Abstract
Antibody 14G7 is protective against lethal Ebola virus challenge and recognizes a distinct linear epitope in the prominent mucin-like domain of the Ebola virus glycoprotein GP. The structure of 14G7 in complex with its linear peptide epitope has now been determined to 2.8 Å. The structure shows that this GP sequence forms a tandem β-hairpin structure that binds deeply into a cleft in the antibody-combining site. A key threonine at the apex of one turn is critical for antibody interaction and is conserved among all Ebola viruses. This work provides further insight into the mechanism of protection by antibodies that target the protruding, highly accessible mucin-like domain of Ebola virus and the structural framework for understanding and characterizing candidate immunotherapeutics.
INTRODUCTION
Ebola hemorrhagic fever is one of the most virulent diseases known, with case fatalities up to 90%. The disease is characterized by a febrile episode, general malaise, and nausea that gradually progress into hemorrhage and shock that are characteristic hallmarks of end-stage disease (30). Ebola virus, an etiological agent of the disease, is a negative-sense RNA virus and belongs to the family Filoviridae (30, 38). The viral genome encodes just seven genes. One of these genes, termed GP, encodes two distinct gene products that share 295 amino acids of N-terminal sequence but have distinct C termini due to a transcriptional editing event (31). The primary gene product (80% of gene transcripts) is termed sGP and is a dimeric, soluble glycoprotein shed abundantly by infected cells. The secondary gene product (20% of transcripts) is termed GP and is the trimeric protein anchored in the viral membrane responsible for virus attachment, fusion, and entry. The viral surface GP is the ideal target for neutralizing antibodies, and yet antibodies produced in natural infections appear to preferentially react with the secreted sGP (24, 25). Even those that cross-react between sGP and GP are likely to be absorbed by the much more abundant sGP and would thus be less available for virus neutralization. Thus, a challenge for design of emergency immunotherapeutics is to find antibodies that are specific for the viral surface GP. One such target is a heavily glycosylated mucin-like domain that is unique to GP and is not shared with sGP.
The unusual mucin-like domains of GP are each 150 amino acids in size and include 5 N-linked and 12 to 15 O-linked glycan addition sites. These heavily glycosylated domains extend from the top and sides of GP and effectively dominate the structure of GP including GP surfaces available for host interaction. They sterically shield host β1-integrin and major histocompatibility complex on the surfaces of infected cells and also shield antibody epitopes lower down on the GP peplomer (14, 28). However, antibody epitopes contained within these projecting mucin-like domains may be better exposed for immune surveillance. Indeed, antibodies belonging to three distinct competition groups that recognize the mucin-like domain of Ebola virus GP have been shown to be protective in mouse models of lethal Ebola virus challenge (37). Studies to determine efficacy in nonhuman primates are ongoing.
The protective efficacy demonstrated by these antibodies and others has raised the possibility of using monoclonal antibodies (MAbs) as a cocktail in both pre- and postexposure treatment of Ebola virus. A structural understanding of how antibodies such as these could protect against Ebola virus challenge will assist in developing and evaluating immune responses elicited by candidate vaccines.
Three epitopes within the mucin-like domain of Ebola virus GP have been delineated based on competitive enzyme-linked immunosorbent assay (ELISA) studies. Antibodies in competition group I are directed against GP residues 401 to 417, those in competition group II are directed against GP residues 389 to 405, and those in competition group III are directed against GP residues 477 to 493 (37). Note that the sequences of these epitopes are specific to Ebola virus (formerly known as Zaire ebolavirus), and the antibodies do not cross-react to other Ebola viruses such as the Sudan, Reston, or Tai Forest viruses (37). Net-N-Glyc and Net-O-Glyc servers predict that these Ebola virus mucin-like domain epitopes are likely unglycosylated (15, 17). A structural understanding of how these antibodies protect and of the conformations that these GP epitopes adopt will be instrumental in development and analysis of immunotherapeutic cocktails.
Any GP containing the heavily glycosylated mucin-like domains has been thus far refractory to crystallization, and this region has had to be deleted from all GPs crystallized (11, 22). Thus, even though we have structural information on the lower core of GP, there is no structural information on the mucin-like domain, save one structure of an antibody bound to a separate mucin-like domain linear epitopes. This crystal structure, of MAb 13F6-1-2, a representative member of competition group I, in complex with its peptide linear epitope (GP residues 401 to 417), has provided insight into its unique structural mechanism of neutralization (23). 13F6-1-2 contains an extremely rare Vλx light chain that is represented by only 0.5% of mouse antibody sequences. The use of this very rare light chain confers noncanonical structures to all three CDRs and may consequently result in the unusual mode of binding of the antibody (23). The GP401-417 peptide, representing the epitope of 13F6-1-2, adopts a linear, extended conformation and binds into a shallow groove in the 13F6-1-2 combining site that extends diagonally across the antibody combining site. Two particular amino acids of GP401-417, Gly P406 and Arg P409, form extensive contacts (eight in all) with 13F6 and are critical for antibody recognition (23).
Delivery of a cocktail of well-characterized protective antibodies against distinct epitopes may provide better protection than delivery of one antibody against a single epitope alone. Here, we extend our structural studies to a separate protective epitope in the mucin-like domain and present the crystal structure of MAb 14G7. MAb 14G7 is the most potent of the MAbs in competition group III and has been crystallized in complex with its distinct, linear Ebola virus GP epitope (residues 477 to 493, GKLGLITNTIAGVAGLI). 14G7 is protective in 90% of mice challenged with Ebola virus when administered 1 day postchallenge (37). The administered dose of 100 μg per rodent is within the therapeutic range of 3 to 5 mg/kg used in humans. Efficacy testing of MAbs 14G7 and 13F6-1-2 in primates is ongoing. Antibodies 14G7 and 13F6-1-2 are specific for Ebola virus, the most virulent of the Ebola viruses, and do not cross-react with other filoviruses.
