Abstract
An HIV-1 vaccine remains elusive, in part because various factors limit the quantity and quality of the antibodies raised against the viral envelope glycoprotein complex (Env). We hypothesized that targeting Env vaccines directly to B cells, by fusing them to molecules that bind and activate these cells, would improve Env-specific antibody responses. Therefore, we fused trimeric Env gp140 to A PRoliferation-Inducing Ligand (APRIL), B-cell Activating Factor (BAFF), and CD40 Ligand (CD40L). The Env-APRIL, Env-BAFF, and Env-CD40L gp140 trimers all enhanced the expression of activation-induced cytidine deaminase (AID), the enzyme responsible for inducing somatic hypermutation, antibody affinity maturation, and antibody class switching. They also triggered IgM, IgG, and IgA secretion from human B cells in vitro. The Env-APRIL trimers induced higher anti-Env antibody responses in rabbits, including neutralizing antibodies against tier 1 viruses. The enhanced Env-specific responses were not associated with a general increase in total plasma antibody concentrations, indicating that the effect of APRIL was specific for Env. All the rabbit sera raised against gp140 trimers, irrespective of the presence of CD40L, BAFF, or APRIL, recognized trimeric Env efficiently, whereas sera raised against gp120 monomers did not. The levels of trimer-binding and virus-neutralizing antibodies were strongly correlated, suggesting that gp140 trimers are superior to gp120 monomers as immunogens. Targeting and activating B cells with a trimeric HIV-1 Env-APRIL fusion protein may therefore improve the induction of humoral immunity against HIV-1.
INTRODUCTION
Despite over 25 years of research, an effective vaccine against HIV-1 remains elusive. Challenges include the remarkable capacity of HIV-1 to limit the induction of effective immune responses and its propensity to escape from those that are generated. So far, neither vaccines aimed at inducing protective antibodies (Abs) nor those intended to stimulate cell-mediated responses have been sufficiently successful (13, 32, 34, 62, 81). Both strategies continue to be pursued, both individually and in combination (2, 60).
The trimeric envelope glycoprotein complex (Env) that is responsible for HIV-1 entry into target cells is the sole target for the induction of neutralizing antibody (nAb) responses. Although an increasing number of broadly active nAbs are now being isolated from HIV-1-infected people, which testifies to the supposition that they can be generated by Env in principle (15, 21, 68, 89, 97, 101, 102, 107), it has not yet been possible to induce such nAbs with Env subunit protein vaccines (13, 14, 32, 34, 51, 62, 76, 81). Multiple immune-evasive properties hinder the ability of engineered Env proteins to induce truly useful nAb responses. These defense mechanisms include surface loops that vary in sequence in response to nAb selection pressure (14, 33, 51). These loops, and also abundant glycan structures, shield the more conserved regions of Env involved in receptor binding (29, 33, 74, 79, 103). HIV-1 particles also bear highly immunogenic nonnative forms of Env that may interfere with responses to functional trimers (23, 43, 67, 75, 77). As a result, most anti-Env antibodies induced during HIV-1 infection or by Env vaccination are nonneutralizing.
Soluble trimeric forms of Env are unstable and thus require modifications to retain an oligomeric structure. Various attempts have been made to overcome this instability and create better mimics of the native complex (33). We have generated cleaved soluble Env gp140 trimers, termed SOSIP gp140, that contain mutations to stabilize the gp120-gp41 and gp41-gp41 interactions (6, 7, 83). Single-particle cryo-electron microscopy (cryo-EM) studies have shown that SOSIP gp140 and the native complex have similar structures and undergo comparable CD4-induced conformational rearrangements (39). SOSIP gp140 is slightly superior to monomeric gp120 at inducing nAb responses (3, 4, 45).
A general constraint of subunit protein and DNA plasmid vaccines is their poor immunogenicity compared to live attenuated or inactivated viral vaccines, which is in part explained by their lack of components such as Toll-like receptor (TLR) activators that provide costimulatory signals. However, HIV-1 Env-based subunit vaccines appear to be rather poor immunogens, even compared to other subunit vaccines such as influenza A virus hemagglutinin (HA) (24). Thus, several high doses of gp120 or gp140 proteins are generally needed to induce moderate binding-antibody titers, and these titers wane quickly, with a short half-life of 30 to 60 days (34). One among several contributory explanations may be interference with dendritic cell (DC) function via the oligomannose N-glycans present on gp120 (1, 49, 59, 92).
The addition of costimulatory molecules can augment the induction of immune responses to poor antigens; covalently linking the costimulatory molecule to the antigen enhances the immunostimulatory effect, because the antigen and “cis-adjuvant” preferentially need to contact the same immune cells, generally antigen-presenting cells (19, 27, 31, 41, 42, 53, 63, 95, 104, 105). The addition of costimulatory molecules also provides an opportunity to skew the immune response in the appropriate and desired direction. A few attempts have been made to conjugate costimulatory molecules to HIV-1 Env (in most cases, monomeric gp120). Heath and colleagues showed that the fusion of gp120 to IFN-γ and/or TNF-α enhanced Env-binding antibody responses in mice, but nAb induction was not measured (61, 72, 73). In a second study, fusion of gp120 to Flt-3 ligand or CTLA4 increased murine Env-specific CD8+ T-cell and binding antibody responses, but nAbs were again not assessed (70, 82). Trimers of gp140 containing an embedded granulocyte-macrophage colony-stimulating factor (GM-CSF) domain induced antibody and T helper responses in mice, which resulted in enhanced neutralization of the highly sensitive SF162 strain (100). Finally, fusing monomeric gp120 and trimeric gp140 to the complement component C3d modestly improved nAb responses in rabbits (12, 37, 50).
The strategies mentioned above were usually intended to improve the targeting and activation of dendritic cells (DCs) and enhance antigen presentation. We have shown that DCs can be activated by fusing trimeric Env to the active domain of CD40 ligand (CD40L; TNFSF5/CD154) (63). CD40L, a tumor necrosis factor (TNF) superfamily (TNFSF) member, is an important immunostimulatory molecule mainly expressed by activated T-helper cells. It stimulates DCs to increase antigen presentation, secrete proinflammatory cytokines, and prime naïve T cells (17, 55, 93, 99). It also induces naive B cells to mature to memory or plasma cells and promotes antibody affinity maturation and IgG or IgA class switching (17, 93, 99).
Targeting an antigen to DC can promote its uptake and processing, thereby facilitating a T-helper-cell response and indirectly aiding the development of a B-cell response. However, the induction of nAb responses requires the presentation of intact antigen to naïve B cells so that complex epitopes can be recognized. We therefore sought to target Env to B cells and activate them directly. B-cell-Activating Factor (BAFF/BLyS/TNFSF13b) and A PRoliferation-Inducing Ligand (APRIL/TNFSF13) resemble CD40L in being homotrimeric type II transmembrane proteins with immunostimulatory functions, in this case, for B cells (26, 56, 58). The principal sources of BAFF and APRIL are innate immune cells such as neutrophils, macrophages, monocytes, DCs, and follicular DCs (FDCs) (20, 46, 54, 56, 58, 85). The two proteins have similar effects on B cells because they share two receptors (BCMA/TNFRSF17 and TACI/TNFRSF13b), although they have different affinities and bind under different circumstances (11, 44, 47, 56). As with CD40L, BAFF and APRIL stimulate B cells to mature into memory cells or antibody-secreting plasma cells in a T-cell-independent manner, as well as activating B-cell-receptor affinity maturation and class switching to IgG or IgA (38, 56, 57). BAFF has also been suggested to counteract the tolerogenic effects of certain antigens, including gp120 (35, 40). These properties of BAFF and APRIL seem particularly advantageous for HIV-1 Env vaccines, because the production of broadly active nAbs may require extensive affinity maturation (89, 91, 107, 108). Furthermore, the promotion of IgA class switching by BAFF and APRIL could help the development of mucosal immune responses that intervene against HIV-1 sexual transmission. Mucosal IgA responses to HIV-1 infection and vaccines are notoriously poor (25, 64, 65, 86, 90, 109). Systems biology approaches have identified the expression of, in particular, the APRIL/BAFF receptor BCMA, but also of APRIL and BAFF themselves, as predictors of nAb responses to yellow fever and influenza vaccines (69, 78). Furthermore, a recent study in mice showed that durable antibody responses that can protect against a lethal influenza challenge critically depend on the APRIL/BAFF receptor TACI (106). Together, these observations support the exploitation of APRIL and BAFF for vaccine targeting.
Here, we investigated whether targeting trimeric HIV-1 Env proteins to B cells via fusion to APRIL, BAFF, or CD40L would improve antibody responses. We found that Env-APRIL, Env-BAFF, and Env-CD40L induce the secretion of IgM, IgA, and IgG from B cells in vitro. Furthermore, Env-APRIL enhanced the antibody response to Env in rabbits, including the induction of nAbs against tier 1 viruses, compared to Env alone. Thus, Env targeting to B cells via fusion to APRIL is a new way to enhance humoral immune responses.
MATERIALS AND METHODS
Plasmids.
We have previously described modifications that improve the stability of soluble, cleaved gp140 trimers based on the R5 subtype B isolate JR-FL (6, 83). The amino acid sequences of gp120 and the gp41 ectodomain were modified as follows (Fig. 1A). We introduced (i) a disulfide bond between residues 501 in gp120 and 605 in gp41 (A501C, T605C) (6); (ii) a trimer-stabilizing substitution in gp41 (I559P) (83); and (iii) a sequence-enhanced site for furin cleavage (RRRRRR) (7). We further modified the JR-FL SOSIP.R6 gp140 construct to include a C-terminal GCN4-based trimerization domain (isoleucine zipper [IZ]) (30, 63). We have shown that this domain further improves trimer stability. A C-terminal octahistidine tag (HHHHHHHH [H8]) was also added. The SOSIP.R6-IZ-H8 construct is referred to here as “Env.”
