Abstract
Filoviruses are enveloped viruses that cause sporadic outbreaks of severe hemorrhagic fever [CDC, MMWR Morb. Mortal. Wkly. Rep. 50:73–77, 2001; Colebunders and Borchert, J. Infect. 40:16–20, 2000; Colebunders et al., J. Infect. Dis. 196(Suppl. 2):S148–S153, 2007; Geisbert and Jahrling, Nat. Med. 10:S110-S121, 2004]. Previous studies revealed that endosomal cysteine proteases are host factors for ebolavirus Zaire (Chandran et al., Science 308:1643–1645, 2005; Schornberg et al., J. Virol. 80:4174–4178, 2006). In this report, we show that infection mediated by glycoproteins from other phylogenetically diverse filoviruses are also dependent on these proteases and provide additional evidence indicating that they cleave GP1 and expose the binding domain for the critical host factor Niemann-Pick C1. Using selective inhibitors and knockout-derived cell lines, we show that the ebolaviruses Zaire and Cote d'Ivoire are strongly dependent on cathepsin B, while the ebolaviruses Sudan and Reston and Marburg virus are not. Taking advantage of previous studies of cathepsin B inhibitor-resistant viruses (Wong et al., J. Virol. 84:163–175, 2010), we found that virus-specific differences in the requirement for cathepsin B are correlated with sequence polymorphisms at residues 47 in GP1 and 584 in GP2. We applied these findings to the analysis of additional ebolavirus isolates and correctly predicted that the newly identified ebolavirus species Bundibugyo, containing D47 and I584, is cathepsin B dependent and that ebolavirus Zaire-1995, the single known isolate of ebolavirus Zaire that lacks D47, is not. We also obtained evidence for virus-specific differences in the role of cathepsin L, including cooperation with cathepsin B. These studies strongly suggest that the use of endosomal cysteine proteases as host factors for entry is a general property of members of the family Filoviridae.
INTRODUCTION
Ebdaviruses and the closely related marburgvirus comprise the family Filoviridae (6, 8, 9, 16). Several lines of recent investigation have elucidated key steps in the pathway for ebolavirus entry into cells. Ebolavirus particles attach to cells through the binding of their glycoprotein (GP) to cell surface receptors or lectins, such as TIM-1 and DC-SIGN, expressed on the plasma membrane (1, 22, 27, 29, 37). Membrane-bound particles are taken up into cells by a macropinocytosis-like mechanism and transported to late endosomes/lysosomes (LE/LY) (20, 30, 31, 34), which contain essential entry host factors. We previously showed that cleavage of ebolavirus Zaire-Mayinga (EBOV-May) GP by endosomal cysteine proteases is required for infection (7). More recent work has revealed a second host factor in LE/LY that is broadly required by filoviruses: Niemann-Pick C1 (NPC1) (5, 10), a multipass transmembrane protein that resides in the limiting membrane (44). According to a recently proposed model, virus GP is cleaved by endosomal cysteine proteases and binds to NPC1 (10).
Several studies have examined the role of protease cleavage in more detail for EBOV-May. They show that cathepsin L functions in concert with cathepsin B to cleave the GP1 subunit of virus GP (7, 35). Structural and functional studies reveal that proteases remove the heavily glycosylated carboxyl-terminal domain of GP1 to expose a more conserved domain that is closely associated with GP2 (12, 19, 25) and that is proposed to contain the binding site for the filovirus receptor (4, 13, 24, 28). Further, we recently showed that cleaved, but not uncleaved, GP1 binds to purified LE/LY membranes in an NPC1-dependent manner and coimmunoprecipitates with NPC1 (10). We identified small molecules that target NPC1, inhibit infection, and block the binding of cleaved GP1 to NPC1-containing membranes (10), strongly suggesting that the conserved N-terminal domain of GP1 is a ligand for NPC1. Taken together, these previous findings suggest a model in which proteolytic cleavage of GP to remove the carboxyl-terminal domain of GP1 and expose its N-terminal domain may be functionally analogous to the role of CD4 binding to HIV gp120 to displace highly variable loops and create the coreceptor-binding site (18). Our recent studies show that NPC1 expression is also essential for infection by virus isolates from the other species that make up the family Filoviridae (23), including the Cote d'Ivoire, Sudan, and Reston ebolaviruses, the new ebolavirus species Bundibugyo (BDBV), and Marburg virus (5, 10). In this study, we tested this model of infection by examining the endosomal cysteine protease requirements for infection by these viruses.
MATERIALS AND METHODS
Cells and cell culture conditions.
Vero, 293T, and mouse embryonic fibroblast (MEF) cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM), 100 μg/ml penicillin-streptomycin, and 2 mM l-glutamine (Invitrogen). 293T and Vero cells were carried in either 10% fetal bovine serum (FBS) or 5% FBS–5% FetalPlex (Gemini). MEF cells were carried in 10% FBS. During virus production and at least 18 h prior to infection, Vero, 293T, and MEF cells were grown in 10% FBS-containing medium. MEF cells were described previously (7). NPC1−/− and NPC1−/− Chinese hamster ovary (CHO) cells stably expressing mouse NPC1 were described previously (10).
Expression plasmids.
Expression plasmids for mouse cathepsin B, mouse cathepsin L, EBOV-May GP, and mucin deletion-containing (ΔMuc) EBOV-May GP were described previously (7). Plasmids encoding ebolavirus Cote d'Ivoire GP, ebolavirus Sudan-Boniface GP, ebolavirus Reston-Pennsylvania GP, and Lake Victoria marburgvirus Musoke GP were obtained from Anthony Sanchez (CDC) and subcloned into pCAGGS. ΔMuc GP constructs were created using PCR to remove ΔMuc EBOV-May GP amino acids (aa) 309 to 489, ΔMuc ebolavirus Cote d'Ivoire GP aa 310 to 489, ΔMuc ebolavirus Sudan GP aa 309 to 490, ΔMuc ebolavirus Reston GP aa 310 to 490, and ΔMuc Bundibugyo (BDBV) GP aa 309 to 489. This PCR added a silent XbaI site at the site of the deletion in each GP. Empty plasmid pCAGGS or a plasmid encoding β-galactosidase was used as a sham vector control.
Chimeric EBOV-May GP1/Reston ebolavirus GP2 and Reston ebolavirus GP1/EBOV-May GP2 were created by restriction digestion of the parental plasmids with XbaI (New England BioLabs) to liberate GP1- and GP2-containing fragments for each virus GP. The fragments were gel purified and ligated using T4 DNA ligase (New England BioLabs). Vectors were verified by DNA sequencing.