MATERIALS AND METHODS
Fab production and purification.
IgG1 14G7-1-2 was elicited by vaccination of BALB/c mice with Venezuelan equine encephalitis virus replicons bearing Ebola virus GP (strain Mayinga, 1976) (37). IgGs were produced via hybridoma culture as previously described (23). Fab fragments of 14G7 were prepared by digesting IgG with 2% pepsin at 37°C for 2 h to yield F(ab′)2, followed by reduction with 5 mM cysteine for 1 h at 37°C to yield Fab′. The reduction was terminated with 50 mM iodoacetamide. Fab fragments were separated from undigested IgG and pepsin by size-exclusion chromatography performed on a Superdex 75 10/300 column buffered with 150 mM NaCl and 10 mM Tris (pH 7.5).
Crystallization.
A linear peptide corresponding to GP residues 477 to 493 (GKLGLITNTIAGVAGLI) was chemically synthesized (Genscript). The 14G7 Fab was initially concentrated to 16 mg/ml and complexed with 20-fold molar excess peptide at 4°C overnight. The complex was further concentrated to 22 mg/ml before crystallization screening via free interface diffusion (Fluidigm Topaz). A single hit in 0.2 M sodium acetate, 0.1 M Bis-Tris propane (pH 6.5), and 20% PEG 3350 was identified and translated to a 1-μl scale at 22°C via hanging-drop method vapor diffusion.
Data collection.
The crystals were cryoprotected with 20% glycerol prior to data collection. Diffraction data extending to 2.8 Å were collected at the Stanford Synchrotron Radiation Laboratory Beamline 11-1 using a MAR 325 charge-coupled device detector and processed using HKL2000 (27). The complex crystallized in space group P21 with unit cell dimensions: a = 89.5 Å, b = 68.3 Å, c = 92.8 Å, and β = 112.4°. The data collection statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics
| Parametera | Value(s)b |
|---|---|
| Space group | P21 |
| Unit cell | a = 89.5 Å, b = 68.3 Å, c = 92.8 Å, α = γ = 90°, β = 112.4° |
| No. of observed reflections | 79,043 |
| No. of unique reflections | 46,342 |
| Resolution range (Å) | 50–2.8 |
| Rsym (%) | 4.3 (20.2) |
| Redundancy | 1.7 (1.7) |
| Completeness (%) | 93.9 (85.1) |
| Avg I/σ(I) | 19.9 (2.2) |
| No. of total atoms | 6,704 |
| No. of peptide atoms | 200 |
| No. of water molecules | 48 |
| Resolution range (Å) | 38–2.8 |
| Rcryst (%) | 23.5 |
| Rfree (%) | 28.7 |
| Avg B values (Å2) | |
| Light chain | 46.2 |
| Heavy chain | 44.9 |
| Peptide | 49.3 |
| Water | 32.3 |
| RMSD | |
| Bond length (Å) | 0.003 |
| Angle (°) | 0.68 |
| Dihedrals (°) | 12.2 |
| Ramachandran favored (%)c | 94.5 (0.7) |
Rsym = Σ |I(k) − <I> |/Σ I(k), where I(k) and <I> represent the scaled intensity values of the individual measurements and the corresponding mean values, respectively. Rcryst = (Σhkl ||Fobs| − k |Fcalc||)/(Σhkl |Fobs|), where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is the same as Rcryst, except that 5% of the reflections are chosen at random and omitted from refinement. RMSD, root mean square deviation.
Values given in parentheses refer to reflections in the outer resolution shell.
That is, the residues in the favored region of Ramachandran plot as validated by molprobity. The outliers are shown in parentheses.
Structure determination and refinement.
The crystal structure of the complex was determined by molecular replacement using Phaser (1–3) with Fab OX108 (26), PDB accession code 3DGG, as a search model since it belongs to the same species and isotype as 14G7 and is 92% identical in sequence. Fab OX108 was divided into two search models, representing its constant and variable domains, respectively, in order to account for possible differences in antibody elbow angle. After molecular replacement, the model was subjected to a round of rigid body and restrained simulated annealing refinement using Phenix. After this first round of refinement, the Rwork and Rfree were 26.1 and 32.3%, respectively. Positive Fo-Fc electron density, which is consistent with a polypeptide bound inside the antibody-combining site, was observed after the first round of refinement and model rebuilding in Phenix and Coot, respectively (13). Water molecules were added in the ensuing rounds of refinement. Automatically derived NCS restraints were applied to the light and heavy chains, as well as the peptide in both complexes in the asymmetric unit. During the last round of refinement, the wxc_scale parameter was adjusted to 0.1. The structural validation of the model was done using both PROCHECK and molprobity (7, 10, 21). Refinement statistics are summarized in Table 1.
Binding studies of 13F6-1-2 and 14G7 GPΔtm.
