Abstract
Orthopoxviruses include the prototypical vaccinia virus, the emerging infectious agent monkeypox virus, and the potential biothreat variola virus (the causative agent of smallpox). There is currently no FDA-approved drug for humans infected with orthopoxviruses. We screened a diversity-oriented synthesis library for new scaffolds with activity against vaccinia virus. This screen identified a nonnucleoside analog that blocked postreplicative intermediate and late gene expression. Viral genome replication was unaffected, and inhibition could be elicited late in infection and persisted upon drug removal. Sequencing of drug-resistant viruses revealed mutations predicted to be on the periphery of the highly conserved viral RNA polymerase large subunit. Consistent with this, the compound had broad-spectrum activity against orthopoxviruses in vitro. These findings indicate that novel chemical synthesis approaches are a potential source for new infectious disease therapeutics and identify a potentially promising candidate for development to treat orthopoxvirus-infected individuals.
INTRODUCTION
The Poxviridae family of DNA viruses includes the orthopoxviruses variola virus (the causative agent of smallpox) and the emerging pathogen monkeypox virus. Naturally occurring smallpox was eradicated through concerted vaccination with the prototypical orthopoxvirus, vaccinia virus, and routine vaccination has since been discontinued (10). The potential use of smallpox as a bioweapon, however, has heightened interest in developing countermeasures (8). In addition, recent reports revealed an increase in human monkeypox cases in Africa over the last 30 years (27), and the first report of human monkeypox in the Western Hemisphere occurred in 2003 (25). Consequently, there is currently renewed interest in effective strategies to treat orthopoxvirus-infected individuals (18).
Orthopoxviruses replicate in the cytoplasm and encode macromolecular machinery for transcription, posttranscriptional mRNA processing, and DNA genome replication (20). Gene expression proceeds by a classical cascade mechanism that is broadly categorized into early, intermediate, and late phases following virus entry into the cell (2). Viral replication occurs in perinuclear factories, and a major mode of transmission of these predominantly intracellular viruses is to adjacent cells via trafficking to the cell membrane or upon infected cell rupture (28). While these core features of the orthopoxvirus life cycle are conserved, host range and virulence factors are divergent (19, 30). Variola virus is an obligate human pathogen causing disease with a mortality rate that can exceed 30%, and it caused an estimated 300 to 500 million deaths in the 20th century (10). Monkeypox has a mortality rate of 1 to 10% and can be transmitted to humans zoonotically from animal reservoirs (21).
There are currently no approved drugs for treatment of orthopoxvirus-infected individuals. Numerous small-molecule inhibitors of orthopoxviruses have been identified; however, many are nucleoside analogs for which selectivity for viral over host enzymes poses developmental challenges (5, 22). Compounds that show promise in preclinical or clinical settings are the cidofovir derivative CMX001 (24), which is a nucleoside analog, and ST-246 (12, 13), which has a unique mechanism of action in preventing viral egress from infected cells. With the majority of inhibitors, including CMX001 and ST-246, viral resistance is achieved rapidly in cell culture (1, 9, 33). As with many antimicrobial strategies, effective treatment is likely to require combination therapy.
We recently generated a series of reporter vaccinia viruses that allow rapid screening of compounds for antiviral activity (7). These viruses were used in a proof-of-principle screen of a diversity-oriented synthesis library. This screen identified a nonnucleoside analog, CMLDBU6128, which could inhibit several orthopoxviruses in vitro. Here we report its identification and the use of newly developed systems to characterize its antiviral effect.
MATERIALS AND METHODS
Cell culture, viruses, and growth curves.
A549 (CCL-85), HeLa (CCL-2), and Vero (CCL-81) cells were obtained from the ATCC. Unless noted otherwise, the vaccinia virus used in this study was strain Western Reserve or a derivative thereof. Recombination strategies, promoter sequences, and insertion sites for reporter viruses are described in detail elsewhere (7). The Venus-A4L virus contains the Venus coding sequence immediately after the A4L ATG start codon in the following sequence: CAATTTTAAAGCCTTAAATGGACTTCTTTAACAAGTTCTC. A flexible glycine-serine linker encoded by the sequence GGTGGAGGCGGTTCA was introduced between the Venus and A4L sequences. Experiments with monkeypox virus strain Zaire 1979 (MPOX), vaccinia virus strain IHDJ (VACV IHDJ), and wild-type cowpox virus (CPX) were performed at USAMRIID under appropriate containment conditions. Virus infections and PFU determinations were performed using standard methods. Infections at a low multiplicity of infection (MOI) utilized an MOI of 0.005; high-MOI infections utilized an MOI of 5 and removal of excess input virus after 1 h. Medium for low-MOI assays was refreshed every 24 h to replenish the compound. The removed medium was saved and added to final cell harvests for total virus yield determination. Cell viability was determined by colorimetric MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay (Invitrogen).
Library screening and compounds.
Generation of the screening library and chemical syntheses were performed by the Center for Chemical Methodology and Library Development at Boston University (CMLDBU), Boston, MA. A549 cells were seeded at 20,000 cells/well in 96-well plates (Corning) the previous day and infected with the late red, early Venus (LREV) virus at an MOI of 10 in the presence of compound. Final compound concentrations were estimated to be approximately 10 μM. At 12 h postinfection, cells were fixed with phosphate-buffered saline (PBS) containing 4% formaldehyde for next-day scoring on a fluorescence plate reader (see below). ST-246 was obtained from SIGA Labs (Corvallis, OR). 1-β-d-Arabinofuranosylcytosine (araC) was purchased from Sigma (C6645). Unless noted otherwise, compound concentrations were as follows: 20 μM CMLDBU6128, 5 μM ST-246, and 1 μg/ml araC.
