Abstract
The core entry machinery of mammalian herpesviruses comprises glycoprotein B (gB), gH, and gL. gH and gL form a heterodimer with a central role in viral membrane fusion. When archetypal alpha- or betaherpesviruses lack gL, gH misfolds and progeny virions are noninfectious. However, the gL of the rhadinovirus murid herpesvirus 4 (MuHV-4) is nonessential for infection. In order to define more generally what role gL plays in rhadinovirus infections, we disrupted its coding sequence in bovine herpesvirus 4 (BoHV-4). BoHV-4 lacking gL showed altered gH glycosylation and incorporated somewhat less gH into virions but remained infectious. However, gL− virions showed poor growth associated with an entry deficit. Moreover, a major part of their entry defect appeared to reflect impaired endocytosis, which occurs upstream of membrane fusion itself. Thus, the rhadinovirus gL may be more important for driving virion endocytosis than for incorporating gH into virions, and it is nonessential for membrane fusion.
INTRODUCTION
The Herpesviridae family contains numerous important pathogens that are classified in three subfamilies (Alpha-, Beta-, and Gammaherpesvirinae). These enveloped viruses enter cells by fusing their envelopes with host cell membranes either by direct fusion at the plasma membrane or by pH-dependent or -independent endocytosis, depending on the virus and on the target cell (4). While most enveloped viruses rely on a single fusogenic protein for entry, herpesviruses are more complex. Indeed, they use a core fusion machinery, composed of glycoprotein B (gB) and the gH/gL heterodimer, that is conserved across the three subfamilies (10). Most of the herpesviruses employ also one or more additional receptor-binding or regulating proteins specific to subfamilies or genera. This complexity explains why herpesvirus entry, and particularly the fusion mechanism, is still poorly understood.
The X-ray structure determinations of herpes simplex virus 1 (HSV-1) and Epstein-Barr virus (EBV) gB (5, 26) suggested that it is a class III viral fusion protein as is glycoprotein G of vesicular stomatitis virus (VSV) (44–45) or baculovirus gp64 (29). However, in contrast to those proteins, gB cannot function on its own and requires the gH/gL heterodimer. This complex is a major target of virus-neutralizing antibodies in several herpesviruses (17, 43), emphasizing its role in virus entry. It has been proposed that gH has features characteristic of class I viral fusogens (14–16). However, the recently published structures of HSV-2 and EBV gH/gL (9, 39) suggested that these proteins do not likely act as cofusogen but rather regulate fusion by gB. These structures also revealed extensive contacts between gL and the N-terminal domain of gH. Interestingly, the sequences of gL and the gH N terminus vary substantially among herpesviruses and it appears that the gH-gL pairs in each herpesvirus have coevolved to form tight complexes with possible specific functions (9).
Bovine herpesvirus 4 (BoHV-4) belongs to the Gammaherpesvirinae subfamily, Rhadinovirus genus, together with the notable human pathogen Kaposi's sarcoma-associated herpesvirus (KSHV). Until recently, little was known about gH and gL in these viruses beyond the fact that they are virion-associated components (33, 41). However, we recently showed that gL was not essential for murid herpesvirus 4 (MuHV-4), another rhadinovirus. MuHV-4 lacking gL both incorporates gH into virions and remains infectious, although it shows some attenuation relative to the wild type (WT) (23). This result is quite surprising compared to results for all alpha- and betaherpesviruses tested to date, in which gL proved to be indispensable (7, 13, 27, 31, 46). This is also intriguing, as rhadinovirus gH/gL is a major neutralization target (17) protected by various antibody evasion mechanisms (24, 36).
In order to know if MuHV-4 gL properties are shared among rhadinoviruses, we disrupted the gL coding exon (ORF47) in the BoHV-4 genome. BoHV-4 lacking gL remained infectious but displayed a growth deficit. This appeared to be associated with impaired cell endocytosis rather than cell binding. Indeed, gL deletion severely altered the trafficking of the virion containing endosomes during entry.
MATERIALS AND METHODS
Cells and virus.
293T (ATCC CRL-11268), Madin-Darby bovine kidney (MDBK; ATCC CCL-22), embryonic bovine lung (EBL; German Collection of Microorganisms and Cell Culture [DSMZ] ACC192), bovine turbinate (BT; ATCC CRL-1390), and embryonic bovine trachea (EBTr; ATCC CCL-44) cells were cultured in Dulbecco's modified Eagle medium (Invitrogen) containing 10% fetal calf serum (FCS), 2% penicillin/streptomycin (Invitrogen), and 1% nonessential amino acids (Invitrogen). The BoHV-4 V.test strain and the V.test BAC G-derived bacterial artificial chromosome (BAC) clone were described elsewhere (22, 49).
Antibodies.
Five mouse monoclonal antibodies (MAbs) raised against BoHV-4 were used in the present study (12). Their specificities were unraveled on 293T cells transfected with the vectors encoding gB-glycophosphatidylinositol (gB-GPI), gH-GPI, or gL-GPI (36). The epitopes depending on the gH-gL heterodimer were reconstituted by coexpressing gH-GPI and gL-GPI (Fig. 1). MAb 35 recognizes gB, as previously stated (34). For some Western blots, we used serum of a rabbit infected intravenously with 108 PFU of the BoHV-4 V.test strain and collected 63 days postinoculation. Lysosome-associated membrane protein 1 (LAMP-1) was detected with rabbit polyclonal antibody (PAb; ab24170; Abcam).
Fig 1.
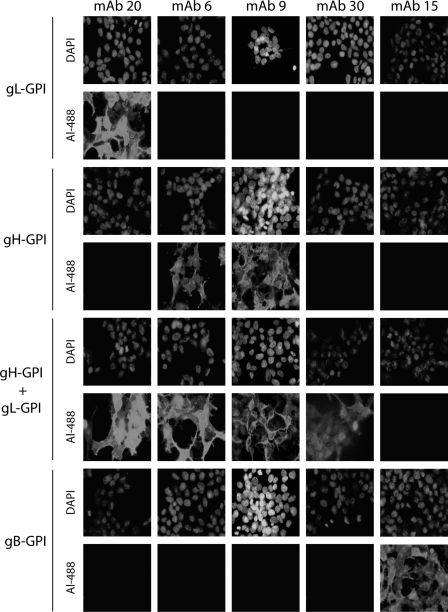
Identification of MAbs recognizing BoHV-4 gL-, gH-, gH/gL-, or gB-dependent epitopes. 293T cells were transfected with the gB or gH extracellular domains or the entire gL fused to a GPI membrane anchor, resulting in gB-GPI, gH-GPI, and gL-GPI, respectively. To reconstitute epitopes depending on the gH-gL heterodimer, we cotransfected the cells with plasmids encoding gH-GPI and gL-GPI. Forty-eight hours after transfection, the cells were fixed and stained with the different MAbs as indicated. Nuclei were counterstained with DAPI.
Indirect immunofluorescent staining of adherent cells.