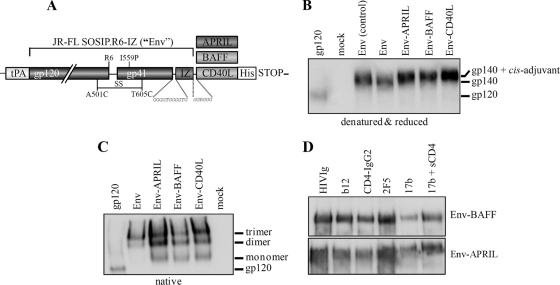
Fig 1.
Chimeric Env-APRIL, Env-BAFF, and Env-CD40L proteins are properly folded. (A) Schematic of the Env, Env-APRIL, Env-BAFF, and Env-CD40L proteins. The clade B JR-FL gp140 protein (amino acids 31 to 681) contains several modifications for stabilization that have been previously described (see Materials and Methods). Codon-optimized sequences encoding the active domains of human and rabbit APRIL, BAFF, or CD40L were added to the gp140 open reading frame at the C terminus. (B) Reducing SDS-PAGE and (C) BN-PAGE analysis of Env, Env-APRIL, Env-BAFF, and Env-CD40L proteins secreted from transiently transfected 293T cells. Recombinant purified JR-FL gp120 (50 ng) and Env from a control transfection were included for comparison. (D) Recognition of Env-APRIL and Env-BAFF by MAbs and CD4. The Env-APRIL and Env-BAFF proteins were immunoprecipitated by CD4-IgG2 and HIVIg MAbs to gp120 (b12) or gp41 (2F5) or via a CD4-induced epitope (MAb 17b) on gp120 in the absence or presence of soluble CD4 (sCD4), followed by reducing SDS-PAGE and Western blot analysis.
To compare trimeric Env with monomeric gp120, we used a variant of SOSIP.R6-IZ-H8 in which the H8 tag was replaced by the epitope for the D7324 antibody (APTKAKRRVVQREKR), generating SOSIP.R6-IZ-D7324 (30). Note that the internal D7324 epitope in the C5 domain of SOSIP constructs is mutated by the introduction of the cysteine at residue 501 that forms the intramolecular disulfide bond with gp41 and that it is also occluded by the proximity of the gp41 subunit; hence, unmodified SOSIP gp140s do not react efficiently with D7324 (6, 83). Because of the addition of C-terminal sequences to SOSIP gp140, SOSIP.R6-IZ-D7324 trimers are poorly cleaved (63).
A SOSIP.R6-IZ-CD40L fusion protein (termed Env-CD40L), based on the human CD40L sequence and described elsewhere (63), served as the template for the new proteins. The rabbit versions of the Env-APRIL, Env-BAFF, and Env-CD40L constructs were generated by replacing the human CD40L moiety with the active domains of rabbit APRIL, BAFF, and CD40L. Codon-optimized rabbit sequences of APRIL, BAFF, and CD40L containing the restriction sites for Asp718I and SfuI were synthesized (Mr. Gene, Regensburg, Germany). The APRIL, BAFF, and CD40L sequences (corresponding to amino acids 115 to 250, 149 to 290, and 119 to 261, respectively) were then cloned C-terminally to SOSIP.R6-IZ by the use of Asp718I and SfuI. Because the amino acid sequence of rabbit CD40L was unknown, we sequenced the gene by amplifying cDNA from New Zealand White rabbit peripheral blood mononuclear cells (PBMCs) (see the supplemental material and Fig. S1 in the supplemental material). The sequence integrity of all clones was confirmed prior to use. The amino acid numbering of SOSIP.R6 gp140 is based on HXB2 Env.
Cell culture and transient transfection.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/ml), and streptomycin (100 μg/ml) as previously described (9). HEK293T cells were transfected using polyethyleneimine (PEI) as described elsewhere (48). Briefly, DNA encoding Env protein was diluted in DMEM (Invitrogen, Breda, The Netherlands) to 1/10 of the final culture volume and mixed with PEI (0.12 mg/ml final concentration). After incubation for 20 min, the DNA-PEI mix was added to the cells for 4 h before replacement with normal culture medium containing 10% FCS (HyClone; Perbio, Etten-Leur, The Netherlands), penicillin, streptomycin, and MEM nonessential amino acids (Invitrogen) (0.1 mM). Culture supernatants were harvested 48 h after transfection. For experiments performed with human B cells, HEK293T cells (3 × 106) were transfected with 10 μg of plasmids encoding Env or fusion proteins by the use of Effectene transfection reagent (Qiagen). Culture supernatants were collected 48 h posttransfection and then incubated overnight with Galanthus nivalis lectin-conjugated agarose beads (Sigma) at 4°C to allow Env binding to the lectin. The beads were then washed three times with phosphate-buffered saline (PBS) and incubated with 1 M methyl α-d-mannopyranoside (Sigma) at 4°C for 2 h. Purified proteins were collected after centrifugation at 10,000 rpm and verified by Western blot analysis.
SDS-PAGE, BN-PAGE, and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), blue native PAGE (BN-PAGE), and Western blot analysis were performed as described elsewhere (83, 87, 88) using the JR-FL V3-specific mouse monoclonal antibody (MAb) PA-1 at a 1:20,000 dilution as an Env probe (96) (Progenics Pharmaceuticals).
Immunoprecipitation assays.
A 100-μl aliquot of 20×-concentrated 293T cell supernatant was incubated overnight at 4°C, with rotation, with MAbs or related reagents (HIVIg, b12, CD4-IgG2, or 2F5 at 4 μg/ml or 17b at 1.5 μg/ml), and, when appropriate, sCD4 (10 μg/ml), in 500 μl of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.2], 150 mM NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, and protease inhibitors [Complete protease inhibitor tablets; Roche, Almere, The Netherlands]). Next, protein G-coated agarose beads (Pierce/Thermo Fisher, Etten-Leur, The Netherlands) were added and incubated for 2 h at 4°C with rotation. The beads were then washed six times with RIPA buffer (supplemented with 0.01% Tween 20), after which the bound proteins were eluted by heating at 100°C for 5 min in 50 μl of 2× SDS-loading buffer containing 100 mM dithiothreitol (DTT). The immunoprecipitates were fractionated by SDS-PAGE (8% polyacrylamide) at 125 V for 1.5 h. Env detection was performed using MAb PA-1 and standard Western blot techniques.
Isolation of human B cells.
Human B cells were isolated from buffy coats of healthy donors obtained from the New York Blood Center. B cells were isolated from peripheral mononuclear cells by the use of B-cell isolation kit II (Miltenyi Biotech). The purity of the sorted B-cell populations was more than 97%, as assessed by CD19 staining. Naïve B cells were isolated from peripheral mononuclear cells by negative selection using naïve B-cell isolation kit II (Miltenyi Biotech).
Ig secretion by human B cells.
Purified B cells (5 × 104) were plated in a 96-well U-bottom plate in 200 μl of complete RPMI 1640 medium containing 10% FBS, 2 mM glutamine, 100 U/ml streptomycin, 100 U/ml penicillin, 1 mM sodium pyruvate, and 10 mM HEPES (all from Invitrogen). The cells were treated with 10 μl of purified Env or Env fusion proteins in the presence of recombinant CD40L (Enzo Life Sciences) (200 ng/ml), interleukin-4 (IL-4) (R&D Systems) (10 ng/ml), and IL-10 (R&D Systems) (200 ng/ml) for 14 days. Culture supernatants were collected for the analysis of immunoglobulin secretion by an enzyme-linked immunosorbent assay (ELISA) (Bethyl Laboratories). The background levels of IgM, IgG, and IgA secretion induced by the stimulation cocktail without Env or fusion proteins were subtracted from the test values. Typically, for each Ig class and in all donors, these background levels were 70 to 140 ng/ml.
AID expression in human B cells.
Purified naïve B cells (2 × 105) were plated in a 96-well U-bottom plate in 200 μl of complete RPMI 1640 medium containing 10% FBS, 2 mM glutamine, 100 U/ml streptomycin, 100 U/ml penicillin, 1 mM sodium pyruvate, and 10 mM HEPES (all from Invitrogen). Cells were treated with 20 μl of purified Env and Env fusion proteins in the presence of IL-4 (10 ng/ml) and IL-10 (200 ng/ml) for 4 days. Cells were washed with PBS twice and collected for real-time PCR. Total RNA from treated naïve B cells was isolated using RNAeasy Mini Spin columns (Qiagen) according to the manufacturer's instructions. cDNA synthesis was carried out for 1 h at 37°C in a 20-μl total volume containing 25 μg/ml random primers, 0.5 mM deoxynucleoside triphosphate (dNTPs), 5 mM DTT, 40 U of RNase inhibitor, and 200 U of murine Moloney leukemia virus (M-MLV) reverse transcriptase in 1× M-MLV buffer (Promega) followed by denaturation at 70°C. cDNA templates were serially diluted and used for real-time PCR. Reaction mixtures were prepared in a 384-well plate in a total volume of 10 μl containing 2× TaqMan gene expression master mix and 0.5 μl of TaqMan FAM (6-carboxyfluorescein) dye-labeled MGBNFQ probe for the quantification of activation-induced cytidine deaminase (AID) (assay no. Hs00757808_m1), and assays were performed using a 7900HT Fast real-time PCR system (Applied Biosystems). Target gene expression was normalized against the level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (assay no. Hs02758991_g1). The threshold cycle (CT) values for ΔCT (ΔCT = CT of target gene − CT of GAPDH) were calculated and converted into arbitrary units by the formula [(2−ΔCt) × 1,000].
Gene gun DNA and protein immunizations.
