entry sites (IRESs) and two open reading frames, encoding two polyprotein precursors (844 and 1,406 amino acids), separated by an intergenic region (IGR) of 588 bases. A dual-luciferase activity assay using DNA and RNA transfection revealed that both IRESs were functional. Quantitative RT-PCR showed that numbers of viral RNAs ranged from 7.55 × 106 to 1.26 × 109 copies/ml of urine and 1.82 × 106 to 4.97 × 1010 copies/ml of fecal sample. This is the first report of the natural occurrence of two functional IRESs in nondicistroviruses. Based on our results, we have proposed a novel species, canine picodicistrovirus (CPDV), to describe this novel member of the picornavirus-like superfamily, which could represent a novel family of viruses.
INTRODUCTION
The picornavirus-like superfamily includes more than 10 families of viruses that are grouped into six clades (26). The genomes of all viruses in this superfamily are characterized by the presence of RNA-dependent RNA polymerase (RdRp), chymotrypsin-like protease, superfamily 3 helicase (S3H), and genome-linked protein (26). Among the families in this picornavirus-like superfamily, Dicistroviridae and Picornaviridae are two families of positive-sense single-stranded RNA viruses with similar gene contents but different genome organizations and hosts. Dicistroviruses are found in arthropods such as shrimps, honey bees, and insect pests of agricultural and medical importance (4, 6, 18, 37, 38, 48). For example, triatoma virus is found in the hematophagous reduviid bug, the vector of Trypanosoma cruzi, which is the cause of Chagas' disease and infects 8 to 11 million people in Latin America (40, 43). Picornaviruses are found in humans and a wide variety of animals, in which they can cause respiratory, cardiac, hepatic, neurological, mucocutaneous, and systemic diseases of various degrees of severity (19, 25, 27, 44, 45, 49, 52). As for the genome organization in these two families of viruses, the region encoding the capsid proteins is located at the 5′ end of the genomes of picornaviruses but downstream from the intergenic region (IGR) for those of dicistroviruses. Furthermore, the genomes of dicistroviruses contain two internal ribosome entry site (IRES) elements, but those of picornaviruses contain only one. It is unknown why these two families of viruses, with such a major difference in their hosts, have similar gene contents. Thus, it is conceivable that there may be a previously undescribed virus, which may be a member of the Dicistroviridae family, a member of the Picornaviridae family, or a member of a previously undescribed family, that possesses a genome structure between those of the Dicistroviridae and Picornaviridae families.
Dogs are well-recognized reservoirs of viruses from at least seven families (42), including highly fatal viruses that can be transmitted to humans, such as the rabies virus. However, no picornaviruses had been reported in dogs until 2011 (24, 36). Therefore, we hypothesized that previously unrecognized picornaviruses may be present in dogs. To test this hypothesis, we carried out a territory-wide molecular epidemiology study in dogs for detection of picornaviruses. In this study, we discovered two novel picornavirus-like viruses in dogs. Complete genome sequencing and comparative analysis showed that one of the viruses formed a cluster phylogenetically related to but distinct from the other genera in the Picornaviridae family. Most interestingly, their genomes possessed two functional IRES elements, a phenomenon present in members of the Dicistroviridae family. One IRES element is at the 5′ untranslated region (UTR) and the other between VP1 and 2A. Based on our results, we proposed a novel species, canine picodicistrovirus (CPDV), to describe this novel member of the picornavirus-like superfamily, which could represent a novel family of viruses.
MATERIALS AND METHODS
Dog surveillance and sample collection.
A total of 368 dogs were captured from 38 different locations in the Hong Kong Special Administrative Region (HKSAR) during 22 months (June 2007 to April 2008 and October 2008 to August 2009) by the Department of Agriculture, Fisheries and Conservation (AFCD), HKSAR. Nasopharyngeal, fecal, urine, and blood samples were collected from these dogs by the Kowloon Animal Management Centre, AFCD, using procedures described previously (28, 29, 31–33, 54, 56, 57).
RNA extraction.
Viral RNA was extracted from nasopharyngeal swabs, fecal swabs, and urine by the use of a viral RNA minikit (QIAgen, Hilden, Germany) and from blood by the use of a QIAamp RNA blood minikit (QIAgen, Hilden, Germany). The RNA was eluted in 60 μl of RNase-free water and was used as the template for reverse transcription-PCR (RT-PCR).
RT-PCR of the 5′ UTR of picornaviruses performed using conserved primers and DNA sequencing.
Picornavirus screening was performed by amplifying a 112-bp fragment of the 5′ UTR of picornaviruses by the use of conserved primers (5′-GGACCCGTGAATGCGGCTAA-3′ and 5′-CACGGAACACCGAAAGTAGT-3′) designed by multiple alignment of the nucleotide sequences of the 5′ UTR of various picornavirus species. Reverse transcription was performed using a SuperScript III kit (Invitrogen, San Diego, CA). The PCR mixture (25 μl) contained cDNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 3 mM MgCl2 and 0.01% gelatin), 200 μM (each) deoxynucleoside triphosphates (dNTPs), and 1.0 U of Taq polymerase (AmpliTaq Gold; Applied Biosystems, Foster City, CA). The mixtures were amplified in 40 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min in an automated thermal cycler (Applied Biosystems, Foster City, CA). Standard precautions were taken to avoid PCR contamination, and no false-positive result was observed in the negative controls.
The PCR products were subjected to gel purification using a QIAquick gel extraction kit (QIAgen, Hilden, Germany). Both strands of the PCR products were sequenced twice with an ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA), using the two PCR primers. The sequences of the PCR products were compared with known sequences of the 5′ UTR of picornaviruses in the GenBank database.
Viral culture.
Four samples, positive for CPDV, were cultured in MDCK (Madin-Darby canine kidney), DH82 (canine macrophage-monocyte), RD (human rhadomyosarcoma), Vero E6 (monkey kidney), and HEL (human embryonic lung fibroblast) cells. Intracerebral, subcutaneous, and intraperitoneal inoculation of suckling mice was also performed.
Genome sequencing of CPDV.
