Abstract
The Epstein-Barr virus (EBV) is highly associated with nasopharyngeal carcinoma (NPC), and it regulates some microRNAs (miRNAs) that are involved in the development of cancer. The role of EBV in the deregulation of cellular miRNAs and how this affects the progression of NPC remain to be investigated. An analysis of the miRNA profile in an EBV-infected cell line revealed that miRNA 203 (miR-203) was downregulated. miR-203 is expressed specifically in epithelial cells. This downregulation of miR-203 was further verified and functionally analyzed. miR-203 was downregulated substantially in epithelial cells and NPC tissues that were latently infected with EBV. Downregulation of miR-203 also occurred during the early stage of EBV infection. Furthermore, the viral oncoprotein, latent membrane protein 1 (LMP1), was responsible for downregulation of miR-203. Removal of the latent EBV genome or suppression of LMP1 resulted in restoration of miR-203 expression. EBV-LMP1 mediated the downregulation of miR-203 at the primary transcript level. E2F3 and CCNG1 were identified as target genes of miR-203. Ectopic expression of miR-203 inhibited EBV-induced S-phase entry and transformation in vivo. Overexpression of the targets overcame the effects of miR-203 mimics on the cell cycle, and the expression of target genes in tumor models was inhibited by miR-203. Inhibitors of Jun N-terminal protein kinase (JNK) and NF-κB blocked miR-203 downregulation. These results imply that EBV promotes malignancy by downregulating cellular miR-203, which contributes to the etiology of NPC.
INTRODUCTION
The Epstein-Barr virus (EBV) is one of the most highly transforming viruses (3) and is associated with several malignancies, including lymphatic Burkitt's lymphoma (BL) and epithelial nasopharyngeal carcinoma (NPC) (54). However, the role of EBV in the development of malignancies, especially in epithelial tumors such as NPC, remains poorly understood. Lack of a suitable EBV-infected epithelial cell line model has been a problem (28, 30, 32). EBV oncogenesis is thought to cause viral latency, which is maintained by the transcription of a limited subset of genes, including EBV nuclear antigen 1 (EBNA1) and latent membrane protein 1 (LMP1). Of the proteins expressed during EBV latent infection, LMP1 is notable for its transforming potential (26). LMP1 activates cell-signaling pathways such as NF-κB, Jun N-terminal protein kinase (JNK), and p38. In addition, EBV-encoded noncoding RNAs (EBERs) and microRNAs (miRNAs) are expressed during latency.
MicroRNAs belong to a class of short, highly conserved noncoding RNAs that are known to suppress the expression of protein-coding genes through imperfect base pairing with the 3′ untranslated regions (UTRs) of target messenger RNAs (mRNAs) (1). miRNAs have been implicated in the control of various biological processes, such as cell proliferation, apoptosis, and differentiation (5, 9). Growing evidence has indicated that deregulation of miRNAs also contributes to tumorigenesis. Recently, deregulation of cellular microRNAs has been reported in NPC, EBV-associated B-cell lymphoma, and gastric carcinoma (11, 29, 44). However, whether and how EBV regulates the aberrant expression of cellular miRNAs in NPC remain to be explored.
The epithelial cell line 293-EBV, which was previously developed in our lab, is stably infected with the whole EBV genome, Maxi-EBV. 293-EBV cells exhibited enhanced proliferation, enhanced S-phase entry during the cell cycle, and malignant potential (32). To further study the contribution of EBV to changes in the biological phenotype, the miRNA profile of 293-EBV cells was examined using microarrays. The expression of approximately 90 miRNAs changed by more than 2.5-fold in 293-EBV compared to EBV-negative 293 cells, and 76.9% of them were downregulated (data not shown). Among the downregulated miRNAs, miRNA 203 (miR-203) was downregulated approximately 3-fold. Because miR-203 is expressed specifically in epithelial cells and is a tumor suppressor (2, 6, 38, 45, 48), it was chosen for further study. Downregulation of miR-203 has been recently described in several types of cancers, particularly epithelial cancers such as esophageal cancer, prostate cancer, and bladder cancer (2, 19, 34, 48). However, it is unknown whether the downregulation of miR-203 is mediated by EBV. Furthermore, the contribution of miR-203 downregulation to the oncogenesis of EBV-associated epithelial cancer remains to be clarified.