EBOV-May and Reston ebolavirus ΔMuc mutations were made using site-directed PCR mutagenesis. The sequences of the primers used were 5′-CTCTAGAGCCTCTGCTAACCA, 5′CATTACAGGTTAGTGAAGTCGACAAACTAGTTTGT, 5′-GTCGACTTCACTAACCTGTAATGT, 5′-ACTCTCAAAGCAACAGATATTGATCAATTGGT, 5′-TCAATATCTGTTGCTTTGAGAGT, 5′-GCTACGCACCTTTTCACTCCTCAACCGTAAGGCAATTGA, 5′-AGTGAAAAGGTGCGTAGCTCA, 5′-CTGAGGACTTACTCAATTCTTAACAGAAAAGCT, 5′-GAATTGAGTAAGTCCTCAGT, 5′-TATACTCGAGCTCGGTACCTCAAACCT, and 5′-GCTAGCTCGAGAAATCAACACA. Chimeric viruses were made as described above.
Infection assays.
Pseudotyped vesicular stomatitis virus (VSV) was created as previously described (7). Pseudotyped VSV particles encoding green fluorescent protein (GFP) (VSVGFP) were added to cells in serial 10-fold dilutions and assayed using fluorescence microscopy or flow cytometry. When using fluorescence microscopy, GFP-positive cells were counted manually and an infectious unit (IU) was defined as one GFP-expressing cell when within the linear range of dilutions. Titer was defined as IU/ml of virus added and was determined using the following formula: (number of GFP-expressing cells × dilution factor)/volume (in ml) of virus added to the well. Two-color flow cytometry was used to determine the relative ratio of infected to uninfected cathepsin B−/− cathepsin L+/+ MEF cells in cells that were cotransfected with an mRFP-expressing plasmid and a sham plasmid, mouse cathepsin B, or mouse cathepsin L. Details of this protocol were described previously (7).
Cathepsin B−/− cathepsin L−/− MEF cell rescue assays were performed with six-well dishes. Sixteen to 20 h after seeding of the dishes, the medium was changed to low (0.2%) serum. After 4 h, cells were transfected using 4 μg total DNA and 10 μl Lipofectamine 2000 (Invitrogen) that were diluted into Opti-MEM (Invitrogen). The plasmids used were pCAGGS (4 μg), mouse cathepsin B (4 μg), mouse cathepsin L (4 μg), and both cathepsin B (2 μg) and cathepsin L (2 μg). Five hours following transfection, cells were washed once with phosphate-buffered saline (PBS) and fresh medium containing 10% FBS was added. Twenty-four hours posttransfection, cells were removed with trypsin-EDTA solution (Invitrogen), spun down, and resuspended in fresh medium and 8,000 cells were added to each well of a 48-well dish. Following an additional 18 h, the medium was changed. Four hours later, pseudotyped VSV encoding luciferase (VSVluc) was added at dilutions within the linear range of infection and sufficient to give signals of 104 to 105 relative luminescence units (RLU) on cells transfected with pCAGGS. Twenty-four hours after the addition of the virus, cells were lysed using standard conditions for the firefly luciferase kit (Promega). Lysates were transferred to a 96-well plate, luciferin reagent was added, and after 1 min of incubation, RLU were measured on an EnVison plate reader (Perkin-Elmer).
Protease inhibitors and protease activity assays.
Protease inhibitors E-64, E-64d, and CA074 (Sigma) were dissolved in dimethyl sulfoxide (DMSO) and dispensed into culture medium immediately before use. The final DMSO concentration in medium was always 1% (vol/vol). Cell monolayers were preincubated with inhibitors or DMSO for 3 to 4 h at 37°C. Viruses were added directly to the culture medium containing DMSO or inhibitors, and infectivities were measured as already described.
The enzymatic activities of cathepsins B and L in acidified lysates (100 mM NaCl, 50 mM Na acetate, 0.5% Triton X-100, pH 5.5) of Vero and MEF cells were assayed as previously described (7, 46). The absence of cathepsin B activity in cathepsin B−/− cathepsin L+/+ MEF cells, the lack of cathepsin B and L activities in cathepsin B−/− cathepsin L−/− MEF cells, and the presence of similar levels of cathepsin L activity in cathepsin B−/− cathepsin L+/+ and wild-type MEF cells were confirmed.
Ebolavirus Sudan infection.
Vero cells were treated with E-64d (300 μM) or vehicle (1% DMSO) for 4 h and then infected with EBOV-May or ebolavirus Sudan-Gulu (multiplicity of infection [MOI], 0.1). After 1 h, the virus inoculums were removed by washing and fresh medium containing E-64d or vehicle was added. Cell supernatant was collected on day 3. RNA was isolated from the supernatant using Virus RNA Extraction kits (Qiagen), and ebolavirus NP RNA was measured using a quantitative reverse transcription (RT)-PCR assay (45). Virus titer was calculated using a standard curve obtained using a virus stock of known titer as determined by plaque assay.
Marburg virus infection.
Vero cells were seeded into six-well plates at a density of 2 × 105/well. One day later, the medium was removed and replaced with fresh medium containing E64d (300 μM). After 4 h, Ebola virus Zaire (MOI, 0.2) or Marburg virus Ci67 (Popp; MOI, 0.2) was added to cells and drug. One hour later, medium containing drug and virus was removed, cells were washed with fresh medium, and fresh medium containing drug was added to each well. Time zero samples were harvested immediately, and remaining samples were incubated for 72 h. Supernatants were harvested for determination of virus by quantitative RT-PCR, and cells were lysed for determination of viral protein by Western blot assay (14).
Protease assay.
VSV particles bearing mucin domain deletion-containing GPs from the indicated viruses were incubated in the presence of 0.2 mg/ml chymotrypsin in reaction buffer (10 mM Tris-HCl, 135 mM NaCl, 1 mM EDTA, pH 7.5). After 1 h, the digestion was stopped by the addition of phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM. Virus particles were deglycosylated by overnight incubation in the presence of peptide N-glycosidase F (PNGase F; New England BioLabs). GP1 digestion products were analyzed under denaturing conditions via immunoblot assay against a conserved N-terminal region of ebolavirus GP1 using polyclonal rabbit anti-ebolavirus GP1 antibody (10).
CHO cell infections.