Three genomes of CPDV were amplified and sequenced using strategies we previously used for complete genome sequencing of other picornaviruses, with the RNA extracted from two fecal and one urine samples as templates (30, 34, 35, 55, 58). The RNA was converted to cDNA by a combined random-priming and oligo(dT) priming strategy. The cDNA was amplified by the use of degenerate primers designed by multiple alignments of the genomes of closely related picornaviruses observed from the initial screening results and additional primers designed on the basis of the results of the first and subsequent rounds of sequencing (see the table in the supplemental material). The 5′ ends of the viral genomes were confirmed by rapid amplification of cDNA ends (RACE) using a SMARTer RACE cDNA amplification kit (Clontech). Sequences were checked manually and assembled to produce final sequences of the viral genomes.
Genome analysis.
The nucleotide sequences of the genomes and the deduced amino acid sequences were compared to those of other picornaviruses. Cleavage sites were predicted by manual inspection of multiple alignments with amino acid sequences of other picornaviruses. Conservation of amino acids in the neighborhood of the cleavage sites was observed and was used to assist the prediction of the cleavage sites. Multiple sequence alignment was performed using MUSCLE 3.8 (10), and the best-fit models of amino acid substitution were chosen based on the Akaike Information Criterion using ProtTest 3.0 (9). The deduced amino acid sequences of 3Dpol and 2C of CPDV and RdRp and S3H of representative viruses from the picornavirus-like superfamily were used for phylogenetic analysis. The maximum-likelihood phylogenetic trees of 3Dpol/RdRp and 2C/S3H were constructed using PhyML 3.0 (16) and the Approximate Likelihood-Ratio Test (aLRT) method (1).
Secondary structure prediction in the 5′ UTR and IGR was performed using the Quikfold server with default settings, except for the use of RNA 3.0 for Energy Rules (http://mfold.rna.albany.edu/?q=DINAMelt/Quickfold) (60). The average evolutionary divergence for each coding region over the three strains of CPDV was estimated by the overall mean distance with p-distance method in MEGA 5 (50).
Preparation of bicistronic plasmids.
To examine the activity of each IRES element, a bicistronic Renilla/firefly dual-luciferase reporter plasmid was constructed on the backbone of pSP-FL+NF and pRL-CMV (Promega, Madison, WI). Firefly luciferase was used as the test reporter and Renilla luciferase as the control reporter. Putative IRES sequences were subcloned into the bicistronic construct containing an upstream Renilla luciferase gene, a downstream firefly luciferase gene, and the studied IRES in the intercistronic space (Fig. 1). Briefly, putative IRES sequences in the 5′ UTR (5′ IRES) and intergenic region (IGR IRES) together with the first 50 amino acids encoding the N terminus of the 2A region were amplified with primers containing the corresponding restriction sites (Table 1) and inserted between the Renilla luciferase and firefly luciferase cistrons, respectively. The bicistronic construct containing the 5′ IRES of hepatitis C virus (HCV) was used as a positive control (51). Ligation products were then transformed into Escherichia coli strain DH5α competent cells. Positive clones were confirmed by restriction fragment analysis and DNA sequencing.
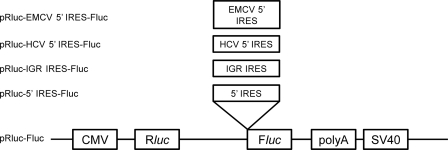
Fig 1.
Schematic representation of the five bicistronic reporter constructs used. pRluc-Fluc, backbone and negative control; pRluc-HCV 5′ IRES-Fluc, positive control; pRluc-EMCV 5′ IRES-Fluc, positive control; pRluc-5′ IRES-Fluc, 5′ IRES of CPDV inserted into pRluc-Fluc backbone; pRluc-IGR IRES-Fluc, intergenic region of CPDV inserted into pRluc-Fluc backbone.
Table 1.
Primers used for amplification, cloning, and sequencing of IRES constructs
In vitro transcription of capped RNA was synthesized using a mMESSAGE mMACHINE kit (Ambion, Austin, Tex). Briefly, bicistronic constructs with T7 promoter were linearized with BamHI followed by assembly of the transcription reaction mix as described by the manufacturer. The reaction mix was incubated at 37°C for 1 h. Following transcription, RNA was treated with TURBO DNase and was further incubated at 37°C for 15 min. RNA transcripts were purified by phenol-chloroform extraction and isopropanol precipitation. The bicistronic construct containing encephalomyocarditis virus (EMCV) IRES was used as a positive control.
Preparation of pGL3-enhancer experimental constructs.
To further examine whether the 5′ IRES and IGR IRES have cryptic promoter activities, each IRES element was excised from the bicistronic constructs at BglII site and subcloned into a pGL3-enhancer vector (Promega, Madison, WI) to yield the final pGL3E/5′ IRES and pGL3E/IGR IRES experimental constructs. The pGL3-enhancer vector lacks a promoter but contains a simian virus 40 (SV40) enhancer located downstream of a firefly luciferase gene. Firefly luciferase was used as the test reporter. A cotransfected plasmid-encoded Renilla luciferase was used as an internal control to normalize the variations between transfection replicates. Empty pGL3-enhancer vector and the vector containing the promoter sequence of the surface gene of hepatitis B virus were used as a negative and a positive control, respectively. Ligation products were then transformed into E. coli strain DH5α competent cells. Positive clones were confirmed by restriction fragment analysis and DNA sequencing.
Cell culture and transient transfection of cloned IRES.
MDCK cells were grown in Dulbecco's modified Eagle medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Grand Island, NY). All cultures were incubated at 37°C in 5% CO2. Cells were seeded 1 day prior to transfection at densities of 0.25 × 105 and 0.5 × 105 per well into 12- and 6-well plates (TPP, Trasadingen, Switzerland), respectively. For each IRES construct, cells were transfected in triplicate in wells of a 12-well plate, with 1 μg of bicistronic DNA per well, using Lipofectamine and Plus reagent and following the instructions of the manufacturer (Invitrogen, Carlsbad, CA). For RNA transfection, cells were transfected in triplicate in wells of a 6-well plate, with 10 μg of capped RNA per well, using the transfection reagents and conditions described above.