In this study, the expression of miR-203 was evaluated in EBV-infected epithelial cells and in poorly differentiated and undifferentiated NPC tissues using sensitive quantitative reverse transcription-PCR (qRT-PCR). miR-203 was downregulated by the same amount in both epithelial cells and NPC tissues. Moreover, the oncoprotein LMP1 was determined to be the viral factor responsible for EBV-mediated downregulation of miR-203. Ectopic expression of miR-203 in EBV-infected cells inhibited EBV-induced S-phase entry and transformation. This approach provided new insight into the interaction between the virus and epithelial cells during cancer development.
MATERIALS AND METHODS
Cell types, plasmids, and miRNAs.
The EBV-infected cell line 293-EBV was previously established by creating a stably transfected cell line (32) that contained the EBV genome, Maxi-EBV (15, 16). The EBV-negative cell line, Lm, was derived from 293-EBV cells from a mixture of 20 clones that had lost the EBV genome during subculture (32). The EBV-transformed lymphocytes (lymphoblastoid cell lines [LCLs]), E14, E18, E20, and E33, were derived from people without cancer. These LCLs were established using the EBV prototype B95-8 strain (46) in our laboratory. The nasopharyngeal carcinoma (NPC) cell lines (HNE1, CNE1, and 6-10B) were EBV negative due to reduplicative subculture (21). Immortalized nasopharynx cells (NP69), LMP1 stably transfected NP69 cells (NP69-LMP1), and NP69 pLNSX cells (NP69-control) (27) were preserved and propagated in our laboratory. NPC cells were cultured in RPMI 1640 medium containing 10% fetal calf serum (FCS). The 293-derived cells were grown in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% FCS. NP69 and NP69-LMP1 cells were cultured in keratinocyte serum-free medium (keratinocyte-SFM; Gibco).
The vector pM-BAC (where BAC is bacterial artificial chromosome) was derived from the Maxi-EBV plasmid after a precise excision of the whole EBV genome using homologous recombination in Escherichia coli (55). The miRNA duplex homologs (mimics) and nonspecific scramble sequences were synthesized and purified by Shanghai Gene-Pharma Co. (Shanghai, China). pcDNA3.1(+) (Invitrogen) was used to construct the LMP2 expression vector. The E2F3 and CCNG1 expression plasmids and the pCMV6-XL control vector were obtained from OriGene.
Tissue samples.
A total of 85 NPC biopsy specimens and 27 normal or nonneoplastic nasopharynx epithelium specimens were obtained from three hospitals that are affiliated with Xiangya Hospital, Central South University. The biopsy samples used in this study were submitted for histopathological diagnosis as poorly differentiated or undifferentiated NPCs and were stored at −80°C for subsequent analysis. Fifty-four tissue samples that were sectioned into serial paraffin sections with eight counterparts were used for in situ hybridization (ISH) analysis to detect miR-203 and/or EBER1 and for immunohistochemical analysis, which was used to detect LMP1. To detect miR-203 using qRT-PCR, 46 NPC biopsy specimens were selected for frozen sectioning (Leica CM 1900), and laser capture microdissection (Leica CTR 6500) was used to obtain pure tumor tissues. All aspects of the study were approved by the local ethics committee.
Nude mice.
Four-week-old specific-pathogen-free (SPF) BALB/c nude mice were purchased from SLAC Laboratory Animal Co., Ltd. (Shanghai, China). All of the animal procedures were performed according to approved protocols and in accordance with recommendations for the proper use and care of laboratory animals.
EBV infection and parallel controls.
NPC cell lines (HNE1, CNE1, and 6-10B) and NP69 cells were infected with recombinant EBV produced from the 293-EBV cells using the transfer infection method (42). Briefly, donor B-lymphocytes were exposed to EBV produced from 293-EBV cells that were induced with the expression plasmids BZLF1 and BALF4 (47) for 3 h at 4°C and then washed well. The acceptor epithelial cells had been previously seeded in 2-ml wells 24 h prior to infection with 3 × 105 cells. After coculture for up to 24 h, the donor cells were removed from the acceptors by washing. After 72 h of coculture, green fluorescent protein (GFP)-positive cells were observed using green fluorescence microscopy. At 7 to 10 days postinfection, the cells were used for the next detection. As an EBV-negative parallel control, the pM-BAC plasmid was transfected into the corresponding cells.
RNA extraction and RT-PCR.