CMLDBU6128 synthesis. (i) 4-Acetyl-3-phenyl-6-(trimethylsilyl)-2,3,7,8-tetrahydro-1H-pyrido[1,2-c]pyrimidin-1-one (3c).
Compound 1c (110 mg, 0.32 mmol, 1.0 equivalent) and auric acid trihydrate (13 mg, 0.03 mmol, 10 mol%) were weighed into a 2-dram reaction vial which was immediately capped, purged, and flushed with argon. Anhydrous 1,2-dichloroethane (3.2 ml; 0.1 M) was introduced by use of a syringe, and the sealed reaction mixture was heated to 80°C for 14 h. The reaction mixture was allowed to cool and directly purified via flash column chromatography (35% to 40% to 50% to 75% ethyl acetate in hexanes). Compound 3c (103 mg; 94%) was isolated as a tan solid.
(ii) 4-Acetyl-3-phenyl-2,3,7,8-tetrahydro-1H-pyrido[1,2-c]pyrimidin-1-one (3a).
Compound 3c (150 mg, 0.44 mmol, 1.0 equivalent) and AgF (112 mg, 0.88 mmol, 2.0 equivalents) were weighed into a foil-wrapped 2-dram reaction vial equipped with a stir bar. The vial was flushed with nitrogen, and 2.2 ml of a 10:2:2:1 mixture of tetrahydrofuran (THF), methanol (MeOH), dimethyl sulfoxide (DMSO), and H2O was added. The vial was capped, and the reaction mixture was stirred at room temperature overnight. The reaction mixture was filtered through a cotton plug, with rinsing with dichloromethane. After solvent removal, the resultant residue was purified by flash column chromatography (20% to 40% to 70% ethyl acetate in hexanes) to afford compound 3a as an orange foam (62 mg; 53%).
Fluorescence plate reader assays.
Well fluorescence was measured on a Tecan Infinite M1000 fluorescence plate reader (Tecan Group Ltd.), using the following excitation/emission wavelengths (in nm, with a 5-nm band-pass length): 515/530 for Venus and 587/610 for mCherry. For endpoint assays, cells and infections were maintained in recommended medium at 37°C and 5% CO2 prior to measurement. For time course assays, cells were seeded and infected in buffered Opti-MEM medium (Gibco). Plates were sealed with optical film and placed in a fluorescence plate reader with the chamber set to 37°C for automated measurement.
Venus mRNA and DNA analyses.
Venus RNA and DNA quantitations were carried out as described in detail elsewhere (K. Dower et al., submitted for publication). Briefly, reverse transcription-PCR (RT-PCR) analysis was performed directly on DNase-treated cell lysates (Ambion) by use of the following Venus TaqMan primer and probe set: AAAGACCCCAACGAGAAGC (forward primer), GTCCATGCCGAGAGTGATC (reverse primer), and 6-carboxyfluorescein (FAM)-TGCTGGAGTTCGTGACCGCC-IBFQ (probe). DNA analysis used this primer and probe set in direct PCR mixtures with cell lysates generated with a filtered home-brewed lysis buffer (10 mM glycine, 1% Triton X-100, pH 2.5, HCl).
35S metabolic labeling.
35S metabolic labeling was carried out using standard methods (4). Gels were stained with Coomassie blue before 35S visualization and measurement using a transilluminator.
Isolation of drug-resistant viruses.
Drug-resistant viruses were generated by serial passage in the presence of compound. The initial infection used 1 × 105 PFU of late Venus (LV) reporter virus in triplicate wells of confluent A549 cells in a 6-well plate (in duplicate for independent selection pools). CMLDBU6128 was added to a 20 μM final concentration, and infections proceeded for 2 days. A portion of the resulting crude virus preparation (∼1/20) was applied to fresh cells in the presence of 20 μM compound. This process was repeated for a total of six passages, with crude virus preparations pooled at passage 2. The final two passages were done with a superinhibitory compound concentration of 40 μM. Clonal isolates from passage 6 were isolated by two plaque purifications.
Viral genome sequencing.
Total DNAs from crude virus preparations were isolated using Qiagen genomic DNA isolation kits. Sequencing libraries were prepared using an Illumina gDNA sequencing kit with 5 μg of DNA input per sample. Libraries were validated and measured by quantitative PCR (qPCR) and then sequenced on an Illumina IIx genome analyzer for 37 cycles (parental virus) or 84 cycles (drug-resistant clones). Resulting reads were mapped to a modified version of the vaccinia virus WR genome (GenBank accession no. NC_006998.1; parental strain) containing the late Venus insertion between J4L and J5R or to the parental consensus sequence (drug-resistant clones), using MAQ (17). Mutations were called using MAQ and then filtered and analyzed using SNiPnfo (not published). Between 47,668 and 5,403,566 sequencing reads were generated for each virus. Of those, between 15,472 and 417,317 mapped to vaccinia virus. Coverage ranged from 5.6× (97.5%) to 89.6× (100%). Mutations in each virus were analyzed separately and then correlated with their clones (e.g., DR1a with DR1b). For each clonal pair, only one coding mutation was detected: V576G in J6R, with an average depth of 33 (DR1), and A954V in J6R, with an average depth of 76 (DR2).
Marker recovery.