To examine the possibility of cryptic promoter activities of the two IRES elements, 1 μg of pGL3-enhancer experimental plasmids was independently transfected into MDCK cells with 10 ng of Renilla luciferase reporter vector as a cotransfection control in triplicate.
Dual-luciferase activity assay.
Luciferase assays were performed as described previously (5, 8). At 24 h after DNA transfection or 8 h after RNA transfection, cells were lysed and analyzed for dual-luciferase activities by the use of a dual-luciferase reporter assay system (Promega, Madison, WI) and an LB9570 microplate luminometer (EG & G Berthold, Bad Wildbad, Germany). Ten microliters of cell lysates was incubated with firefly and Renilla luciferase-specific substrates following the manufacturer's instructions. Relative levels of luciferase activity (Fluc/Rluc) derived from dual-luciferase reporter constructs and pGL3-enhancer experimental constructs were determined by normalizing firefly luciferase activity to Renilla luciferase activity. IRES activity was calculated as the mean of the results of three independent experiments. Statistical analysis was performed using the unpaired Student's t test. P < 0.05 was considered to be statistically significant.
Quantitative RT-PCR.
Quantitative RT-PCR to detect the 5′ UTR of CPDV was performed on the 38 positive fecal and 9 positive urine samples by the use of TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA) with primers 5′-CCGACCAACATCTCATGTCG-3′ and 5′-CCCGGAGGAATTCTGGGTTA-3′ and probe 5′-6-carboxyfluorescein (FAM)-CTGGATGATCTTTTAAGACTC-BHQ1-3′ and a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). The reaction mixture was subjected to thermal cycling at 50°C for 2 min and 95°C for 10 min followed by 50 cycles of 95°C for 15 s and 55°C for 1 min. RNA standards were synthesized by using a MEGAscript T7 kit (Ambion, Austin, TX) according to the manufacturer's instructions. Briefly, an amplicon was produced using primers 5′-CCGACCAACATCTCATGTCG-3′ and 5′-CGCTTCACAGCTCCACGGTCA-3′ and was purified using a QIAquick gel extraction kit (QIAgen, Hilden, Germany). A 0.2-μg volume of the amplicon was mixed with 2 μl each of ATP, GTP, CTP, and UTP, 10× reaction buffer, and enzyme mix in a standard 20-μl reaction mixture. The reaction mixture was incubated at 37°C for 4 h, followed by addition of 1 μl of TURBO DNase, and was further incubated at 37°C for 15 min. The synthesized RNA was purified by lithium chloride precipitation. The concentration of purified RNA was quantified by UV light absorbance. The size of the purified RNA was assessed by running an aliquot of the purified RNA on an agarose gel. pTRI-Xef control template was included as a control for the whole process.
Nucleotide sequence accession numbers.
The nucleotide sequences of the genomes of CPDV have been deposited in the GenBank sequence database under accession no. JN819202 to JN819204.
RESULTS
Dog surveillance and identification of two novel picornavirus-like viruses.
A total of 1,472 nasal, fecal, urine, and blood samples from 368 dogs were obtained from various locations in HKSAR. RT-PCR for a 112-bp fragment in the 5′ UTR of picornaviruses was positive for 49 fecal, 3 nasopharyngeal, and 10 urine samples from 52 dogs. The sequences from the positive specimens fell into two distinct clusters, suggesting the presence of two novel picornavirus-like viruses (n = 47 for the first novel virus and n = 15 for the other “canine picornavirus” [“CanPV”]). The fecal sample from one dog was positive for both viruses. The fecal sample from another dog was positive for the first novel virus, and the urine sample from that dog was positive for “CanPV.” Detailed analysis and characterization of “CanPV” are to be reported elsewhere.
Viral culture.
No cytopathic effect was observed by RT-PCR in any of the cell lines inoculated with the samples that were positive for CPDV. RT-PCR using the culture supernatants and cell lysates after three passages for monitoring the presence of viral replication also showed negative results. No signs of illness such as paralysis were observed in suckling mice within 14 days after inoculation. Brain, muscle, and subcutaneous tissues were negative for CPDV by RT-PCR.
Genome organization and coding potential of CPDV.
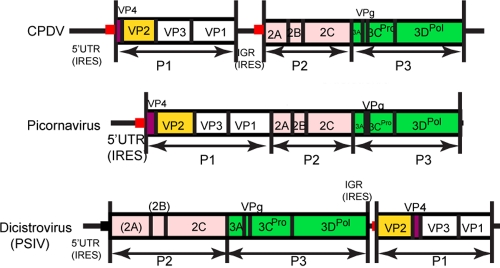
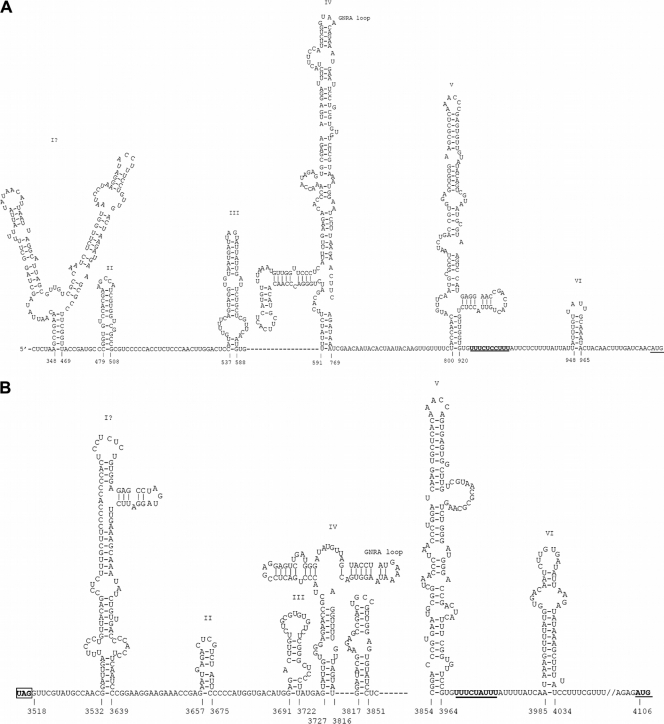
The complete genomes sequenced from the three samples are 8,754 to 8,755 bases, after excluding the polyadenylated tract, and their G + C contents are 42%. The genome possesses a 5′ UTR of 982 bases (Fig. 2). In contrast to the other picornaviruses, the genomes of all three strains of CPDV contain two putative IRES elements and two open reading frames (ORFs) (Fig. 2). Using one strain (209F) as an example, the two ORFs were 2,535 and 4,221 bases in length. These two ORFs encode two polyprotein precursors of 844 and 1,406 amino acids, respectively, separated by a short IGR of 588 bases (from position 3518 to 4105) (Fig. 2). There is 72% nucleotide identity between the 3′ 92 bases of 5′ UTR (from position 813 to 904) and the 3′ 89 bases of IGR (from position 3864 to 3952). These two regions also share 72% and 85% nucleotide identities, respectively, with stem-loop V of the poliovirus IRES, which was previously shown to be strongly conserved in enteroviruses (Fig. 3) (39). Although a putative GNRA loop is present in domain IV of the two IRESs of CPDV, the GNRA loops in the two IRESs of CPDV have predicted structures that differ from the GNRA loop in other type 1 IRESs (Fig. 4). Moreover, the cloverleaf structure immediately upstream from all picornavirus type I IRESs was not identified in the 5′ UTR of CPDV.
