Abstract
Integrase (IN) strand transfer inhibitors (INSTIs) have been developed to inhibit the ability of HIV-1 integrase to irreversibly link the reverse-transcribed viral DNA to the host genome. INSTIs have proven their high efficiency in inhibiting viral replication in vitro and in patients. However, first-generation INSTIs have only a modest genetic barrier to resistance, allowing the virus to escape these powerful drugs through several resistance pathways. Second-generation INSTIs, such as dolutegravir (DTG, S/GSK1349572), have been reported to have a higher resistance barrier, and no novel drug resistance mutation has yet been described for this drug. Therefore, we performed in vitro selection experiments with DTG using viruses of subtypes B, C, and A/G and showed that the most common mutation to emerge was R263K. Further analysis by site-directed mutagenesis showed that R263K does confer low-level resistance to DTG and decreased integration in cell culture without altering reverse transcription. Biochemical cell-free assays performed with purified IN enzyme containing R263K confirmed the absence of major resistance against DTG and showed a slight decrease in 3′ processing and strand transfer activities compared to the wild type. Structural modeling suggested and in vitro IN-DNA binding assays show that the R263K mutation affects IN-DNA interactions.
INTRODUCTION
The high mutation rate of HIV-1 reverse transcriptase (RT) allows the virus to escape pressure through adaptive mutations that include drug resistance mutations that limit the effectiveness of antiretroviral drugs (5, 31, 66, 69, 70). The use of multiple drugs in combination can hamper this process by restraining viral replication, limiting the emergence of resistant strains. The addition of integrase inhibitors to the arsenal of drugs against HIV-1 is important since these inhibitors are active against viruses resistant to other drug classes (16, 49, 63).
The HIV-1 integrase enzyme catalyzes two reactions. The first is 3′ processing, which consists of cleavage of a dinucleotide at both 3′ ends of the reverse-transcribed linear viral DNA and results in the exposure of reactive hydroxyl groups. The second step termed “strand transfer” is carried out through a nucleophilic attack by exposed 3′ hydroxyl groups on host genomic DNA (26, 47). Even though 3′ processing may be a suitable therapeutic target, the integrase inhibitors developed so far are integrase strand transfer inhibitors (INSTIs) that preferentially inhibit strand transfer while only modestly affecting 3′ processing (18, 24, 26). Raltegravir (RAL) was the first INSTI to be approved for therapy in 2007 (64) and is safe and efficient in both treatment-naïve and treatment-experienced subjects (11, 17, 23, 35, 49, 62, 63). Elvitegravir (EVG) is another INSTI currently in advanced clinical trials (10, 12, 77).
Although first-generation INSTIs strongly inhibit HIV-1 replication, they possess only a modest genetic barrier to resistance. Three main resistance pathways have been identified for RAL, involving initial mutations of the N155, Q148, and Y143 residues within IN (11). Both N155 and Q148 confer cross-resistance to EVG (19, 41, 47, 67), while Y143 has been reported to be specific for RAL (44). Numerous secondary mutations confer low levels of resistance against both drugs (reviewed in reference 47). Second-generation INSTIs have been developed, which possess a more robust resistance profile than RAL and EVG (3, 21, 27, 33, 67). These include MK-2048 (3, 4, 20, 67) and dolutegravir (DTG) (3, 20, 33, 45, 46, 61, 67).
Selection studies have shown that MK-2048 can select a G118R resistance mutation (3), and during similar studies with DTG, changes were observed at positions E92, L101, T124, S153, and G193 (33, 57, 59). However, fold changes (FC) in susceptibility were moderate (FC, <2.5) for all of these substitutions; the substitutions at the well-characterized polymorphic positions L101 and T124 did not increase DTG or RAL FC (33, 68). Although no major resistance mutation against DTG has been identified to date, thus far the accumulation of multiple mutations is required to result in an FC of >10, confirming that second-generation INSTIs possess a higher genetic barrier for resistance than their first-generation counterparts (20, 33).
To further investigate this subject, we performed in vitro selections with DTG using viruses of subtypes B, C, and recombinant A/G. The most common mutation selected was R263K, and introduction of R263K into HIV-1 pNL4-3 by site-directed mutagenesis revealed low-level resistance to DTG. In addition, R263K diminished both viral fitness and integration without affecting reverse transcription. Cell-free assays indicated that R263K had minimal effect on DTG susceptibility but decreased integrase 3′ processing and strand transfer activities. Molecular modeling suggests that R263K can trigger structural and catalytic changes within IN that explain its selection by DTG. The use of an IN-DNA binding assay confirmed that R263K partially impairs IN-DNA binding. This is the first characterization of a drug resistance mutation consistently selected in both B and non-B subtype viruses passaged with DTG. This is also the first report of selection with DTG in primary human cells.
MATERIALS AND METHODS
Cells and antiviral compounds.
TZM-bl and PM1 cells were obtained through the NIH AIDS Research and Reference Reagent Program (see Acknowledgments for details). The 293T cell line was obtained from the American Type Culture Collection (CRL-11268). Cells were subcultured every 3 to 4 days in Dulbecco's minimal essential medium (DMEM) for TZM-bl and 293T cells or RPMI medium for PM1 cells supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin and maintained at 37°C and 5% CO2.Umbilical cord blood mononuclear cells (CBMCs) were isolated by Ficoll-Hypaque (GE Healthcare) gradient centrifugation from blood obtained through the Department of Obstetrics, Jewish General Hospital, Montréal, Canada. CBMCs were cultured as previously described (75).
Merck & Co., Inc., Gilead Sciences, and GSK kindly provided RAL and MK-2048, EVG, and DTG, respectively. Compounds were solubilized in dimethyl sulfoxide.
Serial passage experiments.
Selections were performed as previously described (3, 51). Briefly, CBMCs infected with the specified viruses were serially passaged in the presence of increasing concentrations of DTG. Mutations were identified by sequencing the IN region of the pol gene according to the procedure described previously (6). Viral strains 5331, 5326, 12197, 6399, and 4742 are primary isolates.
Generation of replication-competent genetically homogeneous HIV-1.
pNL4-3IN(R263K) was created by site-directed mutagenesis of the wild-type pNL4-3 obtained through the NIH AIDS Research and Reference Reagent Program with the following primers: sense (5′-GTAGTGCCAAGAAAAAAAGCAAAGATCATCAGGG-3′) and antisense (5′-CCCTGATGATCTTTGCTTTTTTTCTTGGCACTAC-3′). The presence of the mutation was confirmed by sequencing. To produce genetically homogenous HIV-1, 16 μg of plasmid coding for pNL4-3 or pNL4-3IN(R263K) was transfected into 293T cells using Lipofectamine 2000 as recommended by the manufacturer (Invitrogen). Fresh medium was added 6 h after transfection. After 48 h, culture supernatants were harvested, centrifuged, and passed through a 0.45-μm filter to remove cellular debris, treated with Benzonase (EMD) to degrade the transfected plasmids, and stored at −80°C for future use. Levels of p24 in culture fluids were measured by enzyme-linked immunosorbent assay (ELISA) (PerkinElmer). Virion-associated RT activity was measured as previously described (75).