VSVluc particles bearing GPs from the indicated ebolaviruses were digested in the presence or absence of chymotrypsin (as described above but not deglycosylated). NPC1−/− CHO cells and NPC1−/− CHO cells stably expressing mouse NPC1 were incubated in the presence or absence of E-64d (300 μΜ). After 4 h, virus particles were added to the cells. Sixteen hours after the addition of virus, luciferase activity was measured as described above.
NPC1 membrane binding assay.
Expression and purification of EBOV-MayΔTM have been described previously (10). An expression vector encoding SUDV GPΔTM (residues 1 to 309 and 491 to 657) that is fused to GCN4 trimerization/His tag was also prepared. Chymotrypsin-cleaved SUDV GPΔTM was created by incubation in the presence of 0.2 mg/ml chymotrypsin in chymotrypsin reaction buffer. After 30 min, the reaction was stopped by the addition of 1 mM EDTA, 1 mM PMSF, and 1× EDTA-free complete protease inhibitor cocktail (Roche). Thermolysin digestion of EBOV-MayΔTM GP was described previously (10). LE/LY were isolated by differential centrifugation and Percoll (Sigma) density gradient centrifugation. LE/LY were disrupted by incubation with methionine methyl ester (Sigma) and used to coat high-binding enzyme-linked immunosorbent assay plates (Corning). Following attachment, unbound LE/LY membranes were removed and plates were blocked with PBS–5% FBS. Bound membranes were incubated with the indicated amounts of native or protease-cleaved trimer in PBS–5% FBS. Unbound GPΔTM protein was removed, membranes were washed, and membrane-bound GPΔTM protein was recovered in SDS loading buffer and analyzed by immunoblot assay using GP1 antiserum as described previously (10).
Cloning of Bundibugyo GP.
TRIzol reagent (Invitrogen)-inactivated ebolavirus Bundibugyo (BDBV) RNA (vRNA) was precipitated using the standard protocol in the TRIzol package insert. First-strand cDNA was created from this vRNA using random primers and SuperScript II (Invitrogen). Frameshifted BDBV GP was amplified via PCR using Phusion HS (New England BioLabs) and primers 5′-TAAATGCATGGTTACATCAGGA, 5′-TGTGAAGTTCTTCTTATTTTCCCAGAAGGC, 5′-TAAGAAGAACTTCACAAAAACCCT, and 5′-TATCTCGAGGACTAGATTAGAGTAGA. The final PCR product was cloned into pCAGGS MCS using NsiI and XhoI. The clone was verified by sequencing. A mucin deletion-containing version of this plasmid was created by PCR with the previous primers, 5′-TCTCCGACATATGGTACCGCAAATCTGCTGACAGGCTCA and 5′-GGTACCATATGTCGGAGAGGTACCGACAGACAGCTCTTCA, and cloning into pCAGGS MCS with NsiI and XhoI. This plasmid was restricted with KpnI and religated to create the mucin (aa 309 to 489) deletion-containing BDBV. Plasmids were verified by sequencing. All restriction enzymes were obtained from New England BioLabs.
RESULTS
Endosomal cysteine proteases are host factors for filovirus entry.
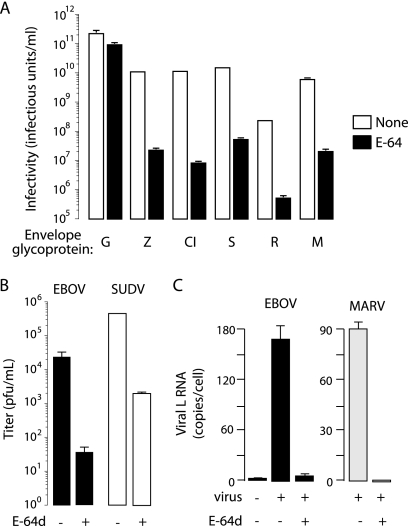
Previous studies show that EBOV-May infection is reduced by >99% by E-64, a highly specific small-molecule inhibitor of endosomal cysteine proteases, including cathepsins L and B (7, 35). We used E-64 as a probe to analyze the host requirements for entry by GPs from other filoviruses. Vero cells were pretreated with E-64 and then challenged with VSV vectors pseudotyped with GPs from EBOV-May, ebolavirus Cote d'Ivoire (CIEBOV), ebolavirus Sudan-Boniface (SUDV), ebolavirus Reston-Pennsylvania (RESTV), and Lake Victoria marburgvirus Musoke (MARV). The titers of VSV particles pseudotyped with EBOV-May, CIEBOV, SUDV, and MARV GPs are 6 × 109 to 1 × 1010 IU/ml, and the titer of VSV RESTV GP particles is 10-fold lower (1.3 × 108 IU/ml) (Fig. 1A). We found that E-64 treatment of target cells reduced infection by these viruses by >99.5% under conditions where VSV G infection was reduced by <50% (Fig. 1A). These findings were confirmed in studies of viruses bearing GPs lacking the mucin-rich domain in GP1. To verify that cysteine proteases are bona fide host factors, we tested the effect of E-64d on the growth of EBOV-May, ebolavirus Sudan-Gulu, and Lake Victoria marburgvirus Musoke in Vero cells. The production of new virus was reduced by more than 99% in the presence of E-64d, as determined by quantitative RT-PCR (Fig. 1B and C). Taken together, the results of these studies indicate that infection by EBOV-May, CIEBOV, SUDV, RESTV, and MARV is dependent on endosomal cysteine proteases sensitive to E-64.
Fig 1.
Endosomal cysteine proteases are host factors for filoviruses. (A) Vero cells were incubated in the presence of E-64 (cysteine protease inhibitor, 300 μM) or vehicle (1% DMSO) for 4 h prior to being challenged with VSV particles encoding GFP (VSVGFP) and bearing GPs from EBOV-May (Z), CIEBOV (CI), SUDV (S), RESTV (R), MARV (M), or VSV (G). After 24 h, infectivity (IU/ml) was determined by manually counting GFP-positive cells using fluorescence microscopy. Data are means ± standard deviations (SD; n = 3). Shown is a representative of four independent experiments. (B) Vero cells were incubated in the presence of E-64d (cysteine protease inhibitor, 300 μM) or vehicle for 4 h prior to being challenged with the filovirus EBOV-May or SUDV (MOI, 0.1). After 3 days, the titer was determined by quantitative RT-PCR. Data are means of three wells ± SD (n = 3). (C) Vero cells were incubated in the presence of E-64d (300 μM) or vehicle for 4 h prior to being challenged with the filovirus EBOV or MARV (MOI, 0.2). After 72 h, copies of the viral L gene per cell were determined by quantitative RT-PCR. Data are means of three wells ± SD (n = 3).
