Abstract
Understanding the role of host factors during lethal influenza virus infection is critical to deciphering the events that determine the fate of the host. One such factor is encoded by the Mx1 gene, which confers resistance to influenza virus infection. Here, we compared pathology and global gene expression profiles in lung tissue from BALB/c (Mx1−) and BALB · A2G-Mx1 mice (Mx1+/+) infected with the fully reconstructed 1918 pandemic influenza virus. Mx1+/+ mice showed less tissue damage than Mx− animals, and pathology and mortality were further reduced by treating the mice with interferon prior to infection. Using global transcriptional profiling, we identified distinct molecular signatures associated with partial protection, complete protection, and the contribution of interferon to the host response. In the absence of interferon treatment, partial protection was characterized by the generation of an acute response with the upregulation of genes associated with apoptosis, reactive oxygen species, and cell migration. Complete protection was characterized by the downregulation of cytokine and chemokine genes previously associated with influenza virus pathogenesis. The contribution of interferon treatment to total protection in virus-infected Mx1+/+ mice was characterized by the altered regulation of cell cycle genes. These genes were upregulated in Mx1+/+ mice treated with interferon but downregulated in the absence of interferon treatment. Our results suggest that Mx1+/+ mice generate a protective antiviral response by controlling the expression of key modulator molecules associated with influenza virus lethality.
INTRODUCTION
The continued emergence of new influenza viruses highlights the need to better understand influenza virus-host interactions and mechanisms of pathogenicity. Such an understanding is necessary to facilitate the development of safe and effective therapeutics and vaccines, critical aspects of preparedness for future pandemics. A clear reminder of the lethal potential of influenza virus infection is the 1918 pandemic, which resulted in over 50 million deaths worldwide (11). In addition, highly pathogenic avian H5N1 influenza viruses continue to circulate in diverse parts of the world (21, 37), and with human infections resulting in a greater than 50% mortality rate, there is considerable concern over the potential for a deadly new pandemic. Here, we sought to compare the host transcriptional responses to the reconstructed 1918 virus in Mx1+/+ mice with the goal of gaining insights into the underlying protective mechanisms that make these animals resistant to lethal influenza virus infection. The expression of the Mx1 gene is under tight transcriptional control of type I and III interferon (26) and codes for a nuclear 72-kDa protein with antiviral properties that controls influenza A virus infection in mice (9, 30).
A study comparing the virulence of influenza A virus in a mouse model of infection (6) found that an unusual influenza virus strain was virulent in Mx1+/+ mice. Interestingly, this virus was well controlled by Mx1+/+ mice treated with interferon before infection, suggesting that rapid virus growth can outcompete the antiviral response. The results from an independent study using mouse infection models suggest that the Mx1 gene protects mice against highly lethal influenza viruses (33). In that study, Mx1 was reported to be a key component of the innate immune system that mediates protection against both zoonotic and human pandemic strains of influenza viruses of high virulence. Another study using a mouse infection model and in vitro experiments showed that the origin of the viral NP gene determines Mx sensitivity and that human influenza viruses acquired adaptive mutations to evade MxA restriction (40).
In the present study, we examined the host response to the 1918 virus in wild-type BALB/c and Mx1+/+ mice; additional findings were obtained from animals treated with interferon before infection. The Mx1+/+ mice were partially protected from lethal 1918 virus infection; however, complete protection was achieved only upon interferon treatment before infection. In contrast, untreated and interferon-treated BALB/c mice succumbed to 1918 virus infection. Global gene expression profiling data revealed distinct molecular signatures associated with partial protection (untreated mice), complete protection (interferon-treated Mx1+/+ mice), and the specific contribution of interferon to complete protection of Mx1+/+ mice. In summary, these findings suggest that mice exhibiting a fully functional Mx1 phenotype are able to generate a strong antiviral state by downregulating key modulator molecules associated with influenza virus lethality.
MATERIALS AND METHODS
Viruses.
The reconstructed 1918 H1N1 virus (32), possessing the A/South Carolina/1/18 hemagglutinin (HA), was previously shown to be highly virulent for both mice and ferrets (15, 31, 32). The 1918 virus was generated utilizing the 12-plasmid reverse-genetics system in a mixture of Madin-Darby canine kidney (MDCK) (ATCC, Manassas, VA) and 293T (ATCC) cells as previously described (31). The titers of virus stocks were determined by plaque assay on MDCK cells, and stocks were maintained in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, Grand Island, NY) supplemented with 10% fetal calf serum (FCS) (HyClone, Logan, UT) and 1% penicillin/streptomycin (Gibco). The reconstructed 1918 virus was grown as previously described (22). All virus challenge experiments were performed under the guidance of the U.S. National Select Agent Program in negative-pressure HEPA-filtered biosafety level 3 enhanced (BSL-3+) laboratories and with the use of a battery-powered Racal HEPA filter respirator (Racal Health and Safety Inc., Frederick, MD) according to Biomedical Microbiological and Biomedical Laboratory procedures (35).
Mouse experiments.
