Abstract
In macrophages, HIV-1 accumulates in intracellular vesicles designated virus-containing compartments (VCCs). These might play an important role in the constitution of macrophages as viral reservoirs and allow HIV-1 to evade the immune system by sequestration in an internal niche, which is difficult to access from the exterior. However, until now, evidence of whether internal virus accumulations are protected from the host's humoral immune response is still lacking. In order to be able to study the formation and antibody accessibility of VCCs, we generated HIV-1 with green fluorescent protein (GFP)-tagged Gag replicating in primary macrophages. Live-cell observations revealed faint initial cytosolic Gag expression and subsequent large intracellular Gag accumulations which stayed stable over days. Taking advantage of the opportunity to study the accessibility of intracellular VCCs via the cell surface, we demonstrate that macrophage internal HIV-1-containing compartments cannot be targeted by neutralizing antibodies. Furthermore, HIV-1 was efficiently transferred from antibody-treated macrophages to T cells. Three-dimensional reconstruction of electron microscopic slices revealed that Gag accumulations correspond to viral particles within enclosed compartments and convoluted membranes. Thus, although some VCCs were connected to the plasma membrane, the complex membrane architecture of the HIV-1-containing compartment might shield viral particles from neutralizing antibodies. In sum, our study provides evidence that HIV-1 is sequestered into a macrophage internal membranous web, posing an obstacle for the elimination of this viral reservoir.
INTRODUCTION
Macrophages are important HIV-1 target cells in vivo (26, 40). They can have a lifespan of several weeks to months, patrol mucosal surfaces, and infiltrate tissues at sites of inflammation and are able to cross the blood-brain barrier. Macrophages are professional antigen-presenting cells (APC) which take up and lyse pathogens to present peptides via the major histocompatibility complex class II (MHC-II) pathway to CD4+ T cells, thereby initiating the adaptive immune response (12). Thus, HIV-1-infected macrophages might play a pivotal role in virus transmission, the establishment of latent reservoirs, and AIDS pathogenesis (8, 16, 23, 26, 40).
HIV-1 productively infects macrophages (11). However, in contrast to T cells, macrophages are infected mainly by isolates using the chemokine receptor type 5 (CCR5) coreceptor, and it has recently been proposed that other nonidentified cellular restrictions are present in macrophages (1). Thus, it is conceivable that HIV-1 might have adapted to macrophage-specific cell biological features (3).
HIV-1 particles are present in large vacuoles of infected macrophages which are referred to as virus-containing compartments (VCCs) (3, 9, 21, 31, 33, 35). These contain typical markers of multivesicular bodies, like MHC-II (35) and the tetraspanins CD63 (33), CD81, CD9, and CD53 (9). From this cumulating data, it has been postulated that in macrophages, in contrast to T cells, HIV-1 accumulates in intracellular vesicles (3). In addition, it has been reported that macrophages harbor infectious HIV-1 over a long period of time (39) and that the virus has evolved strategies to inhibit the acidification of endosomal compartments, thereby preventing viral degradation (21). However, the formation and dynamics of the VCCs are still elusive, and it has also been proposed that these correspond to internally sequestered plasma membrane domains, which are connected to the cell surface via small microchannels (4, 9, 22, 45).
We aimed to reconcile these discrepancies and first studied if VCCs can be formed intracellularly and then investigated if macrophage internal HIV-1 can be targeted by antibodies. For this, we generated R5-tropic HIV-1 with an internal green fluorescent protein (GFP) tag in Gag (18) which is infectious for macrophages and characterized Gag production by time-lapse microscopy. Our results demonstrate that VCCs appear inside HIV-1-infected macrophages and stayed stable for hours and even days. In addition, HIV-1 within VCCs was not accessible to neutralizing antibodies, could be transferred to T cells, and might thus be shielded from the humoral immune response. By correlative fluorescence and transmission electron microscopy (TEM), we characterized the spatial ultrastructure of Gag accumulations. Viral particles were found within an extensive intracellular membranous network which was infrequently connected to the cell surface and was absent from uninfected cells. This membranous web might explain the poor accessibility of VCCs by antibodies and could play a role in the long-term storage of HIV-1 in macrophages.
MATERIALS AND METHODS
Plasmids and proviral constructs.
HIV-1 pUC-NL4-3 Gag internal GFP (iGFP) (18) and the R5-tropic HIV-1 pBR-NL4-3 92th014.12 (32) have been already described. For generation of an R5-tropic HIV-1 Gag-iGFP (GG), we excised and ligated the Gag-iGFP cassette using the single-cutter restriction sites BssHI and AgeI into the pBR-NL4-3 vector backbone. Subsequently, we introduced the R5-tropic V3-loop V92th014.12 (32) with the flanking restriction enzymes NheI and StuI. The same strategy was used to construct an R5-tropic version of pBR-NL4-3 internal ribosome entry site (IRES)-enhanced GFP (eGFP) (36), termed pBR-NL4-3 V92th014.12-IRES-eGFP. To generate pBR-NL4-3 Gag internal cyan fluorescent protein (iCFP), CFP was amplified with the primers Gag-iCFP_MluI (TCGACGCGTATGGTGAGCAAGGGCGAG) and Gag-iCFP_XbaI (ACGTCTAGACTTGTACAGCTCGTCCAT) and ligated into pUC-NL4-3 Gag-iGFP (18). In a second step, the Gag-iCFP fragment was ligated via BssHII and AgeI into the pBR-NL4-3 V92th012.12 backbone. Mem-yellow fluorescent protein (YFP) and CD4-YFP have been described before (2). Plasmids expressing CD81 or transferrin receptor with an N-terminal YFP tag (YFP-CD81 and YFP-transferrin receptor (TfR)) and Gag with a C-terminal CFP tag (Gag-CFP) were constructed by PCR amplification of Gag from pBR-NL4-3 or CD81 and TfR from a Jurkat cDNA library with the following primers: GAG_NheI (CCGCTAGCATGGGTGCGAGAGCGTCGGTATTAAGCGGG), GAG_AgeI (CGACCGGTGCACCTGCTCCTTGTGACGAGGGGTCGCTGC), CD81_XhoI (ATCTCGAGCTATGGGAGTGGAGGGCTGCAC), CD81_EcoRI (CAGAATTCTTATCAGTACACGGAGCTGTTCC), TfR_XhoI (CGCTCGAGCTATGATGGATCAAGCTAG), and TfR_EcoRI (CTGAATTCTTAAAACTCATTGTCAATGTCC). The corresponding ligation procedure has been described elsewhere (2). All PCR-derived inserts were sequenced to confirm sequence identity.