Replication capacity in TZM-bl cells.
Relative replication of the recombinant wild-type and mutant viruses was evaluated using a noncompetitive short-term infectivity assay with TZM-bl cells as previously described (58, 75). Briefly, 20,000 cells per well were seeded into 96-well culture plates (Corning). After 24 h, cells were infected with the indicated amount of virus normalized by p24. Luciferase activity was measured at 48 h postinfection using the luciferase assay system (Promega) and a MicroBeta2 luminometer (PerkinElmer).
qPCR for reverse transcripts and integrated viral DNA.
A quantitative PCR (qPCR) for reverse transcripts and integrated DNA in PM1 T-cell lines was performed as previously described (3), with minor modifications. Briefly, cellular DNA was extracted with a DNeasy blood and tissue kit (Qiagen). PCR was performed with Platinum qPCR SuperMix-UDG (Invitrogen) on a Corbett Rotor-Gene 6000 thermocycler (Corbett). The samples were normalized for their β-globin DNA contents. The cycling conditions were 50°C for 2 min, 95°C for 2 min, and 50 repeats of 95°C for 10 s, 60°C for 10 s, and 72°C for 30 s. For the quantification of reverse transcripts, reactions were performed in the presence of 65 ng of DNA template. Integrated DNA was quantified as previously described (3) in a two-step PCR. Late reverse transcripts, Alu-LTR (where LTR stands for long terminal repeat), and β-globin primers and probes have been described previously (3).
Protein expression and purification.
Escherichia coli strain XL10-Gold ultracompetent cells, Tetr Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac Hte [F′ proAB lacIqZΔM15 Tn10 (Tetr) Amy Camr] (Stratagene), were used for plasmid production while BL21(DE3) Gold cells, F− ompT hsdSB(rB− mB−) dcm gal λ(DE3) (Stratagene), were used for protein expression. Luria-Bertani (LB) broth (Multicell) prepared in Milli-Q water and supplemented with 100 μg/ml ampicillin was used for all bacterial growth. Expression and purification of integrase recombinant proteins were performed as previously described for His-tagged integrase (2) with some modifications. Fractions containing purified integrase as judged by SDS-PAGE were dialyzed into storage buffer (20 mM HEPES, 1 M NaCl, 1 mM EDTA, 5 mM dithiothreitol [DTT], 10% glycerol [pH 7.5]), aliquoted, and stored at −80°C. Protein aliquots could be kept for several months at −80°C without loss of activity.
Protein concentration and identity.
Protein concentrations were measured using an extinction coefficient of 50,420 M−1 cm−1, which was calculated using ProtParam (71) and verified using the Bio-Rad protein assay with bovine serum albumin (BSA; Sigma) as a standard. The BSA concentration was determined by readings at an optical density at 280 nm (OD280) using its published extinction coefficient (48).
DNA substrates for in vitro assays.
The following DNA substrates were used for strand transfer assays: donor LTR DNA sense (A), *5AmMC12-ACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGT-3′ (where *5AmMC12 refers to a reactive amino group attached to the 5′ end of the oligonucleotide using a 12 carbon linker), and antisense (B), 5′-ACTGCTAGAGATTTTCCACACTGACTAAAAG-3′; target DNA sense (C), 5′-TGACCAAGGGCTAATTCACT-3Bio, and antisense (D), 5′-AGTGAATTAGCCCTTGGTCA-3Bio. For 3′ processing assay, primer B was used together with the LTR-3′-sense oligonucleotide (E) *5AmMC12-ACCCTTTTAGTCAGTGTGGAAAATCTCTAGCAGT-BioTEG. The following pairs of oligonucleotides were used for DNA binding assays: LTR-D1, 5′-CTTTTAGTCAGTGTGGAAAATCTCTAGCAGT-3′, and LTR-D2, 5′-RhoR-XN/ACTGCTAGAGATTTTCCACACTGACTAAAAG-3′. To make functional duplexes, equimolar amounts of sense and antisense primers were mixed in a microcentrifuge tube and annealed by heating for 10 min at 95°C and slow cooling to room temperature over a period of 4 h.
Strand transfer activity.
Strand transfer reactions with the wild-type and R263K integrase proteins were carried out using a microtiter plate assay as published (2, 29), with the major difference being the choice of time resolved fluorescence (TRF) over fluorescence for signal detection. Donor DNA LTR oligonucleotide duplex A/B diluted to 100 nM in phosphate-buffered saline (PBS) (pH 7.4) (Bioshop) was covalently linked to Costar DNA-Bind 96-well plates (Corning) by incubation at room temperature for 4 h; negative-control wells lacked any DNA duplex. The plates were blocked with 0.5% BSA in blocking buffer for 18 h. Before use, the plates were washed twice with each of PBS (pH 7.4) and assay buffer (50 mM morpholinepropanesulfonic acid [MOPS; pH 6.8], 50 μg/ml BSA, 50 mM NaCl, 0.15% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 30 mM MnCl2-MgCl2). Purified integrase proteins, diluted in assay buffer supplemented with 5 mM DTT, were added followed by a 1.5-h incubation at 37°C for the 3′ processing reaction. Then INSTIs were added or not added, followed by an additional 1-h incubation at 37°C in the presence of the biotinylated target DNA duplex C/D for the strand transfer reaction to occur. Following the strand transfer reaction, the plates were washed three times with wash buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Tween 20, 2 mg/ml BSA). The plates were then incubated with Eu-labeled streptavidin (PerkinElmer) diluted to 0.025 μg/ml in wash buffer in the presence of 50 μM DTPA (Sigma). After additional washes with the same buffer, Wallac enhancement solution was added (PerkinElmer). The low pH of the enhancement solution caused the charging of conjugated Eu to yield Eu3+ ions. The TRF of Eu3+ in the Wallac enhancement solution was measured on a FLUOstar Optima multilabel plate reader (BMG Labtech) in TRF mode with excitation and emission slits of 355 nm and 612 nm, respectively.
3′ processing activity.
The determination of 3′ processing activity of purified recombinant integrase proteins by microtiter plate assay was also performed. Briefly, 3′-biotinylated DNA duplex E/B was covalently linked to Costar DNA-Bind plates under similar conditions as for strand transfer. Purified IN was added, and after 2 h of incubation, 3′-biotinylated ends cleaved by the enzyme were washed away and the remaining signal measured as described for the strand transfer assay.
Microplate-based assay for IN-DNA binding.
An IN-DNA binding assay was established by modifying a previously described procedure (76). Briefly, various concentrations of IN in PBS (pH 7.4) were loaded into Nunc FluoroNunc microplate wells (200 μl per well) and the plate was incubated at room temperature for 2 h. Unbound proteins were removed by three washes in 200 μl PBS per well (pH 7.4). The plate was then blocked with 200 μl PBS/well containing 2% BSA (wt/vol) for 2 h and washed three times in PBS. A 5′-rhodamine-labeled LTR duplex (20 nM) was then added in 100 μl of binding buffer (20 mM MOPS [pH 7.2], 20 mM NaCl, 5 mM DTT, 7.5 mM MgCl2), and the plate was incubated at room temperature for 1 h. The plate was then washed three times in PBS. After removal of the final wash by inversion, 100 μl PBS was added to each well, and fluorescence was measured in a FLUOstar Optima plate reader (BMG Labtech) at excitation and emission wavelengths of 544 and 590 nm, respectively. All measurements were performed in duplicate, and experiments were repeated at least twice.
