Abstract
The HIV-1 Vpr protein participates in the early steps of the virus life cycle by influencing the accuracy of reverse transcription. This role of Vpr was related to the recruitment of the nuclear form of the uracil DNA glycosylase (UNG2) enzyme into virus particles, but several conflicting findings have been reported regarding the role of UNG2 encapsidation on viral infectivity. Here, we report that the catalytic activity of UNG2 was not required for influencing HIV-1 mutation, and this function of UNG2 was mapped within a 60-amino-acid domain located in the N-terminal region of the protein required for direct interaction with the p32 subunit of the replication protein A (RPA) complex. Importantly, enforced recruitment of overexpressed UNG2 into virions resulted in a net increase of virus infectivity, and this positive effect on infectivity was also independent of the UNG2 enzymatic activity. In contrast, virus infectivity and replication, as well as the efficiency of the viral DNA synthesis, were significantly reduced when viruses were produced from cells depleted of either endogenous UNG2 or RPA p32. Taken together, these results demonstrate that incorporation of UNG2 into virions has a positive impact on HIV-1 infectivity and replication and positively influences the reverse transcription process through a nonenzymatic mechanism involving the p32 subunit of the RPA complex.
INTRODUCTION
HIV-1 Vpr is a small basic protein of 96 amino acids that is specifically incorporated into virus particles through a direct interaction with the p6 C-terminal domain of the Pr55Gag precursor protein (2, 23). Its presence in the core of mature virions is subsequently required during the early steps of the virus life cycle in the newly infected cell. After virus entry, the viral core is released into the cytoplasm, where the viral reverse transcriptase catalyzes the synthesis of viral DNA from RNA. One reported function of Vpr is to influence the accuracy of the reverse transcription process, leading to a modulation of the HIV-1 mutation rate (24, 26). In addition, Vpr displays several other activities, including a perturbation of the cell cycle progression resulting in an arrest at the G2/M transition, an induction of apoptosis, and the transcriptional modulation of host cell genes (23).
Initial studies showed that incorporation of Vpr into virions ensued a significant reduction of mutations introduced by the error-prone reverse transcriptase during viral DNA synthesis, and this activity was associated with its binding to the nuclear form of the uracil DNA glycosylase (UNG2) (9, 26). UNG2 is a base excision repair enzyme that participates in the cellular mechanisms for the specific removal of uracil residues from DNA resulting from misincorporation of dUTPs during replication or cytosine deamination (42). Therefore, the role of UNG2 in DNA repair at the replication fork during chromosomal replication is well established, since UNG2 contains determinants required for interactions with proliferating cell nuclear antigen (PCNA) and the 32-kDa subunit (RPA2) of replication protein A (RPA) (1, 14, 19, 28, 30). In addition, UNG2 plays a specific role in somatic hypermutations and class-switch recombination (CSR) at the immunoglobulin locus of B lymphocytes (42). Interestingly, Vpr is able, via direct interaction with UNG2, to exert a dominant-negative effect on the CSR process when it is ectopically expressed in B cells (3).
Subsequently, we reported evidence indicating that the interaction between Vpr and UNG2 results in the incorporation of the catalytically active form of this cellular enzyme into HIV-1 particles (9). The residue Trp54 of Vpr, which is located in the loop that connects the second and third alpha-helices of the protein, plays a crucial role in this interaction (9, 26, 40, 41), and a Vpr mutant with a substitution at the Trp54 position (i.e., VprW54R) failed to recruit UNG2 into virus particles, even if it was itself correctly incorporated into virions (25, 26). Although the VprW54R mutant was not able to assure the accuracy of reverse transcription, UNG2 expressed as a chimeric protein fused to the C-terminal extremity of the VprW54R mutant (VprW54R-UNG2 fusion) was efficiently incorporated into virions and restored a virus mutation rate equivalent to that measured with wild-type Vpr (9). This demonstrated that the presence of UNG2 in virus particles participated in the maintenance of the integrity of the viral genome by influencing the accuracy of reverse transcription. Other studies also confirmed that UNG2 was efficiently recruited into virus particles (17, 18, 37, 48), indicating that this recruitment might influence the accuracy of the reverse transcription process and had a positive influence on virus replication (9, 17, 37). Interestingly, it has been recently reported that HIV-1 DNA generated in infected macrophages and CD4-positive T cells is heavily uracilated (47). However, the specific role of UNG2 incorporation into virions was also challenged by other studies (18, 40, 48). While the specificity of the interaction between Vpr and UNG2 was not questioned, these studies reported data suggesting that UNG2 had either a detrimental effect on virus replication (40, 48) or was dispensable for virus replication (18). In the model that UNG2 has a detrimental effect on virus replication, the role of Vpr was proposed to induce the proteasomal degradation of UNG2 in virus-producing cells in order to prevent its recruitment into virus particles (39, 40). However, other data have indicated that the Vpr-induced reduction of endogenous UNG2 observed in HIV-1-infected cells was not related to proteasomal degradation (21).
The goal of the present study was to further investigate the contribution of UNG2 virion incorporation on viral mutation and to reevaluate the role of UNG2 on viral infectivity. We report here that incorporation of UNG2 into virions influences viral mutation through a nonenzymatic mechanism and that UNG2 has a net positive impact on viral replication. Indeed, HIV-1 particles produced in cells overexpressing UNG2 resulted in a net increase of virus infectivity, whereas infectivity and replication were significantly reduced when viruses were produced from cells depleted of endogenous UNG2.
MATERIALS AND METHODS
Vectors and expression plasmids.
The HIV-1 vector used for analysis of the HIV-1 mutation rate, as well as plasmids used for expression of the hemagglutinin (HA)-tagged forms of the wild-type (wt) UNG2 protein and UNG2 fused to the C terminus of the VprW54R mutant (VprW54R-UNG2 fusion), have been described previously (9). Plasmids for expression of the VprW54R-UNG2 fusions containing deletions or point mutations within the UNG2 part of the fusion were constructed by PCR-mediated site-directed mutagenesis using specific primers containing the desired mutations, and the PCR products were subcloned into the pAS1B plasmid as described previously (9). The plasmids for the expression of the Flag-tagged and green fluorescent protein (GFP)-tagged forms of the murine UNG2 were kindly provided by Tasuku Honjo and Nasim Begum (Kyoto, Japan) (3). The HIV-1-based packaging vectors pCMVDR8.3 (lacking the env and vpr genes) and pCMVDR8.2 (lacking only the env gene) were kindly provided by Didier Trono (Geneva, Switzerland), while the HIV-1 vector encoding GFP (pHIvec2.GFP) and the plasmids encoding the HIV-1 HXBc2 and YU-2 envelope glycoproteins were described previously (20). The wt infectious clone of the NL4.3 HIV-1 isolate (pNL4.3) has been described (9). The pLKO.1 lentiviral vectors harboring short hairpin RNA (shRNA) targeting either UNG2 or the p32 subunit (RPA2) of RPA were purchased from Sigma.
Cell culture and transfection.
293T cells and HeLa-CD4 cells were grown in Dulbecco minimal essential medium supplemented with 10% fetal calf serum, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml (Invitrogen); shRNA-transduced cells were cultivated with 1 μg of puromycin (Invitrogen/ml). Human monocytes were isolated from blood of healthy donors (Hôpital Saint-Vincent-de-Paul, Paris) by density gradient sedimentation in Ficoll (GE Healthcare), followed by adhesion selection for 2 h at 37°C. After extensive washing, the monocytes were differentiated into macrophages for 8 days in complete culture medium RPMI 1640 supplemented with 10% human serum (from total blood of same blood donors), 100 IU of penicillin/ml, and 100 μg of streptomycin/ml (Invitrogen). All cells were grown at 37°C with 5% CO2. 293T cells were transfected for virus productions and immunoprecipitation assay by the calcium phosphate DNA precipitation technique, as described previously (9, 21, 26). For UNG2 transient expression, HeLa-CD4 cells were transfected with 8 μg of pASB1-UNG2 plasmid using FuGENE HD (Promega) according to the manufacturer's instructions.