Cell culture, transfection, and infection.
293T cells were maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum (FCS; Invitrogen), the antibiotics penicillin and streptomycin, and 1% l-glutamin (PAA). Jurkat-LTRG R5 cells (29) and CemM7 cells which express CD4, CCR5, and CXCR4 and a tat-inducible luciferase and GFP reporter under the control of the HIV-1 long terminal repeat (LTR) (6) were cultured in RPMI 1640 medium (Gibco) supplemented with 10% FCS and antibiotics. For the generation of primary human macrophages, we first isolated peripheral blood mononuclear cells (PBMCs) from the buffy coat by Ficoll gradient centrifugation (36). Next, 15 × 106 PBMCs were seeded in 100- by 15-mm petri dishes with vents (Greiner Bio-One). Four days later, suspension cells were discarded and medium was changed. The adherent cell population was cultured three additional days to differentiate into monocyte-derived macrophages (MDMs). MDMs and PBMCs seeded for differentiation were cultured in macrophage medium: RPMI 1640 (Gibco) supplemented with 4% human AB serum (Sigma), the antibiotics penicillin and streptomycin (PAA), 10 mM nonessential amino acid solution (Invitrogen), sodium pyruvate (Gibco), vitamins (Biochrom), and 1% l-glutamine (PAA). All cells were grown at 37°C and 5% CO2. HIV-1 virus stocks were generated by standard calcium phosphate transfection of 293T cells essentially as described previously (36). For infection of macrophages, Jurkat-LTRG R5 cells, or CemM7 T cells, we quantified HIV-1 stocks by p24 enzyme-linked immunosorbent assay (ELISA) (37) and incubated the cells for 6 h with the indicated amounts of p24. Cells were subsequently washed and cultured with the appropriate medium. If not indicated otherwise, macrophages were infected with 50 ng p24 per 200,000 cells.
Viral replication, infectivity assays, and coculture experiment.
Human macrophages were isolated as described in “Cell culture, transfection, and infection” and infected with 50 ng p24 from R5-tropic HIV-1 NL4-3 Gag-iGFP per 50,000 cells or the corresponding untagged HIV-1. To assess viral spread and replication, aliquots of the infected cell culture supernatants were taken in two-day intervals and stored at −20°C. Virus production was analyzed by the quantification of p24 in the supernatants (37). Infectivity assays were done by infection of Jurkat-LTRG R5 cells with the indicated amounts of p24. Three days postinfection, cells were harvested and the number of GFP-positive cells was determined by standard fluorescence-activated cell sorting (FACS) measurements, essentially as described before (37). Macrophages were generated as described and seeded in a 24-well plate at a density of 50,000 cells/well. Postadherence macrophages were infected with 20 ng p24 from R5-tropic HIV-1 IRES-eGFP and cultured for an additional 6 days. Cells were then extensively washed to remove free virus and incubated with 20 μg/ml anti-TfR or anti-GP120 2G12 antibodies for 1 h at 37°C. After antibody removal, macrophages were cocultured with 100,000 CemM7 cells/well for 10 h. Subsequently, CemM7 cells were collected with cold 5 mM EDTA–phosphate-buffered saline (PBS) and recultured in 500 μl RPMI for up to 4 days.
Immunofluorescence, antibodies, and confocal microscopy.
A total of 100,000 macrophages were seeded in a 12-well plate with added coverslips 24 h before infection. Six days postinfection, cells were washed with PBS and fixed for intracellular staining with 2% paraformaldehyde (PFA) for 30 min at 4°C. Then, cells were permeabilized with 1% Saponin-PBS for 10 min at room temperature (RT). Cells were stained with primary anti-HIV-Gag antibody (Beckman-Coulter; KC57-RD1, mouse derived), broadly neutralizing anti-HIV-1 GP120 antibodies 2G12 (Polymun Scientific) (42), VRC01, and VRC03 (46) or anti-human-CD81 antibody (Ancell; clone 1.3.3.2.2, mouse derived) and anti-human-TfR antibody (Abcam; clone MEM-75, mouse derived) for 30 min at RT and with anti-mouse Alexa Fluor 555-conjugated secondary antibody (Invitrogen; goat derived), anti-human Alexa Fluor 633-conjugated secondary antibody (Invitrogen; goat derived), or anti-mouse phycoerythrin (PE)-conjugated secondary antibody (Invitrogen; goat derived) for 20 min at RT. Between and after staining steps, cells were washed 3 times with PBS for 5 min. For antibody accessibility experiments, nonpermeabilized MDMs were incubated at 37°C for 60 min with a final antibody concentration of 20 μg/ml. At the end of the incubation period, cells were washed and fixed with 2% PFA, and internalized antibody was detected as described above. For antibody internalization at 4°C, macrophages were essentially treated the same, except that all steps were done on ice. Finally, stained macrophages grown on coverslips were mounted on microscope slides using Mowiol mounting solution (2.4 g polyvinylalcohol, 6 g glycerin, 18 ml PBS) and imaged with a Zeiss LSM 510 Meta confocal microscope.
Live-cell microscopy.
Live-cell microscopy experiments were performed with a fully motorized Nikon Ti-Eclipse inverted microscope equipped with the hardware-based perfect focus system and the Nikon DS-Qi1 high-speed monochrome digital camera. If not indicated otherwise, 200,000 differentiated MDMs were seeded in a microdish (35 mm; Ibidi) and infected with HIV-1 GG as described above. During image acquisition, macrophages were kept in a microscope desk-mounted TokaiHit incubation chamber at 37°C and 5% CO2. If not indicated otherwise, the CFI Apochromat 60× total internal reflection fluorescence (TIRF) objective (numerical aperture [NA] 1.49) was used for imaging. Cells were excited with the Nikon Intensilight and the appropriate filters.
Transmission electron microscopy.
For correlative TEM, 100,000 macrophages were grown on culture dishes with imprinted grids (Ibidi) in order to relocalize target cells, preselected by fluorescence microscopy, and infected with HIV-1 GG. Seven days postinfection, epifluorescence and differential inference contrast (DIC) images of the infected macrophages were collected followed by fixation with 2.5% glutaraldehyde (GA) in PBS for 30 min at RT. 293T cells were also fixed with GA and processed for TEM as described below. Subsequently, cells were washed with PBS, postfixed for 30 min with 1% OsO4 in PBS, washed with double-distilled water (ddH2O), and stained with 1% uranyl acetate in water. The samples were gradually dehydrated with ethanol and embedded in Epon resin for sectioning. For pinpoint selection of relocalized samples, single cells were stamped out and sectioned as series parallel to the plain of the culture dish. Ultrathin sections (50 nm thick) were prepared using an Ultracut microtome (Reichert Jung). All sections were counterstained with 2% uranyl acetate and lead citrate. The electron micrographs were obtained with an FEI Eagle 4k HS charge-coupled-device (CCD) camera attached to an FEI Technai G2 20 Twin transmission electron microscope (FEI) at 80 kV.