Data processing.
All cell-free experiments, except when otherwise indicated, were the result of at least 3 sets of experiments, performed in triplicate to yield 9 independent values. To determine the kinetic parameters for target-dependent strand transfer activity, the results of at least 4 sets of triplicate results (n ≥ 12) were fit to a Michaelis-Menten algorithm using GraphPad Prism 4.0 software. Fifty percent inhibitory concentrations (IC50s) were calculated on the basis of 9 independent experiments performed with wild-type B and INR263K enzymes by using the sigmoid dose-response function of the same software. Values of strand transfer activity in the absence of drug for each experiment were determined arbitrarily as 100%. The extent of resistance for individual inhibitors was measured by using an unpaired Student's t test with Welch's correction. Mean IC50s were expressed together with the standard error of the mean (SEM).
Homology modeling.
Homology models of full-length HIV-1 IN intasome and strand transfer complex (STC) were created based on the available partial crystal structures of HIV-1 integrase (8, 25, 40, 72) and the published intasome (26) and STC (39) of the prototype foamy virus (PFV) using the I-TASSER three-dimensional (3D) protein prediction server (56). Model quality was assessed based on root mean square deviation (RMSD) of the global homology structure from the PFV lead templates and available HIV-1 IN using the RCSB PDB Protein Comparison Tool (55). PyMOL (http://pymol.org/, The PyMOL Molecular Graphics System, version 1.3; Schrödinger, LLC) was used for structural visualization and image processing.
RESULTS
Isolation of R263K mutant viruses with DTG.
During previous serial passage experiments with DTG, substitutions were observed at T124A, S153F/Y, and L101I from the HIV-1 lab strain IIIB and E92Q and G193E from the HIV-1 lab strain NL432; both lab strains are subtype B virus (33, 57, 59). To extend these observations, we performed serial passage experiments with DTG using healthy donor primary human CBMCs infected with various subtype B, C, and A/G viruses (Table 1). After 20 weeks, the R263K mutation was observed in all five subtype B (5/5) selections and one of two subtype A/G selections. Of note, only a fraction of the 5326 viral population carried this substitution. During the selections, neither L101I nor T124A appeared. However, the S153Y substitution was observed in combination with R263K for one subtype B virus and S153T was partially detected in one subtype C virus. Importantly, the only subtype B or A/G virus that did not bear the R263K mutation carried the G118R mutation, which we previously characterized as a resistance mutation to MK-2048, another second-generation INSTI (3). The same mutation (G118R) was observed in one subtype C virus. The R263K mutation was detected alone or in combination with other substitutions that varied among virus strains (Table 1). R263K emerged early in culture in both subtype B and A/G viruses. In addition, R263K was the most frequent mutation at weeks 34 and 37, observable in 4/6 subtype B and A/G viruses. At the same time, G118R was present in one subtype C virus and one subtype A/G virus. Various secondary mutations accumulated with R263K and G118R, in particular H51Y, a secondary mutation previously associated with EVG resistance (32, 60). For one subtype B virus, 5326, the initial R263K mutation was lost, and a single substitution S153Y was detected at week 37. Together, these results identified R263K as a common mutation that emerges in the presence of DTG and prompted us to further investigate this substitution.
Table 1.
Serial passage experiments with CBMCs infected with subtype B, A/G, and C HIV-1 viruses in the presence of increasing concentrations of DTGa
| Subtype | Virus | Baseline polymorphisms | Week 20 |
Week 37 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DTG concentration (μM) | Acquired mutations | DTG concentration (μM) | Acquired mutations | |||||||||
| B | 5331 | I72V | 0.05 | R263K | ||||||||
| BK-132 | M154I, V201I | 0.05 | R263K | W243G/W | 0.05 | R263K | E138E/K | |||||
| 5326 | V72I, I203 M | 0.05 | R263K/R | S153Y | R166K/R | 0.05 | S153Y | |||||
| PNL4.3 | I72V, I113V, L234V | 0.05 | R263K | M50I/M, V151I | 0.05 | R263K | M50I, V151I | |||||
| 12197 RAL TI WT for INI | I203 M | 0.01 | R263K | D288E | 0.025 | R263K (week 34) | D288E (week 34) | |||||
| AG | 6399 | V72I, T125A, V201I | 0.025 | G118R | E69E/K | 0.05 | G118R | |||||
| 96USSN20 | V72I, T125A, V201I | 0.1 | R263K | 0.1 | R263K | H51H/Y | ||||||
| C | 4742 | V72I, Q95P, T125A, V201I, I203 M | 0.05 | G118R | 0.05 | G118R | H51Y | |||||
| 96USNG31 | V72I, T125A, V201I | 0.01 | S153S/T | 0.025 | H51Y, G193E/G | |||||||
Baseline polymorphisms and acquired substitutions are indicated.
R263K confers resistance against DTG in vitro.
To study the R263K substitution in the context of a homogeneous genetic background, this mutation was introduced by site-directed mutagenesis into pNL4-3 proviral DNA. The resulting pNL4-3IN(R263K) virus and its wild-type counterpart, pNL4-3, were produced by transfecting proviral DNA into 293T cells. The activity of DTG was tested in TZM-bl cells against pNL4-3 and pNL4-3IN(R263K) (Table 2). Dose-response studies to determine IC50s showed that DTG inhibited wild-type pNL4-3 with an IC50 of 3.273 nM (with a 90% confidence interval of 1.178 to 9.095 nM). The calculated IC50 was 36.69 nM (17.49 to 76.98 nM) for pNL4-3IN(R263K). These values were statistically different according to the extra sum-of-squares F test (P < 0.02). This corresponds to an 11.2-fold decrease in DTG susceptibility in the presence of the R263K mutation.
Table 2.
Effects of the R263K mutation on IC50 and 95% confidence intervals for DTG in TZM-bl cells
| Backbone | Genotype | DTG |
||
|---|---|---|---|---|
| IC50 (nM) | 90% confidence intervals (nM) | FC | ||
| pNL4-3 | wta | 3.273 | 1.178 to 9.095 | |
| INR263K | 36.69 | 17.49 to 76.98 | 11.2 | |
wt, wild type.
Characterization of R263K in cell-based assays.