A BIAcore-2000 surface plasmon resonance spectrophotometer was used to measure the binding avidity of 13F6-1-2 and 14G7 IgGs to soluble GP ectodomain (GPΔtm). The glycoprotein and IgGs were buffer exchanged into HBS-EP (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% polysorbate 20), and the antibodies were immobilized on various channels of CM5 sensor chips using an amine coupling protocol. Streptavidin was immobilized in a similar manner on a different channel as a control. GPΔtm was injected at 10 μl/min for 250 s over the channels at a concentration of 0.275, 0.55, 1.1, and 2.2 μM, respectively. The dissociation was measured by injecting HBS-EP buffer over the channels for a period of ∼600 s. Resulting chromatogram data were fitted to a 1:1 Langmuir binding model using the BIAevaluation 3.0 software to obtain the kinetic binding parameters (Kd).
Neutralization assays.
A biologically contained Ebola virus containing Renilla luciferase instead of the essential viral protein VP30 was rescued and propagated in VeroVP30 cells as described previously (16). EbolaΔVP30-green fluorescent protein virus was diluted in 10% fetal calf serum in minimum essential medium and incubated with the purified MAbs (at the indicated concentrations, diluted into normal mouse serum (Jackson Immunoresearch) in the presence or absence of guinea pig complement (final concentration, 2%; Cedarlane) for 1 h at 37°C. The virus-antibody mixture was transferred to wells of a 96-well plate seeded to confluence with Vero VP30 cells and incubated for 14 h at 37°C. After incubation, the cellular luciferase activity was determined using EnduRen (Promega) as a readout of virus infection. Individual measurements were performed in triplicate, and the percent neutralization was calculated based on virus replication in the presence of a normal mouse IgG negative control.
ELISAs.
Viruses (Ebola virus, strain Mayinga, 1976; Ebola virus, strain Eckron, 1995; Sudan virus, strain Boniface; Tai Forest virus; Reston virus; Marburg virus, strain Musoke; and Ravn virus) were purified by sucrose gradient and irradiated with 6 million rads for 30 min. Recombinant, transmembrane-deleted GPs from Ebola virus (strain Mayinga), Sudan virus (strain Boniface), Bundibugyo virus, and Marburg virus (strain Ci67) were expressed in 293T cells and purified by Ni-NTA affinity chromatography. Plates were blocked for 2 h at room temperature with 3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS). The MAbs 14G7, 13F6-1-2, 42/3.7 (35), and KZ52 were allowed to bind for 1 h at room temperature at 1 and 5 μg/ml in 1% BSA in PBS. A secondary, horseradish peroxidase-conjugated goat anti-mouse antibody diluted 1:2,000 (or goat anti-human for KZ52) was allowed to bind for 1 h at room temperature. Plates were developed with TMB substrate and read at 450 nm.
Protein Data Bank accession code.
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 2Y6S.
RESULTS
14G7 Fab structure.
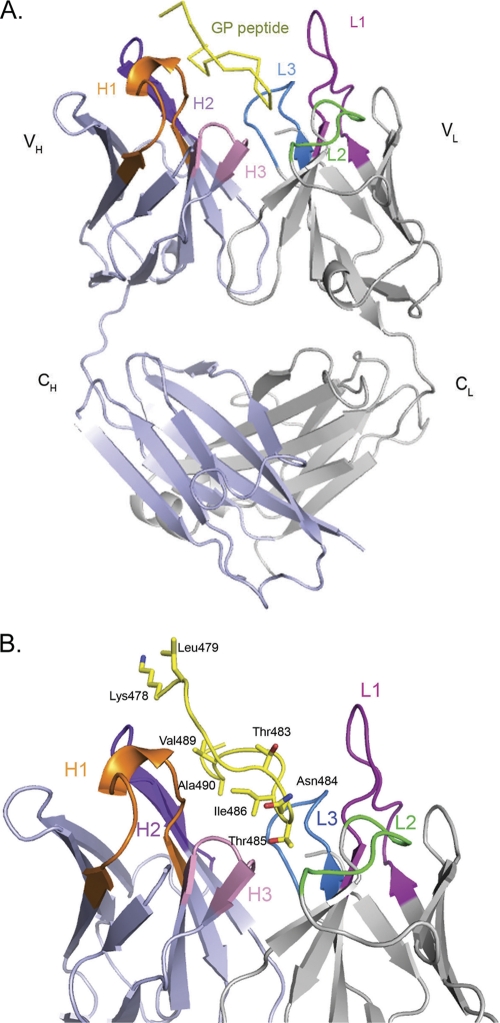
Fab fragments were prepared by pepsin digestion, purified, and incubated overnight with a synthetic peptide representing the GP epitope prior to crystallization. Crystals of the Fab-peptide complex diffract to 2.8-Å resolution, and the structure was determined by molecular replacement. Two copies of the antibody-peptide complex are contained in the crystallographic asymmetric unit. Both complexes within the asymmetric unit are quite similar and, for simplicity, discussions henceforth will focus on one complex unless otherwise noted. Electron density is visible for residues 1 to 126 and residues 134 to 213 of the heavy chain, residues 1 to 217 of the light chain, and residues 478 to 492 of the GP peptide. There is no interpretable electron density for the N- and C-terminal residues of the GP peptide (residues 477 and 493), but previous epitope mapping experiments suggest that these residues are probably not critical for antibody recognition (37). The overall structure of the complex illustrating the bound peptide and the CDRs involved in binding is shown in Fig. 1.
Fig 1.