Homologous recombination was performed as described previously (7). Briefly, HeLa cells were infected with late Venus vaccinia virus at an MOI of 0.1 for 1 h and then transfected with 3 μg of homologous recombination cassette by use of Lipofectamine 2000. To cover the mutation sites, the DR1 cassette included strain WR nucleotides 84589 to 85589 (GenBank accession no. NC_006998.1); the DR2 cassette included nucleotides 85724 to 86724. At 2 h posttransfection, 20 μM CMLDBU6128 was added to each well, and at 24 h posttransfection, fresh drug was added. At 48 h posttransfection, the cells were collected and crude virus preparations were made and serially diluted on Vero cells with medium containing 20 μM CMLDBU6128. The plaques were fixed and counted at 4 days postinfection.
RESULTS
Screening and identification of CMLDBU6128.
We generated single- and multiple-reporter vaccinia viruses with fluorescent proteins under the control of temporally regulated early (C11R), intermediate (G8R), and/or late (F17R) viral promoters (Table 1) that can be used to monitor stages of vaccinia virus gene expression (7). For screening, we used a dual reporter virus that contained late mCherry (red) and early Venus (LREV virus). Compounds were screened for the ability to reduce viral reporter gene expression, determined by well fluorescence, following high-MOI infection of A549 cell monolayers (Fig. 1A). The compound library was developed at CMLDBU and consisted of 2,070 compounds with high stereochemical and positional variation to provide broad coverage of chemical space in a limited screen.
Table 1.
Replication-competent vaccinia virus reporter viruses (based on strain Western Reserve)
| Name | Description | Fluorescent reporter(s) | Viral promoter(s) |
|---|---|---|---|
| PLV | Promoterless Venus | Venus | None |
| EV | Early Venus | Venus | C11R |
| IV | Intermediate Venus | Venus | G8R |
| LV | Late Venus | Venus | F17R |
| LR | Late red | mCherry | F17R |
| LREV | Late red, early Venus | mCherry, Venus | F17R (mCherry), C11R (Venus) |
| TrpV | Triple virus | TagBFP, mCherry, Venus | F17R (TagBFP), G8R (mCherry), C11R (Venus) |
| Venus-A4L | Venus-A4L fusion | Venus-A4L | Endogenous A4L |
Fig 1.
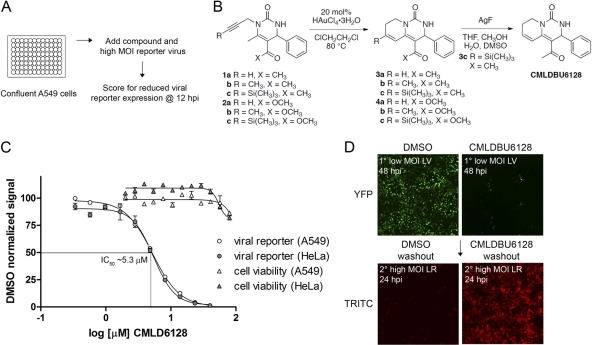
(A) Screen summary. Confluent A549 cells were infected with a fluorescence reporter vaccinia virus at a high MOI. A diversity-oriented synthesis library was screened for compounds that reduced reporter expression at 12 h postinfection. (B) Structure and synthesis of CMLDBU6128, identified from this screen. (C) Late viral reporter expression from high-MOI infections of A549 and HeLa cells (12 h postinfection) and cell cytotoxicity determination (24 h) over a dose range of CMLDBU6128. (D) Sequential low-MOI–high-MOI infection. A549 cells were infected with Venus-expressing reporter virus at a low MOI, and infections proceeded for 48 h (primary infection). Medium exchange was followed by high-MOI infection with an mCherry-expressing reporter virus for 24 h (secondary infection). Images were taken using a 2.5× objective.
None of the library compounds reduced early Venus expression at 12 h postinfection, and the majority also did not prevent late mCherry expression. However, three compounds reduced late mCherry expression to 29 to 56%. Of these, one did not show activity upon resynthesis, and another was antiviral but had a cell-type-specific cytopathic effect. The third, CMLDBU6128, was chosen for further characterization. The structure of the pyridopyrimidone compound CMLDBU6128 and the optimized synthesis scheme that was developed are depicted in Fig. 1B.
CMLDBU6128 is an antiviral compound.
The effect of CMLDBU6128 on a single-reporter virus containing late Venus (LV virus) is shown in Fig. 1C. The 50% inhibitory concentration (IC50) for LV reporter expression at 12 h postinfection was ∼5.3 μM, and the IC90 was ∼10.4 μM, with similar results obtained in both A549 and HeLa cells. Little to no cell cytotoxicity was observed at 24 h for up to 80 μM CMLDBU6128 in either cell type. We therefore chose a compound concentration of 20 μM for subsequent experiments. The noncytotoxic, antiviral properties of CMLDBU6128 are further illustrated in Fig. 1D. A549 cells were infected with LV virus at a low MOI, and the infection was allowed to spread for 48 h. With DMSO but not CMLDBU6128, this resulted in a monolayer of Venus-expressing cells from complete virus transmission. Cell monolayers were then subjected to medium exchange to drug-free conditions and to secondary, high-MOI infection with a single-reporter virus containing late mCherry (late red [LR] virus). Twenty-four hours following secondary infection, mCherry-expressing cells were observed only with the CMLDBU6128-limited primary infection, as vaccinia virus-infected cells do not support superinfection (3, 6). The results show that CMLDBU6128 limits virus spread and that viable, infection-supporting cells remain in this limited infection.