Fig 2.
Genome organization of CPDV, dicistrovirus, and picornavirus.
Fig 3.
(A) Pairwise alignment between the 5′UTR of CPDV and that of poliovirus. (B) Pairwise alignment between the IGR of CPDV and the 5′UTR of poliovirus.
Fig 4.
IRES structures of the 5′ UTR (A) and the IGR (B) in CPDV. The pyrimidine tract and the AUG start codon are underlined.
The P1 (capsid-coding) regions in the genomes of CPDV encode the capsid proteins VP4, VP2, VP3, and VP1 (Table 2). The P1 of CPDV possessed the highest amino acid identity (25.1%) to that of our recently discovered feline picornavirus (34). As with aphthoviruses, cardioviruses, enteroviruses, erboviruses, hepatoviruses, sapeloviruses, senecaviruses, teschoviruses, and tremovirus, the “VP0” of CPDV is predicted to be cleaved into VP4 and VP2, with the presence of a putative cleavage site of E/S based on sequence alignment (Tables 2 and 3).
Table 2.
Coding potential and estimates of average evolutionary divergence of the genomes of CPDV
| Putative protein | Position | No. of amino acids | Avg evolutionary divergence (%) |
|
|---|---|---|---|---|
| Amino acid | Nucleotide | |||
| ORF at amino acids 983-3517 | ||||
| VP4 | M1-E44 | 44 | 1.5 | 7.6 |
| VP2 | S45-Q282 | 238 | 0.8 | 7.1 |
| VP3 | A283-Q532 | 250 | 0.5 | 7.5 |
| VP1 | S533-E844 | 312 | 2.1 | 7.6 |
| ORF at amino acids 4106-8326 | ||||
| 2A | M1-E141 | 141 | 1.2 | 6.3 |
| 2B | S142-E267 | 126 | 1.9 | 8.8 |
| 2C | D268-E608 | 341 | 1 | 9.8 |
| 3A | D609-E695 | 87 | 8.1 | 12.3 |
| 3B | S696-E721 | 26 | 0 | 6.8 |
| 3C | M722-E923 | 202 | 0 | 6.8 |
| 3D | G924-K1406 | 483 | 1.1 | 11.7 |
Table 3.
Comparison of genomic features of CPDV and the different genera in the Picornaviridae family
| Region | Function, conserved motif, or feature (reference[s]) | Generaa |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aphthovirus | Avihepatovirus | Cardiovirus | Enterovirus | Erbovirus | Hepatovirus | Kobuvirus | Parechovirus | Sapelovirus | Senecavirus | Teschovirus | Tremovirus | Unclassified Orthoturdivirus (TV1) | Unclassified Paraturdivirus (TV2, TV3) | Unclassified (Bat picornavirus group 1, Bat picornavirus group 2) | Unclassified (Bat picornavirus group 3) | CPDV | ||
| 5′UTR | Pattern: Yn-Xm-AUG | Y10X15 (FMDV-C) | Y7X65 (DHV1) | Y9X18 (EMCV) | Y9X18 (PV) | Y12X15 (ERBV) | Y9X14 (HAV) | Y7X11 (AiV) | Y7X20 (HPeV2) | Y16-17 X17-18 (SpV1) | N (SVV-001) | Y7X13 (PTV1) | N (AEV) | Y9X19 | Y6X19-34 | N | Y8X19 | 5′ UTR, Y10X50; IGR, Y8X19 |
| IRES (3, 11, 20, 59) | Type II | Type IV | Type II | Type I | Type II | HAV-like | Non-type I to IV | Type II | Type IV | Type IV | Type IV | Type IV | Undefined | Undefined | Type IV | Type I | Undefined | |
| L | Protease (14, 46) | Y† | N† | Y/N† | N† | Y† | N† | Y/N† | N† | Y/N† | Y/N† | Y/N† | N† | Y/N† | Y/N† | Y/N† | Y/N† | N† |
| VP0 | Cleaved into VP4 and VP2 | Y | N | Y | Y | Y | Y | N | N | Y | Y | Y | Y | N | N | Y | Y | Y |
| Myristylation site (7): GXXX[ST] | Y | N | Y | Y | Y | N | Y | N | Y | Y | Y | N | Y | Y | Y | Y | N | |
| VP1 | Motif: [PS]ALXAXETG | N | N | N | Y | N | N | N | N | Y | Y | N | N | N | N | Y | Y | N |
| IGR IRES | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N | Y | |
| 2A | Function (21, 47): | NPGP | NPGP, H-box/NC | NPGP | Chymotrypsin-like protease | NPGP | Unknownb | H-box/NC | H-box/NC in HPeV and LV; NPGP in LV | Chymotrypsin-like protease or unknown§ | NPGP | NPGP | H-box/NC | H-box/NC | Unknown | Chymotrypsin-like protease | Chymotrypsin-like protease | Unknown |
| 2C | NTPase motif (13): GXXGXGKS | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Helicase (15): DDLXQ | Y§ | DDFGQ | Y§ | Y§ | Y§ | DD[LI]GQ | DD[LI]GQ | DD[LA]GQ | Y§ | Y§ | Y§ | Y§ | DDVGQ | Y§ | Y§ | DDVGQ | Y§ | |
| 3Cpro | Catalytic triad (12): H-D/E-C | H-D-C | H-D-C | H-D-C | H-E-C | H-D-C | H-D-C | H-E-C | H-D-C | H-E-C | H-D-C | H-D-C | H-D-C | H-E-C | H-E-C | H-E-C | H-E-C | H-E-C |