Crystal structure of the 14G7 Fab-peptide complex. For clarity, only one of the two essentially identical molecules in the asymmetric unit is shown. (A) Overall structure of the 14G7 Fab in complex with its Ebola virus epitope. (B) Close-up side view of the Fab-peptide complex showing the spatial orientation of the peptide within the Fab pocket. The antibody CDRs L1, L2, L3, H1, H2, and H3 are colored magenta, green, blue, orange, purple, and pink, respectively, while the peptide is in yellow. All of the figures for the present study were generated by using MacPyMol (35).
Rather than containing a rare light chain, as does 13F6-1-2, 14G7 contains a κIII light chain with sequence >90% identical to others of the same class, suggesting that antibodies such as 14G7 may be easier to elicit in vaccination programs. The elbow angle of each 14G7 Fab is 174.3°, similar to the majority of κ chains with elbow angles below 180°. Comparative analysis of the conformations adopted by antibody complementarity-determining regions (CDRs) reveals that they can be broadly categorized into 18 canonical classes (4). Of those in MAb 14G7, CDR H1 belongs to canonical class 1 (class 1/10A), while CDR H2 belongs to class 2 (class 2/10A). Notably, CDR H3 is quite short and has a kinked base. CDRL1 is particularly long (15 residues) and falls under canonical class 5. CDR L2 belongs to canonical class 1/7A, while CDR L3 is very similar to canonical class 1.
The CDR H3 of 14G7 is composed of only 6 amino acids. CDRs with relatively short lengths cannot be clustered under either bulged or nonbulged torso conformations. However, the kinked base formed due to hydrogen bonding between the carbonyl of Phe-100 and the indole group of Trp-103 gives CDR H3 a semblance of a bulged torso. The apex of the H3 is composed of residues Ala-99, Phe-100, Asp-101, and Tyr-102. The entire scaffold is stabilized by hydrogen bond contacts between the carbonyl of Thr-98 and the amide backbone of Asp-101 and another bond between the Phe-100 carbonyl and Trp-103 HNε1.
Conformation of the GP peptide.
The VL-VH interface in the 14G7 antibody combining site forms a 14-Å deep pocket into which Ebola virus GP482-491 binds. The Fab-peptide interaction buries a total of 1,699 Å2 of molecular surface area (801 Å2 on the antibody and 898 Å2 on the peptide). The GP peptide adopts two β turns. The first is a type I β-turn conformation that is formed by residues Thr-483, Asn-484, Thr-485, and Ile-486 and is stabilized by a hydrogen bond between the main chain carbonyl of Thr-483 and the amide nitrogen of Ile-486 (Fig. 2). This β turn causes the peptide to reverse direction and effectively doubles possible contacts with the antibody-combining site. Between this β turn and the next are residues Ala-487 and Gly-488, which extend upward and outward along the heavy chain. The second β turn, a type IV, is formed by residues Val-489, Ala-490, Gly-491, and Leu-492 (Fig. 2). 14G7-1-2 recognizes GP expressed on the surface of live Ebola virus and Ebola virus-infected cells, and hence, it is likely that the double β-turn conformation displayed by the peptide reflects the preferred conformation of this epitope in the full-length, viral surface GP.
Fig 2.
Diagram and stick representation of Fab-peptide complex showing the overall tandem beta turn conformation of the peptide within the Fab cleft. The proximal type I β turn is formed by residues 483TNTI486, while residues 489VAGL492 form the distal type IV β turn. CDR H1 is colored orange, H2 is violet, and H3 is pink. CDR L1 is colored magenta, L2 is green, and L3 is blue.
Of the GKLGLITNTIAGVAGLI peptide bound, contact is made by all of the residues in the central TNTIAGVAGL sequence with a mix of main-chain and side-chain contacts (boldfacing indicates the overlapping region of separate peptides used in epitope identification). Binding of the peptide in the antibody pocket is mediated by seven key hydrogen bonds. Of these, five are mediated by peptide main-chain atoms and two are formed by peptide side chain atoms (Table 2). The main-chain amide of Ile-482 of GP makes a 2.8-Å bond with the carbonyl oxygen of Asp-31 of CDR H1 (Fig. 3A). The type I β turn of the GP peptide (formed by Thr-483, Asn-484, Thr-485, and Ile-486) is anchored to the antibody pocket by three hydrogen bonds: the first between N∂2 of GP Asn 484 and O∂2 of Asp-101 in CDR H3, the second between the O∂ of GP Thr-485 and the carbonyl oxygen of Ala-99 in CDR H3, and the third between the carbonyl oxygen of GP Thr-485 and the imidazole ring of His 38 in CDR L1 (Fig. 3A). In between the type I and IV β turns, the carbonyl oxygen of GP Gly-488 contacts Nη of Arg-100 in CDR L3 (Fig. 3B). The type IV β turn in GP is anchored by a hydrogen bond between the main-chain amide of Ala-490 and the O∂ of Glu-50 (2.8 Å) of CDR H2 (Fig. 3B). In total, the antibody heavy chain forms 70% of the hydrogen bond contacts, and all three CDR regions of the heavy chain make contact with the peptide. Of the light chain, only CDRs L1 and L3 form contact with the GP peptide; L2 does not.
Table 2.