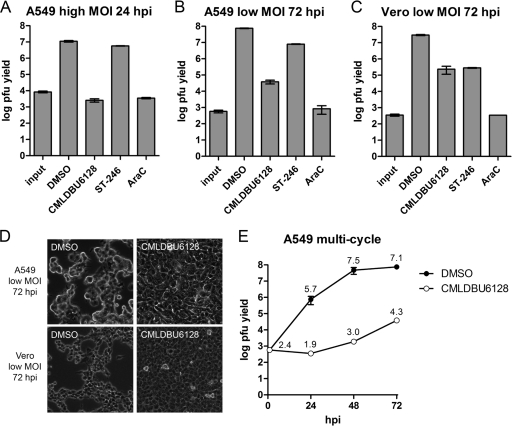
To confirm the antiviral effect more directly, low- and high-MOI viral growth assays using wild-type vaccinia virus strain Western Reserve were performed (Fig. 2). Two reference compounds were included: ST-246, which inhibits the egress of infectious viral progeny, and AraC, a nucleoside analog and inhibitor of DNA replication (31, 33). In high-MOI assays with A549 cells (Fig. 2A), the virus yield was reduced by 3.6 log (CMLDBU6128), 0.3 log (ST-246), and 3.5 log (AraC) at 24 h. In low-MOI assays with A549 cells (Fig. 2B), the virus yield was reduced by 3.3 log (CMLDBU6128), 1.0 log (ST-246), and 4.5 log (AraC) at 72 h. Similar results were obtained in low-MOI assays with Vero cells, indicating that the inhibitory effect was not cell or species specific (Fig. 2C) (1.9-, 2.0-, and 4.9-log reductions with CMLDBU6128, ST-246, and AraC, respectively). Microscopy of the 72-h multicycle endpoints showed intact cell monolayers with the inhibition of low-MOI virus spread by CMLDBU6128 (Fig. 2D). Consistent with inhibition of late viral reporter gene expression, CMLDBU6128 was antiviral in viral proliferation assays. This was further confirmed in a multicycle growth assay, where it reduced infectious progeny formation 24, 48, and 72 h following low-MOI infection of A549 cells (Fig. 2E).
Fig 2.
(A) Single-cycle growth yields on A549 cells. (B and C) Multicycle growth yields on A549 and Vero cells, respectively. (D) Phase-contrast microscopy of cell monolayers of A549 and Vero cells at the endpoint of multistep growth for panels B and C (10× objective). All growth curves were created with vaccinia virus strain Western Reserve. (E) Time course of vaccinia virus growth in the presence of DMSO (filled circles) or CMLDBU6128 (open circles). The log value is listed above each point. Compound concentrations used were as follows: 20 μM CMLDBU6128, 5 μM ST-246, and 1 μg/ml of AraC.
CMLDBU6128 interferes with intermediate and late viral gene expression.
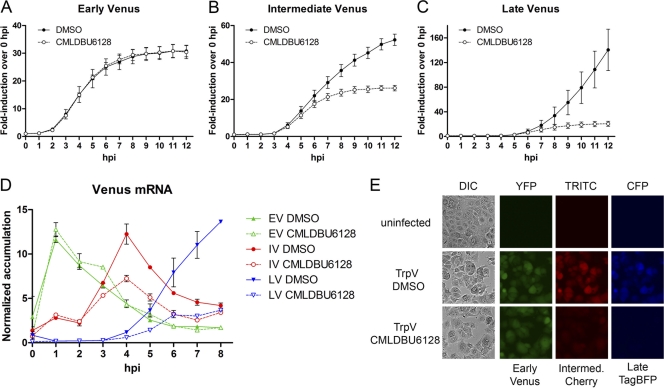
We next tested the effect of CMLDBU6128 on a panel of single-reporter viruses harboring early Venus (EV virus), intermediate Venus (IV virus), or late Venus (LV virus) (Table 1). A fluorescence plate reader was used for automated hourly monitoring of Venus expression after high-MOI infection of A549 cells (Fig. 3). Reporter expression by the canonical promoter elements occurred with the anticipated timing and magnitude, and there was no detectable Venus expression with a virus that contained Venus with no preceding promoter element (promoterless Venus [PLV] virus) (Fig. 3A to C and data not shown). EV reporter expression was unaffected by CMLDBU6128, consistent with the lack of effect on early reporter expression in our initial screen (Fig. 3A). In contrast, IV reporter expression initiated correctly but reached only half-maximal levels (Fig. 3B). A similar but more pronounced defect was observed with LV reporter expression, with maximum fluorescence reduced by 85% (Fig. 3C). Virtually identical results were observed when the compound was added 4 h before virus, as opposed to simultaneously, indicating that these differences were not due to kinetic limitations of drug uptake or action (data not shown).
Fig 3.
(A) A549 cells were infected with early Venus (EV; A), intermediate Venus (IV; B), and late Venus (LV; C) reporter viruses at a high MOI in the presence of DMSO or CMLDBU6128. Viral reporter fluorescence was measured hourly for 12 h. (D) Promoter-dependent Venus mRNA accumulation over a time course of infection with EV, IV, or LV virus. (E) Microscopy of A549 cells after high-MOI infection with TrpV reporter virus, which contains early Venus, intermediate mCherry, and late TagBFP in a single virus. Uninfected and DMSO- or CMLDBU6128-treated cells at 12 h postinfection are shown.