| RNA-binding domain motif (17): KFRDI | N | N | N | Y | N | Y | N | N | Y | Y | N | Y | N | N | NFRDI | KYRDI | N | |
| Motifs (12): GXCG, GXH | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | AMH | Y | Y | |
| 3Dpol | Motif: KDE[LI]R (23) | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Motif: GG[LMN]PSG> (23) | Y | GGMCSG | Y | Y | GALPSG | GSMPSG | Y | Y | Y | Y | Y | GSMPSG | Y | GGMPS[GR] | Y | Y | GAMPSG | |
| Motif: YGDD (23) | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | |
| Motif: FLKR (23) | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | |
Y, yes (present); N, no (absent). Y†, presence of L and L is protease; Y/N†, presence of L but L is not protease; N†, absence of L; Y§, motif DDLXQ is present. FMDV-C, foot-and-mouth disease virus; DHV1, duck hepatitis virus 1; ERDV, equine rhinitis B virus; HAV, hepatitis A virus; AiV, avian influenza virus; HPeV, human parechovirus; SpV, swinepox virus; SVV, simian varicella virus; PTV, Punta Toro virus; AEV, avian encephalomyelitis virus.
Unknown, 2A of HAV, DPV, CPDV, TV2, and TV3 does not contain the characteristic catalytic amino acid residues with chymotrypsin-like proteolytic activity, the NPGP motif, or the H-box/NC motif.
The P2 regions in the genomes of CPDV encode nonstructural proteins 2A, 2B, and 2C (Table 2). The P2 of CPDV possessed the highest amino acid identity (26.0%) to that of the recently discovered canine kobuvirus (24, 36). In similarity to those of turdiviruses 2 and 3, the 2A of CPDV does not possess the characteristic catalytic amino acid residues with chymotrypsin-like proteolytic activity or the NPGP or H-box/NC motifs (Table 3) (21, 47). Moreover, it also does not possess any homology to other known viral or cellular proteins. As with all the other picornaviruses, 2C of CPDV possesses the GXXGXGKS motif for NTP binding (13). In similarity to most picornaviruses, 2C of CPDV possesses the DDLXQ motif for putative helicase activity (15).
The P3 regions in the genomes of CPDV encode 3A, 3B (VPg [small genome-linked protein]), 3Cpro (protease), and 3Dpol (RdRp) (Table 2). The P3 of CPDV possessed the highest amino acid identity (28.7%) to that of our recently discovered turdivirus 3 (55). As with 3Cpro of enteroviruses, kobuviruses, sapelovirus, orthoturdiviruses, and paraturdiviruses, 3Cpro of CPDV contains the catalytic triad of His-Glu-Cys, whereas other picornaviruses contain the catalytic triad of His-Asp-Cys (Table 3) (2). In similarity to other picornaviruses, 3Cpro of CPDV contains the conserved GXCG motif, which has been speculated to form part of the active site of the protease (Table 3) (12). Further structural and mutagenesis studies should delineate whether this is also the active site of 3Cpro of CPDV. As in other picornaviruses, 3Cpro of CPDV contains the conserved GXH motif, which forms part of the substrate binding pocket of the protease (Table 3) (12). As for 3Dpol, in similarity to other picornaviruses, 3Dpol of CPDV contains the conserved KDE[LI]R, YGDD, and FLKR motifs, although the GG[LMN]PSG motif is replaced by the GAMPSG motif in CPDV (Table 3) (23).
The average evolutionary divergence of the coding regions in CPDV ranged from 0% to 8.1% for amino acid differences and from 6.3% to 12.3% for nucleotide differences (Table 2).
Phylogenetic analyses.
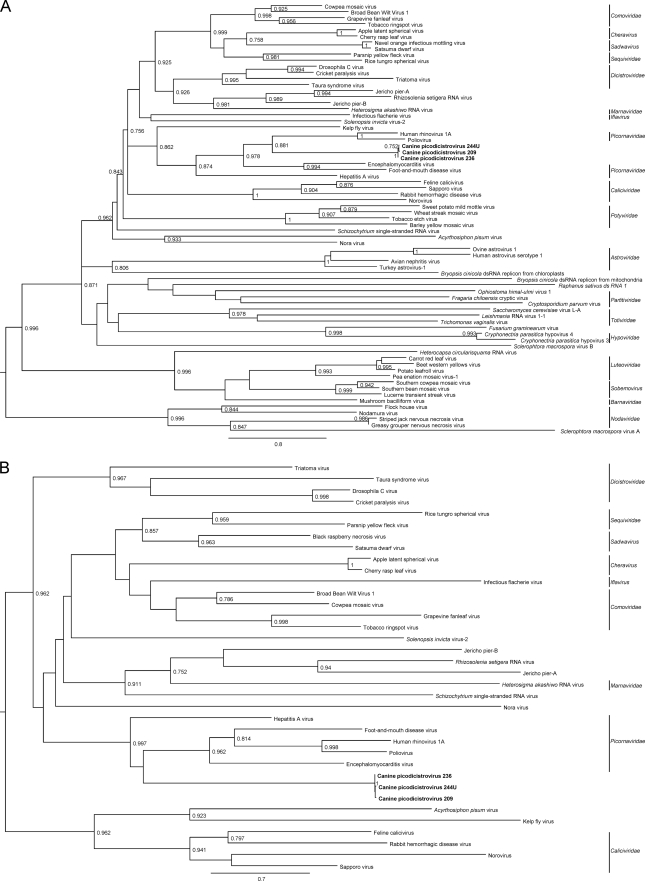
The phylogenetic trees constructed using the amino acid sequences of 3Dpol and 2C of CPDV and RdRp and S3H of other viruses of the picornavirus-like superfamily are shown in Fig. 5. The three strains of CPDV were clustered. Although it seems that they were most closely related to other picornaviruses, there was <35% amino acid identity between the 3Dpol of CPDV and the RdRp of other picornaviruses and <35% amino acid identity between the 2C of CPDV and the helicases of other picornaviruses.