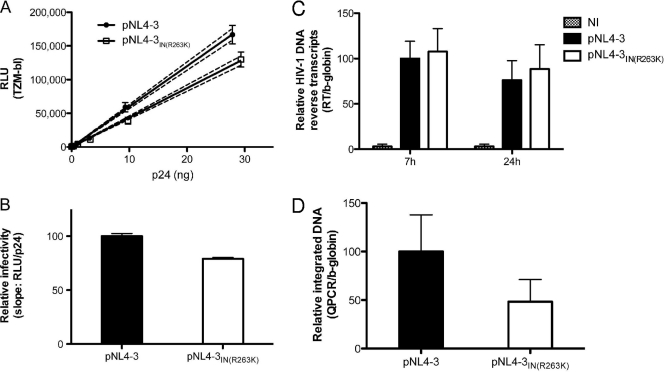
To analyze how R263K impacts viral infectivity, we performed TZM-bl infection assays with increasing amounts of wild-type pNL4-3 and pNL4-3IN(R263K) viruses (Fig. 1A). For both viruses, the measured luciferase activity from TZM-bl cells was directly proportional to the amount of virus, measured in ng of p24. These experiments indicated that R263K slightly decreases viral infectivity, as shown by the reduction in luciferase activity induced by pNL4-3IN(R263K) compared to wild-type pNL4-3. Best-fit linear regressions were created for each virus to determine the slope as an estimate of infectivity (Fig. 1B). This analysis shows that R263K decreases viral infectiousness by approximately 20%.
Fig 1.
The R263K mutation specifically decreases HIV-1 integration. (A and B) The R263K mutation in the IN region decreases HIV-1 pNL4-3 infectivity. (A) Relative luciferase units (RLU) produced by TZM-bl cells 48 h after infection with wild-type and IN(R263K) pNL4-3 viruses. Viral stocks were quantified by p24 ELISA and diluted as indicated. Infections were performed in triplicate for each viral dilution. Results for each triplicate are represented as means ± standard deviations (SD). The calculated linear regressions are shown as solid lines and the 95% confidence intervals as dotted lines. The results presented are representative of three independent experiments. (B) Infectivity of wild-type and R263K mutant viruses represented by the means ± SD of the calculated slopes for three independent TZM-bl infectivity assays (P < 0.01, t test), normalized against the wild-type slope, arbitrarily set at 100%. (C) The R263K mutation does not affect HIV-1 pNL4-3 reverse transcript production. Reverse transcription products were measured by qPCR at 7 h and 24 h after infection of PM1 cells with wild-type and R263K mutant viruses. Infections were performed in duplicate with two different viral stocks for each virus for a total of 4 independent infections for each time point and each virus. qPCRs were performed in duplicate for each sample. Results were normalized for β-globin gene expression and expressed relative to the normalized signal measured for the wild-type virus at 7 h postinfection, arbitrarily fixed at 100% for each set of infections. Results from noninfected cells (NI) are indicated. Means ± SD are shown. (D) HIV-1 pNL4-3 integration is diminished in the presence of the R263K mutation. Integrated DNA was quantified by qPCR in PM1 cells infected with wild-type and R263K pNL4-3 viruses for 72 h. Infections were performed twice in duplicate with two separate viral stocks, for a total of 8 independent infections for each virus. qPCRs were performed in duplicate for each sample. Results were normalized for β-globin gene expression and expressed relative to the signal detected for wild-type virus, arbitrarily set at 100% for each set of infections. Means ± SD are shown.
Since integrase has been found to regulate RT activity (14, 50, 52, 74), we investigated whether the R263K mutation had an effect on reverse transcription. DNA reverse transcripts from pNL4-3 and pNL4-3IN(R263K) were quantified in PM1 cells by qPCR at 7 h and 24 h after infection (Fig. 1C). Both viruses produced similar amounts of reverse transcripts, suggesting that R263K did not affect RT activity. To test the role of integration in decreased infectivity observed with the R263K mutation, we quantified viral integration (Alu-LTR) by qPCR (Fig. 1D), as described previously (3). These experiments showed that the pNL4-3IN(R263K) virus was less efficient at integrating its genome into cellular DNA than the wild-type virus. Since our results indicate that reverse transcription is not affected by the R263K substitution, we conclude that IN carrying this mutation is less active than the wild-type enzyme.
Expression and purification of INR263K.
We next carried out a biochemical characterization of IN carrying the R263K mutation. Although arginine (R) and lysine (K) are hydrophilic, basic, and positively charged at neutral pH, with only small differences in their biochemical properties, substitution from R to K leads to a change from a guanidinium to an amine side chain. To investigate the effect of this change on the biochemical properties of HIV-1 integrase, the R263K mutation was introduced by site-directed mutagenesis into the coding sequence of the subtype B integrase carrying the solubility mutations F185H and C280S. These latter substitutions have been previously shown to increase solubility of recombinant integrase without changing its activity in vitro (2, 4). Both the wild-type and R263K enzymes were expressed fused to a hexahistidine tag and purified simultaneously, as previously described (2, 4). The purity of the recombinant enzymes was shown to be better than 90% as measured by gel analysis, and their identity was confirmed by Western blotting (not shown).
Strand transfer and 3′-processing activities of INR263K.
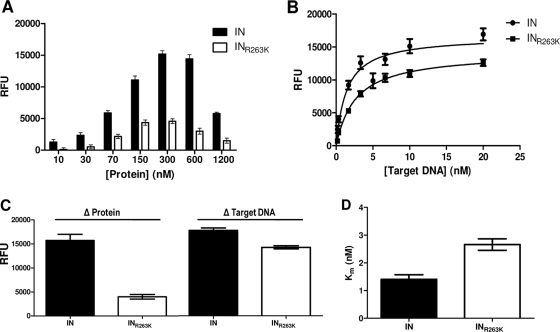
Since cell-based experiments suggest a defect in the activity of integrase carrying the R263K substitution, we analyzed the biochemical properties of the wild-type and variant (INR263K) enzymes in cell-free assays. We measured both 3′-processing and strand transfer activities of the INR263K protein and compared the results to those obtained with wild-type IN protein (Fig. 2). Both assays were performed in microtiter plates in the absence of drug inhibitors. First, the optimal protein concentration for the strand transfer assay was determined by using increasing amounts of wild-type and INR263K recombinant proteins (Fig. 2A). Wild-type IN showed a maximal strand transfer activity at 300 nM, whereas INR263K reached a maximum at 150 nM. INR263K strand transfer activity was lower than the wild type at all protein concentrations.
Fig 2.
Strand transfer activity of purified recombinant wild-type (IN) and R263K (INR263K) integrases. (A) Strand transfer activity expressed in relative fluorescent units (RFU) in the presence of 3 nM target DNA and variable concentrations of purified recombinant protein. (B) Strand transfer activity (RFU) in the presence of 300 nM purified recombinant protein and variable concentrations of target DNA. (C) Calculated maximum strand transfer activities for wild-type IN and INR263K with variable protein or target DNA concentration. The maximum activities in the presence of increasing concentrations of proteins (Δ Protein) were calculated by excluding the higher concentration (1,200 nM). (D) Calculated Michaelis-Menten constant (Km) for purified IN and INR263K.
Similar experiments were also performed in the presence of increasing concentrations of target DNA (Fig. 2B), and saturation curves were generated for each enzyme to determine maximal activity. The results indicate that INR263K has a lower maximal strand transfer activity than does wild-type IN (17,679 ± 503 relative fluorescence units [RFU]/hr for the wild type compared to 14,166 ± 321 RFU/hr for INR263K), corresponding to a 20% decrease in activity. Calculated apparent maximum activities (Vmax) and Michaelis-Menten constants (Km) are summarized in Fig. 2C and D, respectively. The Km was 1.40 nM for the wild type and 2.66 nM for INR263K, confirming the inhibitory effect of the R263K mutation on the integration process.