FACS-FRET.
Flow cyometric measurement of fluorescence resonance energy transfer (FRET) and the according gating strategy were performed as already described (2). Briefly, 300,000 293T cells were seeded in a 6-well plate 12 h before transfection and transfected using the calcium phosphate method. Cells were harvested and assayed 1 day posttransfection for FACS-FRET analysis in a FACS CantoII flow cytometer (BD Bioscience). CFP and FRET were measured by exciting cells with the 405-nm laser, and CFP emission was collected with a standard 450/40 filter, while FRET was measured with a 529/24 filter (Semrock). YFP was excited with the 488-nm laser, and emission was collected with a 529/24 filter (Semrock). At least 10,000 CFP/YFP-positive cells were analyzed for FRET.
Image analysis and software.
Live-cell microscopical sequences were analyzed using the Nikon Elements AR version 3.1 software. Some sequences or z-stacks were processed using the two-dimensional (2-D) real-time deconvolution or three-dimenional (3-D) Huygens-based deconvolution plug-ins or the AutoQuantX2 (MediaCybernetics) software. Generally, both deconvolution softwares yielded the same results. 3-D reconstructions and colocalization analyses were performed with Bitplane Imaris version 6.4.2. In general, images were never modified apart from enhancing contrast and/or brightness. Movies were generated and compressed with the freely available ImageJ version 1.4.2 and VirtualDub version 1.9.8 software packages. Statistical analyses were performed using the GraphPad Prism version 5 software package.
RESULTS
Establishment of a macrophage-tropic HIV-1 with an internal GFP tag in Gag.
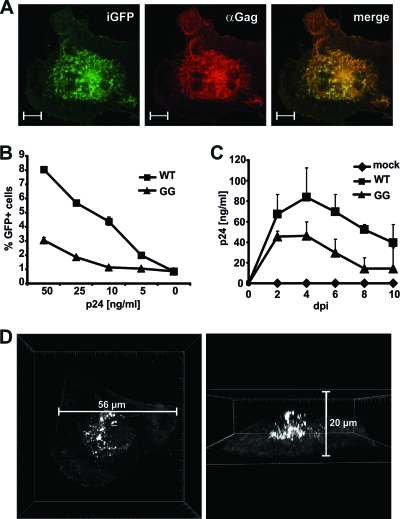
HIV-1 assembly and budding from primary T cells and most cell lines normally occurs from the plasma membrane (PM), whereas macrophages seem to accumulate viral particles intracellularly (see Fig. S1 in the supplemental material) (5, 11, 30, 31). To follow up on Gag production and trafficking in living primary human macrophages, we generated R5-tropic HIV-1 Gag-iGFP (GG). Since GFP expression resembles Gag localization (Fig. 1A) (18, 19), this virus allows for the discrimination of free cytosolic Gag from HIV-1 assembly sites, where GFP and hence Gag accumulate (see Fig. S2 in the supplemental material). Jurkat T cells expressing CCR5 could be infected in a dose-dependent manner, although infectivity was reduced 2- to 3-fold in comparison to the non-GFP-expressing virus (Fig. 1B). Similarly, viral titers were reduced in HIV-1 GG-infected macrophages, but the kinetics of viral replication was comparable to that of wild-type (WT) HIV-1, with peak p24 values at days 4 to 5 postinfection (Fig. 1C). Gag distribution in nonpermeabilized monocyte-derived macrophages (MDMs) by confocal microscopy showed faint GFP expression at the plasma membrane and the cytosol, whereas accumulating GFP and thereby Gag was apparent near the nucleus inside the macrophage (Fig. 1D), confirming the staining pattern in HIV-1-infected MDMs by a Gag-specific antibody (3). In sum, we demonstrate that R5-tropic HIV-1 GG is a useful tool to study the kinetics of Gag expression and movement in primary human macrophages in the context of infectious and replicating HIV-1.
Fig 1.
Characterization of R5-tropic HIV-1 NL4-3 Gag-iGFP (HIV-1 GG). (A) GFP expression resembles Gag distribution in HIV-1 GG-infected macrophages. Four days postinfection (dpi), MDMs were stained with mouse anti-Gag, and subcellular localization of GFP as well as Gag expression were analyzed by confocal microscopy. The scale bar indicates 10 μm. (B) Jurkat LTRG-R5 cells were infected with the indicated amounts (ng) of p24 from either R5-tropic 92th014.12 HIV-1 (WT) or HIV-1 GG, and the percentage of GFP-positive cells was determined 3 days later by flow cytometry. Means and standard deviations (SD) are derived from results from triplicate infections with two independent virus stocks. Similar results were obtained in two other infection experiments. (C) Replication kinetics of HIV-1 WT and HIV-1 GG in macrophages. MDMs were infected with 50 ng of p24, and supernatants were taken in 2-day intervals. Virus in the supernatants was determined by p24 ELISA. The graph shows mean values and SD from one experiment with MDMs infected in triplicates with two independent virus stocks. Similar results were obtained with MDMs from three other donors. (D) Representative 3-D reconstruction of Gag-iGFP distribution in HIV-1 GG-infected MDMs 4 dpi. Z-stacks of infected macrophages were recorded by fluorescence confocal microscopy and reconstructed using Bitplane Imaris 6.4.2. Typically, Gag showed a pattern of intense accumulations near the central nucleus of the macrophage, in contrast to weak expression in the cytosol and at the plasma membrane (PM).
Gag expression and movement in living HIV-1-infected macrophages.