Analysis of HIV-1 mutant frequencies.
The ability of wt or mutated Vpr or VprW54R-UNG2 fusions to complement a vpr-defective HIV-1 was analyzed in a single-cycle replication assay for mutant frequencies, as described previously (9, 26). Briefly, the plasmids for expression of HA-tagged forms of wt or mutated Vpr, VprW54R, or VprW54R-UNG2 were transiently cotransfected with helper packaging plasmids into cells containing a single integrated HIV-1 vector provirus containing the lacZ gene as a mutation target. The viruses produced were then used to infect permissive HeLa cells, which allowed for a determination of the virus mutant frequency per round of replication. Proviral DNA was then purified with the Lac repressor protein as previously described (26) and introduced into Escherichia coli to screen for mutations in the lacZα gene region. The ratio of the number of white plus light blue bacterial colonies to the total number of colonies observed provided the forward mutation rate for a single retroviral replication cycle. A total of 421 colonies were screened, and the average mutant frequency of the vpr-null mutant HIV-1 in the absence of Vpr transcomplementation was 0.15 (63/421) mutant/cycle.
UNG2- and RPA2-depleted cells.
To stably knockdown UNG2 or RPA2 endogenous expression, pLKO.1 lentiviral vectors harboring short hairpin RNA (shRNA) targeting UNG2 or RPA2 were obtained from Sigma. The hairpin consists of a 21-base stem and a 6-base loop. The sense oligonucleotide sequence targeting UNG2 was 5′-GCAGTTGTGTCCTGGCTAAAT-3′, and that for RPA2 was 5′-CAATCAAGCAAGCTGTGGATT-3′. First, vesicular stomatitis virus glycoprotein G (VSV-G)-pseudotyped lentiviral particles (LVPs) harboring shRNA targeting either UNG2 or RPA2 were produced in 293T cells by cotransfecting pLKO1-shRNA, an HIV-1 packaging vector (8.91, lacking the env and auxiliary genes), and a VSV-G expression plasmid. The pLKO.1 vector plasmid expressing shRNA against firefly luciferase (Sigma) was used to produce control vector particles. At 48 h posttransfection, LVPs were pelleted from supernatants by ultracentrifugation (22,000 rpm for 1.5 h at 4°C) and used to transduce 293T or HeLa-CD4 cells. After 24 h, transduced cells were cultured with fresh medium containing puromycin. After selection for 6 to 7 days, the levels of UNG2 or RPA2 were evaluated by Western blotting and immunofluorescence using specific antibodies.
Virus production and infection.
Single-round-infection HIV-1 carrying the GFP gene was produced in 293T cells as follows. Cells were seeded in T75 flasks at a density of ∼2.5 × 106 cells/T75 flask and transfected 16 h later by the calcium phosphate precipitation technique with a DNA mix containing 8 μg of the HIV-1-packaging plasmid (pCMVDR8.3, pCMVDR8.2, or pNL4.3), 4 μg of the HIV-1 vector encoding GFP (pHIvec2.GFP), 2 μg of the plasmid encoding the HIV-1 envelope glycoproteins HXBc2 or YU-2, and 8 μg of the additional plasmid for expression of UNG2 or VprW54R-UNG2 fusions. The cells were washed 6 h later and then cultured in 8 ml of complete medium for 48 h. The supernatant was then collected, filtered through a 0.45-μm-pore-size filter, and ultracentrifuged to pellet viruses as described previously (20). Amounts of CAp24 produced were determined by enzyme-linked immunosorbent assay (ELISA; Innogenetics). For single-round infection experiments, HeLa-CD4 cells or primary monocyte-derived macrophages were seeded into wells of a six-well plate at a density of 105 or 106 cells/well, respectively. After 24 h, infection was performed using 0.5 μg of CAp24 of produced viruses, and the cells were cultured at 37°C for 60 h. Cell samples were then fixed in 1% paraformaldehyde (Sigma-Aldrich) and analyzed on a Cytomix FC500 cytometer (Beckman-Coulter). The percentage of GFP-positive cells was determined by analyzing the data with the RXP analysis software. Viral infectivity was calculated by normalizing the percentage of GFP-positive cells to that obtained in cells infected with wt or Δvpr viruses. To monitor HIV-1 replication kinetic, HeLa-CD4 cells were infected with 0.2 μg of CAp24 of replication-competent virus, and cell culture supernatant was collected 2, 4, 6, and 8 days after infection for CAp24 determination by ELISA.
UNG enzymatic assay.
UNG enzymatic activity incorporated into purified virus particles was assayed as described previously (9), using 1 μg of crude viral protein with the single-stranded DNA oligonucleotide substrate 5′-TTTTTTTTTTTTUTTTTTTTTTTTT-3′.
Quantification of viral DNA.
At 24 h prior to infection, HeLa-CD4 cells were seeded into a six-well plate at a density of 2 × 105 cells/well. Before infection, replication-competent viruses were incubated with DNase I (Roche) for 1 h at 37°C, and 0.5 μg of CAp24 was then used for infection. At 3 h after infection, the viruses were washed off, and the cells were subsequently incubated at 37°C in complete medium supplemented with 0.5 μM saquinavir in order to restrict viral replication to a single cycle. Then, 7 h later, cell samples were collected, and DNA was extracted using a QIAamp DNA blood minikit (Qiagen) according to the manufacturer's protocol. The total level of HIV-1 DNA was quantified via the LightCycler 480 qPCR system (Roche Applied Science) using the following protocols. Briefly, the quantitative PCR for total HIV-1 DNA was carried out using primers targeting the gag region within the HIV-1 genomic sequence in a 10-μl final volume consisting of 2× FastStart DNA Tag polymerase (Roche) and 0.3 μM concentrations of sense MH532 (5′-TGTGTGCCCGTCTGTTGTGT-3′) and antisense MH531 (5′-GAGTCCTGCGTCGAGAGATC-3′) primers (TIB MolBiol). The fluorescent probe primers 5′-LC640-TCTCTAGCAGTGGCGCCCGAACAG-PH and 5′-CCCTCAGACCCTTTTAGTCAGTGTGGAA-FL were used at a concentration of 0.2 μM. Total DNA was expressed as copy numbers per cell, with the DNA template normalized by β-globin gene amplification using a LightCycler control kit DNA (Roche).
Immunoprecipitation and immunoblot analyses.
293T cells were seeded into T75 flasks at a density of 4 × 106 cells and transfected 24 h later with 10 μg of the GFP-tagged murine or human UNG2 expression plasmid in combination with the HA-Vpr expression plasmid. After 24 h, the cells were lysed in NP-40 lysis buffer supplemented with protease inhibitor (Roche) for 30 min under gentle agitation at 4°C, as described previously (7). After centrifugation at 13,200 rpm, immunoprecipitations were carried out on 450 μg of cell lysate proteins by incubation with 3 μg of anti-GFP antibody (7.1/13.1; Roche) and 30 μl of protein A-Sepharose 4B beads (Sigma) for 2 h under gentle agitation at 4°C. Elution from beads was carried out by incubation in 30 μl of 1× Laemmli buffer containing 5% β-mercaptoethanol for 10 min at 95°C. The concentrations of proteins were quantified by Bradford analysis according to the manufacturer's protocol (Bio-Rad), and 25-μg portions of the proteins were then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% acrylamide NuPAGE Novex bis-Tris precast gels (Invitrogen). Immunoprecipitated and cell lysate proteins were then analyzed by Western blotting with anti-HA (3F10; Roche) and anti-GFP (sc-8334; Santa Cruz Biotechnology) antibodies, and secondary horseradish peroxidase (HRP)-coupled antibodies (Sigma). For analysis of the CAp24 contents of viral particles produced in cells overexpressing UNG2 or the VprW54R-UNG2 fusion, purified virions were resuspended in 80 μl of 1× Laemmli buffer containing 5% β-mercaptoethanol and analyzed by SDS-PAGE and Western blotting with anti-p24 antibody (ab9071; Abcam). The incorporation into viral particles of the VprW54R-UNG2 fusions and murine or human UNG2 proteins was analyzed as described previously (9) using anti-Flag (Sigma), anti-HA (3F10; Roche), and anti-p24 (NIH; Abcam) antibodies. For analysis of UNG2 or RPA2 expression in shRNA-transduced or transfected 293T or HeLa-CD4 cells, cell lysate proteins were similarly analyzed by SDS-PAGE and Western blotting using anti-HA (3F10; Roche), anti-Flag (Sigma), anti-UNG2 (Abcam), anti-RPA2 (Abcam), and anti-β-actin antibodies (Sigma).