The pPPI4 plasmid encoding fusion proteins was amplified using DH5α cells and isolated using an EndoFree Plasmid Giga kit (Qiagen, Venlo, The Netherlands). New Zealand White rabbits (four per group) were immunized at weeks 0, 2, 4, and 8 with 125 μg of endotoxin-free DNA, via the abdominal dermis, by the use of gene gun technology. One group was immunized at weeks 0, 2, 4, and 8 with 30 μg of JR-FL gp120 in Imject Alum (40 mg/ml aluminum hydroxide and 40 mg/ml magnesium hydroxide). The injection mixture was made according to the instructions of the manufacturer (Pierce) and delivered subcutaneously at six sites: two each in the shoulder, abdomen, and hind limb. At week 16, all rabbits were injected with 1 ml of PBS containing 30 μg of trimeric JR-FL SOSIP.R6 gp140 protein mixed with 60 μg of Quil A (Brenntag, Frederikssund, Denmark). The injections were performed as follows: 300 μl was injected intradermally (50 μl into each of the 6 sites specified above), 400 μl intramuscularly (200 μl into each hind leg), and 300 μl subcutaneously (into the neck region). Blood samples were obtained at weeks 0, 2, 4, 6, 8, 12, 16, and 18 and from the final bleed at week 20.
Immunizations were carried out under contract by Genovac (Freiburg, Germany) at the facilities of Harlan Winkelmann (Eystrup, Germany). All animals were kept according to DIN EN ISO 9001:2008 standards, the regulations of the German Welfare Act of 19 May 2006 (BGBI I S. 1206), the regulations of the European Union guidelines (86/609/EWG of 24 November 2006), and the European Agreement of 18 March 1986 for the protection of animal trials and other scientific studies using vertebrates (Act of 11 December 1990 [BGBI II S. 1486]). All protocols dealing with animal manipulations were in accordance with guidelines published by FELASA (Federation of European Animal Science Association) and GV-SOLAS (German Society of Laboratory Animal Science) and were reviewed by the Harlan animal care committee. The study was approved by the Landesuntersuchungsamt (Kreis Nienburg/Weser, Germany) (permit 39/30-11-1998).
Env-specific and total immunoglobulin ELISAs.
Anti-gp120 antibody titers were measured by D7324-capture ELISA, essentially as described previously (30). Anti-trimeric gp140 titers were determined using SOSIP.R6-IZ-D7324, which was engineered to contain the D7324 epitope at its C terminus (30, 63). For measuring total serum immunoglobulin levels, goat anti-rabbit IgG (Jackson ImmunoResearch, Newmarket, United Kingdom)–0.1 M NaHCO3 (pH 8.6; 10 μg/ml) was coated overnight onto microplate wells (100 μl/well). After blocking, serially diluted serum was added for ∼2 h. Bound mouse IgG was detected with horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (Jackson ImmunoResearch, Suffolk, England) (1:5,000 [0.2 μg/ml]), followed by quantification in a luminometer. Midpoint titers were calculated using Graphpad Prism (version 5.03) by determining the dilution of the serum at which the optical density was 50% of the maximum value. For quantification, a standard curve using known amounts of purified rabbit IgG (Jackson) was used.
Neutralization assays.
The TZM-bl reporter cell line stably expresses high levels of CD4 and HIV-1 coreceptors CCR5 and CXCR4 and contains the luciferase and β-galactosidase genes under the control of the HIV-1 long-terminal-repeat promoter. The line was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health (John C. Kappes, Xiaoyun Wu, and Tranzyme Inc. [Durham, NC]). Single-cycle infection and inhibition experiments using TZM-bl cells were performed as described using 3-fold serially diluted sera (8, 9, 28). The percentage of neutralization was determined by measuring how much of the luciferase signal was reduced by each serum dilution compared to the results seen when serum was absent (defined as 100%). The 50% (midpoint) neutralization titers were determined. The viruses tested at the Academic Medical Center included the tier 1 virus SF162 and the homologous tier 2 virus JR-FL (Table 1; see also Table 3). The viruses tested at the Duke Central Immunology Laboratory for AIDS Vaccine Research and Development were the tier 1 strains MN, SF162.LS, and BaL.26 and the tier 2 strains JR-FL, 6535.3, QH0692.42, PVO.4, and RHPA4259.7 (Table 2 and data not shown). The sera were heat inactivated (30 min 56°C) before use.
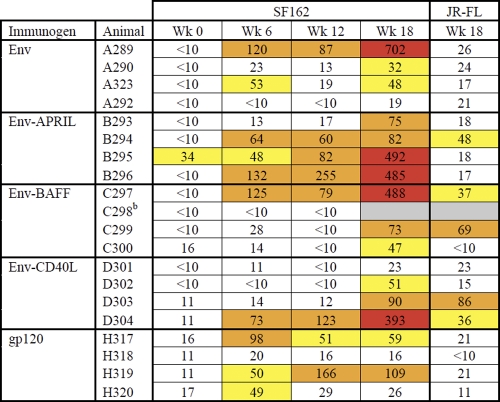
Table 1.
50% neutralization titers against SF162 and JR-FLa
Data are from the Academic Medical Center. The titer data are colored according to the following color scale: yellow, 50% neutralization titers of 30 to 60; orange, 50% neutralization titers of 60 to 300; red, 50% neutralization titers of >300.
b Animal died of unrelated causes between week 12 and week 18.
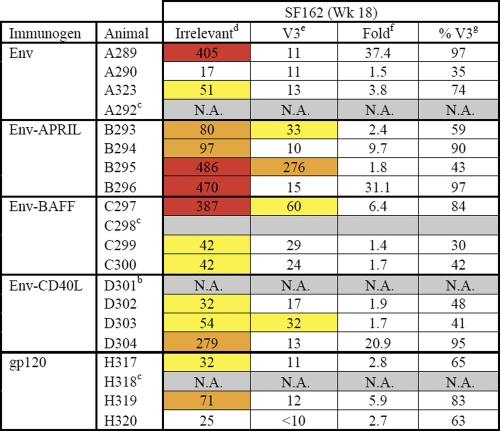
Table 3.
V3-peptide depletion of SF162 nAbsa
Data are from the Academic Medical Center. The titers are colored according to the following color scale: yellow, 50% neutralization titers of 30 to 60; orange, 50% neutralization titers of 60 to 300; red, 50% neutralization titers of >300.
b N.A., not analyzed; sera from these rabbits did not neutralize SF162 potently in earlier experiments (see Table 1).
c Animal died of unrelated causes between week 12 and week 18.
d Values represent 50% neutralization titers in the presence of irrelevant peptide.
e Values represent 50% neutralization titers in the presence of V3 peptides.
f Values represent fold decreases in 50% neutralization titers caused by V3 peptides.
g Values represent percentages of V3-specific neutralization.
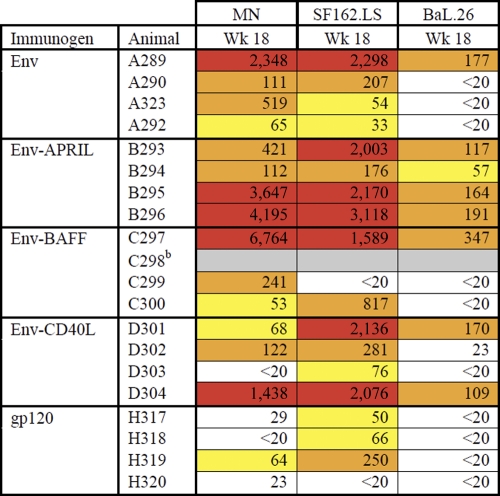
Table 2.
50% neutralization titers against tier 1 virusesa
Data are from the Central Immunology Laboratory. The titers are colored according to the following color scale: yellow, 50% neutralization titers of 30 to 100; orange, 50% neutralization titers of 100 to 1,000; red, 50% neutralization titers of >1,000.
b Animal died of unrelated causes between week 12 and week 18.
IgG depletion experiments.
Serum (120 μl) was mixed with 200 μl of a 50% (vol/vol) slurry of protein G plus agarose (Pierce) and 680 μl of phosphate-buffered saline (pH 7.4). The mixture was incubated overnight at 4°C, and the beads were then washed thoroughly with 1× RIPA buffer containing 0.05% Tween 20 as described for the immunoprecipitation assay. Rabbit IgG was eluted from the beads using 550 μl of IgG elution buffer (Pierce), and the solution was immediately neutralized with 50 μl of 1 M Tris-HCl (pH 9.5). Purified IgG was dialyzed against PBS.
V3 peptide competition experiments.
Rabbit sera were serially diluted and preincubated with a mix of three overlapping peptides (20 μg/ml of each) spanning the JR-FL V3 region (Env V3-1 [NNNTRKSIHIGPGRA], Env V3-2 [SIHIGPGRAFYTTGE], and Env V3-3 [GRAFYTTGEIIGDIR]) or with an unrelated peptide (QAPKPRKQ [60 μg/ml]) for 1 h at room temperature. The peptide-serum mixtures were then tested for HIV-1 neutralization. To control for nonspecific inhibition by the peptides, we also performed neutralization assays using MAb b12 (6 μg/ml) in the presence or absence of the three V3 peptides; they did not inhibit b12 neutralization (data not shown).
BN-PAGE trimer shift assays.
Blue native PAGE (BN-PAGE) analyses were performed as described previously (22, 23, 67). Briefly, virus-like particles (VLPs) bearing wild-type (wt) JR-FL trimers or D368R JR-FL trimers were incubated with MAbs or sera, washed, and gently solubilized in 0.12% Triton X-100–1 mM EDTA–1.5 M aminocaproic acid with a protease inhibitor cocktail containing 4-(2-aminoethyl)benzenesulfonyl fluoride, E-64, bestatin, leupeptin, aprotinin, and sodium EDTA (P-2714; Sigma). An equal volume of 2× sample buffer (100 mM morpholinepropanesulfonic acid [MOPS], 100 mM Tris-HCl [pH 7.7], 40% glycerol, 0.1% Coomassie blue) was added. Samples were loaded onto a 4% to 12% Bis-Tris NuPAGE gel (Invitrogen) and separated at 4°C for 3 h at 100 V. Ferritin (Amersham) was used as a size standard. The gel was then blotted onto a polyvinylidene difluoride membrane that was destained, immersed in blocking buffer (4% nonfat milk–PBS), and probed with a cocktail consisting of MAbs 2G12, b12, E51, 39F, 2F5, 4E10, 7B2, and 2.2B followed by an anti-human Fc alkaline phosphatase conjugate (Jackson) and SigmaFast BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium) substrate (Sigma).
Statistical analyses.