All animal research was conducted according to the guidance of the CDC's Institutional Animal Care and Use Committee and in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. BALB/c mice carrying a defective allele of the Mx1 resistance gene (29) or congenic BALB · A2G-Mx1 mice carrying the functional Mx1 allele (28) (kindly provided by Peter Staeheli, University of Freiburg, Freiburg, Germany), were anesthetized by intraperitoneal injection of 0.2 ml of 2,2,2-tribromoethanol in tert-amyl alcohol (Avertin; Sigma-Aldrich, Milwaukee, WI). One hundred times the 50% lethal dose (LD50), 3.2 × 105 PFU of the reconstructed 1918 virus in 50 μl of infectious virus diluted in phosphate-buffered saline (PBS), was inoculated intranasally. LD50 titers were calculated by the method of Reed and Muench (25).
The replication of the reconstructed 1918 virus in mice (3 animals/group/time point) was examined by determining the virus titers in the lung (12, 24, and 72 h), brain, and spleen (24 and 72 h) following intranasal inoculation of virus. The clarified homogenates were titrated for virus infectivity in eggs from an initial dilution of 1:10 (lung). Additional inoculated mice (6 animals/group, making a total of 24 animals) were followed for morbidity and mortality. In addition, half of the animals were intranasally treated with 10,000 units of recombinant human alpha interferon (IFN-α) A/D (R&D Systems, Minneapolis, MN) before infection with the reconstructed 1918 virus and euthanized 12, 24, and 72 h posttreatment.
Histopathology.
Collected lung sections were fixed by submersion in 10% neutral buffered formalin, routinely processed, and embedded in paraffin. Sections were cut to 5 μm, stained with hematoxylin and eosin (HE), and examined microscopically for histopathological alterations.
RNA preparation and oligonucleotide microarray processing.
For RNA isolation, lungs (3 animals/group/time point) were frozen in individual tubes and stored in solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate, 0.5% sarcosyl, 0.1 M β-mercaptoethanol) as previously described (2). RNA was extracted from mouse lungs using a Qiagen RNeasy Minikit with RNA Later solution (Qiagen, Valencia, CA) according to the manufacturer's instructions. The RNA was further purified using RNeasy columns (Qiagen, Valencia, CA). RNA samples were spectroscopically verified for purity, and the quality of the intact RNA was assessed using an Agilent 2100 Bioanalyzer. For each set of treatment conditions, three of the five RNA samples exhibiting the highest RNA integrity number (RIN) determined using the Bioanalyzer were used for microarray analysis. cRNA probes were generated from each sample by the use of an Agilent one-color LowInput Quick Amp Labeling Kit (Agilent Technologies, Santa Clara, CA). Individual cRNA samples were hybridized to Agilent mouse whole-genome oligonucleotide 4 by 44 microarrays (approximately 39,000 unique mouse genes) according to the manufacturer's instructions. Samples from individual animals were not pooled to enable examination of animal-to-animal variation as part of the data analysis: they included three animals per time point for each virus (60 animals in total). Select samples were hybridized a second time (n = 2 technical replicates) to verify the quality of the process.
Slides were scanned with an Agilent DNA microarray scanner, and the resulting images were analyzed using Agilent Feature Extractor version 8.1.1.1. This software was used to perform image analysis, including the significance of signal and spatial detrending and to apply a universal error model. For these hybridizations, the most conservative error model was applied. Quality control (QC) reports generated from Agilent Feature Extraction software were used to assess data quality for each microarray and to identify outliers. The raw data were then loaded into a custom-designed laboratory information management system.
Microarray analysis and bioinformatics.
Global gene expression in infected lungs was analyzed using GeneData Expressionist, Analyst Module (version 2.2.1); Spotfire DecisionSite for Functional Genomics version 9; and Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Inc., Redwood City, CA). Raw intensity data from all microarrays were normalized by applying the central tendency algorithm with the standard reference set at the 75th percentile of each microarray. One- or two-way analysis of variance (ANOVA) was carried out for various groups of samples stratified by mouse strains and/or time points (see Results for more grouping details). Fixed-effect models were built to identify gene signatures associated with protection phenotypes against 1918 virus infection (see Fig. 3, 5, and 7). The Benjamini-Hochberg algorithm was used to control the false discovery rate (FDR) of multiple testing (1). Primary data are available (http://viromics.washington.edu) in accordance with proposed Minimum Information about a Microarray Experiment (MIAME) standards. Functional and network analysis of statistically significant gene expression changes was performed using IPA. The Benjamini-Hochberg algorithm was applied to correct for multiple testing in determining the probability that each biological function assigned to the genes within each statistical analysis result was due to chance alone.
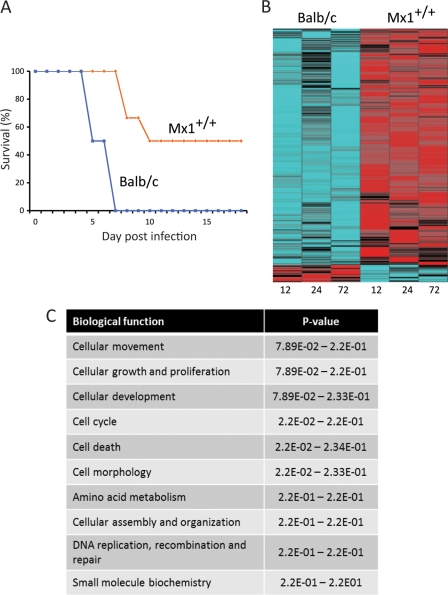
Fig 3.