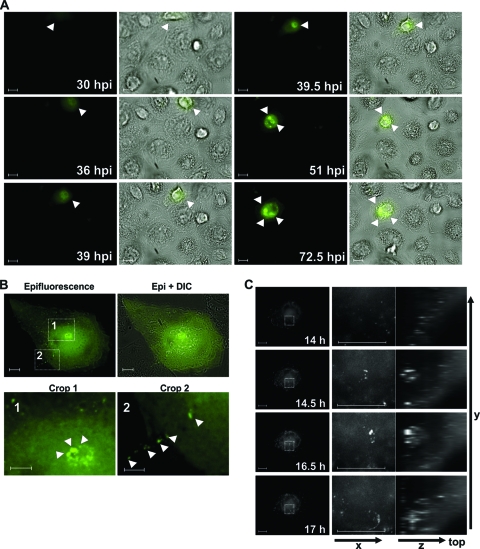
To study the formation of HIV-1 Gag, we performed live-cell microscopy of primary macrophages infected with HIV-1 GG (see Movie S1 in the supplemental material). In order to enable long-term observation of macrophages and to reduce phototoxicity as well as bleaching, we used a temporal resolution of 15 and 30 min, respectively. Cytosolic Gag was detected about 24 to 30 h postinfection (hpi). In this early phase of Gag expression, no viral particle accumulations could be observed, neither at the PM nor in the perinuclear space (Fig. 2A). At 36 to 39 hpi, first Gag accumulations were evident inside the macrophage, which then clustered into large intracellular bodies near the nucleus (Fig. 2A). Gag accumulations fused and divided several times within 30 h of observation but stayed rather stable within a single half-hour observation step (Fig. 2A; see also Movie S1). Higher magnification of regions of Gag expression near the nucleus (monitored as shown in Fig. 2B) revealed that large accumulations consisted of multiple smaller punctate Gag clusters. Although limited by the resolution of fluorescence microscopy, these smaller punctate Gag dots might represent grouped or single virions (Fig. 2B, inlay 1). Furthermore, we observed some trafficking of punctate Gag from the large central accumulation toward the periphery (see Movie S2 in the supplemental material) and transfer to an adjacent macrophage within single 15-min observation steps (Fig. 2B, inlay 2). On the other hand, we could also observe some Gag, although less pronounced, which seems to move from the periphery to more central areas in the macrophage (see Movie S2). Imaging of several macrophages did not reveal a distinct pattern of Gag movement, and faint Gag was detectable at the plasma membrane and in the cytosol, whereas intense accumulations were frequently observed in the perinuclear area (Fig. 2).
Fig 2.
Kinetics of Gag expression in HIV-1 GG-infected MDMs. (A) Macrophages were infected with HIV-1 GG and observed by time-lapse fluorescence microscopy for a period of 4 days. Images were recorded every 30 min with a 40× Plan Fluor objective (Nikon). White arrowheads indicate areas of Gag expression and accumulation. Movie S1 in the supplemental material displays the complete series. (B) Four days postinfection, an HIV-1 GG-infected macrophage was imaged every 15 min for 60 h. Two areas of interest from one representative image are highlighted by a rectangle. Inlay one shows higher magnification of a Gag accumulation near the nucleus, displaying punctate Gag expression. From this area, the smaller punctate Gag accumulations exert dynamic movement (see Movie S2 in the supplemental material). Inset 2 displays an area of cell-to-cell transfer of smaller Gag accumulations (white arrowheads) to an adjacent macrophage. (C) 4-D time lapse of an HIV-1 GG-infected macrophage. Four days postinfection, z-stacks of HIV-1 GG-infected MDMs were collected in 30-min intervals over a period of 24 h. Images depict a 3-h sequence in which a Gag accumulation (squared inset) is generated intracellularly and disperses after a few hours. Z-stacks were deconvolved with a Huygens-based deconvolution algorithm (Nikon NIS Elements AR3.1). The scale bar shows a distance of 10 μm.
Two-dimensional live-cell imaging does not allow for the spatial localization of Gag. Therefore, we expanded our analyses to four-dimensional live-cell imaging and recorded z-stacks of HIV-1 GG-infected macrophages over time (Fig. 2C). Some small punctate Gag accumulations could be detected at the periphery, but also intense GFP appeared inside the macrophage, near the nucleus and apart from the PM. In accordance with our previous results (Fig. 2A), assembled Gag stayed stable over several hours and then dispersed. From these data, we conclude that HIV-1-infected macrophages can accumulate newly formed Gag in internal VCCs.
Macrophage internal VCCs are inaccessible to antibodies.
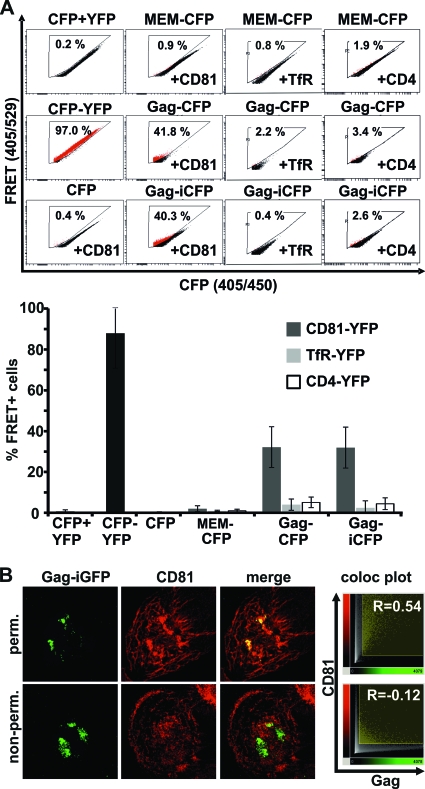
Intracellular accumulation of HIV-1 can be considered an immune evasion mechanism. However, HIV-1-containing compartments might be derived from the PM or connected via small microchannels (4, 9, 44, 45). Therefore, it is still not known if macrophage-residing HIV-1 might be protected from the immune response. To test accessibility of intracellular HIV-1 via the cell surface, we first administered an antibody which detects the assembly compartment. It has been previously reported that the tetraspanin CD81 colocalizes with HIV-1 Gag in macrophages (9), and cumulating data suggest that CD81 may have a role in HIV-1 assembly in general (15, 28). Furthermore, Gag and CD81 coimmunoprecipitated in chronically HIV-1-infected T cells (15). We first tested the association of HIV-1 Gag with CD81 in living cells. For this, we utilized a recently developed FACS-based FRET assay (2). 293T cells transfected with a Gag-CFP expression plasmid or full HIV-1 Gag-iCFP displayed a strong FRET signal with YFP-CD81 (Fig. 3A). To exclude possible false-positive FRET signals, we included a variety of controls. First, we coexpressed Gag-CFP and HIV-1 Gag-iCFP not only with CD81-YFP but also with CD4-YFP and TfR-YFP, which are unrelated membrane proteins. Second, we measured FRET between a membrane-localized control construct (MEM-CFP) and CD81-YFP, CD4-YFP, and TfR-YFP; third, we cotransfected CFP with YFP or CD81-YFP (Fig. 3A). Furthermore, we assessed expression of the Gag-CFP plasmid in macrophages, which also resulted in formation of VCCs (see Fig. S3 in the supplemental material). Thus, HIV-1 Gag alone as well as in the presence of other viral proteins is binding or in close proximity to CD81. This finding is in agreement with a recent study published by the Ono lab (17).