All statistical analyses were performed using GraphPad Prism 5.03. B-cell experiments were analyzed with one-tailed Student t tests. One-tailed Mann-Whitney U tests were used to analyze immunogenicity data. Kruskal-Wallis tests were also performed to compare the same groups, followed by Dunn's multiple comparison test in cases in which the medians were found to be significantly different, unless indicated otherwise. Nonparametric (two-tailed) Spearman tests were performed to determine correlations.
RESULTS
Chimeric Env-APRIL/BAFF/CD40L proteins are expressed efficiently.
To target Env to immune cells, we fused stabilized soluble trimeric gp140 to the globular domains of APRIL, BAFF, or CD40L, intending the immunostimulatory molecules to act as cis-adjuvants. Both human and rabbit versions were made, to match the species used for testing (human cells in vitro, rabbits in immunization studies). The SOSIP.R6-IZ gp140 protein was based on the CCR5-using subtype B primary isolate JR-FL and modified to increase its stability, as described previously (6, 7, 30, 63, 83) (Fig. 1A).
We transiently transfected HEK 293T cells and used reducing SDS-PAGE and Western blots to determine the expression levels of secreted Env-APRIL, Env-BAFF, and Env-CD40L. All the proteins were expressed at comparable levels (Fig. 1B). They were secreted as uncleaved gp140 proteins, because adding C-terminal domains to the gp41 ectodomain (e.g., an isoleucine zipper [IZ] domain and/or a costimulatory molecule) inhibits Env cleavage (63). The slower migration of the fusion proteins is consistent with the extra mass (∼17 kDa per monomer) associated with APRIL, BAFF, and CD40L.
We performed a BN-PAGE analysis to assess whether the presence of the APRIL, BAFF, or CD40L domain affected gp140 trimerization. The unmodified reference gp140 was secreted predominantly in the form of trimers, as described previously (Fig. 1C) (30, 63). The chimeric proteins also formed trimers, although significant amounts of monomers and dimers were also present (Fig. 1C). The APRIL, BAFF, and CD40L moieties therefore do affect trimer formation and/or stability, but do so only modestly.
Chimeric Env-APRIL/BAFF/CD40L proteins bind CD4 and nAbs.
We next studied whether the fusion proteins were folded correctly by immunoprecipitating them with HIVIg, CD4-IgG2, and various MAbs to conformationally sensitive and/or neutralization epitopes (Fig. 1D). HIVIg, CD4-IgG2, and MAb b12 all recognized Env-APRIL and Env-BAFF efficiently. The MPER epitope of MAb 2F5, located adjacent to the trimerization domain and the APRIL/BAFF/CD40L sequences, also precipitated the Env-APRIL and Env-BAFF fusion proteins. MAb 17b directed to a CD4-induced epitope overlapping with the coreceptor-binding site on gp120 bound more strongly in the presence of soluble CD4, indicating that the fusion proteins can undergo CD4-induced conformational changes (Fig. 1D). Similar results were previously obtained with Env-CD40L (63). Overall, these results show that the Env components of the Env-APRIL/BAFF/CD40L fusion proteins are properly folded and present conformationally sensitive neutralization epitopes.
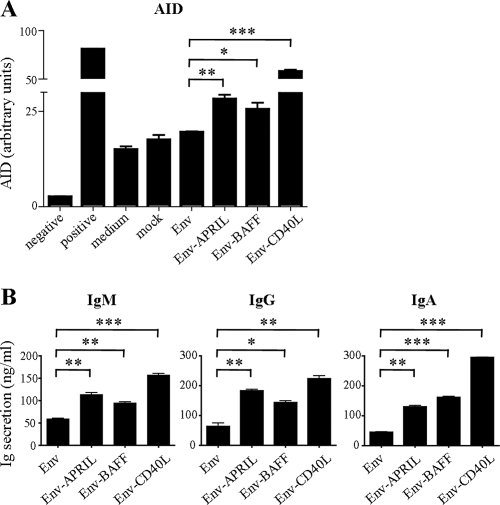
Chimeric Env-APRIL/BAFF/CD40L proteins induce AID expression and immunoglobulin secretion in human B cells.
We next investigated whether the APRIL, BAFF, and CD40L domains of the chimeric fusion proteins were bioactive by testing whether they induced expression of activation-induced cytidine deaminase (AID), the enzyme responsible for initiating somatic hypermutation, antibody affinity maturation, and antibody class switching. B cells treated with Env showed some AID expression, which can be attributed to the IL-4 and IL-10 in the stimulation cocktail. Env-APRIL consistently induced ∼1.6-fold-higher AID levels compared to unconjugated Env. Env-BAFF also upregulated AID levels (by ∼1.5-fold), while Env-CD40L induced ∼3.3-fold-higher AID levels compared to unconjugated Env.
We next tested the capacity of the Env-APRIL/BAFF/CD40L fusion proteins to stimulate immunoglobulin secretion from B cells. Human B cells were incubated with Env or Env fusion proteins, and the levels of IgM, IgG, and IgA after 14 days of culture were determined by ELISA. The unmodified Env protein induced low but detectable levels of IgM, IgG, and IgA secretion from B cells, consistent with previous reports that Env can activate B cells through binding to lectin receptors (Fig. 2B) (40). In comparison, the Env-APRIL, Env-BAFF, and Env-CD40L proteins all stimulated the secretion of significantly greater amounts of IgM, IgA, and IgG (Fig. 2B). These observations are consistent with a report that Env can cooperate with TNF superfamily members to enhance B-cell activation (40). Overall, these studies indicate that the APRIL, BAFF, and CD40L domains of the Env fusion proteins are functional in that they bind and activate human B cells to induce AID expression and antibody secretion.
Fig 2.
Env, Env-APRIL, Env-BAFF, and Env-CD40L induce AID expression and Ig secretion in human B cells. (A) AID mRNA levels in naïve human B cells stimulated with Env, Env-fusion proteins, or control stimuli were measured by real-time PCR. Negative-control B cells did not receive any stimulus (no IL-4 or IL-10). Positive-control cells received CpG-B in addition to the stimulation cocktail. Similar results were obtained with naïve B cells from three different donors. (B) IgM (left panel), IgG (middle panel), and IgA (right panel) levels in the supernatant of human B cells mock stimulated or stimulated with Env or the respective fusion proteins supplemented with a maturation cocktail were measured by ELISA after 14 days. Similar results were obtained with B cells from three different donors. *, **, and *** indicate P < 0.05, P < 0.01, and P < 0.001, respectively, as determined by a one-tailed Student's t test.
Env-APRIL induces stronger anti-Env binding antibody responses in rabbits.
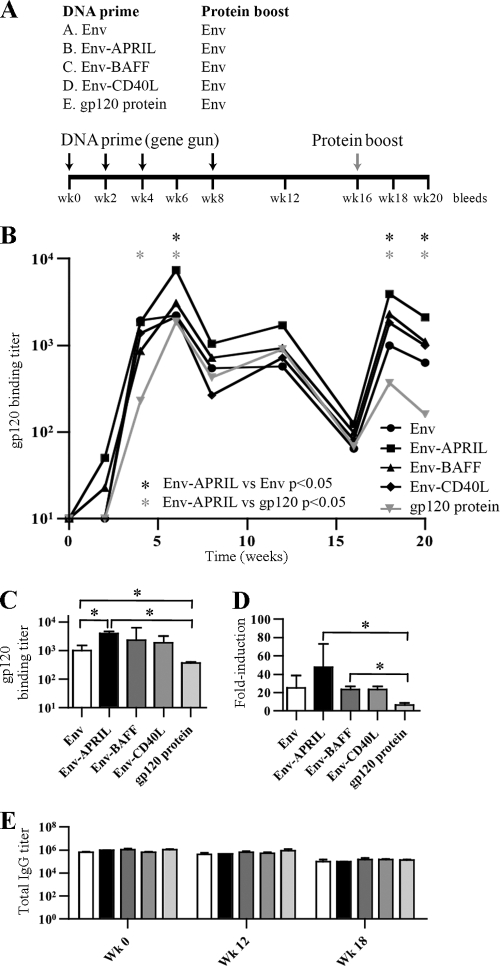
We assessed whether targeting Env to B cells via APRIL, BAFF, or CD40L improves antibody responses in vivo. As outlined in Fig. 3A, groups of 4 rabbits were primed with DNA plasmids encoding unmodified Env (group A), Env-APRIL (group B), Env-BAFF (group C), or Env-CD40L (group D) via gene gun immunization or received monomeric gp120 protein in Imject Alum (group E). This experimental design was chosen because the DNA can be taken up by dermal DCs, which then mature and migrate to draining lymph nodes. There, the Env fusion protein can be expressed in a B-cell-rich environment, providing the opportunity for B cells to be stimulated both by the Env component, through the B-cell receptor (BCR), and by the incorporated costimulator (i.e., APRIL, BAFF, or CD40L). The gp120 protein arm (group E) allows a comparison with immunization regimens that have been used in human clinical trials (32, 76, 80).
Fig 3.
Env-APRIL induces higher antibody titers than Env in rabbits. (A) Rabbit immunization scheme. (B) Midpoint anti-gp120 IgG titers over the course of the immunization experiment as determined by ELISA. The mean values from 4 rabbits per group are given. (C) Midpoint anti-gp120 IgG titers at week 18. The mean values ± standard errors of the means (SEM) derived from 4 rabbits are shown. (D) Fold induction of IgG anti-gp120 titers. The mean relative increases ± SEM of the results from 4 rabbits at week 18 compared to week 16 are given. (E) Total IgG midpoint titers at weeks 0, 12, and 18. Mean values ± SEM are given. * indicates P < 0.05 as determined by a one-tailed Mann-Whitney test.
All the rabbits were boosted with unmodified, cleaved JR-FL SOSIP.R6 gp140 protein in Quil A, a saponin-based adjuvant. The rationale for this choice was 2-fold. First, using the same boosting protein for all groups allowed us to attribute any differences in boosting efficiency to the induction of different memory responses during the priming phase. Second, by boosting with cleaved trimers, we hoped to promote responses that recognize fully processed Env and hence might have neutralization activity.