Mx1+/+ mice are partially protected against 1918 virus infection. (A) Survival plot of a total of 18 mice (9 animals/mouse strain). (B) Heat map illustrating 547 differentially transcribed genes associated with 50% protection of animals compared to the wild-type animals in the absence of interferon treatment; cutoff values were ≥2-fold change and a P value of ≤0.01 (ANOVA), FDR corrected. Blue indicates downregulation. Red indicates upregulation in the activation of genes. (C) Top 10 biological functions assigned to the 547 differentially regulated genes determined by IPA. The P values of the functional categories are shown on the right.
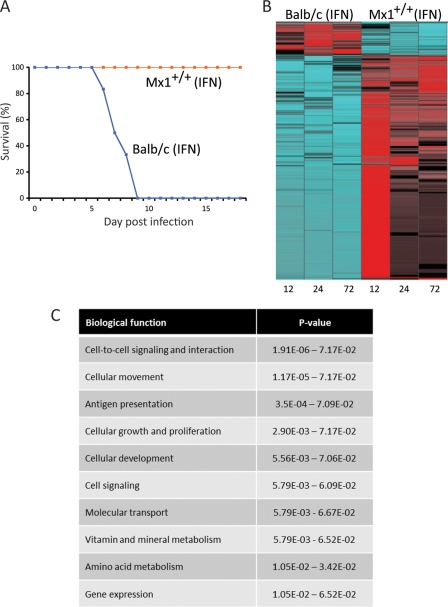
Fig 5.
Interferon-treated Mx1+/+ mice are resistant to lethal 1918 virus infection. (A) Survival plot of a total of 18 mice (9 animals/mouse strain). (B) Heat map illustrating 2,071 differentially transcribed genes associated with complete protection of Mx1+/+ animals compared to the wild-type animals during interferon treatment; cutoff values were ≥2-fold change and a P value of ≤0.01 (ANOVA), FDR corrected. Blue indicates downregulation. Red indecates upregulation in the activation of genes. (C) Top 10 biological functions assigned to the 2,071 differentially regulated genes determined by IPA. The P values of the functional categories are shown on the right.
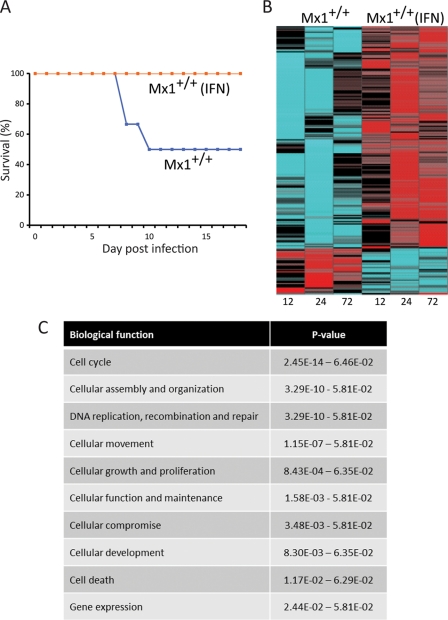
Fig 7.
Contribution of interferon to complete protection. (A) Survival plot of a total of 18 mice (9 animals/treatment). (B) Heat map illustrating 234 differentially transcribed genes that may contribute to the additional 50% difference to achieve complete protection of Mx1+/+ treated versus Mx1+/+ untreated animals; cutoff values were ≥2-fold change and a P value of ≤0.01 (ANOVA), FDR corrected. Blue indicates downregulation. Red indicates upregulation in the activation of genes. (C) Top 10 biological functions assigned to the 234 differentially regulated genes determined by IPA. The P values of the functional categories are shown on the right.
RESULTS
Mx1+/+ mice partially control 1918 virus infection.
Previous studies by our group and others (6, 27, 33) have shown that knock-in mice carrying a fully functional Mx1 gene are resistant to lethal influenza virus infection; however, those studies did not investigate the host response elicited under such experimental conditions. In the present study, our main goal was to understand the contribution of the elicited host response to survival. To achieve this goal, we performed global gene expression analyses on lung tissue from mice infected with the fully reconstructed 1918 virus.
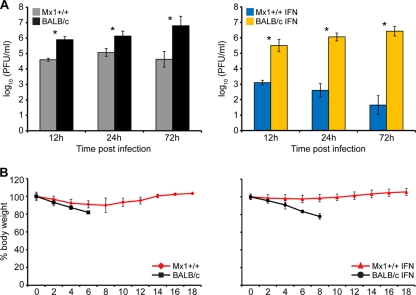
Our initial experiments showed that Mx1+/+ mice were able to reduce the growth of the 1918 virus by 1 log unit at 12 and 24 h postinfection (p.i.) with a reduction of 2 log units by 72 h p.i. compared to the virus growth observed in Mx1− mice (Fig. 1A, left). Administration of interferon to the animals prior to infection resulted in an even greater decrease of 1918 virus growth in Mx1+/+ mice, with 2.5-, 3.5-, and 5-log-unit reductions at 12, 24, and 72 h p.i., respectively. However, there was no significant change in virus growth in BALB/c (Mx1−) mice (Fig. 1A, right); these observations were in agreement with previous studies (33). Although there was a large amount of virus detected in the lungs of infected animals, especially in BALB/c mice, we did not detect viral dissemination to the brain or spleen (data not shown). However, morbidity data showed dramatic differences between the two mouse strains. While BALB/c mice were highly susceptible to 1918 virus infection, as shown by the rapid and steady decrease in body weight, interferon-treated Mx1+/+ mice were insensitive to 1918 infection (as measured by weight loss) throughout the experiment. In the absence of interferon treatment, an initial slight decline followed by a recovery in body weight in Mx1+/+ mice may be observed as a characteristic of interferon treatment (Fig. 1B).