Total RNA was isolated from the cells or tissues using TRIzol reagent (Invitrogen). A reverse transcription reaction was performed using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. Real-time quantitative PCR (qPCR) was performed using an IQ5 Multicolor Detection System (Bio-Rad) with a Hairpin-it miRNAs qPCR Quantification Kit (GenePharma, Shanghai, China) according to the manufacturer's instructions. The primers used for detection of the miRNAs by qRT-PCR were designed based on the miRNA sequences provided by the Sanger Center miRNA Registry. The U6 snRNA was used as an endogenous control. The primers used for the detection of primary transcript of miR-203 (pri-miR-203) were designed and purchased from Applied Biosystems (Hs 03302931-pri; ABI), and β-actin was used as an endogenous control. The following program was used for the RT reaction: 16°C for 30 min, followed by 42°C for 30 min and then 85°C for 10 min. The following program was used for qPCR: 95°C for 3 min followed by 40 cycles of 95°C for 12 s and then 62°C for 35 s. The relative changes in expression were calculated using the 2−ΔΔCT (where CT is threshold cycle) method. Three parallel repeats were performed for each sample in each experiment, and the results were expressed as the mean of three independent experiments. Semiquantitative RT-PCR was also performed to detect the genes, and PCR was performed on a Light-Cycler 1.5 System with a Fermentas Revert Aid First Strand cDNA Synthesis kit and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal control. The primers used were E2F3 (forward, 5′-TCCCGCTTTACTCTTCAGGA-3′; reverse, 5′-GGGTCAGGCACATAGCATTT-3′), CCNG1 (forward, 5′-AAGCCTTCTGTGTTGGCATT-3′; reverse, 5′-GCAGTACGCCCAGAAACAAT-3′), LMP1 (forward, 5′-TGAACACCACCACGATGACT-3′; reverse, 5′-GTGCGCCTAGGTTTTGAGAG-3′), LMP2 (forward, 5′-TCGAAGCTTATGGGGTCCCTAGAAATGGTG-3′; reverse, 5′-CGTGAATTCTTAGCCACACTGTCTGTCTAG-3′), β-actin (forward, 5′-GCATCCCCCAAAGTTCACAA-3′; reverse, 5′-AGGACTGGGCCATTCTCCTT-3′), and GAPDH (forward, 5′-CAAGGTCATCCATGACAACTTTG-3′; reverse, 5′-GTCCACCACCCTGTTGCTGTAG-3′). The following program was used for PCR: an initial denaturation at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 58°C for 40 s, and 72°C for 30 s, with a final step at 72°C for 10 min.
ISH and immunohistochemical analysis.
In situ hybridization (ISH) analysis was used to detect EBV and miR-203 expression. An oligonucleotide probe complementary to EBER1 was digoxigenin labeled at the 3′-terminal end. Oligonucleotide probes complementary to hsa-miR-203 (where hsa indicates Homo sapiens) (product 18097-01; Exiqon) and scramble (normal control [NC]) were digoxigenin labeled at the 5′-terminal end. Tumor specimens were formalin fixed, paraffin embedded, and sectioned. The probes were detected using an enhanced sensitive ISH detection kit (Boster Inc., Wuhan, China) according to the manufacturer's instructions. Negative controls, without probe, were also performed. To examine the expression of LMP1, E2F3, and CCNG1 in tissues, immunohistochemistry analysis was performed. Sectioned specimens were deparaffinized and dehydrated for the analysis. Antibodies to LMP1, E2F3, and cyclin G1 were obtained from Abcam (Hong Kong). Samples that were not exposed to the primary antibody were used as negative controls.
Luciferase reporter assay.