Fig 5.
Maximum-likelihood trees for 3Dpol/RdRp (A) and 2C/S3H (B) of CPDV and representative viruses of the picornavirus-like superfamily. Midpoint rooting was used. aLRT branch support is shown where values are greater than 0.750. The trees are drawn to scale, and scale bars indicate 0.8 (A) and 0.7 (B) amino acid substitutions per site as inferred according to the RtREV+I+G+F model with 4 gamma categories. The three CPDV strains are shown in bold. dsRNA, double-stranded RNA.
Dual-luciferase activity assay.
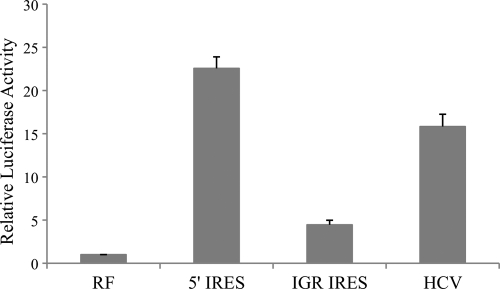
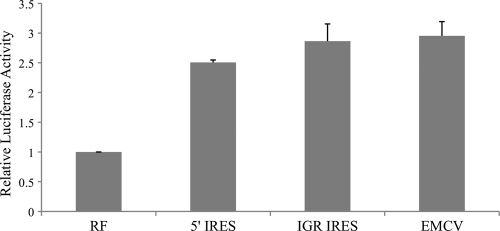
In dual-luciferase experiments employing DNA transfection, the relative luciferase activities of bicistronic vector containing 5′ IRES and the IGR IRES of CPDV were 22.6-fold (P < 0.005) and 4.5-fold (P < 0.01) higher than that of RF-negative control, respectively (Fig. 6), indicating that these two IRES elements, located in the 5′ UTR and intergenic region, were functional in MDCK cells. There was no evidence that this increase in activity was due to splicosomal activity, since the relative luciferase activities of 5′ IRES and IGR IRES were 2.5-fold (P < 0.005) and 2.9-fold (P < 0.005) higher than that of RF-negative control, respectively (Fig. 7) and were comparable to that of EMCV IRES in the RNA transfection assays. However, upon transfection of DNA constructs into 293T cells, the relative luciferase activities of 5′ IRES and IGR-IRES were greatly reduced and were comparable to the amount of activity in the RF-negative control, indicating that these two IRES elements were not functional in the 293T cells (data not shown).
Fig 6.
Luciferase activities of MDCK cells after transfection of bicistronic construct containing 5′ and IGR IRESs. Vertical bars represent means ± standard deviations of the results of three independent experiments. RF (negative control), pRluc-Fluc; 5′ IRES, pRluc-5′ IRES-Fluc; IGR IRES, pRluc-IGR IRES-Fluc; HCV (positive control), pRluc-HCV 5′ IRES-Fluc.
Fig 7.
Luciferase activities of MDCK cells after transfection of in vitro-transcribed capped RNAs. Vertical bars represent means ± standard deviations of the results of three independent experiments. RF, negative control; EMCV, EMCV IRES (positive control).
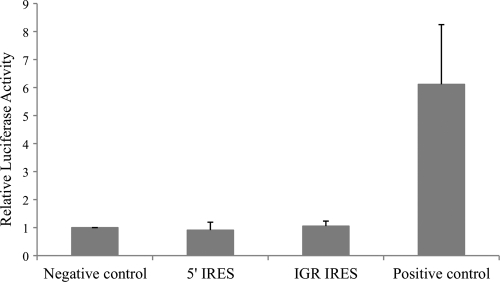
In MDCK cells, the relative luciferase activities of cells transfected with pGL3E/5′ IRES and pGL3E/IGR IRES were comparable to that of the negative control, whereas the luciferase activity of cells transfected with the positive control was 6-fold (P < 0.05) higher than that of negative control (Fig. 8). These results demonstrated that the two IRES elements do not have any cryptic promoter activities.
Fig 8.
Luciferase activities of MDCK cells after transfection of pGL3-enhancer experimental constructs with a cotransfection control. Vertical bars represent means ± standard deviations of the results of three independent experiments. Negative control, empty pGL3-enhancer vector; 5′ IRES, pGL3E/5′ IRES; IGR IRES, pGL3E/IGR IRES; positive control, vector containing promoter of the surface gene of hepatitis B virus.
Quantitative RT-PCR.
Quantitative RT-PCR showed that the amount of CPDV RNA ranged from 7.55 × 106 to 1.26 × 109 copies per ml of urine and 1.82 × 106 to 4.97 × 1010 copies per ml of fecal sample.
DISCUSSION
We report the discovery of a CPDV in dogs that could represent a novel family of viruses in the picornavirus-like superfamily. The genomes of CPDV encode RdRp, chymotrypsin-like protease, S3H, and genome-linked protein, characteristics of members of the picornavirus-like superfamily. In addition, CPDV also exhibited genomic features similar to those of other picornaviruses. With the exception of two IRES elements, the genome organization of CPDV is similar to other picornaviruses, with the characteristic order 5′-VP4, VP2, VP3, VP1, 2A, 2B, 2C, 3A, 3B, 3Cpro, 3Dpol-3′. Phylogenetically, CPDV is more related to other picornaviruses than to members of other families of viruses, as shown by the phylogenetic tree constructed using both RdRp and S3H. On the other hand, P1, P2, and P3 of CPDV possessed very low amino acid identities of <30% to those of all other known picornaviruses. Furthermore, the genomes of CPDV possess two IRES elements, a phenomenon never known for any naturally occurring picornaviruses. The presence of two IRES elements is genuine instead being due to artifacts as a result of the existence of two different viruses in each specimen, because for all three complete genomes of CPDV that we sequenced, we confirmed all assembled sequences using genome-specific primers for PCR across overlapping regions. Based on this unique feature and on the low amino acid identities with other picornaviruses, CPDV could represent a novel family of viruses.