Cells were fixed and permeabilized with acetone 95% for 10 min at −20°C or with paraformaldehyde (4% [wt/vol]) for 10 min on ice and Tween 20 (0.1% [vol/vol]) in phosphate-buffered saline (PBS; anti-LAMP-1 stainings). After the cells were washed with PBS, immunofluorescent staining (incubation and washes) was performed with PBS containing 10% (vol/vol) FCS. Samples were incubated at 37°C for 45 min with the different mouse anti-BoHV-4 MAbs or anti-LAMP-1 rabbit polyserum. After three washes, samples were incubated at 37°C for 30 min with Alexa Fluor 488 or 568 goat anti-mouse (GAM) IgG (2 μg/ml; Invitrogen) or with Alexa Fluor 568 goat anti-rabbit (GARb) IgG (2 μg/ml; Invitrogen). When stated in the figure legends, nuclei were counterstained with DAPI (4,6-diamidino-2-phenylindole) or To-Pro-3 iodide (Invitrogen; 1 μM in PBS). Fluorescence was then visualized with a Nikon Eclipse TE2000-S microscope and a Leica DC300F charge-coupled-device (CCD) camera system or with a Leica true confocal scanner (TCS) SP laser scanning microscope.
Production of BoHV-4 gL STOP and revertant strains.
We disrupted the BoHV-4 V.test strain gL coding sequence (42) (GenBank accession number JN133502; genomic coordinates 60731 to 60309) by introducing stop codons into the coding sequence for the gL signal peptide (gL STOP). BoHV-4 recombinants were produced using BAC cloning and prokaryotic recombination technologies as described before (22). The V.test BAC G plasmid was used as the parental plasmid (22). The V.test BAC G gL STOP was produced using a two-step galactokinase (galK gene product) positive/negative selection in bacteria (52) (Fig. 2). The first recombination process (galK positive selection) consisted of introducing the galK gene in ORF47 (genomic coordinate 60693), coding for gL, resulting in the V.test BAC G ΔORF47 galK plasmid. Recombination was achieved using the gL galK cassette. It consisted of the galK gene flanked by 50-bp sequences corresponding to ORF47 regions (60644 to 60693 and 60694 to 60744 of the BoHV-4 V.test strain genome). This cassette was produced by PCR using the pgalK vector (52) as the template and gL-fwd-galK (5′-ATGGATTGGATAGAAAATCTAAAGTTATTCCGAATGGGCATATAAACAATCCTGTTGACAATTAATCATCGGCA-3′) and gL-rev-galK (5′-ACAATATTAACAATGAGAGATATCTATGTTTTTTGTCTTTTCATGATTTTTCAGCACTGTCCTGCTCCTT-3′) as forward and reverse primers, respectively. The second recombination process (galK negative selection) consisted of replacing the galK sequence with a gL STOP cassette to generate the BoHV-4 V.test strain gL STOP plasmid. The gL STOP cassette consisted of a synthetic double-stranded DNA (Eurogentec) corresponding to genomic coordinates 60742 to 60643 with the introduction of 36 nucleotides coding for in-frame stop codons and restriction sites after genomic position 60692 (Fig. 2). These 36 nucleotides do not insert stop codons in any of the five other frames of the genome. The V.test BAC G gL revertant plasmid was produced similarly from BoHV-4 V.test strain gL STOP plasmid. The first recombination process (galK positive selection) was identical to the one described above. The second recombination process (galK negative selection) consisted of restoring ORF47 to generate a revertant plasmid. This cassette was produced by PCR using the BoHV-4 V.test strain genome as the template and gL-zone-rec-sens (5′-AAGGTACCCATGATTCTATCAGTTGGTGG-3′) and gL-zone-rec-rev (5′-AAGCATGCCAGGAAATTGTGATTGAAGATG-3′) as forward and reverse primers, respectively. Reconstitution of infectious virus from BAC plasmids was obtained by transfection in MDBK cells. The viruses used (BAC+) express enhanced green fluorescent protein (eGFP) from a human cytomegalovirus (HCMV) IE1 promoter located in the BAC cassette (22), so eGFP expression provides a convenient marker of infection. Typically, eGFP is detectable as soon as 8 h postinfection.
Fig 2.
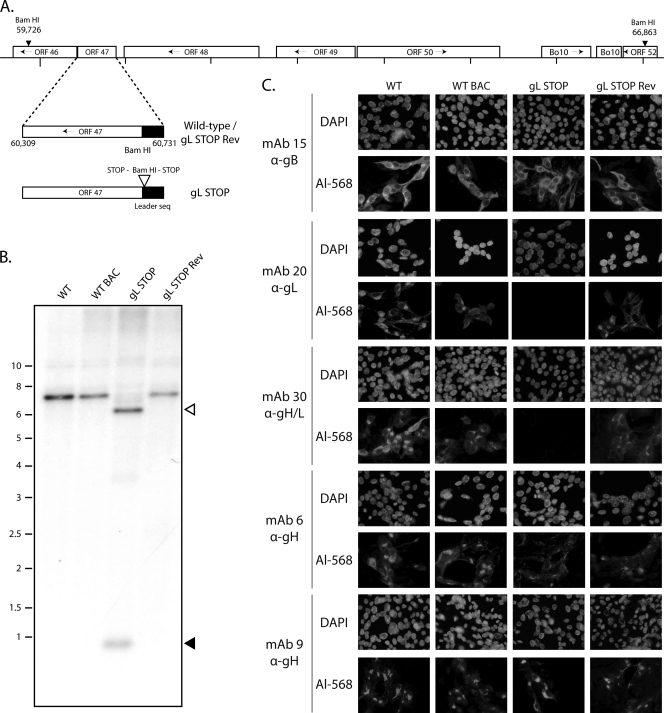
Generation of a gL-deficient BoHV-4 mutant. (A) Schematic representation of the strategy followed to produce the recombinant BoHV-4 strain. The gL-deficient BoHV-4 mutant was derived from a cloned BoHV-4 BAC by the galK counterselection method. The gL coding sequence (ORF47) was disrupted by inserting stop codons near the end of the coding sequence for its predicted signal peptide (gL STOP). The mutation incorporated a new BamHI restriction site. (B) Verification of the molecular structure. Viral DNA was digested with BamHI, resolved by agarose gel electrophoresis, and hybridized with a 32P-labeled probe, corresponding to nucleotides 60087 to 61334. Open arrows show the restriction fragment that contains gL STOP. The 7,137-bp wild-type (WT) band becomes 6,553 bp for the gL STOP mutant. Black arrows show the remaining 620-bp fragment. Marker sizes (in kbp) are indicated on the left. (C) Glycoprotein expression by the gL-deficient BoHV-4 strain. MDBK cells were infected with the WT, the WT BAC, gL STOP, and gL STOP Rev BoHV-4 (MOI of 0.5 PFU per cell). Thirty-six hours postinfection, the cells were fixed, permeabilized, and stained with MAb 15 (gB), MAb 20 (gL), MAb 30 (gH-gL), MAb 6 (gH), or MAb 9 (gH). An Alexa 568-conjugated goat anti-mouse secondary antibody was then used. Nuclei were counterstained with DAPI.
Southern blot.
Southern blot analysis of viral DNA digested with BamHI was performed with probe corresponding to the gL surrounding region. This probe was produced by PCR using the BoHV-4 V.test strain genome as the template and gL-zone-rec-sens and gL-zone-rec-rev as forward and reverse primers, respectively (genomic coordinates 60087 to 61334).
Western blot.