Basic information about hsa-miR-203 was collected from miRBase (http://microrna.sanger.ac.uk/sequences). The sequence and the possible targets are predicted by the online software TargetScan, version 5.0, and Pictar. Two single strands of the 3′ UTR of the target genes that contained the miRNA binding sites were synthesized (accession number NM_001949 for E2F3 and NM_004060 for CCNG1). Then, two single strands of the 3′ UTRs of the target genes, with 5 or 6 bases deleted in the miR-203 binding site, were synthesized as mutant controls. Restriction sites for HindIII and SpeI were located at both ends of the oligonucleotides for further cloning. The following oligonucleotides were used in this study: the 3′ UTR of E2F3 (sense, 5′-CTAGTACAGCAATCTTCCTTAATAGCATTTCAAGCCGTGCCTTCTCCGCAGAAA-3′; antisense, 5′-AGCTTTTCTGCGGAGAAGGCACGGCTTGAAATGCTATTAAGGAAGATTGCTGTA-3′), the 3′ UTR of mutant E2F3 (sense, 5′-CTAGTACAGCAATCTTCCTTAATAGC------AGCCGTGCCTTCTCCGCAGAAA-3′; antisense, 5′-AGCTTTTCTGCGGAGAAGGCACGGCT------GCTATTAAGGAAGATTGCTGTA-3′; dashes indicate the deletion of ATTTCA), the 3′ UTR of CCNG1 (sense, 5′-CTAGTGAGGTTTATGTCAAAAGCAACATTTCACAAATGTACTTTTAAGGCATAA-3′; antisense, 5′-AGCTTTATGCCTTAAAAGTACATTTGTGAAATGTTGCTTTTGACATAAACCTCA-3′), and the 3′ UTR of mutant CCNG1 (sense, 5′-CTAGTGAGGTTTATGTCAAAAGCAAC-----ACAAATGTACTTTTAAGGCATAA-3′; antisense, 5′-AGCTTTATGCCTTAAAAGTACATTTGT-----GTTGCTTTTGACATAAACCTCA-3′; dashes indicate the deletion of ATTTC). The sense and antisense strands were annealed, digested with HindIII and SpeI, and ligated into the pmir-Report luciferase vector (Ambion, Austin, TX). Five luciferase reporters (LRs) were constructed. These constructed plasmids are referred to as LR-blank (no insertion), LR-203/E2F3, LR-203/E2F3m (where m indicates mutant), LR-203/CCNG1, and LR-203/CCNG1m. Cells were seeded into 24-well plates 24 h prior to transfection. The following day, 200 ng of reporter plasmid and 10 pmol of miR-203 mimics were cotransfected into cells using Lipofectamine 2000. Cell lysates were collected 24 h posttransfection and assayed for luciferase activity using a luciferase assay kit (Promega, Madison, WI), and β-galactosidase activity was measured using an enzyme assay kit (Promega). The results were normalized to β-galactosidase activity.
Design of siRNA and Western blotting.
The sequence of the 19-nucleotide (nt) small interfering RNA (siRNA) that was designed to target the LMP1 transcript (36) was 5′-GGAATTTGCACGGACAGGC-3′. The plasmid stably encoding the siRNA targeting LMP1 (siRNA-LMP1) and the negative-control plasmid were synthesized by Shanghai GeneChem Co. Ltd., China. Western blotting was performed as described previously (32). The antibody to β-actin was purchased from Sigma-Aldrich (St. Louis, MO). Antibodies to LMP1, E2F3, and cyclin G1 were the same ones that were used for the immunohistochemical analysis.
Assessment of pathway inhibition.
The JNK inhibitor SP600125, the p38/mitogen-activated protein kinase (MAPK) inhibitor SB203580, or the NF-κB inhibitor BAY11-7082 was used to block specific signaling pathways. Dimethyl sulfoxide (DMSO) was used as a vehicle control. 293-EBV and NP69-LMP1 cells were cultured in 5 μM SP600125, SB203580, or BAY11-7082 for 48 h. After treatment, the cellular proteins were extracted and subjected to SDS-PAGE. Antibodies to phospho-c-Jun (Cell Signaling Technology Co.), phospho-p38 (Santa Cruz), and NF-κB p65 (Abcam) were used for the Western blotting. Total RNA was also isolated from the cells and subjected to qRT-PCR to determine miR-203 expression levels.
Flow cytometry analysis.
Cells were collected, washed twice with phosphate-buffered saline, and fixed in 70% ethanol overnight. Then, cells were centrifuged at 1,500 × g for 8 min, resuspended in 50 μg/ml propidium iodide (Sigma-Aldrich) in phosphate-buffered saline, and immediately analyzed by flow cytometry using a MOFLO XDP flow cytometer (Beckman Coulter, Fullerton, CA). The appropriate forward and side scatter gates were used, and 1 × 105 cells were examined per experiment. Data were analyzed with Modfit software (Verity Software House, Topsham, ME). The values were expressed as the mean and the standard deviation of three independent experiments.
Experiments in nude mice.
To examine the effect of miR-203 on tumor growth in EBV-infected cells in vivo, a xenograft model was used as previously described (32). Three million cells in 0.2 ml of growth medium were subcutaneously injected into four BALB/c nude mice that were 4 weeks old. One week after the inoculation, the first tumor was detected. It was 1 mm in diameter by palpation. At this point, the mice were given an intratumoral injection (sterile insulin syringe; Becton, Dickinson, and Co.) with 1 μg of the miR-203 mimic or NC (scrambled sequence) in 40 μl of sterile diethyl pyrocarbonate (DEPC)-treated water once every 2 days for 5 weeks. When the size of the tumor increased, a multipoint, equal injection was performed for both groups.
Statistical analysis.