This is the first report of the natural occurrence of two functional IRES elements in nondicistroviruses. Although IRES elements were previously reported in a variety of viruses, such as picornaviruses (22, 39) and HCV (51), the natural occurrence of two functional IRES elements in viral families other than the Dicistroviridae has never been previously reported. Notably, bicistronic poliovirus, with two functional IRES elements, had been constructed in vitro by inserting an additional IRES element between P1 and P2, thus breaking the original ORF into two parts (41). In the present study, two IRES elements were observed in all three sequenced strains of CPDV, a naturally occurring virus discovered in dogs in field studies. Both IRES elements were confirmed to be functional by DNA transfection and RNA transfection studies. Our data indicate that both CPDV IRESs are functional and could promote translation of structural and nonstructural polyprotein precursors from the genomic mRNA. However, we cannot exclude the possibility that a subgenomic CPDV mRNA is generated during infection, and further experiments to investigate this should be undertaken when conditions have been identified that allow the virus to be cultured. We speculate that additional members could be present in this possibly novel family of virus. It should be stressed that complete genome sequencing of known and novel picornaviruses has to be performed to detect these additional members and classify the viruses properly. Partial genome sequencing or sequencing just fragments would not be adequate. Otherwise, viruses would be misclassified, as some viruses of different families may possess some similar genomic features as a result of recombination. For example, the presence of a hemagglutinin esterase gene in Betacoronavirus subgroup A is thought to have been resulted from heterologous recombination from influenza C virus (53).
CPDV could represent a missing link between dicistroviruses and picornaviruses. The similarities in gene contents and close phylogenetic relationships between the members of Dicistroviridae and Picornaviridae have made people believe that these two families of viruses could be close relatives. However, there are two major differences in the genome structures of these two families of viruses. First, VP2-VP4-VP3-VP1 is located at the 3′ ends in the genomes of dicistroviruses, but VP4-VP2-VP3-VP1 is located at the 5′ ends in the genomes of picornaviruses. Second, the genomes of dicistroviruses possess two functional IRES elements, one at the 5′ UTR upstream from P2 and the other upstream from VP2, whereas the genomes of picornaviruses possess only one IRES element at the 5′ UTR upstream from VP4. Therefore, multiple steps of translocation and IRES deletion/duplication should be required for dicistroviruses to evolve to become picornaviruses or vice versa. In this study, we showed that the organizations and gene contents of the genomes of CPDV were between those of dicistroviruses and picornaviruses (Fig. 2). Irrespective of the direction of evolution, hypothetical intermediate but undiscovered viruses may still exist or might have become extinct during the course of evolution. Further epidemiology studies and complete genome sequencing of additional picornavirus- and dicistrovirus-like viruses may discover additional intermediate members between these two virus families. Before the era of complete genome sequencing, dicistroviruses and picornaviruses were considered two unrelated families of viruses. After their genomes were sequenced, it was recognized that they were in fact two closely related families with similar gene contents though different genome organizations. The present report illustrates the importance of complete genome sequencing for novel viruses. If sequencing had been restricted to only part of the genome (e.g., RdRp), this unique genome structure of CPDV would not have been discovered.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alan Chi-Kong Wong, Siu-Fai Leung, Chik-Chuen Lay, Thomas Sit, K. F. Chan, Michelle L. Yeung, Byung Mo Hwang, Suet Yee Ng, Patrick I. T. Lau, and Steven D. Benton from the HKSAR Department of Agriculture, Fisheries, and Conservation (AFCD) for facilitation and support and members of the Animal Management Centres of AFCD.
We are grateful for the generous support of Carol Yu, Richard Yu, Hui Hoy, and Hui Ming in the genomic sequencing platform and Eunice Lam for her generous donation on emerging infectious disease research. This work is partly supported by a Research Grant Council grant (HKU 783611 M), University Development Fund and Strategic Research Theme Fund, The University of Hong Kong; The Tung Wah Group of Hospitals Fund for Research in Infectious Diseases; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau; the Providence Foundation Limited in memory of the late Lui Hac Minh; and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the HKSAR Department of Health.