Immunofluorescence analysis.
Transfected or transduced HeLa-CD4 or 293T cells were seeded at low density into a six-well plate containing coverslips. After 24 h, the cells were washed in phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (Sigma-Aldrich) for 20 min at room temperature. The reaction was quenched by subsequent wash of the coverslips with PBS. Coverslips were then incubated for 1 h at room temperature with PBS-bovine serum albumin (BSA) supplemented with 0.1% Triton X-100 (Sigma-Aldrich) and anti-HA antibody (Roche) for UNG2 transient-expression experiments and with anti-UNG2 (PU59 [kindly provided by Geir Slupphaug, Trondheim, Norway]) or anti-RPA2 (Abcam) antibody for shRNA experiments. The cells were then washed twice in PBS-BSA and incubated for 45 min in PBS-BSA supplemented with Alexa 488-coupled anti-rat, Alexa 555-coupled anti-rabbit, or Alexa 488-coupled anti-mouse antibodies (Invitrogen), respectively. Coverslips were then washed twice in PBS-BSA and once in PBS and inverted in mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole; Slow-Fade, Invitrogen). Samples were examined under an epifluorescence microscope (Leica), and acquisition of images was carried out as described previously (21).
RESULTS
Enzymatic activity of UNG2 is not required for modulating HIV-1 mutation.
In order to further explore the specific contribution of UNG2 incorporated into HIV-1 particles for modulation of the viral mutation frequency, we used a mutation rate assay we previously described (9, 25, 26), in which UNG2 was expressed as a chimeric protein fused to the C terminus of the VprW54R mutant. This Vpr variant failed to recruit cellular UNG2 into virions and to influence the virus mutation rate, although it was incorporated as efficiently as the wt Vpr protein (9). In contrast to the 4-fold increase in mutant frequency observed by transcomplementation of a vpr-defective HIV-1 (HIV-1Δvpr) with the VprW54R mutant, the VprW54R-UNG2 fusion gave rise to a virus mutant frequency equivalent to that observed by complementation with the wt Vpr protein (Fig. 1A). This demonstrates that the VprW54R-UNG2 fusion represents a valuable tool for analyzing the direct contribution of UNG2 incorporation to the modulation of the virus mutation rate.
Fig 1.
Analysis of catalytically inactive mutants of UNG2 for modulation of HIV-1 mutation rate. (A and C) The ability of wt or mutated Vpr or VprW54R-UNG2 fusions to complement a vpr-defective HIV-1 (Δvpr) was analyzed in a single-cycle replication assay for mutant frequencies. The plasmids for expression of HA-tagged forms of Vpr, VprW54R, or VprW54R-UNG2 fusions were cotransfected with helper packaging plasmids into cells containing a single integrated HIV-1 vector provirus containing the lacZ gene as a mutation target. The viruses produced were then used to infect permissive HeLa cells, which allowed for a determination of the virus mutant frequency per round of replication. A total of 421 colonies were screened, and the average mutant frequency of the vpr-null mutant HIV-1 in the absence of Vpr transcomplementation was 0.15 (63/421) mutant/cycle. Values are the means of three independent experiments. Error bars represent one standard deviation (SD) from the mean. (B and D) Virion incorporation and enzymatic activity of the VprW54R-UNG2 fusion proteins. 293T cells were cotransfected with a vpr-defective HIV-1-based vector in combination with plasmids for expression of HA-tagged VprW54R-UNG2 mutated fusions. Virions were collected from cell supernatants, and proteins from cell and virion lysates were analyzed by Western blotting with anti-HA and anti-CAp24. In panel B (right side), UNG activity from the VprW54R-UNG2 fusions incorporated into virions was assayed with a 25-bp single-stranded DNA oligonucleotide substrate containing a uracil base at position 13, and apurinic sites were then cleaved by adding NaOH and EDTA and boiling for 30 min. The samples were run on a polyacrylamide denaturing gel. The gel was stained with SYBR Gold, and nucleic acids were visualized with an UV transilluminator. The control lane contains untreated DNA substrate.
Substitutions of residues specifically required for the catalytic activity, recognition of uracil residues, and the binding to the DNA substrate (29) were first introduced in the UNG2 portion of VprW54R-UNG2 to generate the mutated VprW54R-UNG2D154N, -UNG2N181V, and -UNG2N213V fusions, respectively. 293T cells were then cotransfected with the HIV-1Δvpr vector lacking the vpr gene in combination with the plasmids for expression of either wt or one of the mutated VprW54R-UNG2 fusions. As shown in Fig. 1B (left part), all VprW54R-UNG2 fusions were efficiently incorporated into virus particles. A low level of UNG activity could be recovered from virions produced from 293T cells expressing the Vpr-UNG2N181V fusion, as determined by detection of the 12-bp product (Fig. 1B, right panel). In contrast, no UNG activity was detected from virions produced from cells expressing the VprW54R-UNG2D154N and -UNG2N213V mutated fusions. However, these three mutated fusions were still able to complement the vpr-defective HIV-1 as efficiently as Vpr or VprW54R-UNG2 in the mutation rate assay (Fig. 1A). Similarly, fusions containing double- or even triple-point mutations within the UNG2 portion could rescue the defective phenotype of VprW54R to modulate HIV-1 mutant frequency (Fig. 1C) and were efficiently incorporated into virions (Fig. 1D). These data indicate that the catalytic activity of UNG2 is not necessary for the modulation of the HIV-1 mutation rate.
The N-terminal region of UNG2 is required for modulating viral mutation.
In order to determine the region within UNG2 being responsible for the modulation of viral mutant frequency, we then generated several N-terminal and C-terminal deletion mutants of UNG2 (Fig. 2A). Mutants deleted of 30, 90, and 140 amino acids (aa) from the N terminus of UNG2 (Δ30, Δ90, and Δ140), as well as truncation mutants spanning aa 1 to 90 and aa 1 to 140, were again expressed as fusions to the VprW54R mutant in virus-producing cells and tested for mutant frequencies as described above. As shown in Fig. 2B, the VprW54R-UNG2Δ30 deletion mutant, lacking the first 30 aa of UNG2, was still able to restore a mutation rate equivalent to that measured by complementation with wt Vpr or VprW54R-UNG2 proteins. In contrast, the VprW54R-UNG2Δ90 fusion, lacking the first 90 aa, failed to complement the vpr-defective virus despite the fact that it was correctly incorporated into virions (Fig. 2C), suggesting that the N-terminal region of UNG2 between aa 30 and 90 is important for restoration of a relative mutant frequency comparable to the wt VprW54R-UNG2 fusion. Indeed, VprW54R-UNG2-1-90 and -1-140 were able to complement the Δvpr virus as efficiently as the wt fusion (Fig. 2B), showing that expression of the N-terminal region between residues 30 and 90 of the UNG2 protein is sufficient for modulation of the virus mutation rate. Interestingly, this region of UNG2 contains the determinants of the protein required for interaction with the p32 subunit of the RPA trimeric complex (12, 28, 30, 31).