Proteases target virus GP1.
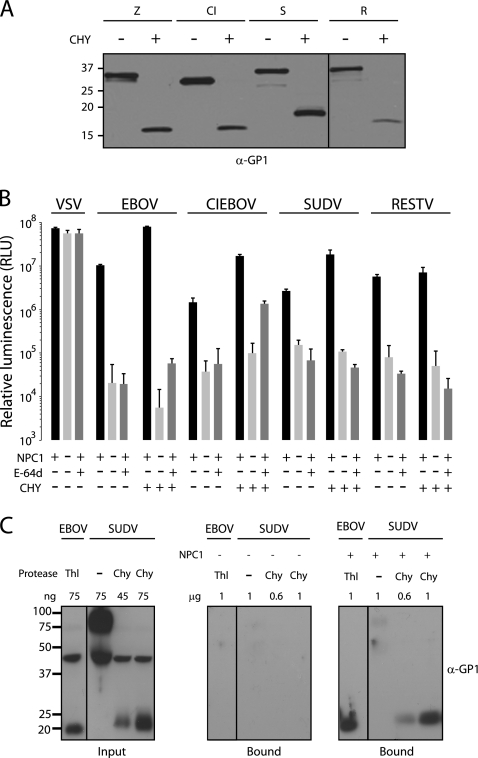
Analysis of the amino acid sequences of GPs from EBOV-May, CIEBOV, SUDV, RESTV, and MARV suggests that the domain structures of GP1 and GP2 are conserved (data not shown), a conclusion that has been recently confirmed for ebolavirus Sudan GP (11, 25). Biochemical studies of EBOV-May indicate that endosomal cysteine proteases remove the heavily glycosylated carboxyl-terminal “cap” domain and expose the N-terminal domain (7, 12, 19, 24, 25, 35) that is the ligand for the essential host factor NPC1 (10). To determine if a protease-sensitive carboxyl-terminal domain in GP1 is conserved among ebolaviruses, we incubated VSV particles pseudotyped with GPs from EBOV-May, CIEBOV, SUDV, and RESTV with chymotrypsin and analyzed the effect on virus infection. Chymotrypsin is a serine endoprotease that faithfully mimics the action of cathepsin L (46). We observed that like chymotrypsin digestion of EBOV-May, chymotrypsin digestion of CIEBOV, SUDV, and RESTV removes the carboxyl-terminal domains of GP1 to create an N-terminal 18- to 20-kDa fragment (Fig. 2A). We analyzed the function of chymotrypsin-cleaved virus particles on CHO cells and found that they are infectious and dependent on both NPC1 and cysteine protease activity (Fig. 2B). We next analyzed the binding of ebolavirus Sudan GP to NPC1 membranes. The source of GP is an ectodomain trimer in which the transmembrane domain is replaced with a GCN4 trimerization domain (SUDVΔTM). We incubated uncleaved and chymotrypsin-cleaved SUDVΔTM GP with purified LE/LY membranes from knockout and NPC1-expressing CHO cells. We found that, like that of EBOV-MayΔTM GP, SUDVΔTM GP binding to membranes is dependent on the cleavage of GP1 and expression of NPC1 (Fig. 2C). Although more work is needed, these findings suggest that at least one function of endosomal cysteine proteases in filovirus infection is to remove the carboxyl-terminal domain of GP1 and expose the NPC1 binding site.
Fig 2.
Function of protease-cleaved ebolavirus GPs. (A) VSV particles bearing GPs from EBOV-May (Z), CIEBOV (CI), SUDV (S), and RESTV (R) were incubated with chymotrypsin (CHY) or reaction buffer alone for 1 h. Virus particles were deglycosylated with PNGase F and analyzed by immunoblot assay using rabbit anti-GP1 antibody. (B) Untreated CHO cells expressing NPC1 or treated with E-64d (300 μM) and NPC−/− CHO cells were challenged with VSV particles encoding luciferase (VSVluc) and bearing GPs from uncleaved or chymotrypsin-cleaved ΔMuc GP from EBOV-May, CIEBOV, SUDV, RESTV, or VSV. After 16 h, infection was measured in RLU. Data are means ± SD (n = 3). (C) SUDVΔTM GP was cleaved with chymotrypsin, and binding to LE/LY membranes from CHO cells expressing NPC1 (right panel) or lacking NPC1 (middle panel) was determined as described in Materials and Methods. Bound proteins were analyzed by immunoblot assay for GP1. Uncleaved SUDVΔTM GP and thermolysin (Thl)-cleaved EBOV-MayΔTM GP were included as controls. Input proteins are shown in the left panel.
Requirement for cathepsin B is not conserved.
EBOV-May infection is strongly dependent on the endosomal cysteine protease cathepsin B (7, 35). To test the hypothesis that cathepsin B activity is required for infection by other filoviruses, we measured the VSV GP-dependent infection of Vero cells treated with the selective cathepsin B inhibitor CA074. We confirmed that GP-dependent EBOV-May infection is reduced as a function of the concentration of CA074 (0 to 80 μM) and is closely correlated with cathepsin B activity (Fig. 3A and B). We observed that like that of EBOV-May, CIEBOV GP-mediated infection is also closely correlated with cathepsin B activity. However, SUDV GP infection was not as sensitive to CA074 inhibition as EBOV-May or CIEBOV GP and neither RESTV nor MARV GP-mediated infection was significantly reduced when cathepsin B activity was inhibited (Fig. 3A and B).
Fig 3.