Virions or infected cells were lysed and denatured by heating (95°C, 5 min) in SDS-PAGE sample buffer (31.25 mM Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 12.5% [wt/vol] glycerol, 0.005% [wt/vol] bromophenol blue, 2.5% [vol/vol] 2-mercaptoethanol). Proteins were resolved by electrophoresis with Mini-Protean Tris-glycine eXtended (TGX) precast 4 to 15% resolving gels (Bio-Rad) in SDS-PAGE running buffer (25 mM Tris-base, 192 mM glycine, 0.1% [wt/vol] SDS) and transferred to polyvinylidene difluoride membranes (Immobilon-P transfer membrane, 0.45-μm pore size; Millipore). The membranes were blocked with 3% nonfat milk in PBS/0.1% Tween 20 and then incubated with primary antibody in the same buffer. Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit IgG PAb or rabbit anti-mouse IgG PAb (Dako Corporation), followed by washing in PBS/0.1% Tween 20, development with enhanced chemiluminescence (ECL) substrate (GE Healthcare), and exposure to X-ray film.
Oligosaccharide digestion.
All reagents were obtained from New England BioLabs (NEB). Samples were denatured in glycoprotein denaturing buffer (0.5% SDS, 40 mM dithiothreitol [DTT]) for 10 min at 100°C and then digested for 3 h at 37°C with 500 NEB units of endo-β-N-acetylglucosaminidase-H (Endo-H) or 250 NEB units of peptide, N-glycosidase F (PNGase F), and/or 250 NEB units of neuraminidase, β1-4 galactosidase, and O-glycanase in G7 reaction buffer (50 mM sodium phosphate, pH 7.5) with 1% NP-40. Reactions were stopped by the addition of Laemmli sample buffer, and proteins were analyzed by immunoblotting as described below.
Virus purification.
BoHV-4 virions grown on MDBK cells were purified as follows. After removal of the cell debris by low-speed centrifugation (1,000 × g, 10 min), virions present in the infected cell supernatant were harvested by ultracentrifugation (100,000 × g, 2 h) through a 30% (wt/vol) sucrose cushion and then centrifuged through two successive 20 to 50% (wt/vol) potassium tartrate gradients in PBS (100,000 × g, 2 h). Virions were finally washed and concentrated in PBS (100,000 × g, 2 h).
Growth curves.
The growth kinetics of mutant and revertant viruses were compared to those of the WT. Cell cultures were infected at a multiplicity of infection (MOI) of 0.01 (multistep assay). After 1 h of adsorption, the cells were washed and then overlaid with minimum essential medium containing 5% FCS. Supernatants of infected cultures or infected cells were harvested at successive intervals, and the amount of infectious virus was determined by plaque assay on MDBK cells.
Flow cytometry.
Cells exposed to eGFP+ viruses were washed in PBS and analyzed directly for green channel fluorescence with a three-laser Becton Dickinson fluorescence-activated cell sorter (FACSAria).
Viral genome detection by real-time PCR.
DNA was purified from infected cells or from 106 purified virions of the different strains using a QIAamp DNA minikit (Qiagen). Real-time PCRs were performed to determine viral and cellular genome copies. Briefly, a 138-bp fragment corresponding to BoHV-4 ORF7 was amplified with the forward primer ORF7 sens (5′-CAGGCAAAAAGTGGCTTCTC-3′) and the reverse primer ORF7 rev (5′-TTGAGGGCCTGGATATTGTC-3′). The PCR products were quantified by hybridization with a TaqMan probe (genomic coordinates 10073 to 10096, 5′-6-FAM-TGACCACCCCACCCATCCATTTTT-BHQ-1-3′, where FAM is 6-carboxyfluorescein and BHQ-1 is Black Hole Quencher-1 dye) and converted to genome copies by comparison with a standard curve of BoHV-4 V.test strain WT DNA, amplified in parallel. PCR amplifications were performed under the following conditions: initial activation of the Taq polymerase (Bio-Rad) at 95°C for 3 min followed by 50 cycles at 95°C for 15 s and 58°C for 40 s. Cellular DNA was quantified in parallel by amplifying part of the bovine actin gene (forward primer, 5′-CGGCATTCACGAAACTACCT-3′; reverse primer, 5′-CAACCGACTGCTGTCACCTTC-3′). The latter PCR products were quantified with Sybr green (Invitrogen), and the copy number was calculated by comparison with a standard curve of cloned bovine actin template. PCR amplifications were performed under the following conditions: initial activation of the Taq polymerase (Bio-Rad) at 95°C for 3 min followed by 45 cycles at 95°C for 30 s, 56°C for 45 s, and 72°C for 45 s. PCR amplifications and fluorescence reactions were carried out with an iCycler system (Bio-Rad).
RESULTS
Identification of gH-specific MAbs.
As demonstrated for other herpesviruses, BoHV-4 gL and gH associate both in virions and in infected cells (33). We have previously characterized monoclonal antibodies recognizing gL alone (MAb 16) or the gH/gL heterodimer (MAb 33) (36). As the absence of gL could interfere with gH stability and recruitment into virions, we first looked for MAbs recognizing a gL-independent gH epitope. Anti-BoHV-4 MAbs were therefore screened for gH, gL, or gH/gL specificity as described in Materials and Methods. Interestingly, similarly to MuHV-4 (17), some epitopes of the gH/gL heterodimer were reconstituted by coexpression of gH-GPI and gL-GPI, suggesting that gL lies close to the virion membrane in the rhadinovirus gH-gL heterodimer. Cells expressing the extracellular part of gB were used as the control (Fig. 1). MAbs 20, 30, and 15 recognized gL, the heterodimer gH/gL, and gB, respectively. MAbs 6 and 9 were shown to recognize gH independently of gL.
Generation of a gL-deficient mutant.
We tested the functional importance of gL for BoHV-4 replication by disrupting ORF47. In the BoHV-4 V.test strain, ORF47 is predicted to be located from genomic coordinates 60731 to 60309 (42). It encodes a predicted 137-amino-acid protein with a 22-amino-acid leader sequence. As a second in-frame ATG is present at genomic coordinates 60701 to 60699, we disrupted this coding sequence by inserting stop codons at genomic coordinate 60692 to generate the BoHV-4 gL STOP strain as described in Materials and Methods. This terminated ORF47 translation 9 amino acids before the end of its predicted signal sequence without any associated deletion (Fig. 2A). A revertant strain, called gL STOP Rev, was finally constructed to validate the gL STOP mutant. The predicted molecular structures of the recombinant strains were confirmed by BamHI restriction mapping (data not shown) and Southern blotting (Fig. 2B) and, further, by DNA sequencing (data not shown).
The gL STOP mutant produced infectious virus after BAC DNA transfection into MDBK cells. Immunostaining of infected cells (Fig. 2C) confirmed the lack of expression of gL-dependent epitopes, as seen with MAb 20 (anti-gL) and MAb 30 (anti-gH/L) staining. In contrast, MAb 6 and 9 stainings showed that gL-independent gH epitopes were normally expressed. Taken together, these results showed that BoHV-4 gL is nonessential.
Characterization of gH in the gL− mutant.