Fig 3.
The Gag assembly compartment in HIV-1 GG-infected MDMs is not accessible to antibodies at 4°C. (A) FACS-FRET of HIV-1 Gag with CD81, TfR, and CD4 in transfected 293T cells. Primary FACS plots show the percentages of cells scoring FRET positive. The graph displays mean values and SD from 4 to 10 independent transfections. (B) Permeabilized or untreated HIV-1 GG-infected macrophages (6 dpi) were stained with anti-CD81 antibody at 4°C to label the HIV-1 assembly compartment and test accessibility toward antibodies. Confocal z-stacks of the macrophages were recorded to display maximum intensity projections and colocalization plots (Imaris 6.4.2; Bitplane). Pearson's correlation coefficient of Gag and CD81 antibody colocalization was also calculated with Imaris 6.4.2. Displayed are results from one out of four independent experiments.
Next, we used an antibody directed against the extracellular region of CD81 to investigate if viral assembly sites in general are accessible to antibodies. Staining was performed at 4°C to reduce membrane movement and phagocytosis. As expected, CD81 strongly colocalized (R = 0.54) with Gag in HIV-1 GG-infected macrophages when cells were fixed and permeabilized prior to staining (Fig. 3B). In contrast to this, only a minor population of Gag colocalized with CD81 (R = −0.12) when living macrophages were incubated with the CD81 antibody over a period of 60 min. Thus, the HIV-1 Gag-containing assembly compartment is largely inaccessible to antibodies at 4°C.
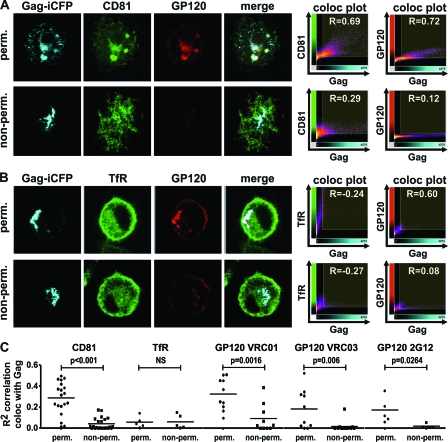
In order to mimic the in vivo situation, we additionally performed antibody feeding experiments at 37°C with anti-HIV-1 GP120 antibodies (Fig. 4). Living infected macrophages were incubated with the same CD81 antibody as used previously, an antibody binding to but not inhibiting the function of the transferrin receptor (TfR), and additionally exposed to three broadly neutralizing antibodies (2G12, VRC01, VRC03) directed against the CD4-binding site of HIV-1 GP120 (42, 46). When macrophages were fixed and permeabilized before staining, CD81 and GP120 showed a pronounced pattern of colocalization with HIV-1 Gag (Fig. 4A, top). Conversely, no colocalization could be observed when living macrophages were exposed to the antibodies for 60 min at 37°C (Fig. 4A, bottom). To control for antibody specificity of Gag and CD81, as well as Gag and GP120 colocalization, we did the same experiment with a monoclonal anti-TfR antibody (Fig. 4B). TfR did not colocalize with Gag in permeabilized or living macrophages, whereas Gag colocalized with GP120 only in the case of permeabilization. These results were confirmed and proven to be statistically highly significant by quantitative assessment of the squared Pearson's correlation coefficient (R2 value) of the colocalizations from 25 imaged macrophages (Fig. 4C). Thus, macrophage internal HIV-1 is protected from recognition by broadly neutralizing antibodies.
Fig 4.
HIV-1 within VCCs is protected from neutralizing antibodies. (A and B) HIV-1 GG-infected macrophages were either fixed and permeabilized or left untreated and incubated at 37°C with anti-CD81 (A) or anti-TfR (B) antibody and broadly neutralizing antibodies targeting GP120 (41, 46) as described in Materials and Methods. Fluorescently labeled secondary antibodies were used to detect sites of antibody binding. (C) Confocal pictures of 25 macrophages were taken at the same intensities in order to be able to quantify and compare fluorescence measurements. The Pearson's correlation coefficient of CD81, TfR, or GP120 antibody colocalizing with HIV-1 Gag was calculated with Imaris 6.4.2 (Bitplane), and the R2 values were plotted in the presented graph. A Student t test was done to assess statistical significance (GraphPad Prism 5).
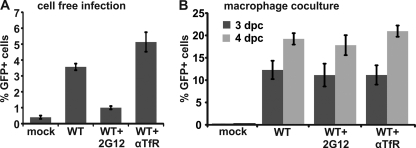
Efficient HIV-1 transfer from antibody-treated macrophages to T cells.
It has been elegantly demonstrated that macrophages transfer internal HIV-1 via virological synapses (VS) to T cells (13, 16). Thus, we set up a functional experiment as described before (16) to test whether treatment of macrophages with GP120 antibody inhibits the efficiency of HIV-1 transfer (Fig. 5). The addition of 20 μg/ml HIV-1 GP120 2G12 antibody (42) suppresses cell-free infection of CemM7 T cells nearly to background levels, whereas TfR antibody had no blocking activity (Fig. 5A). Next, macrophages were infected for 6 days with R5-tropic HIV-1-expressing GFP via an IRES element. Infected macrophages were then extensively washed and incubated for 1 h at 37°C with TfR antibody or 2G12 to potentially neutralize macrophage internal HIV-1. After antibody removal, macrophages were cocultured for 10 h with CemM7 T cells to allow cell-to-cell transfer of HIV-1. CemM7 T cells were then removed and cultured for another 3 to 4 days for productive infection and reporter gene expression. Importantly, preincubation with 2G12 did not suppress the efficiency of macrophage internal HIV-1 transfer to T cells (Fig. 5B). This result is in agreement with our previous observations from the antibody feeding experiments (Fig. 3 and 4) and establishes on a functional level that HIV-1 within macrophages is shielded from antibody neutralization.
Fig 5.
Antibody preincubation does not suppress HIV-1 transfer from macrophages to T cells. (A) A total of 100,000 CemM7 T cells were incubated with 20 ng of p24 from R5-tropic HIV-1 IRES-eGFP and 20 μg/ml of the indicated antibodies for 12 h in a total volume of 200 μl. Then, 500 μl RPMI was added and cells were cultured for two additional days. The number of GFP-positive cells was assessed by FACS measurement. Displayed are mean values and SD from results of three independent infections. (B) A total of 50,000 macrophages were infected with R5 HIV-1 IRES-eGFP and pretreated for 1 h with 20 μg/ml of the indicated antibodies at 37°C. Then, antibodies were removed and macrophages were cocultured for 10 h with CemM7 cells. CemM7 were subsequently collected by 5 mM EDTA-PBS and cultured for another 3 or 4 days. The number of GFP-positive cells was assessed by FACS. The graph shows means and standard errors of the means (SEM) from results of infections of macrophages from three different donors with two independent virus stocks. dpc, days postcoculture.