Cathepsin (Cat) B is not an essential host factor for all filoviruses. (A, B) Effect of cathepsin B selective inhibitor CA074 on cathepsin B and L activities (A) and on infectivity (B) by VSVGFP particles bearing filovirus GPs. Vero cells were incubated in the presence of increasing concentrations of CA074 (0 to 80 μM) or vehicle for 4 h prior to cell lysis or the addition of VSVGFP particles bearing GPs from MARV (M) or ΔMuc GP from EBOV-May (Z), CIEBOV (CI), SUDV (S), or RESTV (R). (A) Cathepsin B and L activities were determined by fluorogenic substrates (n = 2). (B) After 24 h, GFP-positive cells were manually counted using fluorescence microscopy. Infectivities from each of two replicates are shown and are representative of four independent experiments. Data presented were determined as follows: (infection [IU/ml] of CA074-treated cells)/(infection of vehicle-treated cells) × 100%. (C) Wild-type and cathepsin B-deficient (cathepsin B−/− cathepsin L+/+) MEF cells were infected with VSVGFP particles bearing GP from MARV (M) or ΔMuc GP from EBOV-May (Z), CIEBOV (CI), SUDV (S), or RESTV (R) or VSV (G). After 24 h, infectivity was determined as described for panel B. Data presented were determined as follows: (infection [IU/ml] of cathepsin B-deficient cells)/(infection of wild-type MEF cells) × 100%. Data are means ± SD (n = 3). Shown is a representative of three independent experiments. (D) Wild-type and cathepsin B-deficient MEF cells were transfected with expression plasmids encoding mouse cathepsin B, mouse cathepsin L, or a sham plasmid. After 24 h, cells were exposed to VSVGFP particles as described above. After 24 h, the percentage of cells infected was determined using flow cytometry. Data presented were determined as follows: (percent infection of transfected cathepsin B-deficient MEF cells)/(percent infection of wild-type MEF cells) × 100%. Data are means ± SD (n = 3). Shown is a representative of three independent experiments.
As an independent test of the role of cathepsin B in infection, we studied MEF cells derived from cathepsin B knockout (cathepsin B−/−) mice. The pattern of infection of cathepsin B−/− MEF cells by filovirus GPs is closely correlated with the pattern of infection of Vero cells treated with CA074: EBOV-May and CIEBOV GP is 1%, SUDV 7%, RESTV 25 to 40%, and MARV 65 to 99% of infection of wild-type MEF cells (Fig. 3C and D). The introduction of an expression plasmid encoding cathepsin B into cathepsin B−/− MEF cells enhanced infection by each of the viruses to 90 to 150% of the infection of wild-type MEF cells (Fig. 3D), thus confirming that the defect in infection is due to cathepsin B deficiency. These findings demonstrate that the dependence of EBOV-May and CIEBOV on cathepsin B is not a conserved property of all filoviruses.
Cathepsin L.
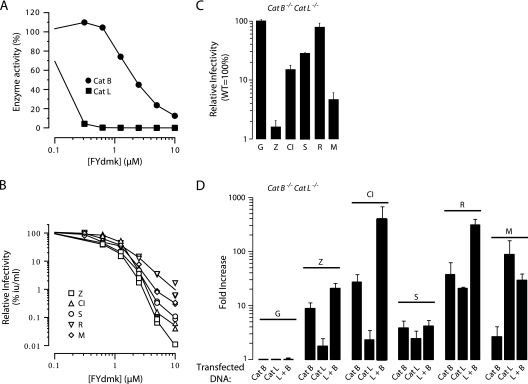
Cathepsin L is an E-64-sensitive endopeptidase that is expressed in cells expressing cathepsin B (15, 17, 32, 40, 42, 43). To investigate the role of cathepsin L in infection by cathepsin B-dependent and -independent viruses, we studied the infection of Vero cells treated with the inhibitor FYdmk. At low concentrations where cathepsin L, but not cathepsin B, activity is blocked (<1 μM), there was no effect of FYdmk on the infection of ebolavirus- or marburgvirus-pseudotyped particles (Fig. 4A and B). However, when the concentration of FYdmk exceeded >1 μM and cathepsins B, and likely other endosomal cysteine proteases, are also inhibited, infection by each virus was reduced, consistent with the results of studies using the general cysteine protease inhibitor E-64. These results indicate that cathepsin L is not essential for either the cathepsin B-dependent or -independent viruses.
Fig 4.
Cathepsin (Cat) L is a host factor for Reston ebolavirus and Marburg virus. (A, B) Effect of the inhibitor FYdmk on cathepsin L and B activities (A) and on the infectivity (B) of VSVGFP particles bearing filovirus GPs. Vero cells were incubated in the presence of increasing concentrations of FYdmk (0 to 10 μM) or vehicle for 4 h prior to cell lysis or the addition of VSVGFP particles bearing GP from MARV (M) or ΔMuc GP from EBOV-May (Z), CIEBOV(CI), SUDV(S), or RESTV (R). (A) Cathepsin B and L activities were determined by fluorogenic substrates (n = 2). (B) After 24 h, GFP-positive cells were manually counted using fluorescence microscopy. Infectivities from two replicates are shown and are representative of four independent experiments. Data presented were determined as follows: (infection [IU/ml] of FYdmk-treated cells)/(infection of vehicle-treated cells) × 100%. (C) Wild-type and cathepsin B/cathepsin L-deficient (cathepsin B−/− cathepsin L−/−) MEF cells were infected with VSVGFP particles bearing GP from MARV (M) or ΔMuc GP from EBOV-May (Z), CIEBOV(CI), SUDV(S), or RESTV (R) or with VSV (G). Infectivity was determined as described in the legend to Fig. 3C. Data are means ± SD (n = 3). Shown is a representative of three independent experiments. (D) Cathepsin B/cathepsin L-deficient MEF cells were transfected with expression plasmids encoding mouse cathepsin B, mouse cathepsin L, both cathepsins B and L, or the vector alone. After 2 days, cells were exposed to VSVluc bearing GP from MARV (M) or ΔMuc GP from EBOV-May (Z), CIEBOV(CI), SUDV(S), or RESTV(R) or to VSV (G). Twenty-four hours later, infectivity was determined by measuring the RLU in cell lysates. Data presented were determined as follows: virus-encoded RLU in cathepsin-transfected cells/RLU in vector-transfected cells. Data are means ± SD (n = 6). Shown is a representative of four independent experiments.