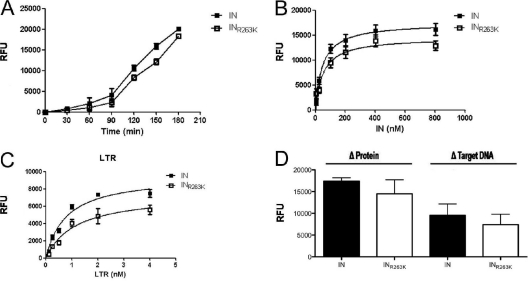
The strand transfer assay measures both integration steps, i.e., 3′ processing and strand transfer of the processed donor DNA to target DNA. To discriminate between the two reactions, we measured 3′-processing activity using the purified recombinant wild-type and INR263K proteins in separate assays. First, 3′-processing activity was measured over time (Fig. 3A). 3′-processing activity increased with time, reaching linear progression between 90 and 160 min. Next, we performed similar experiments in the presence of increasing concentrations of integrase proteins (Fig. 3B). Calculated Vmax values after 2 h were 17,414 ± 277 and 14,489 ± 1,158 RFU for wild-type and INR263K enzymes, respectively. The same assay was also performed with increasing concentrations of donor LTR DNA (Fig. 3C). The Vmax values for the wild type and INR263K were 9,535 ± 946 and 7,404 ± 863 RFU, respectively. Calculated Vmax values are summarized in Fig. 3D. Together, these results show that integrase 3′-processing and strand transfer activities were decreased by approximately 20% when the R263K mutation was present.
Fig 3.
3′-processing activity of purified recombinant wild-type (IN) and R263K (INR263K) integrases. (A) Time-dependent 3′-processing activity expressed in relative units of time-resolved fluorescence (RFU) in the presence of 100 nM protein and 10 nM viral LTR DNA mimic (P < 0.02, paired t test). (B) 3′-processing activity (RFU) after 2 h of incubation with 10 nM target DNA and increasing concentrations of protein. (C) 3′-processing activity (RFU) after 2 h of incubation with 400 nM protein and increasing concentrations of target DNA (LTR). (D) Calculated maximum 3′-processing activities for wild-type IN and INR263K with variable protein or target DNA concentrations.
Effects of the R263K mutation on susceptibility to INSTIs in vitro.
Next, we measured the effects of DTG, EVG, RAL, and MK-2048 on wild-type and INR263K enzymes in vitro (Table 3). Strand transfer assays were performed with the wild-type and mutant IN, in the presence of increasing concentrations of each of these drugs, and the resulting strand transfer activity was measured. Dose-response curves were calculated to determine IC50s. DTG inhibited wild-type and R263K IN with an IC50 of 3.49 nM and 3.74 nM, respectively. The 95% confidence interval for INR263K was wider than that for the wild-type enzyme, with a higher limit that was twice that of wild-type integrase. Our results indicate that R263K confers marginal resistance to EVG (1.8-fold change) with the same trend observed in the 95% confidence interval for the variant enzyme as observed for DTG. In addition, our results show that R263K slightly increases integrase susceptibility to RAL and MK-2048. None of the drugs tested showed any significant inhibition of 3′ processing (not shown), as previously reported (18).
Table 3.
Effects of the R263K mutation on IC50 and 95% confidence intervals for various INSTIs in vitro
| Enzyme | DTG |
RAL |
EVG |
MK-2048 |
||||
|---|---|---|---|---|---|---|---|---|
| IC50 (nM) | 95% confidence intervals (nM) | IC50 (nM) | 95% confidence intervals (nM) | IC50 (nM) | 95% confidence intervals (nM) | IC50 (nM) | 95% confidence intervals (nM) | |
| IN | 3.485 | 2.672 to 4.546 | 10.38 | 7.407 to 14.54 | 1.239 | 0.4682 to 3.278 | 2.578 | 1.923 to 3.455 |
| INR263K | 3.738 | 1.589 to 8.796 | 6.587 | 2.863 to 15.15 | 2.170 | 0.5246 to 8.977 | 1.472 | 0.5283 to 4.104 |
In silico studies of R263K mutant integrase.
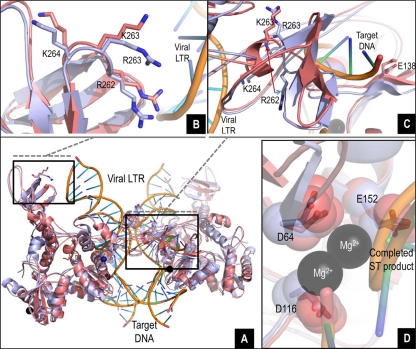
The crystal structures of integrase from the PFV bound to DNA (26, 40) and DTG (27) have recently been elucidated. Other studies demonstrated that the structure of the HIV-1 IN may be analogous to that of PFV (9, 27, 34). Accordingly, we used Protein Data Bank (PDB) identification numbers 3L2R (26), 3S3M (27), and 3OS0 (39) as starting points for structural refinement of wild-type and R263K intasomes, drug-bound intasome, and STC, respectively. Modeled HIV-1 integrase structures showed good global agreement with their template structures and with each other (Fig. 4A). When integrase intasome and STC models were overlaid with the DNA ligands of their respective templates, R263 in wild-type integrase was within 4 Å of the viral LTR fragment in both the intasome (not shown) and STC (Fig. 4A). The R263K mutation brings the R262 and K263 residues closer to the viral LTR mimic (Fig. 4B), possibly affecting LTR binding. The short length of the LTR template prevented proper analysis of these differences in integrase-DNA interactions, but it is possible that this basic rich region, which has already been shown to be important for DNA binding and 3′ processing (15, 38), has more DNA interactions than are obvious from modeling. A more dramatic spatial and orientation shift of R262 relative to its wild-type position was observed in the inner integrase subunits (Fig. 4C), causing steric clashes that translate to the target DNA binding site and the active site periphery. A close-up of one active site shows insignificant differences in the relative positions of the D64D116E152 catalytic residues between the wild type and R263K (Fig. 4D), leading to the hypothesis that the reduction in both integrase activities caused by this mutation may be due to altered DNA substrate interactions. Preliminary docking studies (not shown) indicated a slight variation in the binding mode of DTG to the R263K IN, whereas the position of docked DTG in wild-type subtype B IN was superimposable on the position of DTG in the PFV IN-DTG cocrystal structure. The manner in which the R263K mutation affects DTG binding is the subject of ongoing work.
Fig 4.
In silico studies of the wild-type and R263K integrases. (A to D) Overlay of the wild-type and R263K integrases, intasome, and strand transfer complex models with viral LTR DNA and target DNA. The tetrameric IN structure is composed of the inner and outer subunits. (B) Detailed view (8 Å) of the overlay showing proximity between residue 263 (R or K) in one of the outer subunits and the viral LTR. (C) Detailed view (12 Å) showing the pronounced shift in localization and orientation of residue R262 in the presence of the R263K mutation at the vicinity of the target DNA in one of the inner subunits. (D) Close-up overlay showing the relative positions of the D64D116E152 core catalytic residues in the wild-type and R263K enzymes in the inner subunits.