The Env-APRIL, Env-BAFF, and Env-CD40L proteins all induced higher anti-gp120 titers than unmodified Env (Fig. 3B and C). As shown by a Mann-Whitney test, the differences were significant (P < 0.05) at weeks 6, 18, and 20 for Env-APRIL compared to Env (4-, 4-, and 3-fold, respectively), and at weeks 4, 6, 18, and 20 for Env-APRIL compared to gp120 (8-, 4-, 11-, and 13-fold, respectively). A Kruskal-Wallis test showed that the titers in the five groups differed (P < 0.05). Env-APRIL gave significantly greater gp120-binding titers compared to monomeric gp120 at weeks 18 and 20 (P < 0.05; Dunn's posttest). Similar trends were observed for Env-BAFF and Env-CD40L, but the differences were not statistically significant. To verify whether the cis-adjuvants specifically enhanced the antibody levels for Env only, we also measured total IgG levels (Fig. 3D). At each time point, IgG levels were found to be equal for all sera, indicating that the observed anti-Env titer enhancement was specific.
Measuring the changes in anti-gp120 titer after week 16, when all the animals were boosted with trimeric SOSIP.R6 gp140 proteins, allowed a comparison of the memory responses that had been induced in the priming phase by the different plasmids. The anti-gp120 IgG titers increased by 48-fold between weeks 16 and 18 in the Env-APRIL-primed rabbits, while the corresponding increases in the Env, Env-BAFF, and Env-CD40L groups were 25-, 23-, and 23-fold, respectively (Fig. 3E). In the animals primed with gp120 protein, the titer increase during the same 2-week period was only 6-fold. The differences between Env-BAFF and gp120 and between Env-APRIL and gp120 were significant (P < 0.05) (Fig. 3E). Overall, these data suggest that the Env-APRIL construct induces effective memory responses.
Env-APRIL induces stronger neutralizing antibody responses in rabbits.
Since Env-based immunogens induce mostly nonneutralizing antibodies, anti-gp120 binding antibody titers rarely correlate with neutralization titers. We therefore measured the ability of sera to neutralize HIV-1 SF162 and JR-FL Env-pseudotyped viruses in a single-cycle assay using TZM-bl reporter cells. SF162 is a neutralization-sensitive tier 1 virus (52, 91), whereas JR-FL, from which our Env immunogens were derived, is a relatively resistant tier 2 virus. Neutralization of tier 1 viruses provides a “low bar” measure of functional antibodies. Prebleed sera were inactive against SF162 (50% neutralization < 10), but postimmunization samples from most animals were able to neutralize this virus (Table 1). After the gp140 protein boost at week 18, the highest SF162 neutralization titers were seen in the group primed with Env-APRIL. At the earlier time points (weeks 6 and 12), SF162 was also more frequently neutralized by Env-APRIL sera. Neutralization of JR-FL was observed sporadically in the Env-fusion protein arms but not at all in the Env- or gp120-immunized groups (Table 1). Neutralization results for SF162 and JR-FL were not correlated, indicating that different nAb specificities were responsible.
The Duke Central Immunology Laboratory for AIDS Vaccine Research and Development performed an independent neutralization analysis using the three tier 1 viruses SF162 and MN (both tier 1A) and BaL (tier 1B) and the tier 2 viruses JRFL, 6535.3, QH0692.42, PVO.4, and RHPA4259.7. Although most of the sera were active against the tier 1 viruses (Table 2), no appreciable tier 2 virus neutralization was observed (data not shown). The titers against SF162 determined in the Central Immunology Laboratory were generally considerably higher than those determined at the Academic Medical Center (compare Tables 1 and 2), but the overall patterns were similar. Again, the most consistent and strongest responses to SF162 arose in the Env-APRIL group, with a similar pattern observed for neutralization of the other tier 1A virus, MN. BaL, a tier 1B virus, is more resistant to neutralization than SF162 and MN (91). Accordingly, only a subset of the rabbit sera neutralized BaL and did so at lower titers. Nonetheless, the most consistent anti-BaL responses were again seen in the Env-APRIL group, where 4 of 4 rabbit sera were able to neutralize this virus. In contrast, sera from only 1 of 4 rabbits in each of the Env, Env-BAFF, and Env-CD40L groups neutralized BaL, while no sera from animals immunized with monomeric gp120 had this activity.
Neutralization is mediated by IgG and dominated by V3 specificities.
To assess whether anything other than antibody, such as interferons, chemokines, or factors influencing cell viability, might have affected the neutralization assays nonspecifically, we used protein G-coupled agarose beads to purify IgG from several sera showing potent SF162 neutralization. The depleted sera, the purified IgG recovered from the columns, and the unfractionated sera were all tested against SF162. Depletion of IgG almost completely eliminated the neutralizing activity of the sera, while the purified IgG was as active as the original sera (data not shown).
Tier 1 virus neutralization is often dominated by anti-V3 antibodies (5, 52). To test whether this was so for our sera, we performed SF162 neutralization experiments in the presence of three overlapping V3 peptides or an unrelated, control peptide (Table 3). The 50% SF162 neutralization titers in the presence of the control peptide were comparable to those obtained in the absence of any peptide (compare Tables 1 and 3). In contrast, the 50% neutralization titers were considerably lower when the V3 peptides were present, indicating that V3 specificities constituted a substantial proportion of the total SF162 neutralization activity in the rabbit sera. Some of the sera did still neutralize SF162 to various extents even when the V3 peptides were present, sera B293 and B295 (both Env-APRIL), C297 (Env-BAFF), and D303 (Env-CD40L) being examples. Thus, these rabbits had developed nAb specificities other than ones to V3. In contrast, none of the Env- or gp120-immunized rabbits developed any V3-independent nAbs.
The neutralizing antibody response matures over time.
During natural infection, heterologous nAbs appear only later in the course of infection (36, 66, 84, 98). In our study, the average neutralization titer increased over time, particularly in the Env-APRIL arm (Fig. 4A). To assess whether this reflected an increase in the quantity or quality of nAbs, we divided the 50% SF162 neutralization titers by the 50% gp120 binding titers (Fig. 4B). The resulting ratios increased significantly over time, suggesting that the increase in SF162 neutralization is attributable to qualitative changes in antibodies over time and not just to quantitative increases (i.e., binding antibody titer increases). When the increases in neutralization/binding ratios were compared for the various experimental groups, there was no statistically significant difference (data not shown), implying that the maturation of the neutralizing response was independent of the antigens and cis-adjuvants evaluated.
Fig 4.
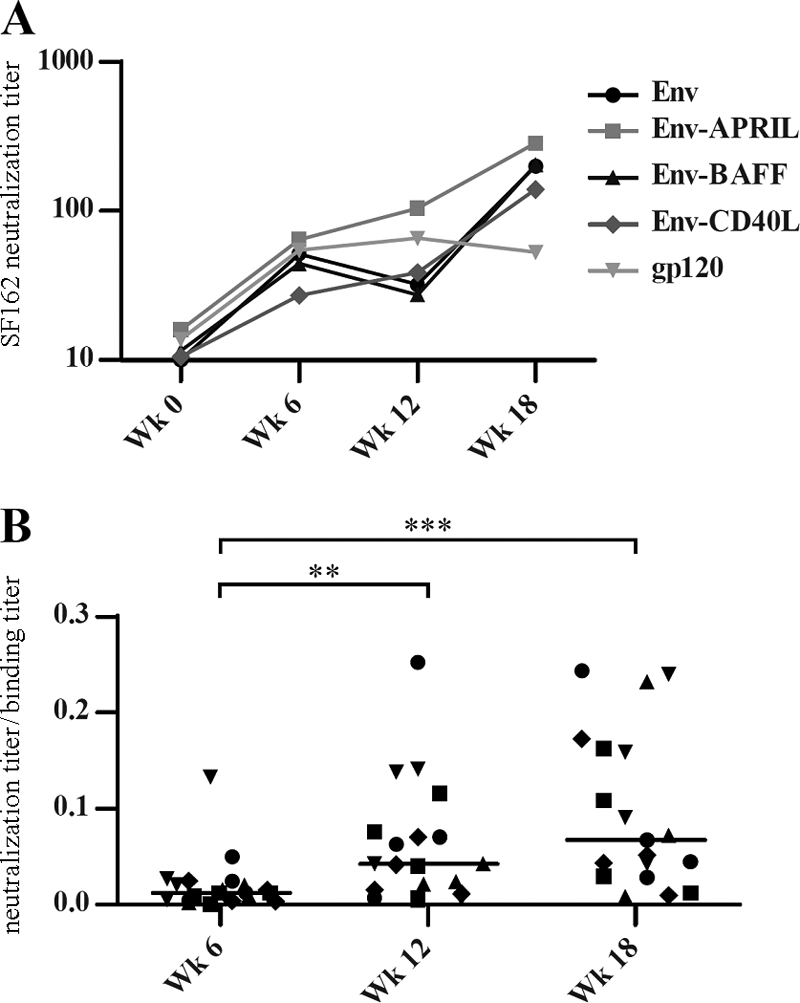
nAb responses mature over time. (A) The mean 50% SF162 neutralization titers are shown for each group. (B) The ratios of the 50% SF162 neutralization titers and the midpoint anti-gp120 titers at weeks 6, 12, and 18 are plotted for each rabbit. ** and *** indicate P < 0.01 and P < 0.001, respectively, as determined by a one-tailed Mann-Whitney test.