Fig 1.
Mx1 competent mice control lethal influenza virus infection. (A) Lung virus titer data from a total of 36 mice (3 animals per time point/treatment/mouse strain). The limit of detection of the assay was 100.95 PFU/ml. (B) Morbidity data from a total of 36 mice (9 mice per group). The error bars indicate standard deviations.
1918 virus infection generates less tissue damage in Mx1+/+ mice.
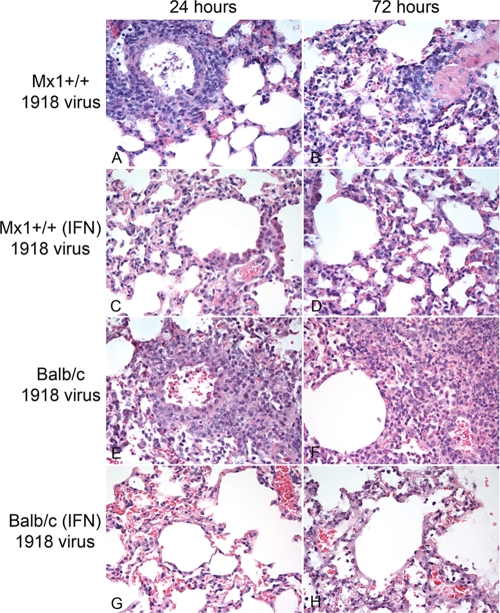
In the absence of interferon treatment, infection with the 1918 virus resulted in lung and airway lesions typical of influenza A virus infection in both Mx1+/+ and BALB/c mice. The lesions varied depending on the individual animals and the area of the lung examined, but in general, the lesions in BALB/c mice were more severe than the lesions observed in the Mx1+/+ mice (Fig. 2). At 24 h p.i., BALB/c mice exhibited moderate to severe necrotizing bronchiolitis and bronchitis. The bronchiolar walls showed vacuolation and loss of epithelium and were thickened by the presence of edema fluid, fibrin, neutrophils, eosinophils, and mononuclear cells. There was perivascular and peribronchiolar infiltration of macrophages, lymphocytes, and plasma cells. At this time point, Mx1+/+ mice exhibited only mild diffuse histiocytic alveolitis with congestion and edema, whereas BALB/c mice had moderate interstitial pneumonia. At 72 h p.i., loss of the bronchial and bronchiolar epithelium was evident, accompanied by severe perivascular and peribronchiolar inflammation, mostly composed of macrophages, but also lymphocytes, plasma cells, and neutrophils. Mild to severe diffuse interstitial pneumonia was present, with thickening of the alveolar walls; presence of inflammatory cells; congestion; and areas of alveolar necrosis, atelectasia, hemorrhage, or consolidation.
Fig 2.
Histopathological changes in tissues from 1918 virus-infected mice. Shown are photomicrographs of lung tissue sections stained with hematoxylin and eosin. (A) Severe necrotizing bronchiolitis and associated peribronchiolar lymphocytic inflammation and congestion in a 1918 virus-infected Mx1+/+ mouse at 24 h p.i. (B) Severe histiocytic-to-lymphocytic alveolitis with some neutrophils, congestion, and edema in a 1918 virus-infected Mx1+/+ mouse at 72 h p.i. (C) Loss of bronchiolar epithelium with minimal peribronchiolar inflammation and moderate histiocytic alveolitis in an interferon-treated Mx1+/+ mouse at 24 h p.i. (D) Moderate histiocytic alveolitis with congestion of the alveolar epithelium in an interferon-treated Mx1+/+ mouse at 72 h p.i. (E) Severe necrotizing bronchiolitis and associated peribronchiolar histiocytic inflammation and congestion in a 1918 virus-infected BALB/c mouse at 24 h p.i. (F) Severe diffuse histiocytic alveolitis with presence of lymphocytes, neutrophils, and plasma cells in a 1918 virus-infected BALB/c mouse 72 h p.i. (G) Moderate histiocytic alveolitis with loss of the bronchial epithelium and congestion in an interferon-treated 1918 virus-infected BALB/c mouse 24 h p.i. (H) Alveolar and bronchial epithelial necrosis with edema and congestion in an interferon-treated 1918 virus-infected BALB/c mouse 72 h p.i.