We analyzed reporter mRNA accumulation to determine if this was the underlying defect in reporter expression. This procedure utilizes quantitative RT-PCR and normalization to infection with the promoterless Venus virus to determine promoter-dependent Venus mRNA accumulation (7). Over an 8-h time course, peak Venus mRNA accumulation for EV, IV, and LV virus infections occurred at 1, 4, and 8 h postinfection, respectively (Fig. 3D). EV reporter mRNA accumulation was unaffected by CMLDBU6128; however, IV and LV reporter mRNA accumulation initiated correctly but failed to reach maximal levels. Peak IV reporter mRNA was reduced by 41%, and peak LV reporter mRNA was reduced by 73%, mirroring the reporter expression defects described above.
The results obtained using these single-reporter viruses were validated by microscopy using a multireporter triple virus (TrpV) which contains early Venus, intermediate mCherry, and late TagBFP (7). A549 cells were infected with TrpV at a high MOI and visualized at 12 h postinfection (Fig. 3E). Consistent with the results described above, early Venus expression was unaffected and intermediate mCherry and late TagBFP expression was reduced uniformly in the infected cell population. Phase-contrast imaging following this high-MOI infection showed that despite this, CMLDBU6128 was unable to prevent the cell morphological changes associated with infection-induced cytopathic events.
CMLDBU6128 does not prevent viral DNA replication or factory formation.
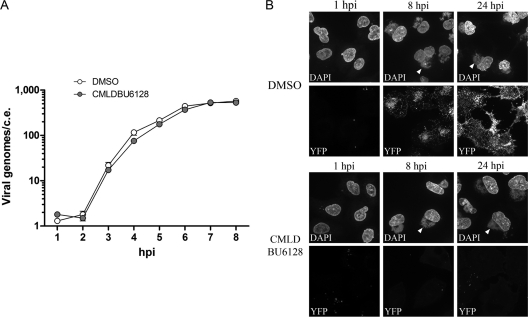
Viral DNA replication resets the transcriptional landscape to postreplicative gene transcription, and consequently, DNA replication inhibitors block intermediate and late viral gene expression (16, 32). To monitor viral DNA replication, we used quantitative PCR to track Venus DNA copy numbers after infection of A549 cells with LV virus. Figure 4A is a log-scale plot of Venus copies per cell equivalent (c.e.) showing that viral DNA replication occurred normally in the presence of CMLDBU6128. The onset of DNA replication was between 2 and 3 h postinfection, and the number of Venus DNA copies/c.e. increased from 1 h postinfection to 8 h postinfection, from 1 to 2 copies to 574 and 529 copies with DMSO and CMLDBU6128, respectively.
Fig 4.
(A) Venus DNA copy number per A549 cell equivalent (c.e.) after high-MOI infection with LV reporter virus. Venus copy numbers were calculated from a standard curve of Venus-containing plasmid. (B) High-magnification microscopy after infection of A549 cells with Venus-A4L virus. DMSO- or CMLDBU6128-treated cells are shown at 1, 8, and 24 h postinfection. Cells were fixed and stained with DAPI before visualization of viral DNA factories (DAPI) and Venus-A4L (yellow fluorescent protein [YFP]). Images were taken using a 63× objective. Arrowheads point to representative perinuclear viral factories.
We next used high-magnification microscopy to determine if viral factory formation occurred normally in the presence of CMLDBU6128. Figure 4B depicts images obtained 1, 8, and 24 h after infection of A549 cells with a vaccinia virus harboring a Venus fusion of the late-expressed viral core protein A4L (Venus-A4L virus). A DAPI (4′,6-diamidino-2-phenylindole)-staining perinuclear viral factory was evident by 8 h and persisted to 24 h postinfection in both DMSO- and CMLDBU6128-treated infections. However, only with DMSO treatment was Venus-A4L visible as a punctate signal in and around viral factories at 8 h postinfection. By 24 h, this signal had spread throughout the cell as Venus-A4L-containing progeny viruses trafficked toward the cell membrane (11, 26). CMLDBU6128-induced defects in intermediate and late gene expression were therefore uncoupled from any gross defects in viral DNA replication or factory formation.
A CMLDBU6128-arrested infection has a general loss of protein synthesis.
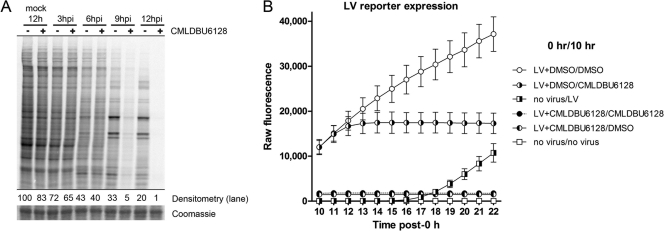
To investigate more globally the effects of CMLDBU6128 on viral gene expression, we used [35S]methionine-cysteine pulse labeling at various times postinfection to monitor active protein translation (Fig. 5A). Cellular protein synthesis in mock-infected cells was unaffected following 12 h of treatment with CMLDBU6128. In control vaccinia virus-infected cells, a characteristic shutoff of host gene expression and transition to viral protein synthesis were observed. CMLDBU6128 did not prevent this host cell shutoff; however, viral protein synthesis was generally reduced. The appearance of viral proteins was diminished slightly at 6 h postinfection and more profoundly at late time points. Despite the virtual absence of active protein synthesis, these infection-arrested cells continued to be viable in metabolic activity assays 12 and 24 h after infection (data not shown).
Fig 5.