The differences between groups were tested using the Student t test or a one-way analysis of variance (ANOVA) test using the SPSS, version 11.0, program. Correlation analysis was performed using the Spearman method in SPSS, version 11.0.
RESULTS
EBV infection downregulates the expression of miR-203 in epithelial cells.
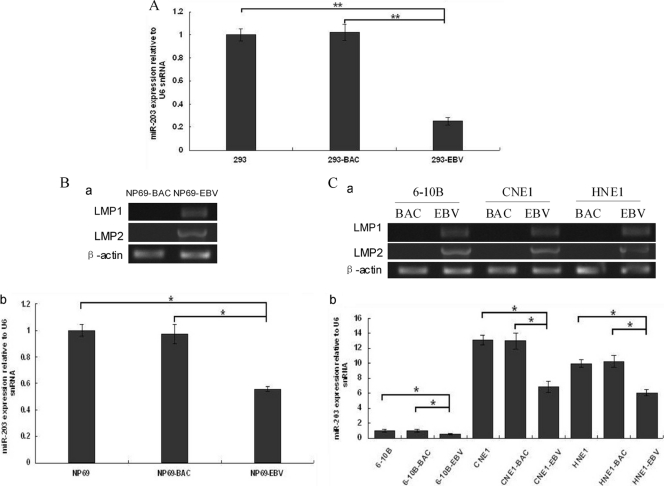
qRT-PCR analysis was performed to verify miR-203 expression in the stably EBV-infected cells, 293-EBV, and miR-203 was found to be downregulated approximately 4-fold (Fig. 1A). Additionally, miR-203 was downregulated in other epithelial or NPC cells, including NP69, 6-10B, CNE1, and HNE1, that were newly infected with EBV (Fig. 1B and C). These results suggested that the downregulation of miR-203 expression is an event that occurs during the early stages of latent EBV infection.
Fig 1.
miR-203 expression in EBV-infected epithelial cells. (A) In EBV stably infected 293-EBV cells, miR-203 was downregulated approximately 4-fold compared with the controls (293 and 293-BAC cells). 293-BAC cells were used as a negative control and were stably transfected with the BAC-based vector pM-BAC, which was established by eliminating the EBV genome from the Maxi-EBV plasmid. (B) Expression of latent genes and miR-203 in immortalized nasopharynx cells (NP69) during the first 7 days of infection using the transfer infection method with infectious EBV particles. The expression of LMP1 and LMP2 genes was determined using semiquantitative RT-PCR analysis (a). The expression of miR-203 was determined using real-time qRT-PCR analysis (b). NP69-BAC cells were used as a negative control and were transfected with the vector pM-BAC. (C) The expression of latent genes and miR-203 in nasopharyngeal carcinoma cells that were reinfected with EBV 10 days postinfection. Expression of LMP1 and LMP2 (a) and of miR-203 (b) are shown. The 6-10B-BAC, CNE1-BAC, and HNE1-BAC cell lines were used as controls and were transfected with the vector, pM-BAC. For panels B and C, the EBV-infected cells were GFP positive under green fluorescence microscopy. miR-203 expression was normalized to that of U6, and the relative levels are shown, with the value of the blank-cell control of each group standardized to 1. The data correspond to the mean values of three independent experiments. A P value of <0.05 (*) or <0.01 (**) was considered to be statistically significant or extremely significant, respectively.
EBV infection is associated with the downregulation of miR-203 in NPC tissues.
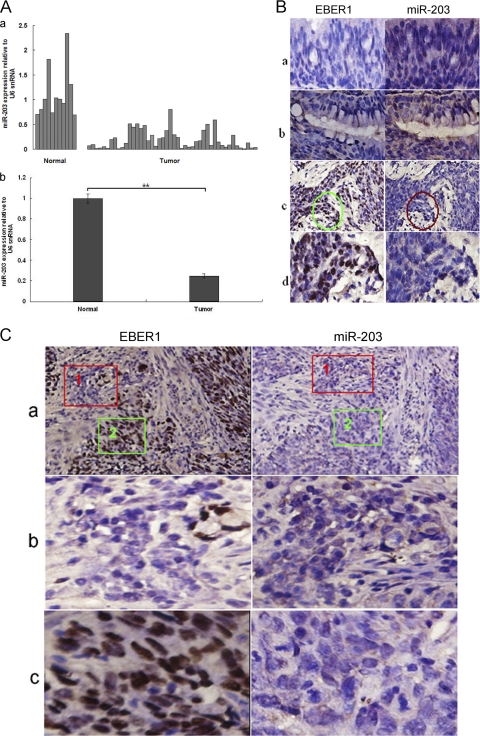
To explore the expression of miR-203 in human samples, miR-203 expression was measured in 46 NPC specimens and 11 normal or nonneoplastic epithelium tissues by qRT-PCR. As shown in Fig. 2A, the average level of miR-203 expression was approximately 4-fold lower in the NPC specimens than in the normal or nonneoplastic tissues (P < 0.01). ISH showed that miR-203 was expressed at a higher level in epithelial tissues and glands adjacent to NPC tissues (EBV-negative) than in NPC tissues (Fig. 2B). In some EBV-positive areas of the NPC tissues, miR-203 expression was almost undetectable. Furthermore, in the areas that were EBV negative (37), miR-203 expression increased (Fig. 2B, rows c and d, and C, row b). Therefore, downregulation of miR-203 was associated with EBV infection in NPC tissues. However, the level of EBV infection and miR-203 downregulation, as judged by the amount of dye, did not directly correspond in every area of the NPC tissues.