Fig 2.
Analysis of deletion mutants of UNG2 for modulation of HIV-1 mutation rate. (A) Schematic representation of the UNG2 deletion mutants expressed as fusions to the VprW54R mutant; amino acids are numbered according to the system of Haug et al. (15). (B) Viruses produced from cells expressing the deleted VprW54R-UNG2 fusions were assayed for mutant frequency phenotype as indicated in Fig. 1A. A total of 356 colonies were screened. and the average mutant frequency of the vpr-null mutant HIV-1 in the absence of Vpr transcomplementation was 0.15 (55/356) mutant/cycle. Values are the means of three independent experiments. Error bars represent one SD from the mean. (C) Virion incorporation of the VprW54R-UNG2 fusion proteins. 293T cells were cotransfected with a vpr-defective HIV-1-based vector in combination with plasmids for expression of HA-tagged VprW54R-UNG2 deleted fusions. Virions were collected from cell supernatants; proteins from cell and virion lysates were analyzed by Western blotting with anti-HA and anti-CAp24.
Incorporation of UNG2 into virus particles positively influences HIV-1 infectivity.
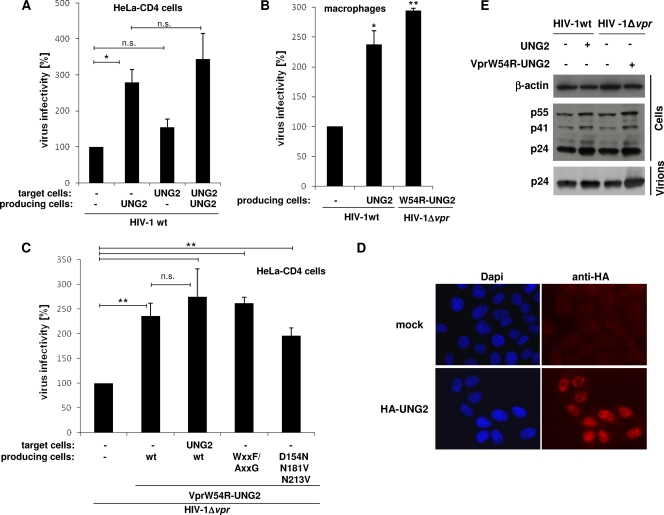
Since the results reported in Fig. 1 and 2 demonstrated that the recruitment of UNG2 into virions was directly responsible for influencing HIV-1 mutation, we then decided to reevaluate the real influence of UNG2 incorporation into HIV-1 particles on virus infectivity. First, the contradictory results reported in the literature about the impact of UNG2 on HIV-1 infectivity and replication (9, 17, 18, 22, 37, 40, 48) were further challenged using a single-round infection assay for the investigation of virus infectivity when UNG2 was overexpressed in either virus-producing cells or target cells. Wild-type reporter viruses carrying the gene encoding GFP were produced in 293T cells overexpressing the HA-tagged form of UNG2, and equivalent amounts of virus, as measured by HIV-1 CAp24 in ELISA, were used to assay virus infectivity on HeLa cells stably expressing the CD4 receptor (HeLa-CD4). As shown in Fig. 3A, overexpression of UNG2 in virus-producing cells had no detrimental effect on virus infectivity, but rather resulted in a 3-fold increase of infectivity of viruses containing UNG2. Moreover, an equivalent 2.5-fold increase of infectivity was measured when viruses produced from UNG2-overexpressing cells were used to infect primary monocyte-derived macrophages (Fig. 3B). In contrast, no significant increase in infectivity was observed when HeLa-CD4 target cells were transfected with HA-UNG2 expression plasmid 1 day prior to infection with wt viruses (Fig. 3A). The inability of UNG2 to affect viral infectivity when overexpressed in target cells was not related to a low level of transfection since we confirmed that almost all HeLa-CD4 cells (∼98%, data not shown) were expressing HA-UNG2, as evidenced by immunofluorescence analysis (Fig. 3D). These results further clarify that UNG2 expression in both virus producing and target cells does not have a negative but has either a supportive effect (virus-producing cells) or no effect (target cells) on virus infectivity.
Fig 3.
Impact of UNG2 overexpression on virus infectivity. (A and C) Wild-type or Δvpr single-round GFP reporter viruses carrying the HIV-1 HXBc2 envelope were produced in 293T cells overexpressing HA-tagged forms of UNG2, or wt or mutated VprW54R-UNG2 fusions, and were used to infect HeLa-CD4 cells overexpressing or not HA-UNG2. Viruses were normalized for CAp24 before infection. The percentages of GFP-positive infected cells were then measured by flow cytometry 60 h later. Viral infectivity was normalized to that of wt (in panel A) or Δvpr (in panel C) viruses produced in cells that did not overexpress UNG2 or VprW54R-UNG2. Values are the means of at least four independent experiments. Error bars represent one SD from the mean. Statistical significance was determined by using the Student t test (n.s., P > 0.05; *, P < 0.05; **, P < 0.01). (B) Wild-type or Δvpr GFP reporter viruses carrying the HIV-1 YU-2 envelope were produced in 293T cells overexpressing HA-tagged forms of either UNG2 or VprW54R-UNG2 and were used to infect primary macrophages derived from blood monocytes from three healthy donors. Viral infectivity was normalized to that of wt viruses produced in cells that did not overexpress UNG2 or VprW54R-UNG2. Values are the means of three independent experiments. Error bars represent one SD from the mean. Statistical significance was determined by using the Student t test (n.s., P > 0.05; *, P < 0.05, **; P < 0.01). (D) Overexpression of UNG2 in HeLa-CD4 target cells. Cells were transfected with the vector for the expression of the HA-tagged form of UNG2 (lower panels) and analyzed 24 h later by indirect immunofluorescence with anti-HA antibody (right panels). Nuclei were stained with DAPI (left panels). Cells were analyzed by epifluorescence microscopy, and images were acquired by using a charge-coupled device camera. (E) Virus maturation of wt and Δvpr HIV-1. 293T cells were cotransfected with wt or vpr-defective HIV-1-based vector in combination with plasmids for expression of HA-tagged forms of UNG2 or VprW54R-UNG2. Virions were collected from cell supernatants; proteins from cell and virion lysates were analyzed by Western blotting with anti-CAp24 and anti-β-actin as a control.
Using the same single-round infectivity assay, we next explored whether the infectivity of vpr-defective viruses could also be influenced through expression of the VprW54R-UNG2 fusion in virus-producing cells. Δvpr viruses containing VprW54R-UNG2 were produced in 293T cells by transfection with the vector for expression of the fusion and then used as previously to infect HeLa-CD4 target cells. As shown in Fig. 3C, Δvpr virus particles produced in cells expressing the VprW54R-UNG2 fusion showed a 2.5-fold increase of infectivity compared to Δvpr viruses. Similarly, a 3-fold increase in virus infectivity was also measured when primary monocyte-derived macrophages were used as target cells (Fig. 3B). Again, overexpression of HA-UNG2 in target cells did not significantly affect infectivity of the Δvpr viruses containing the VprW54R-UNG2 fusion (Fig. 3C). Similarly, VprW54R-UNG2 fusions with point mutations in the catalytic site (VprW54R-UNG2D154N/N181V/N213V) or in the Vpr-binding motif (VprW54R-UNG2WxxF/AxxG) of UNG2 were still able to increase infectivity of Δvpr viruses, as efficiently as the wt VprW54R-UNG2 fusion, when they were expressed in virus-producing cells (Fig. 3C). Therefore, the enzymatic activity of UNG2 and the WxxF motif are not necessary for modulation of virus infectivity. In order to rule out any detrimental effect on virus maturation and release related to UNG2 or VprW54R-UNG2 overexpression in virus-producing cells, cell fractions and purified virus particles were analyzed by Western blotting with anti-CAp24 antibody. As shown in Fig. 3E, overexpression of UNG2 or VprW54R-UNG2 affected neither the expression levels of the Pr55Gag precursor in producer cells nor the release of viral particles in the cell culture supernatant. Taken together, the data reported in Fig. 3 support the notion that incorporation of UNG2 into virions has a positive impact on infectivity of HIV-1 particles.