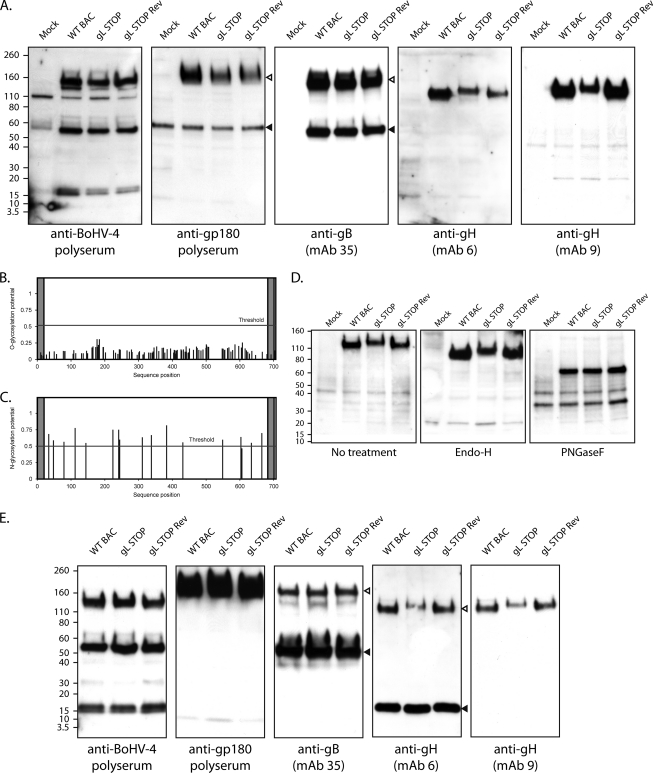
To analyze the influence of the absence of gL on other structural proteins, Western blot analyses of infected cells were performed. Immunoblotting with a polyclonal serum raised against whole virus showed no observable difference between gL STOP and WT or gL STOP Rev strain-infected cells (Fig. 3A). There was also no evidence of a difference in gB or gp180, another viral envelope glycoprotein involved in entry (35). In contrast, while anti-gH MAbs 6 and 9 detected an ∼120-kDa protein in both WT and gL STOP Rev strain-infected cells, they both detected a higher band in gL STOP strain-infected cells.
Fig 3.
Western blot analysis. (A) MDBK cells were mock infected or infected with the WT BAC, gL STOP, and gL STOP Rev BoHV-4 strain (1 PFU/cell). Forty-eight hours later, cells were scraped and immunoblotted for virion components, using either a rabbit serum raised against whole virus (total BoHV-4) or MAbs specific for gp180, gB, and gH, as indicated. In the anti-gp180 blot, open and filled triangles indicate the specific 180-kDa protein and a background band, respectively. In the anti-gB blot, open and filled triangles indicate uncleaved and furin-cleaved gB, respectively. (B) Prediction of O-glycosylation sites for the complete BoHV-4 V.test strain gH protein sequence using the NetOglyc 3.1 algorithm. The shaded regions indicate the signal peptide and transmembrane region. The red line indicates the significance threshold. (C) Prediction of N-glycosylation sites for the complete BoHV-4 V.test strain gH protein sequence using the NetNglyc 1.0 algorithm. The shaded regions indicate the signal peptide and transmembrane region. The red line indicates the significance threshold. (D) MDBK cells were mock infected or infected with the WT BAC, gL STOP, and gL STOP Rev BoHV-4 strain (1 PFU/cell). Forty-eight hours later, cells were scraped and deglycosylated with EndoH or PNGase F and then immunoblotted for gH (MAb 9). (E) Purified virions were subjected to Western blotting with rabbit serum raised against whole virus (total BoHV-4) or MAbs specific for gp180, gB, and gH, as indicated. In the anti-gB blot, open and filled triangles indicate uncleaved and furin-cleaved gB, respectively. In the anti-gH blot (MAb 6), open and filled triangles indicate the specific 120-kDa gH protein and a background band, respectively. The position of an MM standard (kDa) is shown.
This difference could reflect differences in gH glycosylation. BoHV-4 V.test strain gH has no potential O-glycosylation site (Fig. 3B) but 16 potential N-glycosylation sites (Fig. 3C). Moreover, it was previously shown to be sensitive to Endo-H and PNGase F digestion (33). Assuming that oligomer formation between gL and gH takes place before N-glycan trimming, we hypothesized that the absence of gL could alter the gH glycosylation pattern. To establish the contribution of glycans to this difference in apparent molecular mass (MM), we digested infected cell lysates with glycanases. We removed high-mannose, hybrid, and complex N-glycans (37) with PNGase F and/or we removed O-glycans successively with sialidase A, β1-4 galactosidase, and O-glycanase. As expected, O-glycanase treatment did not affect the apparent MM of any gH (data not shown). In contrast, PNGase F treatment reduced the gH apparent MM to approximately 70 kDa in all samples, consistent with the predicted MM for unglycosylated gH. We can therefore conclude that the absence of gL affects BoHV-4 gH N-glycosylation. While PNGase F can cleave any type of N-linked glycans, Endo-H specifically cleaves high-mannose and certain types of hybrid sugars but not complex N-glycans. As described previously, WT virion gH was sensitive to Endo-H treatment (33) and its apparent MM was lower than 110 kDa (Fig. 3D). However, while Endo-H also reduced the apparent MM of gH present in gL STOP-infected cells, the gH MM in this sample was still higher than in WT and gL STOP Rev-infected cells. Taken together, these results show that there are Endo-H-sensitive N-glycans on gH independently of gL. More importantly, they show that gL affects the number and/or the processing of gH complex N-glycans.
The processing of N-linked glycans determines protein homeostasis in the eukaryotic cell (1). The difference in gH N-glycans observed with gL STOP-infected cells could also affect gH stability. Moreover, as shown for other viruses, BoHV-4 gL forms a complex with gH in virions. Therefore, the absence of gL could affect gH stability and recruitment into virions. To analyze whether gL is necessary for virion localization of gH, we performed Western blot analyses of purified virions. While no observable difference between gL STOP and WT or gL STOP Rev virions was observed with an anti-BoHV-4 polyserum or with anti-gp180 or gB antibodies, gH displayed again a higher apparent MM in gL-deficient virions. Moreover, while the abundances of the other structural proteins looked similar for the different viral strains, the amount of gH was slightly lower in gL STOP virions than in the WT and gL STOP Rev. This indicates that BoHV-4 gH is present in the virion in the absence of gL, although its glycosylation and its recruitment efficiency are affected. We next analyzed the consequences of these differences on viral growth.
gL-deficient BoHV-4 mutant shows an in vitro growth deficit.
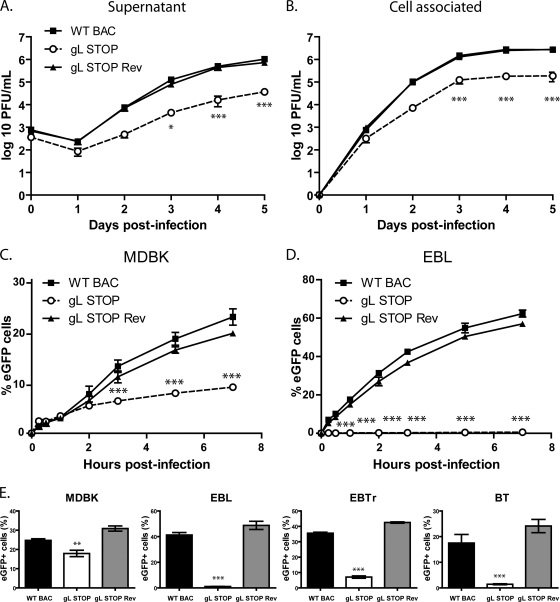
As observed with MuHV-4 (23), the relatively slow spread of viral infection after transfection of the gL STOP BAC DNA into MDBK cells suggested that gL is important for BoHV-4 propagation in vitro. We addressed the role of gL in BoHV-4 lytic infection in vitro by multistep growth curves in MDBK cells (Fig. 4A and B). The results obtained showed an important gL-dependent growth deficit both in the supernatant (Fig. 4A) and in cell-associated virus (Fig. 4B).