The character of the lesions varied in both the virus-infected Mx1+/+ mice and BALB/c mice treated with interferon compared to the mice not treated with interferon. At 24 h p.i., lesions to the bronchiolar and bronchial epithelium were moderate in all mice with, in some cases, loss of the respiratory epithelium, but with minimal or no peribronchial or peribronchiolar inflammation. Alveolar lesions were moderate to severe at this time point, with diffuse alveolar necrosis, hemorrhage, and edema. At 72 h p.i., Mx1+/+ mice had mild to moderate diffuse interstitial pneumonia with alveolar necrosis, and the inflammatory cells were mostly macrophages. In contrast, the BALB/c mice exhibited more severe pathology, with mild to severe necrotizing bronchiolitis and moderate to severe alveolitis with alveolar necrosis, congestion, and edema.
Mx1+/+ mice exhibit increased survival of 1918 virus infection.
Our results indicated that when infected with the 1918 virus, Mx1+/+ mice had lower viral titers and less severe lung lesions than BALB/c mice. In addition, whereas the 1918 virus was uniformly lethal in BALB/c mice, 50% of the Mx1+/+ mice survived infection (Fig. 3A). In order to assess the effects of both genetic background (MX1+/+ and BALB/c) and time points (12, 24, and 72 h) on gene expression, two-way ANOVA was performed using genetic backgrounds and time points as two independent variables. We identified 547 genes that were differentially expressed (Fig. 3B). A large proportion of these genes were downregulated in the BALB/c animals, whereas the same genes were strongly upregulated in the Mx1+/+ mice. The top 10 biological categories in which these genes are involved are shown in Fig. 3C.
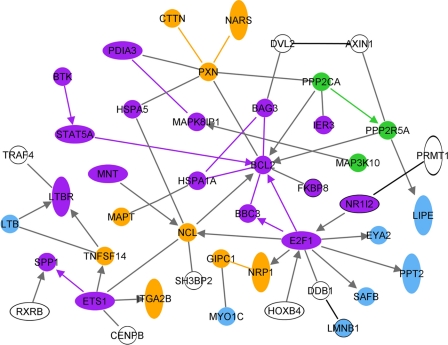
We used IPA to generate a network of genes functionally associated with survival. As annotated in IPA, the resulting network included genes associated with apoptosis, cell migration, connective tissue disorders, and the production of reactive oxygen species (Fig. 4). All of these genes, with the exception of spp1 and Ppp2r5a, were upregulated in the Mx1+/+ mice but were downregulated in BALB/c mice. These results suggest that increased survival of Mx1+/+ mice is associated with the upregulation of these pathways.
Fig 4.
Molecular signature associated with partial protection of Mx1+/+ mice. The biological network shows the direct functional relationship of genes associated with partial protection determined by IPA identified from the 547 differentially regulated genes. This network highlights upregulated and downregulated (SPP1 and PPP2R5A only) genes in Mx1+/+ mice in the absence of interferon treatment. Purple, apoptosis; orange, cell migration; light blue, connective tissue disorder; white, production of reactive oxygen species.
Interferon-dependent host response associated with complete protection.
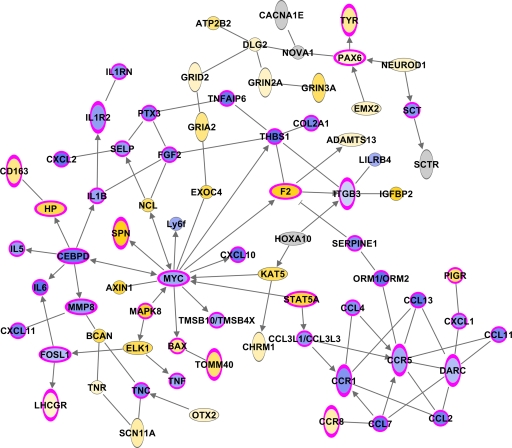
Previous studies by our group and others (6, 33) showed that pretreatment of Mx1+/+ mice with interferon results in increased resistance to influenza virus infection. In the present study, we observed similar results, with all Mx1+/+ mice pretreated with interferon surviving a challenge with a high dose of 1918 virus (Fig. 5A). In this context, we wanted to gain insights into the specific host response required for complete protection. Using a two-way ANOVA approach, we identified 2,071 differentially expressed genes between the Mx1+/+ and BALB/c interferon-treated mice (Fig. 5B). There was a clear anticorrelation in gene expression between the two mouse strains; however, the Mx1+/+ mice showed a strong upregulation of these genes by 12 h p.i. that continued, although to a lesser extent, at later time points. Functional analysis of these genes using IPA showed that the majority of the genes were associated with host physiological functions, such as cell-to-cell signaling and interaction or cellular movement (Fig. 5C). Network analysis of these genes allowed us to identify a set of physically interacting genes that might be responsible for survival (Fig. 6). The most striking feature of this molecular signature was that the Mx1+/+ mice were able to downregulate the expression of inflammatory cytokine and chemokine genes, as well as acute-phase genes. These results suggest that complete survival of mice is linked to the downregulation of key modulator molecules. We also note that these changes in gene expression were not due to differences in the response to interferon, since in the absence of infection, the gene expression signatures of interferon-treated Mx1+/+ and BALB/c mice were similar (see Table S1 in the supplemental material for a complete list of differentially expressed genes).