Depletion of UNG2 in virus-producing cells impairs viral infectivity and replication.
We next sought to further confirm the net positive influence of UNG2 on virus infectivity, and UNG2 expression was then depleted not only in virus-producing 293T cells but also in target HeLa-CD4 cells. In these experiments, we used lentiviruses harboring specific shRNA against UNG2 (shUNG2) and luciferase as a control (shLuc). As evidenced by Western blot analysis of cell lysates from transduced 293T and HeLa-CD4 cells (Fig. 4A), the UNG2 protein band of 39 kDa was efficiently reduced in lysates from shUNG2-transduced cells but not from shLuc-transduced cells. Downregulation of endogenous UNG2 in shUNG2-tranduced HeLa-CD4 cells was also confirmed by immunofluorescence analysis using anti-UNG2 antibody (Fig. 4B). Wild-type HIV-1 was then produced in shUNG2- or shLuc-transduced 293T cells and subsequently used for infection of shUNG2- or shLuc-transduced HeLa-CD4 target cells (Fig. 4C). Interestingly, when viruses were produced in UNG2-depleted cells, a strong decrease in virus infectivity was observed compared to viruses produced in shLuc-treated cells. In contrast, depletion of UNG2 in target cells did not significantly decrease virus infectivity (Fig. 4C), even though downregulation of nuclear UNG2 was efficient in more than 80% of the shUNG2-transduced HeLa-CD4 cells, as determined by immunofluorescence analysis (Fig. 4B). As predicted, virus infectivity was also significantly decreased when UNG2 expression was downregulated in both virus-producing and target cells, indicating the dominant effect of the downregulation of UNG2 in virus-producing cells.
Fig 4.
Impact of UNG2 depletion on virus infectivity. (A) and (B) Depletion of UNG2 in virus-producing (293T) and target (HeLa-CD4) cells. 293T or HeLa-CD4 cells were transduced with lentiviruses expressing shRNA against either UNG2 or luciferase used as a control. In panel A, lysates from shRNA-transduced 293T and HeLa-CD4 cells were analyzed by Western blotting using anti-UNG1/2 and anti-β-actin antibodies. In panel B, shRNA-transduced HeLa-CD4 cells were analyzed by indirect immunofluorescence using anti-UNG1/2 antibody (left part, right panels), and the number of shRNA-transduced cells without nuclear UNG2 staining was quantified over 100 cells (right part). Nuclei were stained with DAPI (left part, left panels). (C) Infectivity of viruses produced in shUNG2-transduced cells. Wild-type GFP reporter viruses were produced in shRNA-transduced 293T cells and then used to infect shRNA-transduced HeLa-CD4 cells as indicated. Viruses were normalized for CAp24 before infection. The percentages of GFP-positive infected cells were then measured by flow cytometry 60 h later. Viral infectivity was normalized to that of viruses produced in shLuc-transduced 293T cells and measured on shLuc-transduced HeLa-CD4 as target cells. (D) Interaction of Vpr with murine UNG2. 293T cells were transfected with plasmids for expression of GFP-tagged murine (mUNG2) or human (hUNG2) UNG2 in combination with the HA-tagged Vpr expression plasmid. Control (mock) corresponds to cells that did not express HA-Vpr. At 24 h after transfection, cell lysates (top and middle panels) were submitted to immunoprecipitation with anti-GFP antibody (bottom panel). Immunoprecipitates were analyzed by Western blotting with anti-GFP (top) and anti-HA (middle and bottom) antibodies. IP, immunoprecipitation. (E) Virion incorporation of mUNG2. shLuc- or shUNG2-transduced 293T cells were cotransfected with a wt HIV-1-based vector in combination with plasmids for expression of Flag-tagged mUNG2. Virions were collected from cell supernatants, and proteins from cell (top panel) and virion (middle and bottom panels) lysates were analyzed by Western blotting with anti-Flag (top and middle) and anti-CAp24 (bottom) antibodies. (F) Infectivity of viruses produced in shUNG2-transduced human cells expressing mUNG2. Wild-type GFP reporter viruses were produced in 293T shRNA-transduced cells expressing or not Flag-tagged mUNG2 and then used to infect HeLa-CD4 cells. Viruses were normalized for CAp24 before infection. The percentages of GFP-positive infected cells were then measured by flow cytometry 60 h later. Viral infectivity was normalized to that of viruses produced in shLuc-transduced cells. Values are the means of at least four independent experiments. Error bars represent one SD from the mean. Statistical significance was determined by using the Student t test (n.s., P > 0.05; *, P < 0.05; **, P < 0.01).
To confirm the specificity of the decrease in virus infectivity observed with viruses produced in UNG2-depleted cells, we checked that this defect was not related to off-target effects of shUNG2-transduction by complementation with the shRNA-insensitive murine form of UNG2 (mUNG2) in shUNG2-transduced human 293T cells. As indicated in Fig. 4D, we determined by coimmunoprecipitation analysis that mUNG2 was able to interact with HIV-1 Vpr as efficiently as the human counterpart (hUNG2). In addition, the shRNA-insensitive mUNG2 version was equally incorporated into HIV-1 particles when produced in either human UNG2-depleted 293T cells or shLuc-transduced control cells (Fig. 4E). When viruses were produced in control cells overexpressing mUNG2 (Fig. 4F), there was a 2.5-fold increase in virus infectivity, which was equivalent to that observed by overexpressing human UNG2 in virus-producing 293T cells (see Fig. 3A). As already reported in Fig. 3C, viruses produced in UNG2-depleted cells were significantly less infectious that viruses produced in control cells (Fig. 4F). In contrast, expression of mUNG2 in shUNG2-transduced virus-producing cells resulted in a restoration of virus infectivity (Fig. 4F). These results confirm that virus production in UNG2-depleted cells results in a specific negative impact on viral infectivity, confirming that incorporation of UNG2 into viral particles is required for maintaining full HIV-1 infectivity.
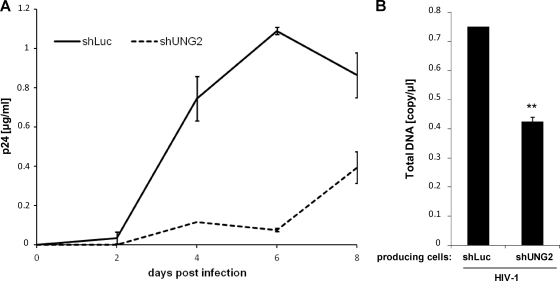
Finally, we evaluated whether UNG2 expression also had a positive influence on virus replication. Wild-type replication-competent HIV-1 was produced in 293T cells and used to infect HeLa-CD4 cells that were depleted of UNG2 (Fig. 5A). Whereas viruses replicated efficiently in shLuc-transduced control cells with a significant CAp24 production detected as soon as 4 days after infection, a strong replication defect could be observed in UNG2-depleted cells with low levels of CAp24 detected 6 days after infection (Fig. 5A). To investigate whether the defect in virus infectivity and replication observed in UNG2-depleted cells was related to a defect in the reverse transcription process, we quantified total viral DNA after infection with wt viruses produced in either UNG2-depleted 293T cells or control shLuc-transduced cells. As shown in Fig. 5B, a significant reduction of the total viral DNA was observed with viruses produced in UNG2-depleted cells compared to the control cells, indicating that UNG2 may affect viral infectivity and replication by directly influencing the number of reverse transcripts produced in the early phase of the viral life cycle.
Fig 5.