Three-dimensional reconstruction of macrophage internal Gag accumulations.
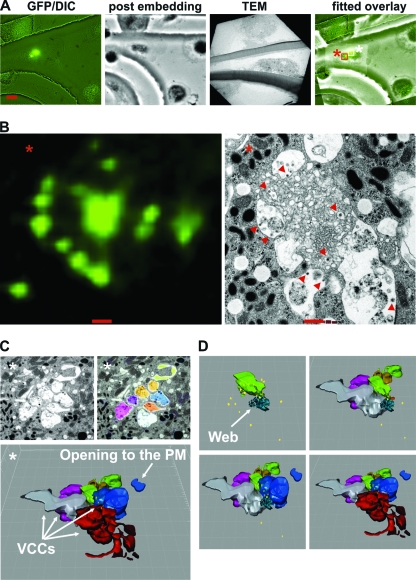
Previous electron microscopical analyses of VCCs indicated that these might be connected to the cell surface via small microchannels (4, 9, 44, 45). However, the spatial expansion of VCCs and their potential association with other cellular structures or organelles are not entirely clear. Therefore, we aimed to reconstruct larger areas of Gag accumulations by cutting serial sections and subsequent EM analyses. Infection rates of primary human macrophages in cell culture vary substantially, with usually 2 to 10% of cells being infected (37, 38) as judged by GFP expression, making it difficult to detect infected cells by TEM. Furthermore, we wanted to specifically assess the ultrastructure of an area harboring multiple Gag accumulations and image the area of prominent Gag expression within the macrophage. In order to identify such a region, we utilized correlative microscopy. For this, macrophages were seeded on culture dishes with imprinted relocalization grids and infected with HIV-1 GG. Seven days later, we took fluorescence images of infected macrophages. Usage of the grid allowed us to reidentify the corresponding region by EM, thus assessing the ultrastructural composition of larger areas of HIV-1 Gag accumulations (Fig. 6A).
Fig 6.
Three-dimensional reconstruction of macrophage internal HIV-1 accumulations. (A) Relocation of an HIV-1 GG-infected macrophage by correlative microscopy. Fluorescence images of infected macrophages grown on a culture dish with a grid were taken to relocate the infected macrophage by TEM 7 dpi. The scale bar shows a distance of 10 μm. (B) Magnification of the area of Gag accumulations, marked with a red square and an asterisk in panel A, and the ultrastructure of this specific region by TEM. Some viral particles are marked with a red arrowhead. The scale bar indicates a distance of 500 nm. (C) 3-D reconstruction of the serial sections displayed in Fig. S4 in the supplemental material. Imaris 6.4.2 (Bitplane) was used to reconstruct the colored vacuoles surrounding the central membranous web region (see also Movie S3 in the supplemental material). Viral particles are depicted as yellow dots. One large VCC, colored in blue, ends up toward a microvillus-enriched region. (D) The membrane web in light green and turquoise (first picture) is surrounded by VCCs. Viral particles (yellow dots) are present inside (see Movie S4 in the supplemental material). Similar structures corresponding to Gag accumulations were observed in two additional macrophages from this donor and in two macrophages from independent donors. We never found comparable structures in uninfected macrophages from the same donors.
Budding and mature viral particles were present in VCCs adjacent to a web of convoluted membranes (Fig. 6B). Notably, we also detected viral particles within the membranous web (Fig. 6B). Concordant with our previous observations, this macrophage harbored at least two sites of GFP accumulation (marked with red and white asterisks). We carried out closer examination of these sites (white asterisk) by analyses of 18 out of 52 50-nm-thick ultrathin serial sections (see Fig. S4 in the supplemental material). These slices were assembled to reconstruct the spatial composition of the VCCs and the associated membranous web in 3-D (Fig. 6C; see also Movie S3 in the supplemental material). The membrane web was in close contact with the surrounding VCCs. Furthermore, viral particles (marked as yellow dots) were present within the web, as well as in the VCCs (Fig. 6D; see also Movie S4 in the supplemental material). Most VCCs were membrane enclosed, separating them from each other. Nevertheless, we identified one vacuole (dark blue) next to the web (light green; turquoise), that opened out toward a microvillus-enriched region near the top of the macrophage (Fig. 6C and D; see also Movie S3 and Fig. S4, section 18, in the supplemental material). Importantly, comparable structures were never observed in multiple sections of uninfected macrophages but confirmed in at least four other HIV-1-infected macrophages of independent donors. Thus, Gag accumulations in HIV-1-infected macrophages represent assembled viral particles within VCCs and an associated complex network of membranes.
The ultrastructure of the HIV-1-containing compartments.
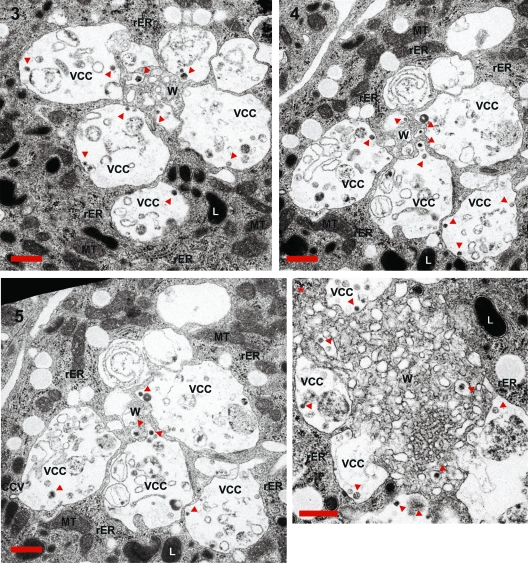
We next wanted to have a more detailed view of the HIV-1 Gag accumulations and carefully analyzed the TEM pictures of the ultrathin sections (Fig. 7). In agreement with previous reports (21, 33), we identified some assembly and budding structures, as judged by the size and morphology of viral particles, at the limiting membrane of VCCs. In contrast, no such structures were observed within the membranous web, which exclusively harbored mature virions. Some mature virions present in the VCCs seemed to be engulfed from the central membranous web (see Fig. 7, slices 4 and 5, as well as Movie S4 in the supplemental material). Although it is difficult to postulate transport features by static TEM pictures, this indicates the possibility that some of the VCC assembled particles might be imported into the membranous web or engulfed by high membrane motility. In sum, the observed compartmentalization of membranes might explain the low accessibility of internal virus toward antibodies, despite the connection of some VCCs with the plasma membrane.