In cathepsin B−/− MEF cells, overexpression of cathepsin L enhanced infection by SUDV, RESTV, and MARV but was unable to overcome the defect imposed by loss of cathepsin B activity for EBOV-May or CIEBOV (Fig. 3D). To determine if cathepsin L is able to support filovirus infection in the absence of cathepsin B, we used fibroblasts obtained from cathepsin B−/− cathepsin L−/− mice. In our initial experiments, we found that infection of these cells by CIEBOV, SUDV, and RESTV closely corresponds to infection of cathepsin B−/− MEF cells and CA074-treated Vero cells (Fig. 4C). However, unlike that of cathepsin B−/− MEF cells and Vero cells treated with a low dose of FYdmk, MARV infection of cathepsin B−/− cathepsin L−/− cells was reduced by >95%. The transfection efficiency of cathepsin B−/− cathepsin L−/− MEF cells was too low to assess the role of cathepsin L and B overexpression using VSVGFP particles. To circumvent this limitation, we utilized pseudotyped VSVluc particles at an MOI of >1, which enhances the sensitivity of the measurement of virus infection. As expected, the expression of cathepsin B was markedly superior to the expression of cathepsin L in supporting infection by EBOV-May and CIEBOV (cathepsin B, 9- to 30-fold; cathepsin L, <3-fold). Remarkably, cathepsin B-dependent infection was markedly increased when cathepsin L was also expressed (Fig. 4D). These findings are consistent with the results of previous studies of EBOV-May (7) and indicate that expression of cathepsin L is not a substitute for the essential role of cathepsin B in EBOV-May and CIEBOV infections. In contrast, expression of either cathepsin L or cathepsin B enhanced the RESTV infection of cathepsin B−/− cathepsin L−/− MEF cells. A consistent observation was that the response of SUDV to overexpression of cathepsin L and/or cathepsin B was not as robust as that of the other viruses. In contrast to the studies of the ebolaviruses, cathepsin L (90-fold) was much more active than cathepsin B (2.5-fold) in supporting MARV infection of these cells (Fig. 4D). Taken together, these findings identify marked virus-specific variation in the effect of cathepsin L expression on filovirus infection.
Mapping of GP determinants of cathepsin B.
EBOV-May and RESTV are closely related but differ in dependence on cathepsin B and therefore provided an opportunity to map the virus determinants of the cathepsin B requirement. To this end, we analyzed virus particles bearing chimeric GPs created by reciprocal exchange of portions of the EBOV-May and RESTV GPs. In our initial experiment, the effect of exchanging GP1 and GP2 was examined. We observed that the titer of EBOV-GP1/RESTV-GP2 particles on Vero cells was similar to that of RESTV and the titer of RESTV-GP1/EBOV-GP2 particles was similar to that of EBOV-May, indicating that the chimeric GPs were functional (Fig. 5A). We observed that neither of the virus particles expressing chimeric GP EBOV/RESTV or RESTV/EBOV were as sensitive as EBOV-May particles to inhibition by CA074 as to inhibition by E-64. These findings indicate that the determinants of cathepsin B dependence are not localized exclusively within either GP1 or GP2. A previous study of EBOV-May-derived viruses selected for resistance to CA074 suggested an explanation. This study showed that single amino acid changes in residues clustered either near the N terminus of GP1 (N40K/S/T, T42A, L43F, or D47V) or in the hr1 segment of GP2 (I584F or K588R) conferred resistance to CA074 (46). Using these findings as a guide, we noted that RESTV GP differs from EBOV-May GP at residue 47 (D→E) in GP1 and residue 584 (I→L) in GP2 (Fig. 5B). To determine if these subtle changes mediate the difference in the behavior of these viruses, we exchanged these residues in EBOV-May and RESTV and tested the effect on sensitivity to CA074. We found that substitution of glutamic acid for D47 or leucine for I584 alone or in combination conferred resistance to CA074 on EBOV-May. However, the introduction of the reciprocal changes E48D and/or L585I into RESTV GP did not confer a requirement for cathepsin B (Fig. 5C and D). These findings indicate that D47 and I584 are necessary for EBOV-May but not sufficient for RESTV to depend on cathepsin B as a host factor for the infection of Vero cells.
Fig 5.
Comparison of cathepsin B requirements for EBOV and RESTV. (A) Virus determinants of cathepsin B dependence. Vero cells were incubated in the presence of CA074 (80 μM), E-64 (300 μM), or the vehicle (1% DMSO) for 3 h prior to the addition of VSVGFP particles bearing ΔMuc EBOV-May GP, RESTV GP, EBOV-May GP1/RESTV GP2, or RESTV GP1/EBOV-May GP2. Infectivity was determined as described in the legend to Fig. 1A. Data are means ± SD (n = 3). Shown are representatives of three independent experiments. (B) Schematic of EBOV-May GP and locations of amino acid residues previously shown to mediate resistance to CA074. Inverted triangles identify residues that mediate resistance. Boxes identify differences in amino acids between EBOV-May and RESTV. The positions of the receptor-binding domain (RBD), the β-13-14 disordered loop, the fusion loop (fl), and heptad repeats 1 and 2 (hr1, hr2) are indicated. Residue numbering relative to EBOV GP. (C and D) Analysis of the effects of D47E and I584L on cathepsin B dependence. The effects of reciprocal substitutions of residues (D47/E48 and I584/L585) that differ between EBOV-May and RESTV GP were measured in native and chimeric GPs from panel A. Infectivity was determined as described in the legend to Fig. 1A. Data are means ± SD (n = 3). Shown are representatives of three independent experiments. n/a, not applicable.
Sequence analysis predicts cathepsin B dependence.
The discovery that the cathepsin B dependence of EBOV-May maps to residues D47 and I584 suggested the possibility that these residues might also predict the protease requirements of other filoviruses. Indeed, we noted that D47 and I584 are conserved in the GPs of EBOV-May and CIEBOV, which are cathepsin B dependent, but not in those of SUDV (E47) and MARV (N47 and L584), which are not. A search of the NCBI database identified 27 partial and complete sequences of GPs obtained from independent isolates classified as EBOV. Only one of the virus isolates, EBOV-1995 (GenBank accession no. ID-AY354458), differed in the cathepsin B-determining residues. Excluding the mucin regions, which are deleted from the GPs in this analysis, the GPs of EBOV-May and EBOV-1995 differ only at residues 47 (D47E) and 544 (I544T), which had not been identified in previous studies of cathepsin B requirements (46). We prepared and analyzed VSV EBOV-May GP-derived particles containing E47 or T544 GP. As expected, these viruses are highly infectious and sensitive to E-64. However, inhibition of cathepsin B by CA074 reduced the titer of T544 GP particles by >99% under conditions where the titer of E47 GP
particles was minimally changed (Fig. 6A). Thus, escape from dependence on cathepsin B was closely correlated with E47 and not T544. Since we began these investigations, an outbreak of hemorrhagic fever occurred in Uganda due to a filovirus that is sufficiently different from EBOV, CIEBOV, RESTV, and MARV to be classified as Bundibugyo (BDBV) (39). Analysis of the BDBV sequence revealed the presence of D47 and I584, thus predicting that BDBV is cathepsin B dependent. Indeed, we found that infection by VSV BDBV GP particles was inhibited by increasing concentrations of CA074 (0 to 80 μM) in a pattern closely correlated with that of EBOV-May (Fig. 6B). In addition, we found that infection by BDBV pseudotypes was blocked by the treatment of cells with E-64 and in cathepsin B−/− MEF cells (data not shown). Thus, in our limited cohort of filoviruses analyzed in this study, the presence of D47 correlated with dependence on cathepsin B for infection.