Fig 2.
Detection of miR-203 and EBV expression in NPC specimens. (A) miR-203 expression analysis in 46 NPC tissues and 11 normal (including nonneoplastic) nasopharynx epithelium tissues as determined by qRT-PCR analysis. The expression of miR-203 in single tissues (a) and the relative level of miR-203 expression as a whole in the tissues (b) are shown. (B) Detection of EBER1 and miR-203 in NPC and the adjacent tissues. Detection was performed using ISH in serial paraffin sections. The noncancerous tissues (a) or gland (b) adjacent to the NPC tissue was negative for the EBV genome (EBER1 ISH) and positive for miR-203. NPC tissues (c) were positive for the EBV genome and negative or weakly positive for miR-203 expression. In row d, magnified images are shown of the circled areas in row c. Original magnifications: ×200 (a, b, and d) and×100 (c). (C) Detection of EBER1 and miR-203 expression in a different specimen of the NPC tissue. Most of the areas in the NPC tissue (a) were positive for the EBV genome and negative or weakly positive for miR-203. Row b shows magnified images of fields 1 in row a that were negative for EBER1 and strongly positive for miR-203. Row c shows magnified images of fields 2 in row a that were positive for EBER1 and negative for miR-203. Original magnifications: ×100 (a) and ×200 (b and c).
CCNG1 and E2F3 are miR-203 target genes.
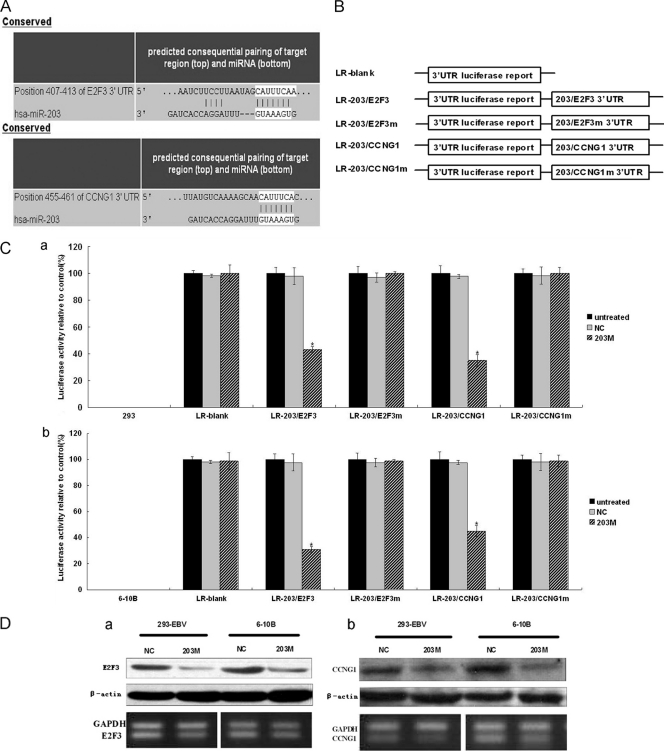
A previous study showed that EBV promoted the G1/S-phase transition in 293-EBV cells, which exhibit enhanced cell proliferation (32). Two genes, E2F3 and CCNG1, were selected from the list of putative miR-203 targets that were predicted by TargetScan and Pictar for further study. The luciferase reporter assay demonstrated that these two genes were targets of miR-203 (Fig. 3). As shown in Fig. 3C, when the 3′ UTRs of E2F3 and CCNG1 were attached to the luciferase gene, the amount of luciferase activity decreased significantly (P < 0.05) in cells transfected with miR-203 mimics (203 M). Furthermore, expression of mutant E2F3 and CCNG1 3′ UTRs restored the luciferase activity. These results verified the effect of miR-203 on the 3′ UTR of E2F3 and CCNG1. To examine the effect of miR-203 on the endogenous expression of E2F3 and CCNG1, two cell lines, 293-EBV and 6-10B, which exhibited a relatively low level of endogenous miR-203 (as shown in Fig. 1A and C), were transfected with miR-203 mimics (203 M), and an obvious decrease in the expression of these two genes was detected at the protein level (Fig. 3D). miR-203 affected the expression of both genes, as detected by semiquantitative RT-PCR, in 293-EBV cells. However, this effect was not so obvious in 6-10B cells (Fig. 3D, panels a and b).