Impact of UNG2 depletion on virus replication and reverse transcription. (A) Virus replication in UNG2-depleted cells. Equivalent amounts of wt replication-competent viruses were produced in 293T cells and then used for infection of shLuc-transduced (plain line) or shUNG2-transduced (dashed line) HeLa-CD4 cells. Aliquots of cell culture supernatant were collected 2, 4, 6, and 8 days after infection for CAp24 quantification. (B) Quantification of viral DNA. Wild-type replication-competent viruses were produced in shRNA-transduced 293T cells as indicated and then used for infection of HeLa-CD4 cells. Viruses were normalized for CAp24 before infection. At 7 h after infection, cell samples were collected and subjected to DNA purification, and the total viral DNA was quantified via quantitative PCR using specific primers for gag. Values are the means of three independent experiments. Error bars represent one SD from the mean. Statistical significance was determined by using the Student t test (n.s., P > 0.05; *, P < 0.05; **, P < 0.01).
Taken together, the data reported in Fig. 4 and 5 show that UNG2 expression has a positive impact on both HIV-1 infectivity and replication.
The p32 subunit of RPA participates in HIV-1 infectivity.
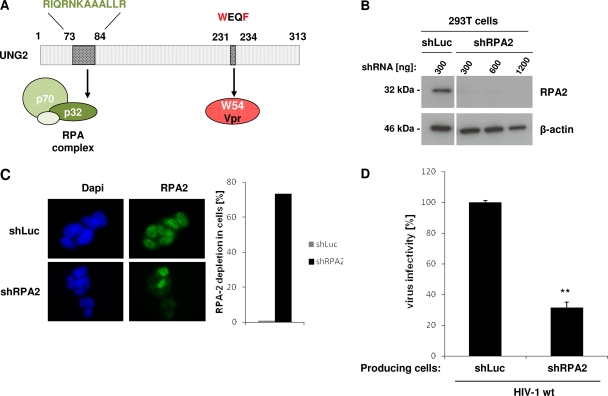
As previously indicated, we mapped and determined that the N-terminal region of UNG2 between residues 30 and 90 was required for modulation of the virus mutation frequency; this region also contains the determinants for interaction with the p32 (RPA2) subunit of the RPA trimeric complex (Fig. 6A) (14, 27, 30, 31). Here, we investigated the potential role of RPA in the modulation of HIV-1 infectivity (Fig. 6). Lentiviruses harboring shRNA against RPA2 (shRPA2) were used for specific depletion in virus-producing 293T cells. Using increasing amounts of lentiviruses for targeting RPA2 (300, 600, or 1,200 ng of CAp24), expression of the protein was successfully depleted for all three concentrations in 293T cells compared to shLuc-transduced control cells, as evidenced both by Western blot and immunofluorescence analyses (Fig. 6B and C, respectively). Intriguingly, when wt HIV-1 was then produced in RPA2-depleted 293T cells and used for infection of HeLa-CD4 cells, we observed that virus infectivity was significantly decreased compared to viruses produced in shLuc-transduced cells (Fig. 6D), suggesting a potential role for RPA2 during HIV-1 infection.
Fig 6.
Impact of RPA2 depletion on virus infectivity. (A) Schematic representation of UNG2 showing the motifs required for interaction with Vpr or with the p32 subunit (RPA2) of the RPA heterotrimeric complex. (B and C) Depletion of RPA2 in virus-producing cells. 293T cells were transduced with lentiviruses expressing shRNA against either RPA2 or Luciferase used as a control. In panel B, lysates from shRNA-transduced cells were analyzed by Western blotting with anti-RPA2 (top) and anti-β-actin (bottom) antibodies. In panel C, shRNA-transduced cells were analyzed by indirect immunofluorescence with anti-RPA2 antibody (left part, right panels), and the number of shRNA-transduced cells without nuclear RPA2 staining was quantified over 100 cells (right part). Nuclei were stained with DAPI (left part, left panels). Cells were analyzed by epifluorescence microscopy, and images were acquired with a charge-coupled device camera. (D) Virus infectivity. Wild-type GFP reporter viruses were produced in shRNA-transduced 293T cells as indicated and then used to infect HeLa-CD4 cells. Viruses were normalized for CAp24 before infection. The percentages of GFP-positive infected cells were measured by flow cytometry 60 h later. Infectivity was normalized to that of viruses produced in shLuc-transduced cells and measured on HeLa-CD4 as target cells. Values are the means of three independent experiments. Error bars represent one SD from the mean. Statistical significance was determined by using the Student t test (n.s., P > 0.05; *, P < 0.05; **, P < 0.01).
DISCUSSION
While several previous studies reported conflicting results regarding the biological relevance of the recruitment of UNG2 into HIV-1 particles (9, 17, 18, 22, 37, 40, 48), our results argue that incorporation of UNG2 into HIV-1 particles is not detrimental for virus infection of target cells but rather has a positive impact on virus replication. These positive effects correlate with a positive influence on the reverse transcription process. While it was recently suggested that UNG2 was specifically required for efficient infection of primary cells with R5 viruses (17), we observed that enforced virion recruitment of UNG2, through overexpression of UNG2 in virus-producing cells, similarly influenced infectivity of X4 and R5 HIV-1 strains in transformed cell lines and primary monocyte-derived macrophages, respectively. Conversely, viral infectivity and spreading were significantly reduced when viruses were produced from or replicated in cells depleted of endogenous UNG2 using specific shRNA, confirming the positive impact of UNG2 incorporation for the full infectivity of HIV-1 particles. In addition, we observed that the p32 subunit of RPA, which has been reported to interact directly with UNG2 (14, 28, 30, 31), may also participate in maintaining HIV-1 infectivity.
While we previously reported that the catalytically active form of UNG2 is incorporated into HIV-1 particles (9), we show here, through substitution of UNG2 residues required for the catalytic activity of the enzyme, recognition of uracil residues, and binding of the protein to the DNA substrate (29), that the uracil excision activity of UNG2 incorporated into virions was required neither for the modulation of the virus mutation rate nor for virus infectivity. These intriguing observations are in agreement with previous reports showing that HIV-1 particles produced in 293T cells overexpressing the specific potent catalytic active-site bacteriophage PBS1 inhibitor (UGI) of human UNG2 were still fully effective for infection of HeLa-CD4 cells or transformed T-cell lines (17, 18, 22).
Interestingly, the genome of numerous viruses from the Poxviridae and Herpesviridae, such as the cytomegalovirus (CMV) and the vaccinia virus, contains an open reading frame coding for a UNG protein with sequence similarities to the mammalian UNG2 (10, 42). In vaccinia virus, the viral UNG is required for efficient virus replication in cultured cells, but viral UNG mutants lacking uracil-removal activity could efficiently replace the wt viral UNG for efficient virus replication in cultured cells (12, 13, 43). Moreover, deletion of UNG in CMV also resulted in a significant decrease of virus replication (35, 36), but again, the uracil-excising activity of CMV UNG did not appear to be important for replication, since poor viral replication was unrelated to the uracil content of input genomes (38). Similarly, our results indicate that the role of the Vpr-mediated incorporation of UNG2 into virus particles for efficient reverse transcription and viral replication may also be different from uracil removal.