Van der Waals contacts and hydrogen bonds formed between Fab 14G7 and GP
| Peptide epitope | Fab residue | Distance (Å) |
|---|---|---|
| Van der Waals contacts | ||
| LysQ478 | TyrH52, ThrH55 | |
| GlyQ480 | ThrH30, AspH31 | |
| LeuQ481 | AspH31 | |
| IleQ482 | AspH31, TyrH32 | |
| AsnQ484 | TyrL53, LeuL54 AspH101 | |
| ThrQ485 | TyrL36, HisL38, LeuL50, TyrL53, AlaL95, AlaH99, PheH100, AspH101 | |
| IleQ486 | TyrL36, ValH33 | |
| AlaQ487 | TyrL36 | |
| GlyQ488 | ArgL100 | |
| ValQ489 | GluH50, TyrH52 | |
| AlaQ490 | LeuL98, TyrH59, GluH50 | |
| GlyQ491 | SerH57 | |
| Hydrogen bond contacts | ||
| ThrQ485 O | HisL38 Nϵ2 | 2.98 |
| GlyQ488 O | ArgL100 Nη2 | 2.95 |
| Ile Q482 N | AspH31 O | 2.87 |
| AlaQ490 N | GluH50 Oϵ2 | 2.75 |
| ThrQ485 Oγ1 | AlaH99 O | 2.30 |
| ThrQ485 Oγ1 | AspH101 Oδ1 | 2.83 |
| LysQ478 N | TyrH52 Oη | 3.34 |
H, L, and Q denote the heavy, light, and peptide chains, respectively.
Fig 3.
14G7 Fab-GP interactions. (A) Hydrogen bonds between the type I β turn of GP and the antibody. Four hydrogen bonds shown here include (i) the main-chain N of Ile-482 to the carbonyl O of Asp-31 of the heavy chain, (ii) a side chain N of Asn-484 to a side chain O of Asp-101 of the heavy chain, (iii) the side chain O of Thr-485 to the carbonyl O of Ala-99 of the heavy chain, and (iv) the carbonyl O of Thr-485 to the imidazole ring of His-38 of the light chain. (B) Hydrogen bond between the type I and IV β turns (the carbonyl O of Gly-488 to Arg-100 of the light chain) and another formed by a residue in the type IV β turn (the main-chain N of Ala-490 to a side chain O of Glu-50 of the heavy chain). Hydrogen bonds are indicated as dashed lines. See Table 2 for a complete list of the corresponding contacts. For clarity, panels A and B are shown in different orientations from that of Fig. 2.
Mutagenesis of the central region of the 14G7 epitope.
The central TIAGV sequence (residues 485 to 489) was contained in adjacent overlapping peptides originally used for epitope identification. We analyzed the contribution of each individual GP residue in the core TIAGV sequence to 14G7 binding in a SPOTS assay by systematically mutating each residue in the epitope to each of the 20 amino acids (Table 3) (9). This analysis indicates that Thr-485 is important for binding and can only be substituted by serine, although the binding affinity for a serine-substituted peptide is reduced. This is in agreement with our structural data that indicate that Thr-485 occupies a pivotal part of the type-I β-turn, which makes intricate interactions with the CDR H3 of 14G7. Table 3 summarizes the contribution of each the amino acids used in the scan. Although key components of the 14G7 epitope are conserved among all Ebola viruses, we report here by ELISA that 14G7 is specific for Ebola virus (formerly known as Zaire ebolavirus) and does not react with other members of the Filovirus family. ELISA using purified, irradiated viruses indicates that 14G7 reacts equivalently with Ebola virus (strain Eckron, 1995) and Ebola virus (strain Mayinga, 1976) but does not react with Sudan virus (strain Boniface), Reston virus, Tai Forest virus, Marburg virus (strain Musoke), or Ravn virus (Fig. 4A). Similarly, ELISA using purified, recombinant GPs indicates that both 13F6-1-2 and 14G7 are specific for Ebola virus GP (strain Mayinga) and do not react with Sudan virus (strain Boniface), Bundibugyo virus, or Marburg virus. MAb 42/3.7 (35) is cross-reactive among Ebola virus, Sudan virus, and Bundibugyo virus GPs (Fig. 4A).
Table 3.
SPOTS analysis of point mutations introduced within the GP477-493 epitopea
| Amino acid | 14G7 epitope binding affinity |
||||
|---|---|---|---|---|---|
| Thr-485 | Ile-486 | Ala-487 | Gly-488 | Val-499 | |
| A | – | – | +++ | – | +++ |
| C | – | – | – | – | + |
| D | – | – | – | +++ | +++ |
| E | – | – | – | – | +++ |
| F | – | – | – | – | – |
| G | – | – | + | +++ | + |
| H | – | +++ | +++ | +++ | +++ |
| I | – | +++ | – | – | +++ |
| K | – | – | – | – | + |
| L | – | + | – | – | + |
| M | – | +++ | – | +++ | +++ |
| N | – | +++ | – | +++ | +++ |
| P | – | – | +++ | + | +++ |
| Q | – | – | – | – | +++ |
| R | – | – | – | – | ++ |
| S | ++ | ++ | +++ | + | +++ |
| T | +++ | +++ | – | – | +++ |
| V | – | +++ | – | – | +++ |
| W | – | – | – | – | – |
| Y | – | – | – | – | ++ |
Fig 4.
(A) Recognition of irradiated viruses by MAb 14G7 and recognition of recombinant viral glycoproteins by MAbs 14G7, 13F6-1-2, 42/3.7, and KZ52. +, Positive binding. –, no binding. Blank lines = not done. Only 14G7 was tested against irradiated virus samples. (B) Neutralization of VP30-deleted, biologically contained Ebola viruses (ΔVP30 EBOV). ΔVP30 EBOV was incubated with either MAb 14G7, 13F6-1-2, 42/3.7, or KZ52 in the presence or absence of guinea pig complement and allowed to attach to Vero E6 cells at 4°C. The cells were washed of unbound material, warmed to 37°C, and plaqued.