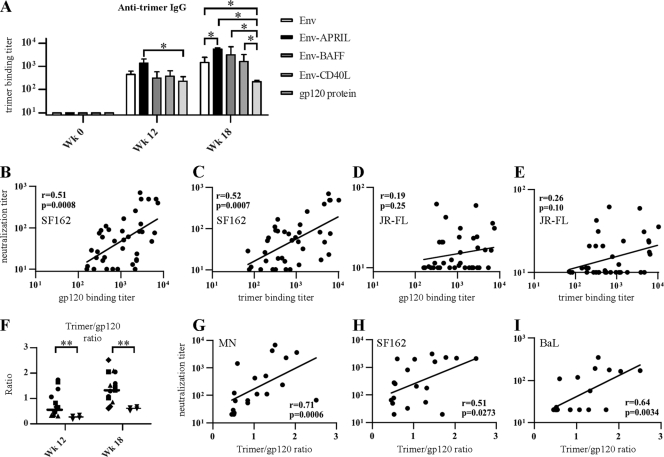
Env-APRIL induces improved anti-trimer responses that correlate with neutralization.
nAbs are uniquely able to recognize the functional, trimeric Env spike. We therefore determined whether the rabbit sera could bind recombinant gp140 trimers, while realizing that such proteins are only imperfect mimics of the native spike. To do so, we used a novel trimer ELISA that employs a construct similar to the Env protein used in the control immunization group. Specifically, the H8 tag of the immunizing Env protein was replaced by a D7324 epitope tag for use in the antigen capture ELISA (30, 63). In this assay, all the week 12 and 18 sera were found to contain Env trimer-reactive antibodies (Fig. 5A). The trimer-binding antibody titers were highest in the Env-APRIL group, and those were significantly greater than the titers determined for the unmodified Env trimer and gp120 monomer groups (which contained the lowest titers). Moreover, unlike the results seen with the other groups, the anti-trimer titers did not increase in the monomeric gp120 group after the Env trimer boost at week 16 (Fig. 5A).
Fig 5.
nAb titers correlate with anti-trimer binding antibody titers. (A) Mean midpoint trimer-binding titers ± SEM as determined by ELISA at weeks 0, 12, and 18. * indicates P < 0.05. (B to E) The 50% SF162 neutralization titers of each individual rabbit (week 12 and 18) are plotted against the midpoint anti-gp120 binding antibody titer (B) and the midpoint anti-trimer binding antibody titer (C). (D and E) Data were determined as described for panels B and C, but only 50% JR-FL neutralization titers are shown. (F) Ratios of the midpoint anti-trimer/anti-gp120 binding antibody titers at weeks 0, 12, and 18. Symbols are the same as described for Fig. 4. ** indicates P < 0.01 as determined by a one-tailed Mann-Whitney test. (G to I) Midpoint neutralization titers for MN (G), SF162.LS (H), and BaL.26 (I) are plotted against the ratio of the midpoint anti-trimer/anti-gp120 binding antibody titers. The Spearman r-values and two-tailed P values are shown.
To investigate whether the presence of anti-trimer antibodies predicted virus neutralization, we plotted the midpoint neutralization titers for SF162 and JR-FL against both the anti-gp120 and anti-trimer binding antibody titers (Fig. 5B to E). There was a strong correlation between the SF162 neutralization titers and both the anti-gp120 and anti-trimer titers (Fig. 5B and C). In contrast, the same correlations were poor and not significant for JR-FL neutralization (Fig. 5D and E).
To gauge what proportions of the antibodies recognize monomers and trimers, we also calculated the ratios of the anti-trimer and anti-gp120 binding antibody titers. There were no significant differences between the Env, Env-APRIL, Env-BAFF, and Env-CD40L groups (data not shown), consistent with the use of the same Env moiety to immunize all the groups. Thus, conjugation of Env trimers to, e.g., APRIL increases the antibody responses reactive with both monomeric gp120 and trimeric gp140, but it does not change the relative proportions of the two categories of Env-reactive antibodies. In contrast, sera from the gp120-primed rabbits contained a significantly lower proportion of trimer-reactive antibodies (Fig. 5E). Thus, the trimer/gp120 binding antibody ratio in this group was 0.26 at week 12, while the average ratio across the trimer-primed rabbits (i.e., the Env, Env-APRIL, Env-BAFF, and Env-CD40L groups combined) was 2.7-fold greater at 0.71 (P < 0.01). Furthermore, the trimer/gp120 binding antibody ratio was ∼2-fold higher in all the groups after the gp140 trimer protein boost at week 16, when the differential between the trimer-primed and gp120-only animals was 2.5-fold (1.27 versus 0.51; P < 0.01). The data suggest that gp140 trimers are more effective than monomeric gp120 at inducing trimer-reactive antibodies.
We next plotted the trimer/monomer binding antibody ratios against the 50% neutralization titers of the tier 1 viruses MN, SF162, and BaL (Fig. 5G to I). For each of these viruses, there was a strong correlation between neutralization and the trimer/monomer ratio. Hence, trimer-reactive antibodies are associated with virus neutralization, at least for tier 1 viruses.
Sera from Env trimer-immunized rabbits recognize the native trimer on virus particles.
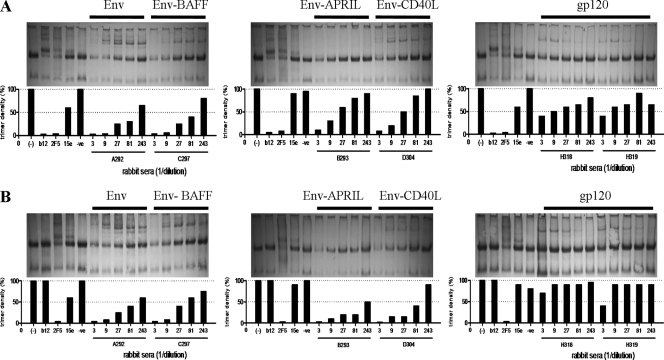
We next investigated whether our sera recognized native trimers on virus particles by using a BN-PAGE trimer shift assay (Fig. 6) (67). In this assay, nAbs b12 and 2F5 efficiently depleted trimers, but the nonneutralizing antibody 15e did not (Fig. 6A). The intensities of the trimer bands are represented by histograms beneath each blot. Thus, the presence of abundant trimer-binding antibodies depleted the trimers, represented by short bars, and the converse occurred when trimer-binding antibodies were infrequent (tall bars). Representative sera from each rabbit group are shown in Fig. 6A. Most sera from animals immunized with Env trimers bound well to native JR-FL trimers (Fig. 6A). In contrast, sera from gp120 recipients recognized the same trimers only poorly (Fig. 6A).
Fig 6.
Sera from gp140-immunized animals recognize native trimers. The binding of representative sera from each group to native JR-FL Env trimers (A) and JR-FL D368R mutant trimers (B) was examined. nAbs b12 and 2F5 and nonneutralizing antibody 15e served as controls. A quantitative evaluation of the trimer band intensity is presented in the bar graphs, where small bars represent efficient trimer binding of the sera and a resulting decrease in the intensity of the trimer band.
To test whether nAbs are directed to the CD4 binding site (CD4bs), we assessed the binding of the same set of sera to D368R-mutant trimers (Fig. 6B). Residue D368 is situated in the CD4 binding loop, and the D-to-R substitution knocks out binding of most CD4BS antibodies, including b12 (5) (Fig. 6; compare b12 binding results in panels A and B). The sera from the Env trimer-immunized rabbits depleted the D368R trimers as effectively as they did the wild-type trimers (Fig. 6). This finding implies that few if any of the antibodies in these sera target the CD4BS, which is consistent with the V3-dominated response (Table 3). The two sera from gp120 recipients bound only very weakly to the D368R trimers, as was observed with the wild-type trimers (Fig. 6).
DISCUSSION
The goal of this study was to improve the humoral response to HIV-1 by targeting trimeric Env to B cells. Most previous immunotargeting strategies have focused on DCs, since these cells play a critical role in the induction of immunity, particularly T-cell responses. Targeting antigens to DCs via lectins such as DEC205 and Clec9A has proven to be useful in this regard (10, 16, 71, 94), although the addition of costimulatory signals such as CD40L may be necessary to overcome tolerance (71), and targeting though CD40L directly may provide an alternative approach (63). While antigen targeting to DCs followed by antigen processing and presentation is beneficial for T-cell responses, B-cell responses require direct contact between naïve B cells and intact antigen. And if the goal is to induce broadly active nAbs, the Env antigen must be conformationally intact. We therefore targeted recombinant Env trimers directly to B cells, via fusion to the sequences of the TNFSF members APRIL, BAFF, and CD40L. Systems biology analyses of the responses to yellow fever and influenza vaccines, as well as a recent mouse study on the important of TACI in durable protection against influenza, support the exploitation of the APRIL/BAFF axis in vaccine design (69, 78, 106).
We showed that all three fusion constructs activate human B cells in vitro and induce the secretion of IgM, IgG, and IgA. However, in the rabbit immunization studies, only the Env-APRIL construct induced significantly better anti-Env antibody responses than Env alone. We do not know why Env-BAFF and Env-CD40L were less effective than Env-APRIL in vivo. One reason may be that APRIL and Env can both bind to heparan sulfate proteoglycans, whereas BAFF and CD40L do not. Another explanation may be that the in vitro experiments do not faithfully mimic the environment and stimuli for B-cell activation in vivo. Furthermore, the in vitro experiments address only the antigen-independent component of B-cell activation by the cis-adjuvant; they do not recapitulate the interplay between activation via APRIL, BAFF, or CD40L and the additional ligation of the BCR by the Env antigen itself. The stronger antibody responses induced by Env-APRIL, compared to the other Env fusion proteins, presumably arose because APRIL was the best activator of B cells after immunization. However, it is also possible that APRIL interacted more efficiently with other cell types, such as DCs. Some DC subsets reportedly express TACI, which could make them sensitive to APRIL but also to BAFF (18, 56). However, if DCs had been the prime targets, we would have expected a stronger response to Env-CD40L, given that CD40L is a very potent activator of these cells (17, 55, 93, 99). Overall, we believe that activation of B cells, and not DCs, was the reason for the improved Ab response induced by Env-APRIL, but this remains to be proved.
One concern with using costimulatory molecules in vaccines is that they might cause nonspecific immune activation. We did not find any evidence that the inclusion of CD40L, APRIL, and BAFF in the Env immunogens caused any nonspecific activation of B cells; there was no increase in total IgG, only in anti-Env antibody titers (Fig. 3). This finding is consistent with the concept underlying cis-adjuvants; the covalent linkage of antigen and adjuvant is intended to activate only those immune cells that also interact with the antigen (19, 27, 31, 41, 42, 53, 63, 95, 104, 105). When antigen and adjuvant are merely mixed, it is plausible that the two components become separated by diffusion, resulting in the antigen losing its costimulator and the adjuvant nonspecifically activating cells that do not encounter the antigen. The strategy of covalent linkage overcomes both these concerns.