It was also reported that despite the absolute requirement of the UNG2 protein for an efficient CSR process in B lymphocytes (42), catalytically inactive mutants of UNG2 were fully proficient in CSR (3–5). These results indicated that the specific function of UNG2 in CSR is not related to the uracil removal activity of the protein and correlate with our observations regarding the specific role of UNG2 for the modulation of the HIV-1 mutation rate. Together, these observations support a model in which both the viral mutation rate and CSR depend on a novel nonenzymatic function of UNG2. As mentioned above, it was previously reported that Ig class switching was drastically inhibited when Vpr was overexpressed in stimulated B cells, indicating that Vpr had a dominant-negative effect on CSR (3). However, Vpr mutants defective for interaction with UNG2, such as the VprW54R mutant, failed to influence CSR. Interestingly, it was also reported that mutations in the conserved UNG2 motif (i.e., WxxF), which is required for Vpr binding to UNG2 (8, 9), blocked CSR without affecting its uracil removal activity, indicating that this motif is equally essential for the role of UNG2 in CSR (3, 5). Overall, it was suggested that the exogenous Vpr competes with an unknown Vpr-like host protein required for a novel enzymatic-independent function of UNG2 in CSR (3, 5). Therefore, the WxxF motif within UNG2 is required for binding to the Vpr-like factor in order to ensure its activity during CSR (3, 5). In the current study, the UNG2 WxxF motif per se was not necessary for enhancement of HIV-1 infectivity, but this motif was strictly required for interaction with Vpr in virus producing cells in order to recruit UNG2 into virions (9), where it would be subsequently required for efficient reverse transcription and viral replication in target cells. Indeed, the VprW54R-UNG2 fusion with substitutions in the WxxF motif of UNG2 (VprW54R-UNG2WxxF/AxxG) was still able to significantly increase infectivity of Δvpr viruses as efficiently as the wt VprW54R-UNG2 fusion.
We furthermore found that the determinants of UNG2 participating in the accuracy of the reverse transcription process are located within a 60-aa region between the N-terminal residues 30 and 90 of the UNG2 protein. It is noteworthy that the same N-terminal region of the UNG2 protein is required for the specific role of UNG2 in CSR in B lymphocytes (3, 5), suggesting that this region, which is neither required for uracil-removal activity nor Vpr binding (29, 41), is involved in a new function of UNG2 shared by CSR and HIV-1 mutation modulation processes. Interestingly, this N-terminal region of UNG2 also contains determinants of the protein required for interaction with the p32 subunit of the RPA trimeric complex (14, 28, 30, 31). RPA has been reported to be essential for the repair of double-strand breaks by homologous recombination (32, 44) and for postreplicative base-excision repair (BER) (11, 31). Through tight binding of RPA to single-stranded DNA (6), it is thought that RPA mediates the coordinated assembly of the DNA repair machinery at sites of DNA damage (16, 46). This function is attributed to the two larger subunits of RPA (RPA-32/RPA2 and RPA-70), while the smaller subunit RPA-14 is believed to serve a structural purpose in the RPA heterotrimer. RPA and UNG2 do colocalize in replication foci (31) and RPA has been demonstrated to be essential for the role of UNG2 in rapid postreplicative removal of uracil in single-strand DNA at the replication fork (14, 28, 30, 31). Given the close functional relationship between UNG2 and RPA for BER, our initial findings indicated a potential role for RPA during HIV-1 infection. Indeed, when viruses were produced in RPA2-depleted cells, virus infectivity was significantly reduced, suggesting that RPA2 might be involved in the infection process, probably through direct interaction with UNG2 and independently of its enzymatic activity. Intriguingly, the same identified N-terminal region within UNG2 has been reported to bind to proliferating cell nuclear antigen (PCNA), a DNA elongation factor that is involved in various processes such as DNA replication and repair (19, 33, 34, 45). Since it was demonstrated that UNG2, RPA2, and PCNA colocalized within replication foci (31), this implies an evident functional relationship between these proteins. Further analyses are therefore needed to investigate the potential recruitment of these additional factors of the DNA repair system for HIV-1 infectivity and replication.
We initially reported data indicating that the interaction between Vpr and UNG2 in virus-producing cells led to the incorporation of the enzyme into viral particles in order to restrict the errors introduced by the reverse transcriptase during viral DNA synthesis in the target cells (9, 26), but the specific role of UNG2 incorporation into virions was subsequently questioned by other studies (18, 40, 48). While our previous studies with the UNG2-binding deficient VprW54R mutant demonstrated the importance of UNG2 recruitment for efficient replication in cell types with low endogenous UNG2 levels (e.g., nondividing cells), our UNG2 depletion experiments reported in the present study in actively dividing HeLa-CD4 cells using shRNAs directed against UNG2 further support the critical requirement of UNG2 recruitment into virions. We now strengthen our previous conclusion by demonstrating that viral infectivity was significantly reduced when viruses were produced in UNG2-depleted cells and was restored in the same cells overexpressing the shRNA-insensitive murine form of UNG2. Moreover, the low infectivity of HIV-1 particles produced in UNG2-depleted cells correlated with a reduced amount of viral DNA generated during the reverse transcription process in the target cells. In contrast, enforced virion recruitment of UNG2, through overexpression of UNG2 in virus-producing cells, similarly influenced HIV-1 infectivity in transformed cell lines and primary monocyte-derived macrophages. Using similar small interfering RNA (siRNA) strategies targeting UNG2 for the production of viruses in UNG2-depleted cells, such as the 293T and HeLa-CD4 MAGI-CCR5 cell lines, or primary macrophages, Priet et al. (37) and Jones et al. (17) both confirmed that the recruitment of UNG2 into viral particles has a positive influence on the reverse transcription process and is required for efficient viral infectivity or replication in HeLa-CD4 MAGI-CCR5 cells or in primary macrophages used as target cells. In contrast, studies by Schröfelbauer et al. (40) and Yang et al. (48) suggested a model in which incorporation of UNG2 into viral particles would have a detrimental effect on reverse transcription by introducing abasic sites into viral DNA in regard to uracil residues resulting from cytosine deamination by the cytidine deaminase APOBEC3G. While Schröfelbauer et al. did not directly question the specific role of UNG2 in the antiviral activity of APOBEC3G (40), Yang et al. reported data indicating that the antiviral activity of overexpressed APOBEC3G was partially affected when viruses were produced in UNG2-depleted 293T cells using an siRNA strategy (48). The data reported in the latter study are in apparent contradiction to results reported by us and others by producing viruses in UNG2-depleted cells which expressed or not APOBEC3G (17, 37) but also with other reports showing that APOBEC3G-mediated restriction of HIV-1 in human cells was independent of UNG (18, 22). Since Yang et al. (48) did not examine in their siRNA strategy used to analyze the role UNG2 if viruses produced in UNG2-depleted cells overexpressing APOBEC3G could be rescued through expression of a RNAi-insensitive form of UNG2, additional investigations are thus required to further understand this apparent contradiction regarding the role of UNG2 virion incorporation for the antiviral activity of APOBEC restriction factors. Finally, Kaiser and Emerman (18) reported data indicating that UNG2 had neither a positive nor a negative impact on HIV-1 infectivity, but viruses were produced in a UNG2-defective B-lymphoid cell line, and the different cell culture systems used to produced virus particles might explain the discrepancy between this report and data reported by us and others (17, 37).
In conclusion, the results reported here support the model in which incorporation of UNG2 into virions has a positive impact on HIV-1 infectivity and replication and positively modulates the virus mutation rate through a nonenzymatic mechanism. Intriguingly, direct interaction between UNG2 and RPA2 might participate in this not yet completely understood mechanism and further investigations are thus needed to elucidate how incorporation of UNG2 is able to influence the accuracy of the reverse transcription process. This could also help to elucidate the novel molecular function of UNG2 in CSR in B lymphocytes.
ACKNOWLEDGMENTS
We thank Geir Slupphaug (Trondheim, Norway), Tasuku Honjo and Nasim Begum (Kyoto, Japan), and Didier Trono (Geneva, Switzerland) for the generous gift of reagents.