Fig 6.
Molecular signature associated with complete protection of Mx1+/+ mice. The biological network shows the direct functional relationship of genes associated with complete protection determined by IPA identified from the 2,071 differentially regulated genes. This network highlights the upregulation (yellow) and downregulation (blue) of genes in Mx1+/+ interferon-treated mice. Genes associated with the inflammatory response are outlined in pink.
Specific contribution of interferon to survival.
Because treatment of Mx1+/+ mice with interferon prior to 1918 virus infection increased survival from 50% to 100% (Fig. 7A), we next investigated the contribution of interferon to differences in the host transcriptional response to infection. Using two-way ANOVA, we identified 234 differentially expressed genes between these two experimental conditions (Fig. 7B), and the top 10 functional categories assigned to these genes are indicated in Fig. 7C.
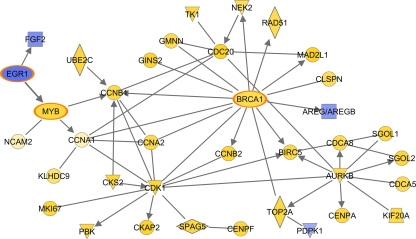
Network analysis identified functionally related genes associated with survival. All of the genes depicted in this specific network were related to cell cycle regulation, including cyclins (CCNA1, CCNA2, CCNB1, and CCNB2), cell cycle DNA damage check point regulation (BRCA1, CDK1, CKS2, TOP2A), and cell division cycle-associated genes (CDCA5, CDCA8) (Fig. 8). A large proportion of these genes were upregulated in the interferon-treated animals but were downregulated in the absence of interferon, suggesting that tight control of this specific set of genes involved in cell cycle regulation is important to elicit a response capable of providing complete protection. Although during our previous studies we found that the expression of some cell cycle-related genes was differentially regulated upon influenza virus infection, the extent and functional network of cell cycle genes associated with IFN pretreatment of Mx1+/+ mice challenged with the 1918 virus is a novel finding.
Fig 8.
Molecular signature of the contribution of interferon to complete protection. The biological network shows the direct functional relationship of cell cycle regulation genes associated with the specific contribution of interferon to survival determined by PA identified from the 234 differentially regulated genes. This network highlights the upregulation (yellow) and downregulation (blue) of genes in the Mx1+/+ interferon-treated mice.
DISCUSSION
In this study, we observed that Mx1+/+ mice were able to partially control 1918 virus growth and that this control was highly enhanced by pretreatment of the animals with interferon. In contrast, BALB/c mice were not able to control virus growth in the presence or absence of interferon treatment. We were not able to detect viral dissemination to brain or spleen in either mouse strain. Histopathological analysis indicated that Mx1+/+ animals had less severe lung damage than was present in BALB/c mice, and pathology was further reduced by interferon pretreatment. These results indicate that the ability of Mx1+/+ mice to control virus growth is linked to reduced disease severity and lethality. This is consistent with earlier studies using a highly pathogenic mouse-adapted virus (6) or the 1918 pandemic strain (33). Nevertheless, because pretreatment of Mx1+/+ mice with interferon was required for full protection, additional elements of the host response are required. We therefore used gene expression profiling to obtain a picture of the global host transcriptional response to infection under each experimental condition.
Elicited host response in untreated mice.
Comparison of the transcriptional responses of Mx1+/+ and BALB/c mice showed a striking anticorrelation in the response to 1918 virus infection. Our results suggest that under these experimental conditions, the normal interferon response is not enough to fully potentiate the protective nature of the Mx1 protein. One of the initial consequences during influenza virus infection is the rapid virus-mediated antagonism of the host innate response to avoid the generation of a strong antiviral state (8). This is accomplished by several tactics, such as inhibition of the RIG-I signaling pathway (7, 17, 19), dysregulation of gene expression (18, 23), downregulation of type I and II interferon receptor signaling (20, 24, 34), inhibition of PKR (12, 14) and OAS (14), and modulation of apoptosis (5, 13, 16, 36, 38, 39). Consequently, on one hand, the 1918 virus antagonizes the innate response, but on the other hand, Mx1 competent animals are able to partially control virus growth. This decrease in virus growth, however, was not enough to fully protect all Mx1+/+ animals.
Our functional genomics analysis performed on lungs from 1918 virus-infected Mx1+/+ mice allowed us to identify a specific molecular signature associated with partial protection of mice. The novelty of this finding is the altered regulation of specific acute-response components, such as genes associated with apoptosis (BCL2, BAG3, BBC3, HSPA1A, SPP1, ETS1, BTK, STAT5A, LTBR, PDIA3, HSPA5, IER3, MAPK8IP1, MNT, FKBP8, and NR1I2) and cell migration (TNFSF14, ITGA2B, MAPT, NCL, GIPC1, NRP1, PXN, CTTN, and NARS). We also observed the upregulation and direct functional relationship of genes associated with connective tissue disorders (LTB, MYO1C, SAFB, EYA2, PPT2, LIPE, and LMNB1) and genes involved in the production of reactive oxygen species in macrophages (PPP2CA, PPP2R5A, and MAP3K10). This functional signature illustrates a striking dichotomy in the gene expression profiles during the course of the experimental infection, where Mx1+/+ mice elicited a steady upregulation of an acute-response environment that likely generated an antiviral state that partially protected the 1918 virus-infected animals.