The gp140 trimer-encoding DNA constructs were better than monomeric gp120 at inducing antibodies that recognize trimeric Env, as demonstrated by the higher trimer/monomer binding antibody ratio (Fig. 5F). Furthermore, this higher trimer/monomer ratio correlated with neutralization (Fig. 5G to I). We cannot completely exclude the possibility that antigen delivery during the priming phase (gp140 DNA versus gp120 protein) is a contributory variable, but we do not favor this explanation. The correlation between ELISA trimer/monomer binding ratios and the SF162 neutralization titers is consistent with the need for HIV-1 vaccines to elicit antibodies capable of interacting with functional Env on virions, although there are two caveats. First, the correlations were found only with the neutralization-sensitive tier 1 isolates SF162, MN, and BaL (neutralization of tier 2 viruses was too sporadic to be analyzed in this way). Second, the soluble JR-FL trimers used in the ELISA experiments are imperfect mimics of the functional trimers found on JR-FL virions. To address the latter point, we also studied binding of the rabbit sera to virion-associated trimers, noting that binding titers of HIV-1-infected human sera to such trimers reportedly correlate well with nAb titers (22). All our trimeric Env immunogens elicited Abs reactive with native JR-FL trimers more efficiently than the gp120 monomer. Overall, we conclude that trimeric Env immunogens are superior to monomeric gp120 at inducing trimer-reactive antibodies and, against tier 1 viruses, neutralizing antibodies.
As noted above, conjugating Env to APRIL increased both the anti-gp120 binding Ab and SF162 nAb titers compared to Env alone, but the two measures of immunogenicity were correlated. Hence, the improvement is in the quantity of anti-Env antibodies produced and not in their quality (i.e., Env-APRIL did not induce a specific increase in only virus-neutralizing antibodies). This outcome was not unexpected, as the Env antigen components were similar in the Env and Env-APRIL immunogens, which should result in similar antibody specificities. Considering the natural role of APRIL in affinity maturation (38, 56, 57), it is possible that fusion to APRIL could also improve the quality of the antibody response by inducing antibodies with the same specificity but higher affinity, but we did not assess this point in vivo. Ideally, APRIL should be fused to an Env immunogen that optimally induces antibodies against conserved neutralization epitopes. Improving the design and delivery of Env trimers so that tier 2 viruses are also neutralized remains a major goal of HIV-1 vaccine research.
Overall, the concept of targeting antigens to B cells by the use of cis-adjuvants could be useful for Env-based subunit vaccines and also for vaccines directed at other pathogens. Because TNFSF members recognize a variety of cell types and the nature of the response can be adjusted by selecting different superfamily members, this approach has flexibility. Targeting B cells in this way may be a useful addition, or an alternative, to antigen-targeting strategies aimed at DCs, in particular when the goal is to improve humoral immune responses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dennis Burton and James Robinson for reagents. The technical assistance from Kenneth Kang is greatly appreciated.
This research was supported in part by the AIDS Fonds (the Netherlands) grants 2005021 (B.B.) and 2008013 (R.W.S.) and by NIH grants R37 AI36082 (J.P.M.), P01 AI82362 (W.C.O.), and AI58763 and AI84714 (J.M.B.), the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (J.M.B.; CAVD-VIMC grant 38619), and the Torrey Pines Institute's AIDS and Infectious Disease Science Center. R.W.S. is a recipient of Veni and Vidi fellowships from the Netherlands Organization for Scientific Research (NWO), a Mathilde Krim research fellowship from the American Foundation for AIDS Research (amfAR), and a Starting Investigator grant from the European Research Council (ERC-StG-2011-280829-SHEV).
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Banerjee K, et al. 2009. Enzymatic removal of mannose moieties can increase the immune response to HIV-1 gp120 in vivo. Virology 389:108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barouch D. H., Korber B. 2010. HIV-1 vaccine development after STEP. Annu. Rev. Med. 61:153–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beddows S, et al. 2007. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 360:329–340 [DOI] [PubMed] [Google Scholar]
- 4. Beddows S, et al. 2005. Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 79:8812–8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Binley JM, et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Binley JM, et al. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Binley JM, et al. 2002. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J. Virol. 76:2606–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bontjer I, et al. 2009. Optimization of human immunodeficiency virus type 1 envelope glycoproteins with V1/V2 deleted, using virus evolution. J. Virol. 83:368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bontjer I, et al. 2010. Stabilized HIV-1 envelope glycoprotein trimers lacking the V1V2 domain, obtained by virus evolution. J. Biol. Chem. 285:36456–36470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boscardin SB, et al. 2006. Antigen targeting to dendritic cells elicits long-lived T cell help for antibody responses. J. Exp. Med. 203:599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bossen C, et al. 2008. TACI, unlike BAFF-R, is solely activated by oligomeric BAFF and APRIL to support survival of activated B cells and plasmablasts. Blood 111:1004–1012 [DOI] [PubMed] [Google Scholar]
- 12. Bower JF, Yang X, Sodroski J, Ross TM. 2004. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J. Virol. 78:4710–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buchbinder SP, et al. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burton DR, et al. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236 [DOI] [PubMed] [Google Scholar]
- 15. Burton DR, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024–1027 [DOI] [PubMed] [Google Scholar]
- 16. Caminschi I, et al. 2008. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood 112:3264–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caux C, et al. 1994. Activation of human dendritic cells through CD40 cross-linking. J. Exp. Med. 180:1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang SK, Mihalcik SA, Jelinek DF. 2008. B lymphocyte stimulator regulates adaptive immune responses by directly promoting dendritic cell maturation. J. Immunol. 180:7394–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen TT, Tao MH, Levy R. 1994. Idiotype-cytokine fusion proteins as cancer vaccines. Relative efficacy of IL-2, IL-4, and granulocyte-macrophage colony-stimulating factor. J. Immunol. 153:4775–4787 [PubMed] [Google Scholar]
- 20. Chu VT, Enghard P, Riemekasten G, Berek C. 2007. In vitro and in vivo activation induces BAFF and APRIL expression in B cells. J. Immunol. 179:5947–5957 [DOI] [PubMed] [Google Scholar]
- 21. Corti D, et al. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS One 5:e8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crooks ET, et al. 2008. Relationship of HIV-1 and SIV envelope glycoprotein trimer occupation and neutralization. Virology 377:364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crooks ET, Tong T, Osawa K, Binley JM. 2011. Enzyme digests eliminate nonfunctional Env from HIV-1 particle surfaces, leaving native Env trimers intact and viral infectivity unaffected. J. Virol. 85:5825–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daly LM, et al. 2005. Innate IL-10 promotes the induction of Th2 responses with plasmid DNA expressing HIV gp120. Vaccine 23:963–974 [DOI] [PubMed] [Google Scholar]
- 25. De Milito A, et al. 2004. Mechanisms of hypergammaglobulinemia and impaired antigen-specific humoral immunity in HIV-1 infection. Blood 103:2180–2186 [DOI] [PubMed] [Google Scholar]
- 26. Dillon SR, Gross JA, Ansell SM, Novak AJ. 2006. An APRIL to remember: novel TNF ligands as therapeutic targets. Nat. Rev. Drug Discov. 5:235–246 [DOI] [PubMed] [Google Scholar]
- 27. Eckl-Dorna J, Batista FD. 2009. BCR-mediated uptake of antigen linked to TLR9 ligand stimulates B-cell proliferation and antigen-specific plasma cell formation. Blood 113:3969–3977 [DOI] [PubMed] [Google Scholar]
- 28. Eggink D, et al. 2009. Detailed mechanistic insights into HIV-1 sensitivity to three generations of fusion inhibitors. J. Biol. Chem. 284:26941–26950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eggink D, Melchers M, Sanders RW. 2007. Antibodies to HIV-1: aiming at the right target. Trends Microbiol. 15:291–294 [DOI] [PubMed] [Google Scholar]
- 30. Eggink D, et al. 2010. Lack of complex N-glycans on HIV-1 envelope glycoproteins preserves protein conformation and entry function. Virology 401:236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faulkner L, et al. 2001. IL-2 linked to a peptide from influenza hemagglutinin enhances T cell activation by affecting the antigen-presentation function of bone marrow-derived dendritic cells. Int. Immunol. 13:713–721 [DOI] [PubMed] [Google Scholar]
- 32. Flynn NM, et al. 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191:654–665 [DOI] [PubMed] [Google Scholar]
- 33. Forsell MN, Schief WR, Wyatt RT. 2009. Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr. Opin. HIV AIDS 4:380–387 [DOI] [PubMed] [Google Scholar]
- 34. Gilbert PB, et al. 2005. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J. Infect. Dis. 191:666–677 [DOI] [PubMed] [Google Scholar]
- 35. Goodnow CC, Sprent J, Fazekas de St Groth B, Vinuesa CG. 2005. Cellular and genetic mechanisms of self tolerance and autoimmunity. Nature 435:590–597 [DOI] [PubMed] [Google Scholar]
- 36. Gray ES, et al. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Green TD, Montefiori DC, Ross TM. 2003. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. J. Virol. 77:2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Groom JR, et al. 2007. BAFF and MyD88 signals promote a lupuslike disease independent of T cells. J. Exp. Med. 204:1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harris A, et al. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He B, et al. 2006. HIV-1 envelope triggers polyclonal Ig class switch recombination through a CD40-independent mechanism involving BAFF and C-type lectin receptors. J. Immunol. 176:3931–3941 [DOI] [PubMed] [Google Scholar]
- 41. Heath AW, Devey ME, Brown IN, Richards CE, Playfair JH. 1989. Interferon-gamma as an adjuvant in immunocompromised mice. Immunology 67:520–524 [PMC free article] [PubMed] [Google Scholar]
- 42. Heath AW, Playfair JH. 1990. Conjugation of interferon-gamma to antigen enhances its adjuvanticity. Immunology 71:454–456 [PMC free article] [PubMed] [Google Scholar]