This study was supported in part by INSERM, the CNRS, the Université Paris-Descartes, the French National Agency for AIDS Research (ANRS), Sidaction (S.B.), and National Institutes of Health grant GM56615 (L.M.M.). C.G. is an ANRS fellowship recipient.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Ali SI, Shin J-S, Bae S-H, Kim B, Choi B-S. 2010. Replication protein A 32 interacts through a similar binding interface with TIPIN, XPA, and UNG2. Int. J. Biochem. Cell Biol. 42:1210–1215 [DOI] [PubMed] [Google Scholar]
- 2. Andersen JL, Le Rouzic E, Planelles V. 2008. HIV-1 Vpr: mechanisms of G2 arrest and apoptosis. Exp. Mol. Pathol. 85:2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Begum NA, et al. 2007. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J. Biol. Chem. 282:731–742 [DOI] [PubMed] [Google Scholar]
- 4. Begum NA, et al. 2004. Uracil DNA glycosylase activity is dispensable for immunoglobulin class switch. Science 305:1160–1163 [DOI] [PubMed] [Google Scholar]
- 5. Begum NA, et al. 2009. Further evidence for involvement of a noncanonical function of uracil DNA glycosylase in class switch recombination. Proc. Natl. Acad. Sci. U. S. A. 106:2752–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bochkarev A, Bochkareva E, Frappier L, Edwards AM. 1999. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 18:4498–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bouchet J, et al. 2011. Inhibition of the Nef regulatory protein of HIV-1 by a single-domain antibody. Blood 117:3559–3568 [DOI] [PubMed] [Google Scholar]
- 8. BouHamdan M, et al. 1998. Diversity of HIV-1 Vpr interactions involves usage of the WXXF motif of host cell proteins. J. Biol. Chem. 273:8009–8016 [DOI] [PubMed] [Google Scholar]
- 9. Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. 2004. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J. Biol. Chem. 279:28419–28425 [DOI] [PubMed] [Google Scholar]
- 10. Chen R, Wang H, Mansky LM. 2002. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J. Gen. Virol. 83:2339–2345 [DOI] [PubMed] [Google Scholar]
- 11. DeMott MS, Zigman S, Bambara RA. 1998. Replication protein A stimulates long patch DNA base excision repair. J. Biol. Chem. 273:27492–27498 [DOI] [PubMed] [Google Scholar]
- 12. De Silva FS, Moss B. 2008. Effects of vaccinia virus uracil DNA glycosylase catalytic site and deoxyuridine triphosphatase deletion mutations individually and together on replication in active and quiescent cells and pathogenesis in mice. Virol. J. 5:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Silva FS, Moss B. 2003. Vaccinia virus uracil DNA glycosylase has an essential role in DNA synthesis that is independent of its glycosylase activity: catalytic site mutations reduce virulence but not virus replication in cultured cells. J. Virol. 77:159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hagen L, et al. 2008. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 27:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haug T, Skorpen F, Lund H, Krokan HE. 1994. Structure of the gene for human uracil-DNA glycosylase and analysis of the promoter function. FEBS Lett. 353:180–184 [DOI] [PubMed] [Google Scholar]
- 16. Iftode C, Daniely Y, Borowiec JA. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141–180 [DOI] [PubMed] [Google Scholar]
- 17. Jones KL, et al. 2010. X4 and R5 HIV-1 have distinct post-entry requirements for uracil DNA glycosylase during infection of primary cells. J. Biol. Chem. 285:18603–18614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaiser SM, Emerman M. 2006. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 80:875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ko R, Bennett SE. 2005. Physical and functional interaction of human nuclear uracil-DNA glycosylase with proliferating cell nuclear antigen. DNA Repair (Amst.) 4:1421–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laguette N, Benichou S, Basmaciogullari S. 2009. Human immunodeficiency virus type 1 Nef incorporation into virions does not increase infectivity. J. Virol. 83:1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langevin C, et al. 2009. Human immunodeficiency virus type 1 Vpr modulates cellular expression of UNG2 via a negative transcriptional effect. J. Virol. 83:10256–10263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langlois M-A, Neuberger MS. 2008. Human APOBEC3G can restrict retroviral infection in avian cells and acts independently of both UNG and SMUG1. J. Virol. 82:4660–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Rouzic E, Benichou S. 2005. The Vpr protein from HIV-1: distinct roles along the viral life cycle. Retrovirology 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mansky LM. 1996. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology 222:391–400 [DOI] [PubMed] [Google Scholar]
- 25. Mansky LM, et al. 2001. Interaction of human immunodeficiency virus type 1 Vpr with the HHR23A DNA repair protein does not correlate with multiple biological functions of Vpr. Virology 282:176–185 [DOI] [PubMed] [Google Scholar]
- 26. Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. 2000. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 In vivo mutation rate. J. Virol. 74:7039–7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mer G, Bochkarev A, Chazin WJ, Edwards AM. 2000. Three-dimensional structure and function of replication protein A. Cold Spring Harbor Symp. Quant. Biol. 65:193–200 [DOI] [PubMed] [Google Scholar]
- 28. Mer G, et al. 2000. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell 103:449–456 [DOI] [PubMed] [Google Scholar]
- 29. Mol CD, et al. 1995. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell 80:869–878 [DOI] [PubMed] [Google Scholar]
- 30. Nagelhus TA, et al. 1997. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J. Biol. Chem. 272:6561–6566 [DOI] [PubMed] [Google Scholar]
- 31. Otterlei M, et al. 1999. Post-replicative base excision repair in replication foci. EMBO J. 18:3834–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park MS, Ludwig DL, Stigger E, Lee SH. 1996. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 271:18996–19000 [DOI] [PubMed] [Google Scholar]
- 33. Prelich G, Kostura M, Marshak DR, Mathews MB, Stillman B. 1987. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature 326:471–475 [DOI] [PubMed] [Google Scholar]
- 34. Prelich G, et al. 1987. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature 326:517–520 [DOI] [PubMed] [Google Scholar]
- 35. Prichard MN, Duke GM, Mocarski ES. 1996. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J. Virol. 70:3018–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prichard MN, et al. 2005. Human cytomegalovirus uracil DNA glycosylase associates with ppUL44 and accelerates the accumulation of viral DNA. Virol. J. 2:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Priet S, et al. 2005. HIV-1-associated uracil DNA glycosylase activity controls dUTP misincorporation in viral DNA and is essential to the HIV-1 life cycle. Mol. Cell 17:479–490 [DOI] [PubMed] [Google Scholar]
- 38. Ranneberg-Nilsen T, et al. 2008. Characterization of human cytomegalovirus uracil DNA glycosylase (UL114) and its interaction with polymerase processivity factor (UL44). J. Mol. Biol. 381:276–288 [DOI] [PubMed] [Google Scholar]
- 39. Schröfelbauer B, Hakata Y, Landau NR. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. U. S. A. 104:4130–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schröfelbauer B, Yu Q, Zeitlin SG, Landau NR. 2005. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 79:10978–10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Selig L, et al. 1997. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 71:4842–4846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sousa MML, Krokan HE, Slupphaug G. 2007. DNA-uracil and human pathology. Mol. Aspects Med. 28:276–306 [DOI] [PubMed] [Google Scholar]
- 43. Stanitsa ES, Arps L, Traktman P. 2006. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 281:3439–3451 [DOI] [PubMed] [Google Scholar]
- 44. Sugiyama T, New JH, Kowalczykowski SC. 1998. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U. S. A. 95:6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takasaki Y, Deng JS, Tan EM. 1981. A nuclear antigen associated with cell proliferation and blast transformation. J. Exp. Med. 154:1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61–92 [DOI] [PubMed] [Google Scholar]
- 47. Yan N, O'Day E, Wheeler LA, Engelman A, Lieberman J. 2011. HIV DNA is heavily uracilated, which protects it from autointegration. Proc. Natl. Acad. Sci. U. S. A. 108:9244–9249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang B, Chen K, Zhang C, Huang S, Zhang H. 2007. Virion-associated uracil DNA glycosylase-2 and apurinic/apyrimidinic endonuclease are involved in the degradation of APOBEC3G-edited nascent HIV-1 DNA. J. Biol. Chem. 282:11667–11675 [DOI] [PubMed] [Google Scholar]