Interferon-dependent host response is associated with complete protection.
Analysis of the global transcriptional responses of interferon-treated Mx1+/+and BALB/c mice challenged with the 1918 virus revealed a striking difference in the expression of 2,071 genes. We found that the majority of these genes were downregulated in BALB/c mice but upregulated in the Mx1+/+ animals. Using a functional analysis approach, we identified a molecular signature associated with complete protection of Mx1+/+ mice. The most relevant feature of this signature was the downregulation of genes involved in hypercytokinemia and hyperchemokinemia, including interleukin 6 (IL-6), IL-1β, tumor necrosis factor (TNF), CCL4, CCL12, CCR1, CCR5, CXCL10, and IL-1RN genes. These results are particularly interesting in the context of our previous studies, where we have shown that lethal influenza virus infection of mice is associated with strong upregulation of the inflammatory response, which includes multiple cytokines and chemokines (3). These results suggest that Mx1 exerts its strongest role against influenza virus infection by targeting and downregulating an excessive cytokine response, which in mice lacking a functional Mx1 is highly upregulated to the detriment of the host.
Specific contribution of interferon treatment to survival.
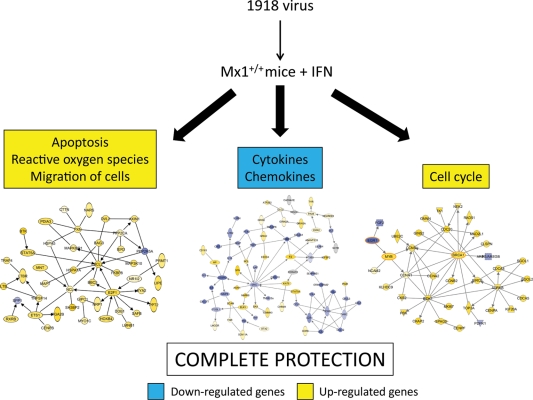
Our experimental approach allowed us to identify the genes responsible for complete protection that were not activated in the absence of interferon pretreatment of Mx1+/+ mice. Functional analysis of the 234 differentially regulated genes between the treated and untreated Mx1+/+ animals allowed us to identify a molecular signature associated with complete protection upon interferon treatment. At first glance, this functional network illustrates the upregulation of a large proportion of genes related to cell cycle regulation (CCNA1, CCNA2, CCNB1, CDK1, CKAP2, CKS2, CDC20, CDCA8, CDCA5, BIRC5, PBK, AURKB, SPAG5, MKI67, TK1, UBE2C, MYB, and BRCA1). These results suggest that complete protection of mice required the activation of a specific set of cell cycle-related genes that were downregulated in the untreated Mx1+/+ animals, which elicited only partial protection. Our previous studies of nonhuman primates and mice infected with highly pathogenic viruses have also shown the differential regulation of cell cycle-related genes (3, 4), suggesting that this is an important mechanism for host survival during lethal influenza virus infection. This is interesting, because it has been shown that influenza virus infection induces cell cycle arrest, which promotes a favorable environment for influenza virus protein expression (10). Consequently, the activation of cell cycle regulatory genes counters this mechanism by promoting host cell proliferation. Our results depict a scenario where complete protection of the Mx1+/+ animals was achieved only when there was sustained upregulation of genes related to apoptosis, reactive oxygen species, migration of cells, cytokines, chemokines, and the cell cycle, suggesting that modulation of the activities of these genes is essential for the generation of a strong antiviral state that protects all interferon-treated Mx1+/+ mice from 1918 virus infection (Fig. 9).
Fig 9.
Model illustrating our understanding of the host response necessary for complete protection against 1918 virus infection in Mx1 competent mice pretreated with interferon. The graph depicts the events leading to complete protection of Mx1+/+ animals under the described experimental conditions. Yellow indicates the upregulation of genes. Blue indicates the downregulation of genes.
In summary, our results suggest that a functional Mx1 is not sufficient to protect all mice from challenge with the fully reconstructed 1918 virus. We identified molecular signatures associated with partial protection (Mx1+/+ mice), complete protection (Mx1+/+ mice pretreated with interferon), and the specific contribution of interferon treatment to the host response. Partial protection of untreated mice was characterized by the generation of an acute-response environment, whereas complete protection was characterized by the downregulation of inflammatory cytokines and chemokines. The contribution of interferon to total protection in the 1918 virus-infected Mx1+/+ mice was characterized by the differential upregulation of cell cycle-related genes in the Mx1+/+ treated mice, which were downregulated in the absence of interferon treatment. Thus, novel therapies aimed at regulating the cytokine and chemokine environment may be of particular benefit in treating highly pathogenic influenza virus infection.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded in part by Public Health Service grant P01AI058113 from the National Institute of Allergy and Infectious Diseases.
We thank Pavel Sova (Department of Microbiology, University of Washington) for helpful discussions and reviewing the manuscript.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the funding agency.