Fig 3.
Identification of the target genes of miR-203 and the effect of miR-203 mimics on those targets. (A) Base pairing between miR-203 and the target genes, E2F3 and CCNG1, was predicted by TargetScan software. (B) Schematic representation of the five reporter constructs, named LR-blank, LR-203/E2F3, LR-203/E2F3m, LR-203/CCNG1, and LR-203/CCNG1m, respectively, where “m” indicates the mutant construct with 5 or 6 bases deleted at the predicted binding site of the gene 3′ UTRs. LR, luciferase reporter. (C) Each of the constructed luciferase reporter plasmids was cotransfected with the indicated material (untreated; NC, scramble; 203 M, miR-203 mimic) into the 293 (a) or 6-10B NPC (b) cells. Luciferase activities were measured after 48 h, and β-galactosidase was used for normalization of the difference of transfection efficiencies. The result showed that miR-203 combined with the predicted sites at the 3′ UTRs of the two genes, resulting in decreased luciferase activity. Data were confirmed in three experiments. A P value of <0.05 (*) was considered to be statistically significant. (D) The effect of miR-203 mimics on the expression of endogenous E2F3 and cyclin G1. The 293-EBV cell line or the NPC-derived cell line, 6-10B, was treated with 203 M or NC; after 48 h of the treatment, E2F3 (a) and cyclin G1 (b) at both protein and mRNA levels were assayed by Western blotting (upper panels) or semiquantitative RT-PCR (lower panels) analysis.
EBV mediates miR-203 downregulation at the level of the primary transcript, and LMP1 is an essential viral factor responsible for miR-203 downregulation.
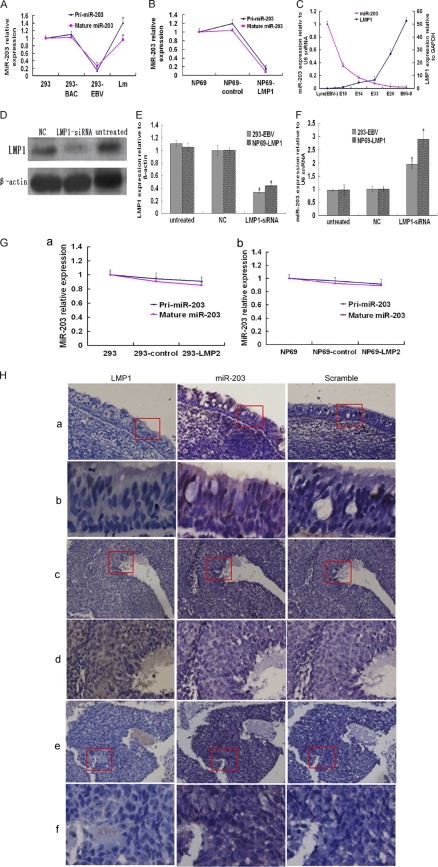
Detection of miR-203 expression levels at both the primary and the mature transcript levels demonstrated that EBV downregulated both levels equally, which implied that EBV downregulated the primary transcript of miR-203 (pri-miR-203) (Fig. 4A), which then resulted in downregulation of the mature miR-203 transcript. Moreover, loss of EBV from the 293-EBV (Lm cells) cells resulted in an almost complete restoration of miR-203 expression (Fig. 4A).
Fig 4.