Fig 6.
Analysis of cathepsin B dependence of EBOV-1995 and BDBV. (A) VSVGFP particles pseudotyped with GP from EBOV-May containing E47 or T544 from EBOV-1995 were prepared, and dependence on cathepsin B was analyzed. Vero cells were incubated in the presence of CA074 (80 μM), E-64 (300 μM), or the vehicle for 3 h prior to the addition of VSVGFP bearing ΔMuc GP from EBOV-May D47E or I544T or wild-type EBOV-May GP. Infection was measured as described in the legend to Fig. 1A. Data are means ± SD (n = 3) Shown is a representative of three independent experiments. (B) Effect of cathepsin B selective inhibitor CA074 on infection by BDBV GP. Vero cells were incubated in the presence of increasing concentrations of CA074 (0 to 80 μM) or the vehicle for 3 h prior to the addition of VSVluc particles bearing ΔMuc GP from EBOV-May (Z), BDBV (B), or RESTV (R). After 6 h, cells were lysed and relative luminescence (RLU) was measured. Data are presented as follows: (virus-encoded luciferase activity of cells treated with CA074)/(luciferase activity of cells treated with vehicle) × 100%. Shown are representatives of three independent experiments. n/a, not applicable.
DISCUSSION
Over the past 6 years, significant progress has been made in understanding how filoviruses gain entry into cells and a model of infection based on this progress has been proposed (10). EBOV-May particles attach to lectins on the cell surface and are taken up by macropinocytosis into vesicles that are transported to LE/LY-containing endosomal cysteine proteases and NPC1 (5, 10, 20, 31, 32, 34, 43). One function of endosomal cysteine proteases is to cleave the carboxyl-terminal “cap” region of GP1 from the mushroom-shaped GP protruding from the virus membrane and expose the stalk containing the protease-resistant N-terminal domain in GP1 that is the ligand for the NPC1 protein (4, 10–12, 19, 24, 25, 28). A second function may be to biochemically destabilize the GP, thereby sensitizing it to triggering for viral membrane fusion (3, 46). Further studies are needed to determine the roles of NPC1 (5, 10) and additional events, including further cleavage of GP1 in this process (3, 11, 21, 35, 46).
In this report, we provide evidence that key aspects of this scheme are conserved among other filoviruses. The new findings show that as in EBOV-May, the carboxyl-terminal domain of SUDV GP1 is a substrate for proteolytic cleavage, that cleaved particles are infectious and NPC1 dependent, and that the protease-resistant N-terminal domain of GP1 binds to purified LE/LY membranes in an NPC1-dependent manner. These findings are consistent with the recent report showing that the domain organization of EBOV-May is conserved in SUDV (11). Moreover, we find that the sensitivity of the C-terminal domain of GP1 to cleavage and the dependence of cleaved particles on NPC1 is conserved among other ebolaviruses. Consistent with this view, we find that isolates from each species of Filoviridae are E-64 sensitive, including the growth of EBOV-May, SUDV, and MARV. While more work needs to be done, including studies of other proteases and MARV, the results of the experiments described in this report, coupled with alignment of primary amino acid sequences of GPs which indicate that the domain structure is likely to be conserved (data not shown), suggest that cleavage and binding are key steps in the filovirus entry pathway. In this model, endosomal cysteine proteases are required for the efficient removal of the carboxyl-terminal domain of GP1 to expose the NPC1 binding domain. Endosomal cysteine proteases may mediate the additional steps necessary for orderly deployment of the virus membrane fusion activity (21, 35, 46). Indeed, two recent reports suggest that cysteine protease cleavage of the β-13-14 loop in GP1 promotes release of the GP2 fusion peptide (3, 11). The studies in this report provide the basis for future studies to compare the virus requirements for specific proteases to cleave GP1, to bind to NPC1, and to release the GP2 fusion peptide.
The endosomal cysteine proteases are a family of 11 acid-dependent proteases that reside in LE/LY (15, 17, 32, 40–43). Little is known about the roles individual members of this family play during filovirus infection. Previously, we showed that EBOV-May requires cathepsin B, but not cathepsin L, for entry (7). We confirm this finding and show that CIEBOV has the same requirement. Additional studies are needed to determine if the virus's sensitivity to the absence of cathepsin B activity is due to a specific requirement for the double-chain isoform of cathepsin B (36). Although cathepsin L is not essential for any of the filoviruses studied, we found that it works in concert with cathepsin B to enhance the infection of EBOV-May, CIEBOV, and RESTV. Furthermore, cathepsin L activity is required for MARV infection of MEF cells but not Vero cells, suggesting that the role of cathepsin L may be shared by other cysteine proteases with endopeptidase activity. Remarkably, RESTV infection is sensitive to E-64 but is not sensitive to the loss of cathepsins B and L. This suggests that one or more additional E-64-sensitive proteases can support RESTV infection.
The substrate specificity of endosomal cysteine proteases is governed largely by the accessibility of the polypeptide chain to the active site of the protease and not by a strong preference for specific sequence motifs (32, 41, 42). Thus, one consequence of the extensive variation in the sequence of the carboxyl-terminal domain of GP1 is that it may alter the repertoire of cysteine proteases that are able to cleave GP1. Therefore, the presence of multiple proteases with overlapping substrate preferences in late endosomes and lysosomes of host cells may provide for redundancy in the conditions for cleavage of GP and might explain the virus-specific differences in dependence on cathepsins B and L observed in our studies. One advantage of this scheme may be that effective cleavage of the GP1 cap and/or fusion peptide release is maintained in the presence of selective pressure from host immune recognition for sequence diversification, analogous to the function of variable loops in HIV gp120 (2, 18). In this model, adaptation to loss of cathepsin B activity by a change to a single amino acid residue (i.e., D47 or I584) may provide a means for a rapid response to changes in endosomal cysteine protease expression between hosts or cell types. Sequence polymorphisms that specifically predict host factor preference have been identified in other virus envelope GPs, including SARS GP N479K interactions with human or civet ACE2 receptor, influenza virus HA1 interactions with α-2 glycan linkage in human or α-2-6 glycan linkage in avian sialic acid, and HIV gp120 binding to receptor CXCR4 and/or CCR5 (2, 26, 33, 38). Analysis of the filoviruses identified in future outbreaks will provide further tests of the utility of using the D47/I584 polymorphisms in GP in determining virus preference for host endosomal cysteine proteases.