(A) [35S]methionine labeling of cells to visualize active translation. Densitometry values are provided normalized to 12-h mock-infected cells (−). A representative slice of the Coomassie blue-stained gel is also shown. (B) Time course analysis of Venus expression in A549 cells subjected to various treatments. The legend indicates the treatments given at 0 h and 10 h. Reporter fluorescence was measured hourly for 12 h after the initial 10-h treatment (until 22 h). “LV” indicates high-MOI infection with the late Venus reporter virus. For example, LV+DMSO/CMLDBU6128 indicates an initial treatment of high-MOI LV reporter virus in the presence of DMSO, followed 10 h later by medium removal and replacement with CMLDBU6128-containing medium.
Because vaccinia virus gene expression occurs in a cascade mechanism, with a requirement of preceding gene expression products for subsequent gene expression, we tested whether delayed drug addition could inhibit established late gene expression. We also tested whether drug withdrawal from an initially inhibited infection would alleviate inhibition. The LV reporter virus was used to allow for quantitative analysis (Fig. 5B). Cells received an initial 0-h treatment followed by medium removal and a second treatment at 10 h, and LV reporter fluorescence was then measured hourly for 12 h (from 10 h to 22 h). Unimpeded infections showed high Venus expression at 10 h, which continued to increase over the time course. CMLDBU6128 addition to an unimpeded infection at 10 h resulted in a flattening of Venus expression within 2 h of addition, showing that the compound could inhibit late gene expression following diffusion into infected cells.
In the complementary experiment, a CMLDBU6128-arrested infection had significantly reduced Venus expression at 10 h, as seen above. However, drug removal at this time did not result in any detectable rise in Venus expression. Indeed, the level of Venus expression following drug removal was indistinguishable from that with sustained drug inclusion. For comparison, infection of A549 cells with LV virus at the 10-h time point showed a clear rise in reporter expression over the course of this analysis. The results indicate that CMLDBU6128 can inhibit late gene expression even after it is established. They also suggest that a CMLDBU6128-arrested infection may be irretrievably defunct.
Mutations in the large subunit of the viral RNA polymerase bypass inhibition.
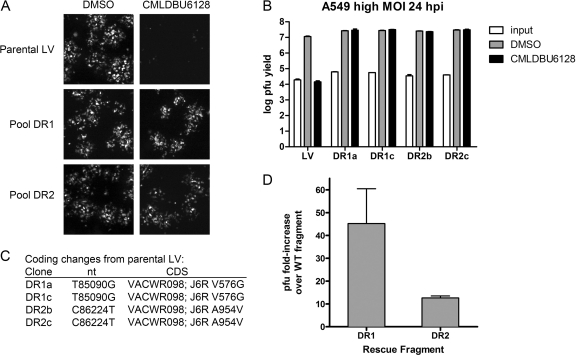
We next determined whether drug-resistant virus could be obtained through serial passage of LV virus in the presence of CMLDBU6128. After six serial passages, this resulted in clear drug resistance, as seen in low-MOI viral spread assays of two independent selection pools (Fig. 6A, drug-resistant pools DR1 and DR2). Two clones from each of these pools were randomly chosen and plaque purified to yield clones DR1a, DR1c, DR2b, and DR2c. These viruses were completely resistant to CMLDBU6128 in single-cycle growth assays (Fig. 6B).
Fig 6.
(A) Fluorescent foci from low-MOI infection of A549 cell monolayers with parental LV virus or two selection pools (DR1 and DR2) in the absence or presence of CMLDBU6128. Images were taken using a 2.5× objective. (B) Single-cycle growth yields for A549 cells infected with four plaque-purified viruses: two from pool DR1 (DR1a and DR1b) and two from pool DR2 (DR2b and DR2c). (C) Coding changes identified by whole-genome sequencing. DR1a and DR1b had an identical J6R V576G mutation, and DR2b and DR2c had an identical J6R A954V mutation. (D) Marker rescue experiments using fragments containing the DR1 (J6R V576G) mutation or DR2 (J6R A954V) mutation. The graph shows fold increases in viral progeny following 2 days of incubation with CMLDBU6128 relative to mock rescue using the corresponding fragment from the wild-type virus.
A stock of parental LV virus and all four clones were sequenced by Illumina whole-genome sequencing. Compared to the reference GenBank sequence for vaccinia virus strain Western Reserve, the parental LV virus had four potential differences: G1091A, A1092G, T194371C, and G194372T. These fell within terminal repeat regions of the viral genome and were likely mapping errors. In addition, all four drug-resistant viruses contained a silent mutation (G185627T; VACWR208 gene, amino acid P140P), which we presume was acquired by LV virus in an intermediate passage. Each drug-resistant pool yielded a single, different coding change in J6R, the gene encoding the large subunit of the viral RNA polymerase. DR1a and DR1c had a T85842G mutation, which encodes a V576G mutation, and DR2b and DR2c had a C86976T mutation, which encodes an A954V mutation (Fig. 6C).
We confirmed that these unique DR1 and DR2 mutations could confer drug resistance in a marker rescue experiment (Fig. 6D). A549 cells were infected with wild-type virus and subsequently transfected with an ∼1-kb PCR fragment surrounding either the J6R V576 or J6R A954 region generated from the corresponding drug-resistant isolate or wild-type control virus. CMLDBU6128 was added to these infections, and viral harvest was performed 2 days later. Viral yield assays showed that rescue with the DR1 fragment resulted in a 40-fold increase in virus yield. Rescue with the DR2 fragment resulted in a 10-fold increase in yield. Taken together, these data indicate that either a V576G mutation or an A954V mutation in J6R can confer resistance to CMLDBU6128.