Fig 7.
Ultrastructural analyses of macrophage internal HIV-1 accumulations. Serial sections 3, 4, and 5 and the slice corresponding to the other Gag accumulation present in the same macrophage (compare Fig. 6 and Fig. S4 in the supplemental material) were magnified to identify ultrastructural features of the sites of Gag accumulation. Some viral particles are marked with red arrows. CCV, clathrin-coated vesicle; L, lysosome; MT, mitochondria; rER, rough endoplasmic reticulum; VCC, virus-containing compartment; W, membranous web. The scale bar indicates a distance of 500 nm.
DISCUSSION
In this study, we investigate the formation of VCCs and their accessibility via cell surface-administered antibodies in living macrophages by the use of an infectious fluorescently labeled HIV-1 genome. HIV-1 Gag-iGFP (HIV-1 GG) is attenuated but retains replicative capacity and infectivity by the insertion of GFP between the p17 matrix and the p24 capsid of Gag (18). In contrast to other HIV-1 constructs, which essentially also carry GFP at the same location (22, 27), the chromophore is flanked by two HIV-1 protease cleavage sites, thus relieving GFP during maturation (18). While a similar virus has been elegantly used to study transfer and trafficking of HIV-1 across synapses of electroporated T cells (19), its practical use has been hampered by the fact that it spreads poorly in most cell lines and primary cells (18). Our R5-tropic HIV-1 GG is infectious for CCR5-expressing cells and spreads and replicates in macrophages, although with approximately 2-fold-reduced efficiency compared to that of the corresponding unlabeled HIV-1. Importantly, since replication kinetics and peak viral titers were similar to those of untagged HIV-1, we are confident that HIV-1 GG resembles the temporal and spatial Gag distribution of wild-type HIV-1. This is important, since until now, the only tagged HIV-1 genome showing replication and spread in primary macrophages was an HIV-1 AD8 variant with a tetracysteine (TC) tag (13). While this is a useful and elegant tool, especially since the TC tag does have only a minor impact on virus infectivity, the need for biarsenic labeling hampers the possibility to image the formation of newly synthesized Gag.
We followed up on Gag synthesis from immediately after infection to up to 4 days. In contrast to T cells or 293T cells (18, 19, 22), the Gag precursor was not found predominantly at the plasma membrane but appeared in large perinuclear accumulations, which stayed stable for days (Fig. 2). HIV-1-containing compartments (VCCs) in macrophages have been described before (9, 21, 31, 33, 35). However, their origin and fate remained elusive, because of the lack of a dynamic view on them (3). Based on their temporal appearance in HIV-1-infected macrophages (Fig. 2) and the presence of budding profiles within membrane-enclosed VCCs (our study and, e.g., references 21 and 33), we conclude that these accumulations can be formed internally. Importantly, this does not exclude the possibility that viral assembly sites are initiated at the PM and are internalized before the completion of release. In this context, it is noteworthy that we also observed faint Gag at the PM (Fig. 1 and 2) and smaller punctate Gag in close vicinity to the PM (Fig. 2C), which could resemble PM assembly or assembly sites at the beginning of internalization (22, 45).
We also do not dismiss the possibility that some of the VCCs in macrophages might be connected to the PM, e.g., by small microchannels (4, 9, 44, 45). Our results are in agreement with a recent study by Welsch and colleagues (44). Using electron tomography and EM with stereology, they described a complex membranous network which harbors viral particles. These VCCs, which were designated “HIV-1 assembly and storage compartments,” were connected to the cell surface by tubelike structures with insufficient diameter for virion passage. Therefore, it was postulated that the nature of the virus-holding compartment may aid in antibody evasion of HIV-1 (44). For the first time, our study provides experimental proof that macrophage internal HIV-1 is indeed inaccessible to surface-administered neutralizing antibodies. These results are based on the usage of four different antibodies, either targeted against CD81, a component frequently found in VCCs (our study and, e.g., reference 9) or targeted against HIV-1 GP120. Notably, all three GP120 antibodies are directed against the CD4 binding site of GP120 and were described as broadly neutralizing with no cross-reactivity against host self or other antigens (42, 46). Furthermore, antibody accessibility experiments were performed at 4°C and 37°C. The 4° condition is for low membrane motility; thus, existing openings toward the PM should stay stable. In contrast, at 37°C we expect to have endocytic uptake and presumably transient openings of VCCs toward the cell surface. Collectively, these results provide evidence that VCCs can be shielded from antibodies via the cell surface. This is particularly important, because macrophages are considered viral reservoirs, transmit HIV-1 via the blood-brain barrier (14, 23, 26), and harbor infectious HIV-1 over several weeks (39).
Cell-to-cell spread of HIV-1 is far more efficient than infection by cell-free virus (7, 10, 34) and occurs via so-called virological synapses (VS) (25). Macrophages have also been reported to transmit HIV-1 via VS to T cells (16, 43), and Freed and colleagues, using a TC-tagged HIV derivative, showed rapid transfer of internal VCC from macrophages toward VS and T cells (13). The Sattentau lab reported that formation of the VS needs CD4 and to a smaller extent the chemokine coreceptor on the target cell (16). In addition, transmitted virus was sensitive toward fusion inhibitors and antibodies that bind CD4 and block interaction with GP120 (16, 24). Interestingly, cell-free infection and cell-to-cell transfer could be suppressed by antibodies targeting the HIV-1 glycoprotein, but this effect was demonstrated only for transfer between T cells (24). Our findings suggest that GP120 antibody-pretreated macrophages efficiently transfer internal HIV-1 to T cells (Fig. 5). This is in agreement with a model in which HIV-1 at least in part assembles into an internal niche. In this scenario, a large proportion of GP120 is not exposed at the plasma membrane of macrophages, a finding which is supported by our surface GP120 staining of HIV-1-infected macrophages (Fig. 4). In contrast, assembly in T cells precedes transport of GP120 to the plasma membrane, which could then be accessible to neutralizing antibodies.