ACKNOWLEDGMENTS
This work was supported by grants U54 AI057159 and R01 CA104266 to J.C. and PIDS-Sanofi-Pasteur fellowship K12-HD052896 and 5K08AI079381 to J.M. K.C. was supported by a postdoctoral fellowship from the New England Research Center of Excellence in Biodefense and Emerging Infectious Diseases (NERCE/BEID). C.F. was supported by the Postgraduate Research Participation Program at the U.S. Army Medical Research and Material Command administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAMRMC. M.C. was supported by the Fonds de la Recherche en Sant é du Quebéc.
We thank Scott Aoki, Anna Bruchez, Brenna Hill, and Daniel Douek for assistance.
The opinions, interpretations, conclusions, and recommendations in this report are ours and are not necessarily endorsed by the U.S. Department of Defense, the U.S. Department of the Army, or the U.S. Department of Health and Human Services.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Alvarez CP, et al. 2002. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 76:6841–6844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arthos J, et al. 1989. Identification of the residues in human CD4 critical for binding HIV. Cell 57:469–481 [DOI] [PubMed] [Google Scholar]
- 3. Brecher M, et al. 2012. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J. Virol. 86:364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brindley M, et al. 2007. Ebola virus glycoprotein 1: identification of residues important for binding and postbinding events. J. Virol. 81:7702–7709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carette JE, et al. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Centers for Disease Control and Prevention 2001. Outbreak of Ebola hemorrhagic fever Uganda, August 2000-January 2001. MMWR Morb. Mortal. Wkly. Rep. 50:73–77 [PubMed] [Google Scholar]
- 7. Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Colebunders R, Borchert M. 2000. Ebola haemorrhagic fever—a review. J. Infect. 40:16–20 [DOI] [PubMed] [Google Scholar]
- 9. Colebunders R, et al. 2007. Marburg hemorrhagic fever in Durba and Watsa, Democratic Republic of the Congo: clinical documentation, features of illness, and treatment. J. Infect. Dis. 196(Suppl. 2):S148–S153 [DOI] [PubMed] [Google Scholar]
- 10. Côté M, et al. 2011. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 477:344–348 (Letter.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dias J, et al. 2011. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 18:1424–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dube D, et al. 2009. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 83:2883–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dube D, et al. 2010. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc. Natl. Acad. Sci. U. S. A. 107:16637–16642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fabozzi G, Nabel C, Dolan M, Sullivan N. 2011. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J. Virol. 85:2512–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujishima A, et al. 1997. The crystal structure of human cathepsin L complexed with E-64. FEBS Lett. 407:47–50 [DOI] [PubMed] [Google Scholar]
- 16. Geisbert TW, Jahrling PB. 2004. Exotic emerging viral diseases: progress and challenges. Nat. Med. 10:S110–S121 [DOI] [PubMed] [Google Scholar]
- 17. Guncar G, Pungercic G, Klemencic I, Turk V, Turk D. 1999. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 18:793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hood CL, et al. 2010. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J. Virol. 84:2972–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunt CL, Kolokoltsov AA, Davey RA, Maury W. 2011. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J. Virol. 85:334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaletsky R, Simmons G, Bates P. 2007. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 81:13378–13384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kondratowicz AS, et al. 2011. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. U. S. A. 108:8426–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuhn JH, et al. 2010. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 155:2083–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuhn JH, et al. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 281:15951–15958 [DOI] [PubMed] [Google Scholar]
- 25. Lee JE, et al. 2008. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li F, Li W, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868 [DOI] [PubMed] [Google Scholar]
- 27. Lin G, et al. 2003. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J. Virol. 77:1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manicassamy B, Wang J, Jiang H, Rong L. 2005. Comprehensive Analysis of ebola virus GP1 in viral entry. J. Virol. 79:4793–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marzi A, et al. 2006. The signal peptide of the ebolavirus glycoprotein influences interaction with the cellular lectins DC-SIGN and DC-SIGNR. J. Virol. 80:6305–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mulherkar N, Raaben M, de la Torre JC, Whelan SP, Chandran K. 2011. The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology 419:72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nanbo A, et al. 2010. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 6(9):e1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otto H-H, Schirmeister T. 1997. Cysteine proteases and their inhibitors. Chem. Rev. 97:133–172 [DOI] [PubMed] [Google Scholar]
- 33. Ribeiro RM, Hazenberg MD, Perelson AS, Davenport MP. 2006. Naïve and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: implications for therapy. J. Virol. 80:802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saeed MF, Kolokoltsov A, Albrecht T, Davey R. 2010. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 6:e1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schornberg K, et al. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174–4178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schornberg K, et al. 2009. α5β1-integrin controls ebolavirus entry by regulating endosomal cathepsins. Proc. Natl. Acad. Sci. U. S. A. 106:8003–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Simmons G, et al. 2003. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115–123 [DOI] [PubMed] [Google Scholar]
- 38. Stevens J, et al. 2006. Structure and receptor specificity of hemagglutinin from an H5N1 influenza virus. Science 312:404–410 [DOI] [PubMed] [Google Scholar]
- 39. Towner JS, et al. 2008. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4:e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turk B, Turk D, Turk V. 2000. Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta 1477:98–111 [DOI] [PubMed] [Google Scholar]
- 41. Turk D, Podobnik M, Kuhelj R, Dolinar M, Turk V. 1996. Crystal structures of human procathepsin B at 3.2 and 3.3 Angstroms resolution reveal an interaction motif between a papain-like cysteine protease and its propeptide. FEBS Lett. 384:211–214 [DOI] [PubMed] [Google Scholar]
- 42. Turk D, Guncar G. 2003. Lysosomal cysteine proteases (cathepsins): promising drug targets. Acta Crystallogr. D Biol. Crystallogr. 59(Pt. 2):203–213 [DOI] [PubMed] [Google Scholar]
- 43. Turk V, Turk B, Turk D. 2001. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 20:4629–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vance JE, Peake KB. 2011. Function of the Niemann-Pick type C proteins and their bypass by cyclodextrin. Curr. Opin. Lipidol. 22:204–209 [DOI] [PubMed] [Google Scholar]
- 45. Weidmann M, Mühlberger E, Hufert F. 2004. Rapid detection protocol for filoviruses. J. Clin. Virol. 30:94–99 [DOI] [PubMed] [Google Scholar]
- 46. Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. 2010. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 84:163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]