CMLDBU6128 has activity against multiple orthopoxviruses.
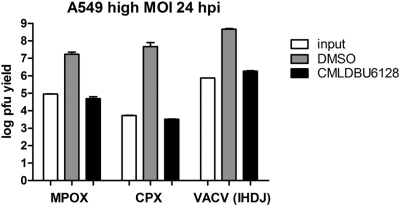
J6R is highly conserved among orthopoxviruses, with >96% identity between vaccinia, monkeypox, cowpox, and variola viruses. This suggested that CMLDBU6128 might act as a broad-spectrum inhibitor. We tested the effect of CMLDBU6128 on orthopoxviruses other than vaccinia virus strain Western Reserve. The results of single-cycle growth assays of monkeypox virus Zaire 1979 (MPOX), cowpox virus Brighton red (CPX), and vaccinia virus strain IHDJ (VACV IHDJ), using A549 cells, are shown in Fig. 7. CMLDBU6128 inhibited the replication of all of these viruses, reducing viral yields by 2.5 log (MPOX), 4.0 log (CPX), and 2.4 log (VACV IHDJ). The replication of these viruses was also inhibited on Vero cells (data not shown). These findings are consistent with drug targeting of a conserved orthopoxvirus function and demonstrate that CMLDBU6128 has broad-spectrum efficacy against orthopoxviruses.
Fig 7.
Single-cycle growth yields for A549 cells infected with monkeypox virus Zaire 1979 (MPOX), wild-type cowpox virus (CPX), or vaccinia virus (VACV) IHDJ in the absence or presence of CMLDBU6128.
DISCUSSION
We have identified a novel antiorthopoxvirus compound through screening of a diversity-oriented synthesis library. Using a series of recently developed systems, we provide evidence that the pyridopyrimidinone compound CMLDBU6128 interferes with productive viral intermediate and late gene expression. This drug phenotype shares similarities with those for the structurally unrelated inhibitors isatin β-thiosemicarbazone (IBT) and distamycin A, although the accumulated evidence suggests that different mechanisms are at play (2, 15, 23). The presence of CMLDBU6128 at the onset of infection caused a defect partway through intermediate gene expression; however, established late gene expression could also be inhibited, and drug removal did not result in a rescue of late gene expression. We speculate that the latter effect is due to the virtually complete host cell shutoff during arrested infection, which may not allow for a restart of the highly orchestrated events required for viral gene expression. Taken together, these observations are promising for a therapeutic effect, as the inhibitory window does not appear to be narrow and there is the potential for persistent inhibition beyond peak drug exposure.
Structurally, CMLDBU6128 does not resemble a nucleoside analog, although drug resistance mutations map to the viral RNA polymerase large subunit gene J6R. One explanation may be that the resistance mutations could alter interactions between J6R and other polymerase subunits. In this model, resulting changes in the polymerase complex would allow for productive transcription despite the presence of CMLDBU6128, depending on the different requirements for temporal gene expression (2). However, an alternative explanation is that CMLDBU6128 is a general inhibitor of viral transcription but that early transcription, which is believed to occur within the viral core shortly after entry, is physically inaccessible to drug (14, 29). The observed effects of CMLDBU6128 on stage-specific transcription would then be due to drug inaccessibility of viral cores or early factories. These possibilities are under investigation, as are additional studies aimed to help elucidate the drug's mechanism of action.
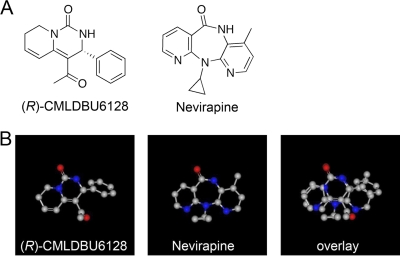
Given these findings, we note that significant structural similarity exists between CMLDBU6128 and nevirapine, an approved nonnucleoside reverse transcriptase inhibitor (NNRTI) of HIV (Fig. 8A). By modeling, CMLDBU6128 appears to be similar to nevirapine: it is a planar 3-ring compound with a single major side group located on the center ring, and the two compounds show 73% overlap when aligned (Fig. 8B). The similarity of these compounds and the disparity between heteromeric HIV reverse transcriptase and multimeric vaccinia virus RNA polymerase may indicate that CMLDBU6128 is a general inhibitor of vaccinia virus transcription, potentially targeting J6R function directly. Absent any biochemical data, this remains speculative. However, this structural class of compounds may represent a chemical space for further exploration in the targeting of viral polymerases.
Fig 8.
(A) Chemical structures of the (R)-enantiomer (R)-CMLDBU6128 (left) and of nevirapine (right). (B) Computationally generated overlay of (R)-CMLDBU6128 and nevirapine. The overlay was generated using the OpenEye Scientific Software shape similarity comparison program ROCS.
In summary, we have identified a small-molecule inhibitor with broad-spectrum activity against orthopoxviruses. The discovery of this compound in a library with increased chemical diversity indicates that such libraries can yield promising compounds for the development of new infectious disease therapeutics.
ACKNOWLEDGMENTS
We thank Robert Jordan (SIGA Technologies, Corvallis, OR) for providing ST-246, Aaron Beeler (Center for Chemical Methodology and Library Development, Boston University) for chemistry support, Mohamed Ragaa Mohamed (Grant McFadden laboratory) for advice on recombinant virus construction, and Arthur J. Goff, Kenny Lin, and Carly Wlazlowsky for guidance on biosafety level 3 experiments. We thank OpenEye Scientific Software for the use of their suite of products.