R263K impairs IN-DNA binding in vitro.
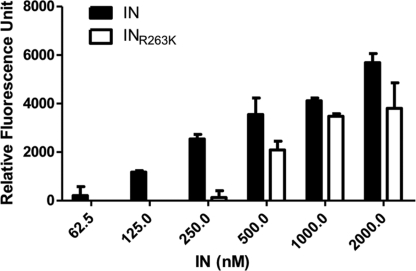
To confirm our in silico studies, we performed in vitro IN-DNA binding assays using increasing concentrations of both wild-type IN and R263K IN (Fig. 5). DNA binding was detected within a range of 62.5 to 2,000 nM purified IN. The results show that IN-DNA binding was significantly reduced in experiments performed with R263K as opposed to wild-type IN. These experiments confirm that the R263K mutation decreases IN-DNA binding under cell-free conditions.
Fig 5.
DNA binding activity of purified recombinant wild-type (IN) and R263K (INR263K) integrases. DNA binding is expressed in relative fluorescence units (RFU) in the presence of 20 nM viral LTR DNA mimic and increasing concentrations of IN protein.
DISCUSSION
To extend previous studies (33, 57, 59), we performed in vitro selection experiments with DTG using viruses of several subtypes, B, C, and A/G. The inclusion of non-B viruses in this study was necessary since such variants represent more than 90% of the HIV-1 pandemic (70). It is important to determine the role that subtype-specific polymorphisms might play in the emergence of DTG-resistance (69).
Our serial passage study did not lead to the isolation of highly resistant viruses, in agreement with previous selection studies (33, 57, 59). Our results show that subtype A/G and C viruses are not more likely to develop resistance to DTG than subtype B viruses. Furthermore, we report the selection of several mutations with DTG in vitro, with R263K being the most common after 20 weeks of culture, i.e., 6/9 viruses. R263K was present in the selections with all five subtype B (5/5) viruses and 1 of 2 A/G viruses (Table 1). At weeks 34 to 37, this mutation was detected in 3 of 4 B subtypes and 1 of 2 A/G subtypes. R263K was absent in the subtype C selections, although not enough viruses may have been tested. The partial R263K substitution identified at week 20 with the subtype B 5326 virus was replaced by S153Y after 37 weeks (Table 1). S153Y was previously documented during serial passage experiments with DTG (33). Whether resistance or fitness accounts for the change from R263K to S153Y is under investigation.
Initially, R263K was identified as a secondary resistance mutation for EVG, arising in the presence of the major resistance mutation T66I (32) and has also been reported in patients treated with RAL (6). Other selection studies with EVG did not show the emergence of R263K (60). Most resistance mutations emerge from within the catalytic core domain (amino acids [aa] 50 to 212). Of more than 33 amino acids involved in resistance to INSTIs, only 2 (6%) are located in the C-terminal region of integrase. Hence, R263K, which is also located in this region, is an unusual mutation. We have shown that IN carrying R263K is less active than wild-type IN, resulting in a decrease in infectiousness (Fig. 1A and B), which correlates with a specific decrease in viral DNA integration (Fig. 1C and D). These results agree with biochemical studies that confirmed that INR263K is impaired in regard to integration (Fig. 2). In fact, each infectivity in TZM-bl cells as well as both 3′ processing and strand transfer activities with variable DNA substrates were all reduced by approximately 20% if the R263K mutation was present (Fig. 1 to 3). Although R263K was shown in a previous study to be devoid of significant impact on DNA binding on its own, the presence of three mutations, i.e., R262D/R263V/K264E, severely impaired the ability of integrase to bind and cleave DNA (38). In contrast, our results show that DNA binding is inhibited in the presence of the single R263K mutation. This discrepancy may be due to the fact that previous workers used only a minimal DNA binding region to test for DNA binding (IN210–270) (38). In addition, our data show that both 3′-processing and strand transfer activities were impeded by the R263K mutation (Fig. 2 and 3), highlighting the importance of the R263 residue for integrase activity. In silico studies (Fig. 4) support this observation, since R263K can mediate structural changes in the HIV-1 IN intasome and strand transfer complex, ultimately impacting on integrase catalytic activities. Indeed, structural modeling of the HIV-1 intasome and strand transfer complex suggests that R263K causes a deviation in the spatial positioning and orientation of R262 in both the inner and outer subunits of IN. In the inner subunit, this deviation could possibly affect IN-target interactions and hence strand transfer. The coordination of Mg2+ by D64D112E152 does not appear to be significantly affected by the R263K mutation.
Structural modeling might also shed light on experimental differences between cell-based and biochemical cell-free assays. Despite its selection in cell culture with DTG (Table 1) and a modest level of resistance to DTG in cell-based assays (Table 2), R263K did not confer significant levels of resistance to DTG in cell-free biochemical assays (Table 3). The difference between cell culture and biochemical assays could be due to the regulation of biological events that are not measured in vitro with purified recombinant proteins. For example, the presence of cellular cofactors that are important but nonessential for integrase activity in vitro and that are not included in the strand transfer assay could play an important role. Furthermore, the polypeptide 262-RRKAK-266 has recently been identified as important for integrase nuclear import (30). Whether interaction with cellular cofactors and nuclear localization contribute to susceptibility to INSTIs is unknown. We previously identified another substitution, G118R, which also translates into a decrease in integrase activity and confers resistance to MK-2048 (3). In this regard, integrase activities with other DTG-associated mutations (33) remain to be investigated. Importantly, neither these mutations nor R263K confers major resistance to DTG in cell culture or in vitro, confirming the potency of this drug. Interestingly, it is possible that second-generation INSTIs may favor the selection of R263K in subtype B viruses whereas G118R may be favored in subtype C.
A number of secondary mutations, M50I, H51Y, E138K, V151I, and D288E, were coselected with R263K in the various cultures (Table 1). The absence of homogeneity in these secondary mutations suggests that they do not result in a major resistance pathway, although combination of these substitutions with R263K by site-directed mutagenesis will be necessary to confirm this point. M50I and D288E are natural polymorphisms detected in clinical isolates (http://hivdb.stanford.edu/, Stanford University HIV drug resistance database). V151I is a polymorphic substitution that has been selected by several INSTIs, without having significant effects on RAL and EVG susceptibility (28, 36, 42, 43). H51Y is an E92Q-linked secondary mutation selected in the presence of EVG in vitro and in patients (43, 60). In addition to R263K, our study shows that G118R was also occasionally selected with DTG (Table 1). We have previously shown that G118R is selected in vitro by MK-2048 and confers resistance to this INSTI (3). Here, we also show that G118R emerged with subtype C and A/G viruses selected with DTG. No subtype B virus displayed this mutation over the course of the selection. Despite this, G118R was the second most common substitution observed during in vitro selection with DTG. From these studies and others (27), it appears that G118R confers low-level resistance to both MK-2048 and DTG in vitro. We have previously shown that E138K is a secondary mutation that increases resistance to MK-2048 and partially restores fitness to viruses carrying the G118R mutation (3). Although susceptibility to DTG is not affected by E138K alone, the double mutation E138K/Q148K has been shown to decrease DTG efficiency (FC = 19) (33). We have shown that E138K can also emerge in the presence of the preexisting R263K mutation. The role of these secondary mutations on viral fitness and resistance is still under investigation.