- 43. Herrera C, et al. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 77:1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ingold K, et al. 2005. Identification of proteoglycans as the APRIL-specific binding partners. J. Exp. Med. 201:1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kang YK, et al. 2009. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine 27:5120–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kern C, et al. 2004. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood 103:679–688 [DOI] [PubMed] [Google Scholar]
- 47. Kimberley FC, et al. 2009. The proteoglycan (heparan sulfate proteoglycan) binding domain of APRIL serves as a platform for ligand multimerization and cross-linking. FASEB J. 23:1584–1595 [DOI] [PubMed] [Google Scholar]
- 48. Kirschner M, et al. 2006. The production of cleaved, trimeric human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein vaccine antigens and infectious pseudoviruses using linear polyethylenimine as a transfection reagent. Protein Expr. Purif. 48:61–68 [DOI] [PubMed] [Google Scholar]
- 49. Klasse PJ, Sanders RW, Cerutti A, Moore JP. 17 April 2011, posting date How can HIV-type-1-Env immunogenicity be improved to facilitate antibody-based vaccine development? AIDS Res. Hum. Retroviruses doi:10.1089/aid.2011.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koch M, Frazier J, Sodroski J, Wyatt R. 2005. Characterization of antibody responses to purified HIV-1 gp120 glycoproteins fused with the molecular adjuvant C3d. Virology 340:277–284 [DOI] [PubMed] [Google Scholar]
- 51. Kwong PD, Wilson IA. 2009. HIV-1 and influenza antibodies: seeing antigens in new ways. Nat. Immunol. 10:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li M, et al. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lim YS, et al. 1998. Vaccination with an ovalbumin/interleukin-4 fusion DNA efficiently induces Th2 cell-mediated immune responses in an ovalbumin-specific manner. Arch. Pharm. Res. 21:537–542 [DOI] [PubMed] [Google Scholar]
- 54. Litinskiy MB, et al. 2002. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat. Immunol. 3:822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma DY, Clark EA. 2009. The role of CD40 and CD154/CD40L in dendritic cells. Semin. Immunol. 21:265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mackay F, Schneider P. 2009. Cracking the BAFF code. Nat. Rev. Immunol. 9:491–502 [DOI] [PubMed] [Google Scholar]
- 57. Mackay F, Schneider P. 2008. TACI, an enigmatic BAFF/APRIL receptor, with new unappreciated biochemical and biological properties. Cytokine Growth Factor Rev. 19:263–276 [DOI] [PubMed] [Google Scholar]
- 58. Mackay F, Schneider P, Rennert P, Browning J. 2003. BAFF AND APRIL: a tutorial on B cell survival. Annu. Rev. Immunol. 21:231–264 [DOI] [PubMed] [Google Scholar]
- 59. Martinelli E, et al. 2007. HIV-1 gp120 inhibits TLR9-mediated activation and IFN-{alpha} secretion in plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 104:3396–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mascola JR, Montefiori DC. 2010. The role of antibodies in HIV vaccines. Annu. Rev. Immunol. 28:413–444 [DOI] [PubMed] [Google Scholar]
- 61. McCormick AL, Thomas MS, Heath AW. 2001. Immunization with an interferon-gamma-gp120 fusion protein induces enhanced immune responses to human immunodeficiency virus gp120. J. Infect. Dis. 184:1423–1430 [DOI] [PubMed] [Google Scholar]
- 62. McElrath MJ, et al. 2008. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet 372:1894–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Melchers M, et al. 2011. A stabilized HIV-1 envelope glycoprotein trimer fused to CD40 ligand targets and activates dendritic cells. Retrovirology 8:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mestecky J, et al. 2004. Paucity of antigen-specific IgA responses in sera and external secretions of HIV-type 1-infected individuals. AIDS Res. Hum. Retroviruses 20:972–988 [DOI] [PubMed] [Google Scholar]
- 65. Mestecky J, Moldoveanu Z, Smith PD, Hel Z, Alexander RC. 2009. Mucosal immunology of the genital and gastrointestinal tracts and HIV-1 infection. J. Reprod. Immunol. 83:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mikell I, et al. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moore PL, et al. 2006. Nature of nonfunctional envelope proteins on the surface of human immunodeficiency virus type 1. J. Virol. 80:2515–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Muster T, et al. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nakaya HI, et al. 2011. Systems biology of vaccination for seasonal influenza in humans. Nat. Immunol. 12:786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nayak BP, Sailaja G, Jabbar AM. 2003. Enhancement of gp120-specific immune responses by genetic vaccination with the human immunodeficiency virus type 1 envelope gene fused to the gene coding for soluble CTLA4. J. Virol. 77:10850–10861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nchinda G, et al. 2008. The efficacy of DNA vaccination is enhanced in mice by targeting the encoded protein to dendritic cells. J. Clin. Invest. 118:1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nimal S, Heath AW, Thomas MS. 2006. Enhancement of immune responses to an HIV gp120 DNA vaccine by fusion to TNF alpha cDNA. Vaccine 24:3298–3308 [DOI] [PubMed] [Google Scholar]
- 73. Nimal S, McCormick AL, Thomas MS, Heath AW. 2005. An interferon gamma-gp120 fusion delivered as a DNA vaccine induces enhanced priming. Vaccine 23:3984–3990 [DOI] [PubMed] [Google Scholar]
- 74. Pantophlet R, Burton DR. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24:739–769 [DOI] [PubMed] [Google Scholar]
- 75. Parren PW, Sattentau QJ, Burton DR. 1997. HIV-1 antibody—debris or virion? Nat. Med. 3:366–367 [DOI] [PubMed] [Google Scholar]
- 76. Pitisuttithum P, et al. 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 194:1661–1671 [DOI] [PubMed] [Google Scholar]
- 77. Poignard P, et al. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Querec TD, et al. 2009. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 10:116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Reitter JN, Means RE, Desrosiers RC. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679–684 [DOI] [PubMed] [Google Scholar]
- 80. Rerks-Ngarm S, et al. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 81. Robb ML. 2008. Failure of the Merck HIV vaccine: an uncertain step forward. Lancet 372:1857–1858 [DOI] [PubMed] [Google Scholar]
- 82. Sailaja G, Husain S, Nayak BP, Jabbar AM. 2003. Long-term maintenance of gp120-specific immune responses by genetic vaccination with the HIV-1 envelope genes linked to the gene encoding Flt-3 ligand. J. Immunol. 170:2496–2507 [DOI] [PubMed] [Google Scholar]
- 83. Sanders RW, et al. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 76:8875–8889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Scapini P, Bazzoni F, Cassatella MA. 2008. Regulation of B-cell-activating factor (BAFF)/B lymphocyte stimulator (BLyS) expression in human neutrophils. Immunol. Lett. 116:1–6 [DOI] [PubMed] [Google Scholar]
- 86. Schäfer F, et al. 2002. Lack of simian immunodeficiency virus (SIV) specific IgA response in the intestine of SIV infected rhesus macaques. Gut 50:608–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Schägger H, Cramer WA, von Jagow G. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220–230 [DOI] [PubMed] [Google Scholar]
- 88. Schägger H, von Jagow G. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223–231 [DOI] [PubMed] [Google Scholar]
- 89. Scheid JF, et al. 2011. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333:1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Schneider JA, et al. 2007. Mucosal HIV-binding antibody and neutralizing activity in high-risk HIV-uninfected female participants in a trial of HIV-vaccine efficacy. J. Infect. Dis. 196:1637–1644 [DOI] [PubMed] [Google Scholar]
- 91. Seaman MS, et al. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J. Virol. 84:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shan M, et al. 2007. HIV-1 gp120 mannoses induce immunosuppressive responses from dendritic cells. PLoS Pathog. 3:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shreedhar V, et al. 1999. Dendritic cells require T cells for functional maturation in vivo. Immunity 11:625–636 [DOI] [PubMed] [Google Scholar]
- 94. Soares H, et al. 2007. A subset of dendritic cells induces CD4+ T cells to produce IFN-gamma by an IL-12-independent but CD70-dependent mechanism in vivo. J. Exp. Med. 204:1095–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tighe H, et al. 2000. Conjugation of protein to immunostimulatory DNA results in a rapid, long-lasting and potent induction of cell-mediated and humoral immunity. Eur. J. Immunol. 30:1939–1947 [DOI] [PubMed] [Google Scholar]
- 96. Trkola A, et al. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187 [DOI] [PubMed] [Google Scholar]
- 97. Trkola A, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. 2009. Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. AIDS 23:2405–2414 [DOI] [PubMed] [Google Scholar]
- 99. van Kooten C, Banchereau J. 2000. CD40-CD40 ligand. J. Leukoc. Biol. 67:2–17 [DOI] [PubMed] [Google Scholar]
- 100. van Montfort T, et al. 2011. A chimeric HIV-1 envelope glycoprotein trimer with an embedded granulocyte-macrophage colony-stimulating factor (GM-CSF) domain induces enhanced antibody and T cell responses. J. Biol. Chem. 286:22250–22261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Walker LM, et al. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Walker LM, et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 104. Wille-Reece U, et al. 2005. HIV Gag protein conjugated to a Toll-like receptor 7/8 agonist improves the magnitude and quality of Th1 and CD8+ T cell responses in nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 102:15190–15194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wille-Reece U, Wu CY, Flynn BJ, Kedl RM, Seder RA. 2005. Immunization with HIV-1 Gag protein conjugated to a TLR7/8 agonist results in the generation of HIV-1 Gag-specific Th1 and CD8+ T cell responses. J. Immunol. 174:7676–7683 [DOI] [PubMed] [Google Scholar]
- 106. Wolf AI, et al. 2011. Protective antiviral antibody responses in a mouse model of influenza virus infection require TACI. J. Clin. Invest. 121:3954–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Wu X, et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wu X, et al. 2011. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science 333:1593–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Xu W, et al. 2009. HIV-1 evades virus-specific IgG2 and IgA responses by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat. Immunol. 10:1008–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.