K.D. and C.M.F. were supported by the Postgraduate Research Participation Program and the U. S. Army Research and Medical Command administered by the Oak Ridge Institute for Science and Education (ORISE). S.S. acknowledges funding from the NIH (P41 GM086180).
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Andrei G, et al. 2006. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 80:9391–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Broyles SS. 2003. Vaccinia virus transcription. J. Gen. Virol. 84:2293–2303 [DOI] [PubMed] [Google Scholar]
- 3. Christen L, Seto J, Niles EG. 1990. Superinfection exclusion of vaccinia virus in virus-infected cell cultures. Virology 174:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Connor JH, Lyles DS. 2002. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 76:10177–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Clercq E. 2001. Vaccinia virus inhibitors as a paradigm for the chemotherapy of poxvirus infections. Clin. Microbiol. Rev. 14:382–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doceul V, Hollinshead M, van der Linden L, Smith GL. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science 327:873–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dower K, Rubins KH, Hensley LE, Connor JH. 2011. Development of vaccinia reporter viruses for rapid, high content analysis of viral function at all stages of gene expression. Antiviral Res. 91:72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drazen JM. 2002. Smallpox and bioterrorism. N. Engl. J. Med. 346:1262–1263 [DOI] [PubMed] [Google Scholar]
- 9. Farlow J, Ichou MA, Huggins J, Ibrahim S. 2010. Comparative whole genome sequence analysis of wild-type and cidofovir-resistant monkeypoxvirus. Virol. J. 7:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fenner F. 1982. A successful eradication campaign. Global eradication of smallpox. Rev. Infect. Dis. 4:916–930 [DOI] [PubMed] [Google Scholar]
- 11. Huang CY, et al. 2008. A novel cellular protein, VPEF, facilitates vaccinia virus penetration into HeLa cells through fluid phase endocytosis. J. Virol. 82:7988–7999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huggins J, et al. 2009. Nonhuman primates are protected from smallpox virus or monkeypox virus challenges by the antiviral drug ST-246. Antimicrob. Agents Chemother. 53:2620–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jordan R, et al. 2009. ST-246 antiviral efficacy in a nonhuman primate monkeypox model: determination of the minimal effective dose and human dose justification. Antimicrob. Agents Chemother. 53:1817–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kates J, Beeson J. 1970. Ribonucleic acid synthesis in vaccinia virus. I. The mechanism of synthesis and release of RNA in vaccinia cores. J. Mol. Biol. 50:1–18 [DOI] [PubMed] [Google Scholar]
- 15. Katz E, Margalith E, Winer B, Goldblum N. 1973. Synthesis of vaccinia virus polypeptides in the presence of isatin-beta-thiosemicarbazone. Antimicrob. Agents Chemother. 4:44–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keck JG, Baldick CJ, Jr, Moss B. 1990. Role of DNA replication in vaccinia virus gene expression: a naked template is required for transcription of three late trans-activator genes. Cell 61:801–809 [DOI] [PubMed] [Google Scholar]
- 17. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McFadden G. Killing a killer: what next for smallpox? PLoS Pathog. 6:e1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McFadden G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moss B. 2007. Poxviridae: the viruses and their replication, p 2905–2947 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 21. Parker S, Nuara A, Buller RM, Schultz DA. 2007. Human monkeypox: an emerging zoonotic disease. Future Microbiol. 2:17–34 [DOI] [PubMed] [Google Scholar]
- 22. Prichard MN, Kern ER. 2005. Orthopoxvirus targets for the development of antiviral therapies. Curr. Drug Targets Infect. Disord. 5:17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prins C, Cresawn SG, Condit RC. 2004. An isatin-beta-thiosemicarbazone-resistant vaccinia virus containing a mutation in the second largest subunit of the viral RNA polymerase is defective in transcription elongation. J. Biol. Chem. 279:44858–44871 [DOI] [PubMed] [Google Scholar]
- 24. Quenelle DC, et al. 2007. Synergistic efficacy of the combination of ST-246 with CMX001 against orthopoxviruses. Antimicrob. Agents Chemother. 51:4118–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reed KD, et al. 2004. The detection of monkeypox in humans in the Western Hemisphere. N. Engl. J. Med. 350:342–350 [DOI] [PubMed] [Google Scholar]
- 26. Rietdorf J, et al. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992–1000 [DOI] [PubMed] [Google Scholar]
- 27. Rimoin AW, et al. 2011. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 107:16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roberts KL, Smith GL. 2008. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 16:472–479 [DOI] [PubMed] [Google Scholar]
- 29. Rosemond-Hornbeak H, Moss B. 1975. Inhibition of host protein synthesis by vaccinia virus: fate of cell mRNA and synthesis of small poly(A)-rich polyribonucleotides in the presence of actinomycin D. J. Virol. 16:34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seet BT, et al. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377–423 [DOI] [PubMed] [Google Scholar]
- 31. Taddie JA, Traktman P. 1993. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutation localized to the 3′-5′ exonuclease domain. J. Virol. 67:4323–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vos JC, Stunnenberg HG. 1988. Derepression of a novel class of vaccinia virus genes upon DNA replication. EMBO J. 7:3487–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang G, et al. 2005. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J. Virol. 79:13139–13149 [DOI] [PMC free article] [PubMed] [Google Scholar]