Footnotes
Published ahead of print 21 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Benjamini Y, Hochberg Yosef. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57:289–300 [Google Scholar]
- 2. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 3. Cilloniz C, et al. 2010. Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J. Virol. 84:7613–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cilloniz C, et al. 2009. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 5:e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ehrhardt C, et al. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grimm D, et al. 2007. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. U. S. A. 104:6806–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo Z, et al. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36:263–269 [DOI] [PubMed] [Google Scholar]
- 8. Hale BG, Albrecht RA, Garcia-Sastre A. 2010. Innate immune evasion strategies of influenza viruses. Future Microbiol. 5:23–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haller O, Frese M, Kochs G. 1998. Mx proteins: mediators of innate resistance to RNA viruses. Rev. Sci. Tech. 17:220–230 [DOI] [PubMed] [Google Scholar]
- 10. He Y, et al. 2010. Influenza A virus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 84:12832–12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson NP, Mueller J. 2002. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76:105–115 [DOI] [PubMed] [Google Scholar]
- 12. Katze MG, et al. 1988. Influenza virus regulates protein synthesis during infection by repressing autophosphorylation and activity of the cellular 68,000-Mr protein kinase. J. Virol. 62:3710–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurokawa M, Koyama AH, Yasuoka S, Adachi A. 1999. Influenza virus overcomes apoptosis by rapid multiplication. Int. J. Mol. Med. 3:527–530 [DOI] [PubMed] [Google Scholar]
- 14. Li S, Min JY, Krug RM, Sen GC. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13–21 [DOI] [PubMed] [Google Scholar]
- 15. Maines TR, et al. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79:11788–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mao H, et al. 2009. Influenza virus directly infects human natural killer cells and induces cell apoptosis. J. Virol. 83:9215–9222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mibayashi M, et al. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000 [DOI] [PubMed] [Google Scholar]
- 19. Opitz B, et al. 2007. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 9:930–938 [DOI] [PubMed] [Google Scholar]
- 20. Pauli EK, et al. 2008. Influenza A virus inhibits type I IFN signaling via NF-kappaB-dependent induction of SOCS-3 expression. PLoS Pathog. 4:e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peiris JS, et al. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perrone LA, et al. 2009. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J. Virol. 83:5726–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plotch SJ, Bouloy M, Ulmanen I, Krug RM. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847–858 [DOI] [PubMed] [Google Scholar]
- 24. Pothlichet J, Chignard M, Si-Tahar M. 2008. Cutting edge: innate immune response triggered by influenza A virus is negatively regulated by SOCS1 and SOCS3 through a RIG-I/IFNAR1-dependent pathway. J. Immunol. 180:2034–2038 [DOI] [PubMed] [Google Scholar]
- 25. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 26. Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Salomon R, et al. 2007. Mx1 gene protects mice against the highly lethal human H5N1 influenza virus. Cell Cycle 6:2417–2421 [DOI] [PubMed] [Google Scholar]
- 28. Staeheli P, Dreiding P, Haller O, Lindenmann J. 1985. Polyclonal and monoclonal antibodies to the interferon-inducible protein Mx of influenza virus-resistant mice. J. Biol. Chem. 260:1821–1825 [PubMed] [Google Scholar]
- 29. Staeheli P, Grob R, Meier E, Sutcliffe JG, Haller O. 1988. Influenza virus-susceptible mice carry Mx genes with a large deletion or a nonsense mutation. Mol. Cell. Biol. 8:4518–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. 1986. Mx protein: constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44:147–158 [DOI] [PubMed] [Google Scholar]
- 31. Tumpey TM, et al. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77–80 [DOI] [PubMed] [Google Scholar]
- 32. Tumpey TM, et al. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315:655–659 [DOI] [PubMed] [Google Scholar]
- 33. Tumpey TM, et al. 2007. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J. Virol. 81:10818–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uetani K, et al. 2008. Influenza A virus abrogates IFN-gamma response in respiratory epithelial cells by disruption of the Jak/Stat pathway. Eur. J. Immunol. 38:1559–1573 [DOI] [PubMed] [Google Scholar]
- 35. Wilson DE, Chosewood LC. 2009. Laboratory biosafety level criteria, p 30–59 In Wilson DE, Chosewood LC. (ed), Biosafety in microbiological and biomedical laboratories, 5th ed Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 36. Xing Z, et al. 2009. Differential regulation of antiviral and proinflammatory cytokines and suppression of Fas-mediated apoptosis by NS1 of H9N2 avian influenza virus in chicken macrophages. J. Gen. Virol. 90:1109–1118 [DOI] [PubMed] [Google Scholar]
- 37. Yuen KY, et al. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471 [DOI] [PubMed] [Google Scholar]
- 38. Zhirnov OP, Klenk HD. 2007. Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis 12:1419–1432 [DOI] [PubMed] [Google Scholar]
- 39. Zhirnov OP, Konakova TE, Wolff T, Klenk HD. 2002. NS1 protein of influenza A virus down-regulates apoptosis. J. Virol. 76:1617–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zimmermann P, Manz B, Haller O, Schwemmle M, Kochs G. 2011. The viral nucleoprotein determines Mx sensitivity of influenza A viruses. J. Virol. 85:8133–8140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.