Affinity of anti-mucin antibodies for their epitopes.
Surface plasmon resonance studies reveal that 14G7 binds to trimeric, transmembrane-deleted Ebola virus GP (GPΔtm; strain Mayinga) ectodomain with a Kd of 103 ± 47 nM and 13F6-1-2 binds to GPΔtm with a Kd of 57.5 ± 51 nM. The higher affinity of 13F6-1-2 correlates with structural observations. Crystal structures of the two antibodies in complex with their linear peptide epitopes reveal that 13F6-1-2 makes 19 hydrogen bonds and 138 nonbonded interactions with GP404-414 (distance cutoff for nonbonded interactions is 4 Å), while 14G7 makes 7 hydrogen bonds and 101 nonbonded interactions with GP477-493.
Anti-mucin antibodies do not neutralize well in vitro.
Although antibodies 13F6-1-2 and 14G7 have acceptable affinity for recombinant GP, bind infected cells and irradiated virions, and can be protective in rodent models in vivo (37), these antibodies are unable to neutralize biologically contained Ebola viruses well in vitro (Fig. 4B). MAb 42/3.7 binds well and is cross-reactive among GPs of the Ebolavirus genus but, similarly, does not neutralize. In contrast, antibody KZ52, which is known to bind the base of GP (22), is able to neutralize viruses in vitro with 50% neutralization occurring between 0.1 and 1 μg/ml.
DISCUSSION
Effective immunotherapy against Ebola viruses will require recognition of well-exposed epitopes on GP, and likely, recognition of those that are unique to the relatively rare viral GP over the more abundant sGP. Epitopes in the projecting mucin-like domains of GP may be well exposed and are certainly unique to GP. This region differs in sequence among the five species of the Ebolavirus genus, and antibodies against this region are typically species specific (28). However, within this region, the residues Gly-480 and Thr-485 are strictly conserved (Fig. 5). The unusual conservation of these residues indicates that they may be important for the structural integrity of the mucin-like domain and/or play a functional role. Thr-485 forms the apex of the first β turn and its hydroxyl group forms hydrogen bonds with two of the six CDRs of the 14G7. Indeed, of the central TIAGV sequence that forms the type I β turn, Thr-485 is the least tolerant of substitutions, accepting only a Ser, although with a lower affinity.
Fig 5.
Multiple sequence alignment of the 14G7 Ebola virus epitope performed using CLUSTAL W. Gly-480 and Thr-485 (boldface red text) are completely conserved across the five species of the Ebolavirus genus. Of these two, Thr-485 is critical for 14G7 binding. The portion of the sequence visualized in the crystal structure is marked by a bracket. The residues that comprise the most significant region of interaction are marked by asterisks and are also important for antibody contact. Zaire denotes Ebola virus (formerly known as Zaire ebolavirus).
Although components of its epitopes are completely conserved, 14G7 is nonetheless specific for Ebola virus (formerly known as Zaire ebolavirus) over other members of the Ebolavirus genus such as Sudan, Reston, Bundibugyo, and Tai Forest viruses (28) (Fig. 4A). Hence, the species specificity must arise from the additional GP residues in the epitope.
Of the central TIAGV sequence, Thr-485 is the least tolerant of substitutions and is completely conserved across all Ebola viruses. The next residue, Ile-486, is Ile in Ebola virus and Tai Forest virus, but Val in Sudan virus and Reston virus. However, a Val substitution is allowed by 14G7. Similarly, Val-489 is conserved across Ebola virus, Reston virus, and Tai Forest virus, but is an Ile in Sudan virus, although an Ile substitution at position 489 is tolerated by 14G7. Hence, the Ebola virus specificity of at least the central portion of the 14G7 epitope appears to be determined by the remaining two residues: Ala-487 and Gly-488. Ala-487 in Ebola virus is replaced by Arg, Thr, and Ser in Reston, Tai Forest, and Sudan viruses, respectively (Fig. 5). Arg and Thr substitutions at position 487 are not tolerated by 14G7. Ser (as in Sudan virus) is tolerated. Gly-488 is conserved among Ebola, Tai Forest, and Sudan viruses but is a Lys in Reston. A Lys at position 488 is not tolerated by 14G7. The failure of 14G7 to bind Sudan virus GP must then lie in residues outside the core TIAGV sequence.
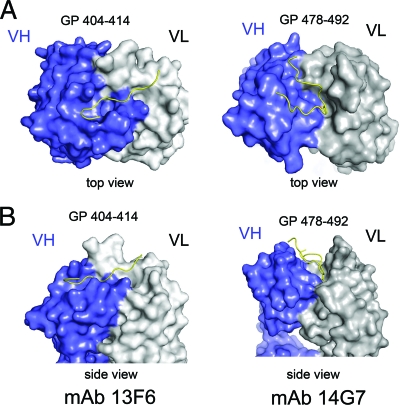
The mode of binding of Fab 14G7 is completely different from that of the other mucin-like domain antibody 13F6-1-2. The epitope of 13F6-1-2, GP residues 404 to 414, is predicted to lack secondary structure and, indeed, the peptide adopts an extended conformation, binding into a straight, shallow groove, approximately 28 Å long, 10 Å wide, and 9 Å deep, that reaches diagonally across the antibody combining site (Fig. 6). The difference in the mode of binding probably lies in the noncanonical CDRs encoded by the extremely rare mouse Vλx light chain. In contrast, 14G7 is encoded by much more typical heavy and light chain genes, and the resulting binding groove of 14G7 is centrally located. In addition, the binding groove is quite deep: specifically, 14 Å deep, 13 Å wide, and 26 Å long. Further, the linear epitope of 14G7 forms a tandem β turn rather than an extended conformation (Fig. 6).