It is tempting to speculate that the complex membrane architecture of VCCs observed by us and Welsch and colleagues (44) prevents antibody recognition of macrophage internal HIV-1. How HIV-1 induces the formation of the membranous web and which are its major constituents remain elusive. Deneka et al. reported colocalization of VCCs with the tetraspanins CD81, CD53, and CD9 (9). We confirm the presence of CD81 in VCCs. Furthermore, the data of our FRET assay implies that HIV-1 Gag itself binds to the cytoplasmic region of CD81 (Fig. 3A). This is a finding which is corroborated by data from Gag-CD81 coimmunoprecipitation experiments (15). Interestingly, the tetraspanins CD9, CD63, CD81, and CD82 have been suggested as gateways for HIV-1 in HeLa cells (28) and T cells (15, 20). Since tetraspanins are membrane associated and build homo- as well as heteromers (41), it is tempting to speculate that Gag itself by the interaction with CD81 and maybe other members of the tetraspanin family might directly be involved in the formation of the membranous web.
In sum, this study emphasizes the potential importance of HIV-1-infected macrophages for viral immune evasion and therefore AIDS pathogenesis. It is crucial to further elucidate the nature of the membranous structures serving as HIV-1 storage compartments and hideouts in order to target the macrophage-residing HIV-1 reservoir.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Holstermann for technical assistance, B. Chen for the generous gift of the pUC-NL4-3 Gag-iGFP provirus, and P. Benaroch for helpful discussions.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: JLTRG-R5 (catalog number 11586) from O. Kutsch and gp120 monoclonal antibodies VRC01 and VRC03 from J. Mascola.
This work was funded by grants to M. Schindler from the Stiftung für Neurovirale Erkrankungen (NVE) and the Heinrich Pette Institute, which is a member of the Leibniz Gemeinschaft (WGL) and is supported by the Free and Hanseatic City of Hamburg and the Federal Ministry of Health. H. Koppensteiner is a stipendiary of the Leibniz Center for Infection (LCI) graduate school.
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ayinde D, Maudet C, Transy C, Margottin-Goguet F. 2010. Limelight on two HIV/SIV accessory proteins in macrophage infection: is Vpx overshadowing Vpr? Retrovirology 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banning C, et al. 2010. A flow cytometry-based FRET assay to identify and analyse protein-protein interactions in living cells. PLoS One 5:e9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benaroch P, Billard E, Gaudin R, Schindler M, Jouve M. 2010. HIV-1 assembly in macrophages. Retrovirology 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bennett AE, et al. 2009. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog. 5:e1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bieniasz PD. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brandt SM, Mariani R, Holland AU, Hope TJ, Landau NR. 2002. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 277:17291–17299 [DOI] [PubMed] [Google Scholar]
- 7. Carr JM, Hocking H, Li P, Burrell CJ. 1999. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology 265:319–329 [DOI] [PubMed] [Google Scholar]
- 8. Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 7:798–812 [DOI] [PubMed] [Google Scholar]
- 9. Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 177:329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dimitrov DS, et al. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 67:2182–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gendelman HE, et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gordon S, Taylor PR. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953–964 [DOI] [PubMed] [Google Scholar]
- 13. Gousset K, et al. 2008. Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathog. 4:e1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gras G, Kaul M. 2010. Molecular mechanisms of neuroinvasion by monocytes-macrophages in HIV-1 infection. Retrovirology 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grigorov B, et al. 2009. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Groot F, Welsch S, Sattentau QJ. 2008. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111:4660–4663 [DOI] [PubMed] [Google Scholar]
- 17. Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. 2011. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J. Virol. 85:9749–9766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hubner W, et al. 2007. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81:12596–12607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hubner W, et al. 2009. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323:1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jolly C, Sattentau QJ. 2007. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J. Virol. 81:7873–7884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. 2007. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe 2:85–95 [DOI] [PubMed] [Google Scholar]
- 22. Jouvenet N, et al. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4:e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Douce V, Herbein G, Rohr O, Schwartz C. 2010. Molecular mechanisms of HIV-1 persistence in the monocyte-macrophage lineage. Retrovirology 7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin N, et al. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J. Virol. 84:3516–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McDonald D, et al. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300:1295–1297 [DOI] [PubMed] [Google Scholar]
- 26. Montaner LJ, et al. 2006. Advances in macrophage and dendritic cell biology in HIV-1 infection stress key understudied areas in infection, pathogenesis, and analysis of viral reservoirs. J. Leukoc. Biol. 80:961–964 [DOI] [PubMed] [Google Scholar]
- 27. Muller B, et al. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78:10803–10813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173:795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ochsenbauer-Jambor C, Jones J, Heil M, Zammit KP, Kutsch O. 2006. T-cell line for HIV drug screening using EGFP as a quantitative marker of HIV-1 replication. Biotechniques 40:91–100 [DOI] [PubMed] [Google Scholar]
- 30. Ono A. 2009. HIV-1 assembly at the plasma membrane: Gag trafficking and localization. Fut. Virol. 4:241–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Orenstein JM, Meltzer MS, Phipps T, Gendelman HE. 1988. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J. Virol. 62:2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Papkalla A, Munch J, Otto C, Kirchhoff F. 2002. Nef enhances human immunodeficiency virus type 1 infectivity and replication independently of viral coreceptor tropism. J. Virol. 76:8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pelchen-Matthews A, Kramer B, Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Phillips DM, Tan X, Perotti ME, Zacharopoulos VR. 1998. Mechanism of monocyte-macrophage-mediated transmission of HIV. AIDS Res. Hum. Retroviruses 14(Suppl. 1):S67–S70 [PubMed] [Google Scholar]
- 35. Raposo G, et al. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718–729 [DOI] [PubMed] [Google Scholar]
- 36. Schindler M, et al. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067 [DOI] [PubMed] [Google Scholar]
- 37. Schindler M, et al. 2010. Vpu serine 52 dependent counteraction of tetherin is required for HIV-1 replication in macrophages, but not in ex vivo human lymphoid tissue. Retrovirology 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schindler M, Wildum S, Casartelli N, Doria M, Kirchhoff F. 2007. Nef alleles from children with non-progressive HIV-1 infection modulate MHC-II expression more efficiently than those from rapid progressors. AIDS 21:1103–1107 [DOI] [PubMed] [Google Scholar]
- 39. Sharova N, Swingler C, Sharkey M, Stevenson M. 2005. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J. 24:2481–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stevenson M. 2003. HIV-1 pathogenesis. Nat. Med. 9:853–860 [DOI] [PubMed] [Google Scholar]
- 41. Tarrant JM, Robb L, van Spriel AB, Wright MD. 2003. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 24:610–617 [DOI] [PubMed] [Google Scholar]
- 42. Trkola A, et al. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Waki K, Freed EO. 2010. Macrophages and cell-cell spread of HIV-1. Viruses 2:1603–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Welsch S, Groot F, Krausslich HG, Keppler OT, Sattentau QJ. 2011. Architecture and regulation of the HIV-1 assembly and holding compartment in macrophages. J. Virol. 85:7922–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Welsch S, et al. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.