Fig 4.
A lack of gL causes a growth deficit associated with a defect in entry. (A and B) MDBK cells were infected with wild-type (WT BAC), gL STOP, and gL STOP Rev BoHV-4 strains in 6-well cluster dishes at an MOI of 0.01 PFU per cell. Supernatant of infected cultures (A) and infected cells (B) were harvested at different times after infection, and the amount of infectious virus was determined by plaque assay on MDBK cells. These plaques were visualized with eGFP and counted. The data presented are the averages of triplicate measurements ± standard deviations and were analyzed by two-way analysis of variance (ANOVA) and Bonferroni posttests (*, P < 0.05; ***, P < 0.001). For supernatants, time zero postinfection is retitration of the inocula to ensure that similar amounts of virus were put on the cells. (C and D) MDBK (C) or EBL (D) cells were exposed to eGFP-expressing wild-type (WT BAC), gL STOP, or gL STOP Rev BoHV-4 strains (0.4 PFU/cell) for the times indicated and then washed with PBS and cultured overnight in the presence of phosphonoacetic acid (PAA; 300 μg/ml). Eighteen hours after the virus was added to the cells, viral infection was assayed by flow cytometry for eGFP expression. The data presented are the averages of triplicate measurements ± standard deviations and were analyzed by two-way ANOVA and Bonferroni posttests (*, P < 0.05; ***, P < 0.001). (E) MDBK, EBL, BT, and EBTr cells were exposed to eGFP-expressing wild-type (WT BAC), gL STOP, or gL STOP Rev BoHV-4 strains (0.4 PFU/cell) and cultured overnight in the presence of PAA (300 μg/ml). Eighteen hours after the virus was added to the cells, viral infection was assayed by flow cytometry for eGFP expression. The data presented are the averages of triplicate measurements ± standard errors of the means and were analyzed by one-way ANOVA and Bonferroni posttests (**, P < 0.01; ***, P < 0.001).
Similar to other herpesviruses, BoHV-4 gL is likely to be involved in entry. We tested viral entry by incubating MDBK (Fig. 4C) or EBL (Fig. 4D) cells with the different viral strains (0.4 PFU/cell) for various times before washing the cells with PBS. The numbers of infected cells (eGFP+) were determined by flow cytometry 18 h later. The gL STOP mutant appeared slow to enter in comparison to results for the WT and gL STOP Rev strains. This entry deficit was particularly important with EBL cells (Fig. 4D); less than 1% of gL STOP-infected EBL cells expressed eGFP even after 24 h of exposure to the virus (Fig. 4E). Similar results were observed with bovine turbinate (BT) and embryonic bovine trachea (EBTr) cells (Fig. 4E).
A lack of gL causes a postbinding deficit.
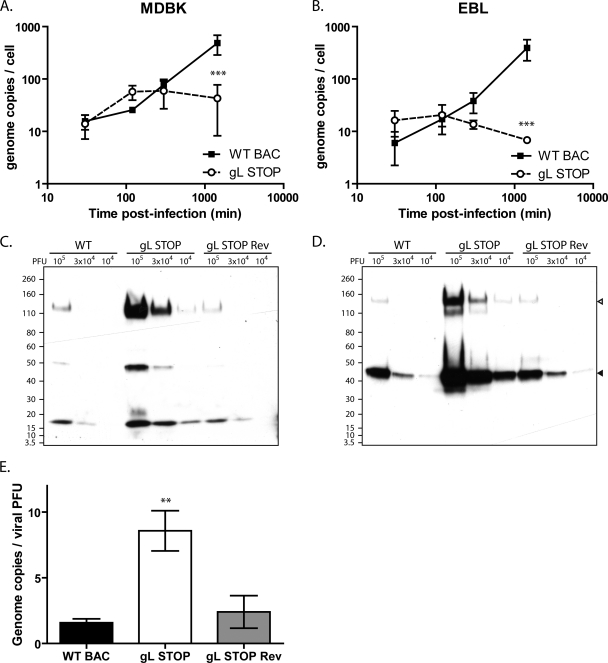
The gL-dependent entry deficit could be due to impaired cell binding or impaired cell penetration of virions. We first investigated cell binding by quantifying the number of viral genomes associated with the cells after various times of infection. Surprisingly, we did not observe reduced binding of gL STOP virions at early time points after infection either in MDBK (Fig. 5A) or in EBL (Fig. 5B) cells—quite the contrary. The apparent better binding of gL STOP virions observed at these early time points could be explained by the noninfectious-to-infectious particle ratio that was approximately 10-fold higher for gL− viruses than for gL+ viruses. Indeed, when protein contents of purified viruses are compared (Fig. 5C and D), viral protein contents for the same numbers of PFU are about 10-fold higher for gL− virus stocks than for the WT and Rev. Genome-to-PFU ratios confirmed that there were proportionally more noninfectious particles in gL− stocks than in gL+ stocks (Fig. 5E). In other words, we underestimate the number of gL STOP virions when standard plaque assay titration is used. At later time points, genome copy numbers increased significantly in WT-infected cells (Fig. 5A and B), reflecting likely viral DNA replication. In contrast, we did not observe a similar increase in gL STOP-infected cells. Taken together, these results suggest that the entry deficit of gL STOP virions is mainly postbinding.
Fig 5.
Binding analysis of the BoHV-4 gL STOP strain. (A and B) MDBK (A) or EBL (B) cells were exposed to WT BAC, gL STOP, or gL STOP Rev BoHV-4 strains (0.4 PFU/cell) for the times indicated and then washed with PBS. Directly after washing, the cells were scraped and DNA was extracted. Cellular and viral genome copies were determined as described in Materials and Methods. The data presented are the averages of triplicate measurements ± standard deviations and were analyzed by two-way ANOVA and Bonferroni posttests (***, P < 0.001). (C and D) WT BAC, gL STOP, or gL STOP Rev BoHV-4 virus stocks were compared for viral protein contents per PFU by immunoblotting with anti-BoHV-4 polyserum (C) or anti-gB MAb 35 (D). In the anti-gB blot, open and filled triangles indicate uncleaved and furin-cleaved gB, respectively. (E) WT BAC, gL STOP, or gL STOP Rev BoHV-4 virus stocks were compared for genome content per PFU by quantitative PCR as described in Materials and Methods. The data presented are the averages from triplicate DNA extractions and measurements ± standard deviations and were analyzed by one-way ANOVA and Bonferroni posttests (**, P < 0.01).