Fig 6.
Comparison of the binding mode of the two structurally characterized antibodies against the Ebola virus mucin-like domain. (A) Top view of the 14G7 and 13F6 peptide epitopes. In the 13F6-peptide complex the peptide lies diagonally across Fab pocket, while in the 14G7 complex the peptide antigen defines a wide S-shape conformation that centrally occupies both Fab light and heavy chain CDRs. (B) Side view of the Fab complexes. The GP401-414 epitope lies in a shallow groove of MAb 13F6-1-2, while the GP487-493 epitope lies in a deep groove of MAb 14G7.
Tandem β turns are also adopted by the V3 loop of the HIV-1 glycoprotein gp120 when bound by antibody (12, 33), and binding of a β turn by an antibody allows for a greater number of interactions per residue of the epitope (29, 34). For the Ebola viruses, recognition of this type of structure facilitates tight binding of the short peptide sequences sandwiched between the multiple glycans in the mucin-like domain of GP.
For many viruses, glycosylation plays a major role in immune evasion by masking antibody epitopes (19, 20, 36). However, although the mucin-like domain as a whole is heavily glycosylated, the particular epitopes recognized by 14G7 and 13F6-1-2 within it are not glycosylated. Thus, these two protective antibodies have identified regions of the mucin-like domain that are accessible and not masked with sugars.
Although the antibodies bind recombinant GP with 50 to 100 nM affinity and also recognize GP on viral surfaces, they do not neutralize virus infection well in vitro (Fig. 4B). Curiously, however, they can be protective in vivo. The reasons for this apparent paradox result from the nature of the particular binding sites on GP recognized by these antibodies, as well as unique remodeling of the filovirus GP that occurs upon viral entry.
Neutralization is an in vitro event in which binding alone of the antibody to viral antigen inactivates the virus or its antigen by blocking receptor or coreceptor binding sites or physically preventing conformational rearrangements required for fusion, as examples. The apparent failure of anti-mucin antibodies such as 13F6-1-2 and 14G7 to neutralize despite their ca. 50 to 100 nM affinities suggests that even the added 150-kDa steric bulk of an IgG linked to the 150-kDa GP monomer is insufficient to prevent attachment or entry. The insensitivity of Ebola virus GP to binding of anti-mucin-domain antibodies likely results from (i) the possible ability of multiple host factors (with multiple binding sites) to mediate initial virus attachment and subsequent entry into the endosome by macropinocytosis or another mechanism and (ii) a required receptor-binding event does not occur until the mucin-like domain is removed by host proteases in the endosome (5, 8, 32). Antibodies such as 13F6-1-2 and 14G7 that remain bound to GP upon entry into the endosome would be removed from the virus upon this cleavage event.
Although these antibodies do not neutralize infection well in vitro, they have been shown to confer passive immunity in pre- and postexposure challenge in rodents (37). In these scenarios, it is likely the ability of antibodies such as 14G7 to recruit immunological effector functions that confers protection and, in rodent models, such protection is sufficient. Because this type of antibody is able to confer protection in vivo in these models, mucin-like domain antibodies could be considered as components of cocktails. Certainly, testing in nonhuman primates of different combinations of antibodies, against a variety of structural epitopes on GP, will be required in order to identify the best combinations for passive immunotherapy.
The mucin-like domains of the filovirus GPs render them different from other viral glycoproteins. The glycoproteins of HIV-1 have glycosylated, variable loops, but these are smaller in size and demonstrate extensive variation among currently circulating HIV-1. In contrast, although sequences of the mucin-like domains differ among the separate viruses in the genus (Ebola virus versus Sudan virus, for example), they remain relatively consistent within a single virus for decades. For example, the mucin-like domains of 1976 Ebola virus, strain Mayinga, and 2005 Ebola virus, strain Etoumbi, are 91% identical overall, with all residues in both the 13F6 and 14G7 epitopes remaining completely conserved for 29 years. An antibody against the Ebola virus mucin-like domain could thus be expected to remain relevant for current field use. Second, filovirus GPs undergo extensive proteolytic remodeling during entry and the mucin-like domains are stripped off in the endosome prior to fusion (6, 32). As a result, antibodies against these sites would likely be most functional outside the cell or on the cell surface. Third, the required receptor-binding event appears to occur only after proteolytic remodeling in the endosome (5, 8, 18). Hence, antibodies that target the receptor-binding sites may not recognize viral-surface GP and thus may not be present in the endosome for neutralization. Fourth, these mucin-like domains of Ebola virus are extremely large. They comprise more than half the molecular weight of GP and dominate the size, structure, and appearance of GP and are thus likely to dominate initial interactions with host attachment and immune factors (Lee et al., in preparation). Indeed, antibodies against these regions are well represented among those elicited by vaccination studies.
The crystal structure and accompanying biochemical and functional analysis presented here highlights 14G7 as an example of the kinds of antibodies that are likely elicited by GP in natural infection or by immunization. The massive size, genetic stability, and possible immunodominance of the mucin-like domains, the loss of these domains during entry, and the likely appearance of a functional receptor-binding site only after the protein is proteolytically remodeled in the endosome together mean that development antibodies against Ebola virus may be a more complex problem than originally thought. Antibodies against multiple different sites on GP will need to be considered both alone and in combination for postexposure prophylaxis.