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 2. Bazan JF, Fletterick RJ. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. U. S. A. 85:7872–7876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belsham GJ. 2009. Divergent picornavirus IRES elements. Virus Res. 139:183–192 [DOI] [PubMed] [Google Scholar]
- 4. Bonning BC, Miller WA. 2010. Dicistroviruses. Annu. Rev. Entomol. 55:129–150 [DOI] [PubMed] [Google Scholar]
- 5. Chan CP, et al. 2006. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 80:9279–9287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen YP, Siede R. 2007. Honey bee viruses. Adv. Virus Res. 70:33–80 [DOI] [PubMed] [Google Scholar]
- 7. Chow M, et al. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482–486 [DOI] [PubMed] [Google Scholar]
- 8. Choy EY, Kok KH, Tsao SW, Jin DY. 2008. Utility of Epstein-Barr virus-encoded small RNA promoters for driving the expression of fusion transcripts harboring short hairpin RNAs. Gene Ther. 15:191–202 [DOI] [PubMed] [Google Scholar]
- 9. Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernández-Miragall O, Lopez de Quinto S, Martinez-Salas E. 2009. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 139:172–182 [DOI] [PubMed] [Google Scholar]
- 12. Gorbalenya AE, Donchenko AP, Blinov VM, Koonin EV. 1989. Cysteine proteases of positive strand RNA viruses and chymotrypsin-like serine proteases. A distinct protein superfamily with a common structural fold. FEBS Lett. 243:103–114 [DOI] [PubMed] [Google Scholar]
- 13. Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. 1989. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 17:4713–4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gorbalenya AE, Koonin EV, Lai MM. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gorbalenya AE, Koonin EV, Wolf YI. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145–148 [DOI] [PubMed] [Google Scholar]
- 16. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 17. Hämmerle T, Molla A, Wimmer E. 1992. Mutational analysis of the proposed FG loop of poliovirus proteinase 3C identifies amino acids that are necessary for 3CD cleavage and might be determinants of a function distinct from proteolytic activity. J. Virol. 66:6028–6034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hanzlik TN, Zeddam JL, Gordon KHJ, Christian PD. 1999. A new view of small RNA viruses of insects. Pestic. Outlook 10:22–26 [Google Scholar]
- 19. Harvala H, Simmonds P. 2009. Human parechoviruses: biology, epidemiology and clinical significance. J. Clin. Virol. 45:1–9 [DOI] [PubMed] [Google Scholar]
- 20. Hellen CU, de Breyne S. 2007. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 81:5850–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hughes PJ, Stanway G. 2000. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 81:201–207 [DOI] [PubMed] [Google Scholar]
- 22. Jang SK, Pestova TV, Hellen CU, Witherell GW, Wimmer E. 1990. Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme 44:292–309 [DOI] [PubMed] [Google Scholar]
- 23. Kamer G, Argos P. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 12:7269–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kapoor A, et al. 2011. Characterization of a canine homolog of human Aichivirus. J. Virol. 85:11520–11525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klein J. 2009. Understanding the molecular epidemiology of foot-and-mouth-disease virus. Infect. Genet. Evol. 9:153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925–939 [DOI] [PubMed] [Google Scholar]
- 27. Krous HF, Langlois NE. 2010. Ljungan virus: a commentary on its association with fetal and infant morbidity and mortality in animals and humans. Birth Defects Res. A Clin. Mol. Teratol. 88:947–952 [DOI] [PubMed] [Google Scholar]
- 28. Lau SK, et al. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 84:2808–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lau SK, et al. 2010. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J. Virol. 84:11385–11394 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Lau SK, et al. 2011. Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 85:8819–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lau SK, et al. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lau SK, et al. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lau SK, et al. 2010. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology 404:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lau SK, et al. 2012. Identification of a novel feline picornavirus from the domestic cat. J. Virol. 86:395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lau SK, et al. 2007. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 45:3655–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L, et al. 2011. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 92:2534–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lightner DV. 1996. Epizootiology, distribution and the impact on international trade of two penaeid shrimp viruses in the Americas. Rev. Sci. Tech. 15:579–601 [DOI] [PubMed] [Google Scholar]
- 38. Lightner DV, et al. 1997. Risk of spread of penaeid shrimp viruses in the Americas by the international movement of live and frozen shrimp. Rev. Sci. Tech. 16:146–160 [DOI] [PubMed] [Google Scholar]
- 39. Malnou CE, Poyry TA, Jackson RJ, Kean KM. 2002. Poliovirus internal ribosome entry segment structure alterations that specifically affect function in neuronal cells: molecular genetic analysis. J. Virol. 76:10617–10626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marti GA, et al. 2009. Prevalence and distribution of parasites and pathogens of triatominae from Argentina, with emphasis on Triatoma infestans and Triatoma virus TrV. J. Invertebr. Pathol. 102:233–237 [DOI] [PubMed] [Google Scholar]
- 41. Molla A, Jang SK, Paul AV, Reuer Q, Wimmer E. 1992. Cardioviral internal ribosomal entry site is functional in a genetically engineered dicistronic poliovirus. Nature 356:255–257 [DOI] [PubMed] [Google Scholar]
- 42. Murphy FA, Gibbs EPJ, Horzinek MC, Studdert MJ. 1999. Veterinary virology, 3rd ed Academic Press, San Diego, CA [Google Scholar]
- 43. Muscio OA, La Torre JL, Scodeller EA. 1988. Characterization of Triatoma virus, a picorna-like virus isolated from the triatomine bug Triatoma infestans. J. Gen. Virol. 69(Pt. 11):2929–2934 [DOI] [PubMed] [Google Scholar]
- 44. Reuter G, Boros A, Pankovics P. 2011. Kobuviruses—a comprehensive review. Rev. Med. Virol. 21:32–41 [DOI] [PubMed] [Google Scholar]
- 45. Rhoades RE, Tabor-Godwin JM, Tsueng G, Feuer R. 2011. Enterovirus infections of the central nervous system. Virology 411:288–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roberts PJ, Belsham GJ. 1995. Identification of critical amino acids within the foot-and-mouth disease virus leader protein, a cysteine protease. Virology 213:140–146 [DOI] [PubMed] [Google Scholar]
- 47. Ryan MD, Flint M. 1997. Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 78(Pt. 4):699–723 [DOI] [PubMed] [Google Scholar]
- 48. Scotti PD, Longworth JF, Plus N, Croizier G, Reinganum C. 1981. The biology and ecology of strains of an insect small RNA virus complex. Adv. Virus Res. 26:117–143 [DOI] [PubMed] [Google Scholar]
- 49. Solomon T, et al. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 10:778–790 [DOI] [PubMed] [Google Scholar]
- 50. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Whitton JL, Cornell CT, Feuer R. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3:765–776 [DOI] [PubMed] [Google Scholar]
- 53. Woo PC, Huang Y, Lau SK, Yuen KY. 2010. Coronavirus genomics and bioinformatics analysis. Viruses 2:1804–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Woo PC, et al. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Woo PC, et al. 2010. Comparative analysis of six genome sequences of three novel picornaviruses, turdiviruses 1, 2 and 3, in dead wild birds, and proposal of two novel genera, Orthoturdivirus and Paraturdivirus, in the family Picornaviridae. J. Gen. Virol. 91:2433–2448 [DOI] [PubMed] [Google Scholar]
- 56. Woo PC, et al. 2009. Comparative analysis of complete genome sequences of three avian coronaviruses reveals a novel group 3c coronavirus. J. Virol. 83:908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Woo PC, et al. 2005. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J. Infect. Dis. 192:1898–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yip CC, Lau SK, Woo PC, Chan KH, Yuen KY. 2011. Complete genome sequence of a coxsackievirus A22 strain in Hong Kong reveals a natural intratypic recombination event. J. Virol. 85:12098–12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu Y, et al. 2011. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 30:4423–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.