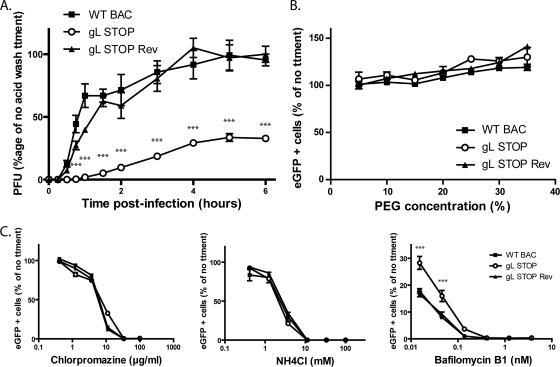
We tested penetration of the different viral strains in MDBK cells (Fig. 6A). The results obtained showed that after 4 h, most of the WT and gL STOP Rev virions had penetrated the cells. In contrast, after 6 h, most of the gL STOP virions were still sensitive to an acid wash, indicating that they were still at the cell surface. These results showed that the gL STOP mutant appeared to be mainly impaired in cell penetration. This deficit could not be overcome by forcing fusion of these cell surface blocked virions. Indeed, polyethylene glycol (PEG) treatment did not significantly enhance gL− virion entry compared to that of gL+ virions (Fig. 6B).
Fig 6.
The entry deficit of the BoHV-4 gL STOP strain is linked with a defect in penetration. (A) MDBK cells were exposed to eGFP-expressing wild-type (WT BAC), gL STOP, or gL STOP Rev BoHV-4 strains (200 PFU/well) for 2 h on ice to allow virus binding but block virus penetration before being washed three times with ice-cold PBS (pH 7.4) to remove unbound virions. After incubation at 37°C for the indicated times, the cells were washed with isotonic acid (pH 3) buffer and overlaid with complete medium containing 0.6% medium-viscosity carboxymethylcellulose (Sigma). At 96 h postinfection, viral plaques (eGFP positive) were counted. The data presented are the averages of triplicate measurements ± standard deviations and were analyzed by two-way ANOVA and Bonferroni posttests (***, P < 0.001). (B) PEG-induced fusion of gL− virions. Cells were infected with eGFP-expressing WT BAC, gL STOP, or gL STOP Rev BoHV-4 strains (37°C, 0.5 PFU/cell). After 2 h, the cells were washed in PBS, and membrane fusion of cell surface virions was induced by PEG treatment at different concentrations. Eighteen hours after the virus was added to the cells, viral infection was assayed by flow cytometry for eGFP expression. Results are expressed as percentages of the levels seen with no PEG treatment. The data presented are the averages of triplicate measurements ± standard deviations and were analyzed by two-way ANOVA and Bonferroni posttests. No statistical difference was observed between viral strains. (C) Dependence of WT and gL-deficient virions on endocytosis and lysosomal acidification. MDBK cells were incubated for 1 h with drugs as shown and then exposed to eGFP+ WT BAC, gL STOP, or gL STOP Rev BoHV-4 virions (0.5 PFU per cell). Drug treatment was maintained throughout the course of infection. Infection rates were determined by a flow cytometric assay of viral eGFP expression after 18 h. The data presented are the averages of triplicate measurements ± standard errors of the means and were analyzed by two-way ANOVA and Bonferroni posttests (***, P < 0.001).
gL-deficient virions still require endocytosis but are less sensitive to inhibitors of endosomal acidification.
Gammaherpesviruses have been reported variously to fuse with the plasma membrane or with endosomal membranes (2, 17, 40). Wild-type BoHV-4 infection of MDBK cells was blocked by inhibitors of clathrin-mediated endocytosis or lysosomal acidification (Fig. 6C). As gL STOP virions display a penetration deficit, we tested their sensitivity to the same drugs. Surprisingly, infection by gL-deficient virions remained as sensitive to chlorpromazine as WT or gL STOP Rev virions (Fig. 6C). NH4Cl treatment, which mainly blocks endocytosis as previously observed (19), gave similar results (Fig. 6C). Interestingly, gL STOP virions were less sensitive to bafilomycin B1, which specifically blocks endosome acidification through inhibition of vacuolar-type H+-ATPase, than those of WT or gL STOP Rev strains (Fig. 6C). gL-deficient virions therefore appeared to still require endocytosis but to be less dependent on low pH.
gL-deficient virions show abnormal distribution during entry.
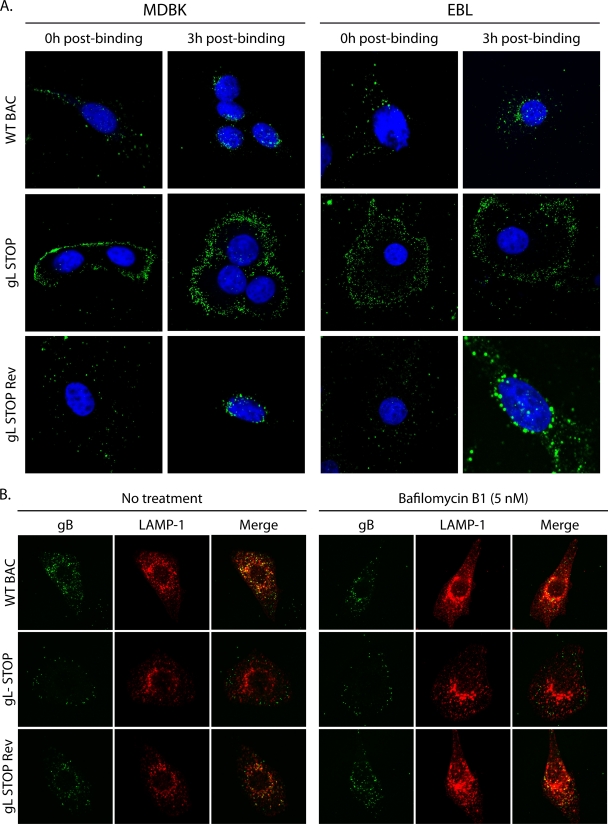
We finally tracked the entry of gL STOP virions into MDBK and EBL cells by immunofluorescence (Fig. 7) with MAb 35 that recognizes gB (34). We bound WT, gL STOP, and gL STOP Rev virions to MDBK or EBL cells for 3 h at 4°C before washing off any unbound virion. We then analyzed virion localization either directly or after incubation at 37°C for 3 h. As previously shown (Fig. 5), we did not observe any gL-related binding deficit in either MDBK or EBL cells (Fig. 7A, 0h post-binding). After incubation at 37°C, most of the endosomes containing WT or gL STOP Rev proteins reached the nuclear margin. In contrast, gL STOP virions remained scattered mainly near the plasma membrane (Fig. 7A, 3h post-binding). We observed that WT BoHV-4 virions, like MuHV-4 virions (19), reach LAMP-1+ endosomes even when fusion was blocked by bafilomycin B1 (Fig. 7B). In contrast, gL− virions remained scattered throughout the cytoplasm but failed to progress to LAMP-1+ late endosomes (Fig. 7B). This result suggests that the entry deficit associated with the absence of gL is downstream of cell binding but upstream of membrane fusion.
Fig 7.
gL-deficient virions show altered glycoprotein distribution during entry. (A) MDBK or EBL cells were exposed to WT BAC, gL STOP, or gL STOP Rev BoHV-4 strains (3 h, 4°C, 10 PFU/cell) and then washed in PBS and either fixed immediately (0 h postbinding) or incubated first (3 h, 37°C) to allow virion endocytosis (3 h postbinding). The cells were then permeabilized and stained for BoHV-4 glycoprotein antigen (MAb 35, anti-gB). Nuclei were counterstained with To-Pro-3. Equivalent data were obtained in three further experiments. (B) BoHV-4 virions were bound to MDBK cells (3 h, 4°C, 10 PFU/cell) with or without bafilomycin (5 nM). The cells were then washed in PBS to remove unbound virions and further incubated with or without drug (3 h, 37°C) to allow virion endocytosis. The cells were then fixed, permeabilized, and stained for gB plus LAMP-1.