ACKNOWLEDGMENTS
We thank Xiaoping Dai (The Scripps Research Institute) and the staff of SSRL Beamline 11-1 for assistance with data collection and Gene Olinger (U.S. Army Research Institute for Infectious Disease) for helpful discussions.
We acknowledge support of NIAID R01 AI 067927 (E.O.S.), the Skaggs Institute for Chemical Biology (E.O.S.), Canadian Institutes for Health Research Postdoctoral Fellowship (J.E.L.), K99 Pathway to Independence from NIAID/NIH (J.E.L.), National Institutes of Health (NIH) R01 AI055519 and membership within and support from the Region V ‘Great Lakes’ Regional Center for Excellence for Biodefense and Emerging Infectious Disease Research (RCE) Program (NIH award U54 AI057153) (Y.K.), and DTRA K.K0001_07_RD_B (J.M.D.).
Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Army.
Footnotes
Published ahead of print 14 December 2011
This article is manuscript 20945 from The Scripps Research Institute.
REFERENCES
- 1. Adams PD, et al. 2010. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adams PD, et al. 2004. Recent developments in the Phenix software for automated crystallographic structure determination. J. Synchrotron Radiat. 11:53–55 [DOI] [PubMed] [Google Scholar]
- 3. Adams PD, et al. 2002. Phenix: building new software for automated crystallographic structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 4. Al-Lazikani B, Lesk AM, Chothia C. 1997. Standard conformations for the canonical structures of immunoglobulins. J. Mol. Biol. 273:927–948 [DOI] [PubMed] [Google Scholar]
- 5. Carette JE, et al. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen VB, et al. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cote M, et al. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cushman I. 2008. Utilizing peptide SPOT arrays to identify protein interactions. Curr. Protoc. Protein Sci. Chapter 18:Unit 18 [DOI] [PubMed] [Google Scholar]
- 10. Davis IW, et al. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic acids Res. 35:W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dias JM, et al. 2011. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 18:1424–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ding J, et al. 2002. Crystal structure of a human rhinovirus that displays part of the HIV-1 V3 loop and induces neutralizing antibodies against HIV-1. Structure 10:999–1011 [DOI] [PubMed] [Google Scholar]
- 13. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 14. Francica JR, et al. 2010. Steric shielding of surface epitopes and impaired immune recognition induced by the Ebola virus glycoprotein. PLoS Pathog. 6:e1001098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gupta R, Jung E, Brunak S. 2004, posting date Prediction of N-glycosylation sites in human proteins. http://www.cbs.dtu.dk/services/NetNGlyc
- 16. Halfmann P, et al. 2008. Generation of biologically contained Ebola viruses. Proc. Natl. Acad. Sci. U. S. A. 105:1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Julenius K, Molgaard A, Gupta R, Brunak S. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O glycosylation sites. Glycobiology 15:153–164 [DOI] [PubMed] [Google Scholar]
- 18. Kaletsky RL, Simmons G, Bates P. 2007. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 81:13378–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karlsson-Hedestam GB, et al. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6:143–155 [DOI] [PubMed] [Google Scholar]
- 20. Kwong PD, Wilson IA. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 10:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 22. Lee JE, et al. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JE, et al. 2008. Complex of a protective antibody with its Ebola virus GP peptide epitope: unusual features of a V lambda x light chain. J. Mol. Biol. 375:202–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maruyama T, et al. 1999. Recombinant human monoclonal antibodies to Ebola virus. J. Infect. Dis. 179(Suppl 1):S235–S239 [DOI] [PubMed] [Google Scholar]
- 25. Maruyama T, et al. 1999. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J. Virol. 73:6024–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nettleship JE, et al. 2008. A pipeline for the production of antibody fragments for structural studies using transient expression in HEK 293T cells. Protein expression and purification. 62:83–89 [DOI] [PubMed] [Google Scholar]
- 27. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 28. Reynard O, et al. 2009. Ebolavirus glycoprotein GP masks both its own epitopes and the presence of cellular surface proteins. J. Virol. 83:9596–9601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rini JM, Schulze-Gahmen U, Wilson IA. 1992. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science 255:959–965 [DOI] [PubMed] [Google Scholar]
- 30. Sanchez A, Geisbert TW, Feldmann H. 2007. Filoviridae: Marburg and Ebola viruses, p 1409–1440 In Knipe DM. (ed), Fields virology. Lippincott/Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 31. Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. 1996. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. U. S. A. 93:3602–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schornberg K, et al. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stanfield R, et al. 1999. Dual conformations for the HIV-1 gp120 V3 loop in complexes with different neutralizing Fabs. Struct. Fold Des. 7:131–142 [DOI] [PubMed] [Google Scholar]
- 34. Stanfield RL, Fieser TM, Lerner RA, Wilson IA. 1990. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 Å. Science 248:712–719 [DOI] [PubMed] [Google Scholar]
- 35. Takada A, et al. 2003. Identification of protective epitopes on Ebola virus glycoprotein at the single amino acid level by using recombinant vesicular stomatitis viruses. J. Virol. 77:1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang CC, et al. 2009. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. U. S. A. 106:18137–18142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson JA, et al. 2000. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287:1664–1666 [DOI] [PubMed] [Google Scholar]
- 38. Zampieri CA, Sullivan NJ, Nabel GJ. 2007. Immunopathology of highly virulent pathogens: insights from Ebola virus. Nat. Immunol. 8:1159–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]