ACKNOWLEDGMENTS
We thank the Canadian Institutes of Health Research (CIHR), the Canadian Foundation for AIDS Research (CANFAR), and ISTP Canada for support. P.K.Q. is a recipient of a CAHR/CIHR doctoral scholarship. T.M. is a recipient of a BMS/CTN postdoctoral fellowship. D.N.S. is a recipient of a CIHR doctoral scholarship.
We are grateful to Daniela Moisi and Ilinca Ibanescu for sequencing, to Susan P. Colby-Germinario for p24 quantification, to Richard D. Sloan for fruitful discussions, and to Daniel A. Donahue for assistance with Alu-LTR qPCR.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc. (13, 53, 54, 65, 73), PM1 from Marvin Reitz (37), pNL4-3 from Malcolm Martin (1), and HIV-1 HXB2 integrase antiserum (no. 758) from Duane P. Grandgenett (7, 22).
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Adachi A, et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bar-Magen T, et al. 2010. HIV-1 subtype B and C integrase enzymes exhibit differential patterns of resistance to integrase inhibitors in biochemical assays. AIDS 24:2171–2179 [DOI] [PubMed] [Google Scholar]
- 3. Bar-Magen T, et al. 2010. Identification of novel mutations responsible for resistance to MK-2048, a second-generation HIV-1 integrase inhibitor. J. Virol. 84:9210–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bar-Magen T, et al. 2009. Comparative biochemical analysis of HIV-1 subtype B and C integrase enzymes. Retrovirology 6:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanco JL, Varghese V, Rhee SY, Gatell JM, Shafer RW. 2011. HIV-1 integrase inhibitor resistance and its clinical implications. J. Infect. Dis. 203:1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brenner BG, et al. 2011. Subtype diversity associated with the development of HIV-1 resistance to integrase inhibitors. J. Med. Virol. 83:751–759 [DOI] [PubMed] [Google Scholar]
- 7. Bukrinsky MI, et al. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. U. S. A. 90:6125–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen JC, et al. 2000. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl. Acad. Sci. U. S. A. 97:8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cherepanov P, Maertens GN, Hare S. 2011. Structural insights into the retroviral DNA integration apparatus. Curr. Opin. Struct. Biol. 21:249–256 [DOI] [PubMed] [Google Scholar]
- 10. Cohen C, et al. 2011. Randomized, phase 2 evaluation of two single-tablet regimens elvitegravir/cobicistat/emtricitabine/tenofovir disoproxil fumarate versus efavirenz/emtricitabine/tenofovir disoproxil fumarate for the initial treatment of HIV infection. AIDS 25:F7–F12 [DOI] [PubMed] [Google Scholar]
- 11. Cooper DA, et al. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355–365 [DOI] [PubMed] [Google Scholar]
- 12. DeJesus E, et al. 2006. Antiviral activity, pharmacokinetics, and dose response of the HIV-1 integrase inhibitor GS-9137 (JTK-303) in treatment-naive and treatment-experienced patients. J. Acquir. Immune Defic. Syndr. 43:1–5 [DOI] [PubMed] [Google Scholar]
- 13. Derdeyn CA, et al. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dobard CW, Briones MS, Chow SA. 2007. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J. Virol. 81:10037–10046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dolan J, Chen A, Weber IT, Harrison RW, Leis J. 2009. Defining the DNA substrate binding sites on HIV-1 integrase. J. Mol. Biol. 385:568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donahue DA, et al. 2010. Stage-dependent inhibition of HIV-1 replication by antiretroviral drugs in cell culture. Antimicrob. Agents Chemother. 54:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eron JJ, et al. 2010. Switch to a raltegravir-based regimen versus continuation of a lopinavir-ritonavir-based regimen in stable HIV-infected patients with suppressed viraemia (SWITCHMRK 1 and 2): two multicentre, double-blind, randomised controlled trials. Lancet 375:396–407 [DOI] [PubMed] [Google Scholar]
- 18. Espeseth AS, et al. 2000. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc. Natl. Acad. Sci. U. S. A. 97:11244–11249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goethals O, et al. 2008. Resistance mutations in human immunodeficiency virus type 1 integrase selected with elvitegravir confer reduced susceptibility to a wide range of integrase inhibitors. J. Virol. 82:10366–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goethals O, et al. 2011. Resistance to raltegravir highlights integrase mutations at codon 148 in conferring cross-resistance to a second-generation HIV-1 integrase inhibitor. Antiviral Res. 91:167–176 [DOI] [PubMed] [Google Scholar]
- 21. Goethals O, et al. 2010. Primary mutations selected in vitro with raltegravir confer large fold changes in susceptibility to first-generation integrase inhibitors, but minor fold changes to inhibitors with second-generation resistance profiles. Virology 402:338–346 [DOI] [PubMed] [Google Scholar]
- 22. Grandgenett DP, Goodarzi G. 1994. Folding of the multidomain human immunodeficiency virus type-I integrase. Protein Sci. 3:888–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grinsztejn B, et al. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 369:1261–1269 [DOI] [PubMed] [Google Scholar]
- 24. Grobler JA, et al. 2002. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl. Acad. Sci. U. S. A. 99:6661–6666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hare S, et al. 2009. Structural basis for functional tetramerization of lentiviral integrase. PLoS Pathog. 5:e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hare S, et al. 2011. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol. Pharmacol. 80:565–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hazuda DJ, et al. 2004. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. U. S. A. 101:11233–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hazuda DJ, Hastings JC, Wolfe AL, Emini EA. 1994. A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res. 22:1121–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jayappa KD, Ao Z, Yang M, Wang J, Yao X. 2011. Identification of critical motifs within HIV-1 integrase required for importin alpha3 interaction and viral cDNA nuclear import. J. Mol. Biol. 410:847–862 [DOI] [PubMed] [Google Scholar]
- 31. Johnson VA, et al. 2010. Update of the drug resistance mutations in HIV-1: December 2010. Top. HIV Med. 18:156–163 [PubMed] [Google Scholar]
- 32. Jones G, et al. 2007. In vitro resistance profile of HIV-1 mutants selected by the HIV-1 integrase inhibitor, GS-9137 (JTK-303), abstr. 627. 14th CROI, Conference on Retroviruses and Opportunistic Infections, Los Angeles, CA [Google Scholar]
- 33. Kobayashi M, et al. 2011. In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob. Agents Chemother. 55:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krishnan L, et al. 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 107:15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lennox JL, et al. 2010. Raltegravir versus efavirenz regimens in treatment-naive HIV-1-infected patients: 96-week efficacy, durability, subgroup, safety, and metabolic analyses. J. Acquir. Immune Defic. Syndr. 55:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Low A, et al. 2009. Natural polymorphisms of human immunodeficiency virus type 1 integrase and inherent susceptibilities to a panel of integrase inhibitors. Antimicrob. Agents Chemother. 53:4275–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lusso P, et al. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lutzke RA, Vink C, Plasterk RH. 1994. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 22:4125–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maertens GN, Hare S, Cherepanov P. 2010. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature 468:326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V. 1998. Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J. Mol. Biol. 282:359–368 [DOI] [PubMed] [Google Scholar]