DISCUSSION
Most enveloped viruses devote just one protein to cell binding and membrane fusion. Herpesviruses are more complex and engage cells via multiple glycoproteins. gL is one of the three glycoproteins, the others being gH and gB, that form the core entry machinery conserved for the Herpesviridae family. All known functions of gL are directly associated with its dimerization with gH. Indeed, gL is a chaperone for gH, and X-ray structures revealed a close association of the two proteins (9, 39). It is therefore not surprising that, in Alpha- and Betaherpesvirinae, virions lacking gL misfold gH (28, 30) and are not able to initiate membrane fusion at entry (28, 30–31).
Our results show that rhadinoviruses behave differently. Similar to MuHV-4 (23), BoHV-4 lacking gL remained infectious, suggesting that it folds gH correctly, although some differences in gH N-glycosylation were observed (Fig. 3). This difference in the apparent MM of gH is conserved after Endo-H treatment but not after PNGase treatment, which indicates that there are more complex N-glycans on gH in the absence of gL and/or that these complex N-glycans are processed to a greater extent. Interestingly, similar qualitative differences in gH were also observed with pseudorabies virus (PRV) virions (31), for which N-glycans on gH appeared to be processed to a greater extent in the absence of gL. This difference could be explained by the fact that gL might impair the trimming of some gH N-glycans to the complex form in the Golgi apparatus. Similar to PRV gH (31), BoHV-4 gH was still incorporated into virions in the absence of gL, although recruitment efficiency was probably lower than in WT or gL STOP Rev virions (Fig. 3E). However, while some PRV gL-independent infectivity was only restored after repeated passages and selection of genomic rearrangement involving gH and gD (32), no similar compensatory mutations were observed in either MuHV-4 (23) or BoHV-4, suggesting that rhadinovirus gH-gL complexes are different. Moreover, KSHV gH also folds correctly independently of gL and is transported to the plasma membrane (25), suggesting that all rhadinoviruses have similar properties.
Although BoHV-4 gL appeared to be nonessential, gL-deficient virions displayed a growth deficit (Fig. 4) associated with a default in entry (Fig. 4C to E). The gH/gL heterodimer has been shown to bind glycosaminoglycans (GAGs) in both KSHV (25) and MuHV-4 (20). This translated to reduced binding of gL-deficient MuHV-4 virions on some cell types (23). In contrast, we did not observe any binding deficit for the BoHV-4 gL STOP mutant in the tested cell lines (Fig. 5). Herpesviruses commonly express multiple heparan-sulfate (HS) binding proteins (48). Among the rhadinoviruses, MuHV-4 binds to HS via gp70 (18) and gH/gL (20); KSHV does so via K8.1 (6, 51), gH/gL (25), its complement control protein (38), and possibly gB (3). In BoHV-4, binding to HS has been shown for gp8 (50). Moreover, it has been proposed that BoHV-4 gp180 regulates virion tropism through GAG interaction and gp180-deficient virions display a binding deficit (35). Finally, KSHV gH alone binds to GAGs (25). This redundancy could explain why BoHV-4 gL STOP virions did not have any observable binding deficit. Similarly, in MuHV-4, the binding deficit associated with the absence of gL was cell type dependent (20).
In contrast, we showed that the BoHV-4 gL-dependent entry deficit is linked with impaired cell penetration of virions (Fig. 6 and 7). Indeed, while WT virions were quickly internalized after cell binding, gL-deficient virions remained stacked at the cell surface for much longer time (Fig. 6A). This result suggests that BoHV-4 gL triggers virion endocytosis. However, infection by gL-deficient virions still remained sensitive to inhibitors of endocytosis (Fig. 6C), and glycoproteins of gL STOP virions were still internalized (Fig. 7), indicating that fusion does not occur at the cell surface. Interestingly, gL STOP virions were less sensitive to inhibitors of endosome acidification than those of WT or gL STOP Rev (Fig. 6C). This could reflect the requirement of a low pH for gL dissociation from gH as has been proposed for MuHV-4 (21). The absence of gL would therefore allow premature membrane fusion and capsid release outside low-pH endosomes.
The absence of gL could also result in a fusion deficit. However, the PEG-induced fusion experiment would have, at least partially, rescued gL− virions, as observed for PRV gL− mutants (31). That was not the case (Fig. 6B). A fusion deficit also does not explain why endosomes containing glycoproteins of gL− virions do not migrate to the nuclear margin, as observed for WT and gL STOP Rev infection in either EBL or MDBK cells (Fig. 7A). Moreover, fusion blockage of WT virions, as observed with bafilomycin-treated cells, is not associated with a defect in endosome migration in contrast to what is observed with gL− virions (Fig. 7B). Taken together, these results show that the major part of the gL− virion defects appeared to reflect impaired endocytosis and altered endosome migration, which occur upstream of membrane fusion itself.
During entry, the crowded and highly packed cytoplasm inevitably creates a big challenge for trafficking of herpesvirus particles (8). Rhadinoviruses could bypass this problem by eliciting endocytosis and endosome migration to the nuclear margin. While complexing cellular surface receptors is likely to be sufficient to induce endocytosis, endosome targeting probably requires virus signaling to the cell. As endosomes containing glycoproteins of gL-deficient BoHV-4 virions show abnormal distribution after incubation at 37°C (Fig. 7), we propose that BoHV-4 gL could be involved in such signaling. Treatment with chlorpromazine (Fig. 6C) shows that both gL+ and gL− virions enter the cells through clathrin-mediated endocytosis. However, the process is much slower in the absence of gL (Fig. 7A). One hypothesis could therefore be that interaction of some gL-dependent epitope with a cell surface receptor induces de novo formation of clathrin-coated pits as observed for some other viruses, such as influenza A virus (47) or VSV (11). In the absence of gL, virions could be internalized by random ongoing endocytic activities. That would explain why they remain at the cell surface for longer periods of time (Fig. 6A) but still require endocytosis (Fig. 6C). As the ongoing endocytic activities are certainly different between cells, this could explain the differences that we observed in gL− virion infectivity between cells.
This potential role of gL could, at least partially, explain why the gH/gL heterodimer is a major neutralization target in rhadinoviruses (17), although HS binding is redundant. Moreover, it would explain why gH/gL targeted neutralization of MuHV-4 WT virions did not affect virus binding or membrane fusion but blocked the accumulation of perinuclear capsids (D. Glauser and P. G. Stevenson, unpublished results).
Taken together, our results show that gL is nonessential in BoHV-4 although it is involved in viral entry. We propose that one of the major functions of gL in rhadinoviruses is to trigger virion endocytosis.
ACKNOWLEDGMENTS
C.L., B.M., and L.G. are Research Fellows and Research Associates of the “Fonds de la Recherche Scientifique—Fonds National Belge de la Recherche Scientifique” (F.R.S.-FNRS), respectively. P.G.S. is a Wellcome Trust Senior Clinical Fellow (GR076956MA).
This work was supported by the following grants: ARC “GLYVIR,” starting grant from the University of Liège (D-09/11), and an Incentive Grant for Scientific Research from the F.R.S.-FNRS (F.4510.10).
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Aebi M, Bernasconi R, Clerc S, Molinari M. 2010. N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35:74–82 [DOI] [PubMed] [Google Scholar]
- 2. Akula SM, et al. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) infection of human fibroblast cells occurs through endocytosis. J. Virol. 77:7978–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Akula SM, Pramod NP, Wang FZ, Chandran B. 2001. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology 284:235–249 [DOI] [PubMed] [Google Scholar]
- 4. Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84:12292–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birkmann A, et al. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583–11593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowman JJ, Lacayo JC, Burbelo P, Fischer ER, Cohen JI. 2011. Rhesus and human cytomegalovirus glycoprotein L are required for infection and cell-to-cell spread of virus but cannot complement each other. J. Virol. 85:2089–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chandran B. 2010. Early events in Kaposi's sarcoma-associated herpesvirus infection of target cells. J. Virol. 84:2188–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chowdary TK, et al. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat. Struct. Mol. Biol. 17:882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat. Rev. Microbiol. 9:369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5:e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dubuisson J, et al. 1989. Production and characterization of monoclonal antibodies to bovid herpesvirus-4. Vet. Microbiol. 19:305–315 [DOI] [PubMed] [Google Scholar]
- 13. Dunn W, et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galdiero S, et al. 2006. Analysis of synthetic peptides from heptad-repeat domains of herpes simplex virus type 1 glycoproteins H and B. J. Gen. Virol. 87:1085–1097 [DOI] [PubMed] [Google Scholar]
- 15. Gianni T, Martelli PL, Casadio R, Campadelli-Fiume G. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha-helix with attributes of an internal fusion peptide, positionally conserved in the herpesviridae family. J. Virol. 79:2931–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gianni T, Piccoli A, Bertucci C, Campadelli-Fiume G. 2006. Heptad repeat 2 in herpes simplex virus 1 gH interacts with heptad repeat 1 and is critical for virus entry and fusion. J. Virol. 80:2216–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gill MB, et al. 2006. Murine gammaherpesvirus-68 glycoprotein H-glycoprotein L complex is a major target for neutralizing monoclonal antibodies. J. Gen. Virol. 87:1465–1475 [DOI] [PubMed] [Google Scholar]
- 18. Gillet L, Adler H, Stevenson PG. 2007. Glycosaminoglycan interactions in murine gammaherpesvirus-68 infection. PLoS One 2:e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gillet L, Colaco S, Stevenson PG. 2008. Glycoprotein B switches conformation during murid herpesvirus 4 entry. J. Gen. Virol. 89:1352–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gillet L, Colaco S, Stevenson PG. 2008. The murid herpesvirus-4 gH/gL binds to glycosaminoglycans. PLoS One 3:e1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gillet L, Colaco S, Stevenson PG. 2008. The murid herpesvirus-4 gL regulates an entry-associated conformation change in gH. PLoS One 3:e2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gillet L, et al. 2005. Development of bovine herpesvirus 4 as an expression vector using bacterial artificial chromosome cloning. J. Gen. Virol. 86:907–917 [DOI] [PubMed] [Google Scholar]
- 23. Gillet L, May JS, Colaco S, Stevenson PG. 2007. Glycoprotein L disruption reveals two functional forms of the murine gammaherpesvirus 68 glycoprotein H. J. Virol. 81:280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gillet L, Stevenson PG. 2007. Antibody evasion by the N terminus of murid herpesvirus-4 glycoprotein B. EMBO J. 26:5131–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hahn A, et al. 2009. Kaposi's sarcoma-associated herpesvirus gH/gL: glycoprotein export and interaction with cellular receptors. J. Virol. 83:396–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heldwein EE, et al. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217–220 [DOI] [PubMed] [Google Scholar]
- 27. Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hutchinson L, et al. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM. 2008. The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat. Struct. Mol. Biol. 15:1024–1030 [DOI] [PubMed] [Google Scholar]
- 30. Kaye JF, Gompels UA, Minson AC. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J. Gen. Virol. 73:2693–2698 [DOI] [PubMed] [Google Scholar]
- 31. Klupp BG, Fuchs W, Weiland E, Mettenleiter TC. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J. Virol. 71:7687–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Klupp BG, Mettenleiter TC. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lomonte P, et al. 1997. Analysis of the biochemical properties of, and complex formation between, glycoproteins H and L of the gamma2 herpesvirus bovine herpesvirus-4. J. Gen. Virol. 78:2015–2023 [DOI] [PubMed] [Google Scholar]
- 34. Lomonte P, et al. 1997. Glycoprotein B of bovine herpesvirus 4 is a major component of the virion, unlike that of two other gammaherpesviruses, Epstein-Barr virus and murine gammaherpesvirus 68. J. Virol. 71:3332–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Machiels B, et al. 2011. The bovine herpesvirus 4 Bo10 gene encodes a nonessential viral envelope protein that regulates viral tropism through both positive and negative effects. J. Virol. 85:1011–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Machiels B, et al. 2011. Antibody evasion by a gammaherpesvirus o-glycan shield. PLoS Pathog. 7:e1002387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maley F, Trimble RB, Tarentino AL, Plummer TH., Jr 1989. Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Anal. Biochem. 180:195–204 [DOI] [PubMed] [Google Scholar]
- 38. Mark L, Lee WH, Spiller OB, Villoutreix BO, Blom AM. 2006. The Kaposi's sarcoma-associated herpesvirus complement control protein (KCP) binds to heparin and cell surfaces via positively charged amino acids in CCP1-2. Mol. Immunol. 43:1665–1675 [DOI] [PubMed] [Google Scholar]
- 39. Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc. Natl. Acad. Sci. U. S. A. 107:22641–22646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller N, Hutt-Fletcher LM. 1992. Epstein-Barr virus enters B cells and epithelial cells by different routes. J. Virol. 66:3409–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naranatt PP, Akula SM, Chandran B. 2002. Characterization of gamma2-human herpesvirus-8 glycoproteins gH and gL. Arch. Virol. 147:1349–1370 [DOI] [PubMed] [Google Scholar]
- 42. Palmeira L, Machiels B, Lete C, Avanderplasschen Gillet L. 2011. Sequencing of bovine herpesvirus 4 V.test strain reveals important genome features. Virol. J. 8:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peng T, et al. 1998. Structural and antigenic analysis of a truncated form of the herpes simplex virus glycoprotein gH-gL complex. J. Virol. 72:6092–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roche S, Bressanelli S, Rey FA, Gaudin Y. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187–191 [DOI] [PubMed] [Google Scholar]
- 45. Roche S, Rey FA, Gaudin Y, Bressanelli S. 2007. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science 315:843–848 [DOI] [PubMed] [Google Scholar]
- 46. Roop C, Hutchinson L, Johnson DC. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rust MJ, Lakadamyali M, Zhang F, Zhuang X. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shukla D, Spear PG. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 108:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thiry E, Pastoret PP, Dessy-Doizé C, Hanzen C, Calberg-Bacq CM. 1981. Herpesvirus in infertile bull's testicle. Vet. Rec. 108:426. [DOI] [PubMed] [Google Scholar]
- 50. Vanderplasschen A, Bublot M, Dubuisson J, Pastoret PP, Thiry E. 1993. Attachment of the gammaherpesvirus bovine herpesvirus 4 is mediated by the interaction of gp8 glycoprotein with heparinlike moieties on the cell surface. Virology 196:232–240 [DOI] [PubMed] [Google Scholar]
- 51. Wang FZ, Akula SM, Pramod NP, Zeng L, Chandran B. 2001. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 75:7517–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]