- 41. Marinello J, et al. 2008. Comparison of raltegravir and elvitegravir on HIV-1 integrase catalytic reactions and on a series of drug-resistant integrase mutants. Biochemistry 47:9345–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Markowitz M, et al. 2007. Rapid and durable antiretroviral effect of the HIV-1 integrase inhibitor raltegravir as part of combination therapy in treatment-naive patients with HIV-1 infection: results of a 48-week controlled study. J. Acquir. Immune Defic. Syndr. 46:125–133 [DOI] [PubMed] [Google Scholar]
- 43. McColl D. J, et al. 2007. Resistance and cross-resistance to first generation integrase inhibitors: insights from a phase 2 study of elvitegravir (GS-9137), abstr. 9. 16th International HIV Drug Resistance Workshop, Barbados, West Indies [Google Scholar]
- 44. Métifiot M, et al. 2011. Elvitegravir overcomes resistance to raltegravir induced by integrase mutation Y143. AIDS 25:1175–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Min S, et al. 2011. Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of dolutegravir as 10-day monotherapy in HIV-1-infected adults. AIDS 25:1737–1745 [DOI] [PubMed] [Google Scholar]
- 46. Min S, et al. 2010. Pharmacokinetics and safety of S/GSK1349572, a next-generation HIV integrase inhibitor, in healthy volunteers. Antimicrob. Agents Chemother. 54:254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mouscadet JF, Delelis O, Marcelin AG, Tchertanov L. 2010. Resistance to HIV-1 integrase inhibitors: a structural perspective. Drug Resist. Updat. 13:139–150 [DOI] [PubMed] [Google Scholar]
- 48. National Committee for Clinical Laboratory Standards 1979. NCCLS standard. National Committee for Clinical Laboratory Standards, Villanova, PA [Google Scholar]
- 49. Nguyen BY, et al. 2011. Raltegravir: the first HIV-1 integrase strand transfer inhibitor in the HIV armamentarium. Ann. N. Y. Acad. Sci. 1222:83–89 [DOI] [PubMed] [Google Scholar]
- 50. Nishitsuji H, et al. 2009. Augmentation of reverse transcription by integrase through an interaction with host factor, SIP1/Gemin2 is critical for HIV-1 infection. PLoS One 4:e7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oliveira M, Brenner BG, Wainberg MA. 2009. Isolation of drug-resistant mutant HIV variants using tissue culture drug selection. Methods Mol. Biol. 485:427–433 [DOI] [PubMed] [Google Scholar]
- 52. Oz I, Avidan O, Hizi A. 2002. Inhibition of the integrases of human immunodeficiency viruses type 1 and type 2 by reverse transcriptases. Biochem. J. 361:557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J. Virol. 83:8289–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prlic A, et al. 2010. Pre-calculated protein structure alignments at the RCSB PDB website. Bioinformatics 26:2983–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sato A, et al. 2009. In vitro passage of drug resistant HIV-1 against a next generation integrase inhibitor (INI), S/GSK1349572, abstr. H-932. 49th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC), San Francisco, CA [Google Scholar]
- 58. Schader SM, Colby-Germinario SP, Schachter JR, Xu H, Wainberg MA. 2011. Synergy against drug-resistant HIV-1 with the microbicide antiretrovirals, dapivirine and tenofovir, in combination. AIDS 25:1585–1594 [DOI] [PubMed] [Google Scholar]
- 59. Seki T, et al. 2010. S/GSK1349572 is a potent next generation HIV integrase inhibitor and demonstrates a superior resistance profile substantiated with 60 integrase mutant molecular clones, abstr. 555. 17th CROI, Conference on Retroviruses and Opportunistic Infections, San Francisco, CA [Google Scholar]
- 60. Shimura K, et al. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 82:764–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Song I, et al. 2011. Effects of etravirine alone and with ritonavir-boosted protease inhibitors on the pharmacokinetics of dolutegravir. Antimicrob. Agents Chemother. 55:3517–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Steigbigel RT, et al. 2008. Raltegravir with optimized background therapy for resistant HIV-1 infection. N. Engl. J. Med. 359:339–354 [DOI] [PubMed] [Google Scholar]
- 63. Steigbigel RT, et al. 2010. Long-term efficacy and safety of raltegravir combined with optimized background therapy in treatment-experienced patients with drug-resistant HIV infection: week 96 results of the BENCHMRK 1 and 2 phase III trials. Clin. Infect. Dis. 50:605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Summa V, et al. 2008. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J. Med. Chem. 51:5843–5855 [DOI] [PubMed] [Google Scholar]
- 65. Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J. Virol. 82:12585–12588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Turner D, Wainberg MA. 2006. HIV transmission and primary drug resistance. AIDS Rev. 8:17–23 [PubMed] [Google Scholar]
- 67. Van Wesenbeeck L, et al. 2011. Cross-resistance profile determination of two second-generation HIV-1 integrase inhibitors using a panel of recombinant viruses derived from raltegravir-treated clinical isolates. Antimicrob. Agents Chemother. 55:321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vavro C, et al. 2010. Polymorphisms at position 101 and 124 in the HIV-1 integrase (IN) gene: lack of effects on susceptibility, abstr. H-935. 50th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC), Boston, MA [Google Scholar]
- 69. Wainberg MA. 2004. HIV-1 subtype distribution and the problem of drug resistance. AIDS 18(Suppl 3):S63–S68 [DOI] [PubMed] [Google Scholar]
- 70. Wainberg MA, Zaharatos GJ, Brenner BG. 2011. Development of antiretroviral drug resistance. N. Engl. J. Med. 365:637–646 [DOI] [PubMed] [Google Scholar]
- 71. Walker JM. 2005. The proteomics protocols handbook. Humana Press, Totowa, NJ [Google Scholar]
- 72. Wang JY, Ling H, Yang W, Craigie R. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wei X, et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilkinson TA, et al. 2009. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J. Biol. Chem. 284:7931–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu HT, et al. 2011. Compensation by the E138K mutation in HIV-1 reverse transcriptase for deficits in viral replication capacity and enzyme processivity associated with the M184I/V mutations. J. Virol. 85:11300–11308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang ZR, et al. 2003. Fluorescent microplate-based analysis of protein-DNA interactions. I. Immobilized protein. Biotechniques 35:980–982, 984,, 986 [DOI] [PubMed] [Google Scholar]
- 77. Zolopa AR, et al. 2010. Activity of elvitegravir, a once-daily integrase inhibitor, against resistant HIV type 1: results of a phase 2, randomized, controlled, dose-ranging clinical trial. J. Infect. Dis. 201:814–822 [DOI] [PubMed] [Google Scholar]