LMP1 downregulated miR-203 expression in cells and NPC tissues. (A) The effect of EBV latent infection on miR-203 expression at the primary and mature transcript levels as determined by qRT-PCR analysis. There was a complete restoration of miR-203 expression in the Lm cells that had lost EBV. These data came from experiments different from those shown in Fig. 1A. (B) miR-203 expression of primary and mature transcripts in immortalized nasopharynx cells stably transfected with LMP1 (NP69-LMP1). The NP69-control was a negative control stably transfected with the corresponding vector, pLNSX. In the 293-EBV (A) and NP69-LMP1 cells (B), miR-203 was downregulated. (C) The relative expression of LMP1 and miR-203 in EBV-transformed LCLs showed a perfect negative correlation by Spearman analysis using SPSS, version 11.0 (r = −0.92, P < 0.01). LMP1 and miR-203 expression were normalized to that of GAPDH and U6, respectively. (D) Western blot analysis of LMP1 in 293-EBV cells at 72 h posttransfection with LMP1-siRNA. (E) The expression of LMP1 after treatment with LMP1-siRNA as measured by qRT-PCR analysis. (F) The expression of miR-203 after treatment with LMP1-siRNA as measured by qRT-PCR analysis. miR-203 expression was normalized to that of U6, and LMP1 expression was normalized to that of β-actin. Values from the qRT-PCR analysis are expressed as the mean of three independent experiments (A, B, C, E, and F). *, P < 0.05; **, P < 0.01. (G) Overexpression of LMP2 had no significant effect on the expression of miR-203 in 293 (a) and NP69 (b) cells. (H) Detection of LMP1 by immunohistochemical analysis and miR-203 by ISH in the NPC or NPC-adjacent tissues. The tissue samples were cut into serial paraffin sections. “Scramble” was used as a negative control for ISH detection. The magnified images in rows b, d, and f show the corresponding fields within the boxes in rows a, c, and e, respectively. Original magnifications: ×100 (a, c, and e) and ×200 (b, d, and f).
LMP1 is the first EBV protein to be identified as an oncoprotein. To investigate whether there was a relationship between LMP1 and miR-203 downregulation, a cell line stably transfected with LMP1, NP69-LMP1, was used to monitor miR-203 expression. We found that miR-203 expression in NP69-LMP1 cells was dramatically decreased at both the primary and mature transcript levels (Fig. 4B), as was previously observed in 293-EBV cells (Fig. 4A). Furthermore, there was a negative correlation between miR-203 expression and LMP1 expression in EBV-transformed lymphocytes (LCLs) (r = −0.92) (Fig. 4C). To further investigate the role of LMP1 in regulating the expression of miR-203, LMP1-siRNAs were transfected into 293-EBV and NP69-LMP1 cells, and a 5-fold decrease in LMP1 expression was observed (Fig. 4D and E). Repression of LMP1 significantly increased miR-203 expression in both cell lines (P < 0.05) (Fig. 4F). In addition, LMP2 did not mediate deregulation of miR-203 primary or mature transcripts (Fig. 4G). The correlation between the expression of LMP1 and miR-203 was further determined in additional NPC or noncancer specimens. As shown in Fig. 4H, miR-203 expression is obvious in the tissues adjacent to NPC tissues where LMP1 is not expressed, and miR-203 expression was downregulated in most areas of the NPC tissues that were positive for LMP1 expression. However, some areas in the NPC tissue that were LMP1 negative expressed larger amounts of miR-203.
The LMP1-mediated JNK and NF-κB pathways are involved in miR-203 downregulation.
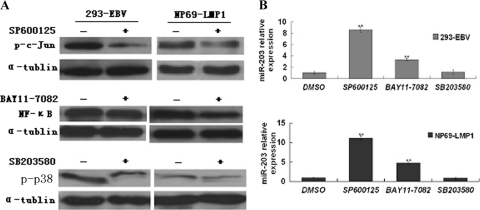
LMP1 has three C-terminal-activating region (CTAR) domains. Both CTAR1 and CTAR2 influence the transcript factor NF-κB and the p38 mitogen-activated protein kinase (MAPK) pathways. CTAR2 is the key effector that triggers c-Jun N-terminal kinase (JNK) and activates NF-κB (17). To determine whether the pathways mediated by LMP1 were involved in LMP1-driven miR-203 downregulation, inhibitors of the NF-κB, JNK, and p38 pathways were used. The JNK inhibitor significantly abolished downregulation of miR-203 (Fig. 5). Additionally, inhibition of NF-κB restored miR-203 expression. However, the p38 inhibitor had no obvious effect on miR-203 expression. In addition, the transfection of an expression plasmid containing an LMP1 mutant lacking CTAR3 had no effect on miR-203 expression in the 293-EBV and NP69-LMP1 cell lines (data not shown). These results suggest that CTAR1 and -2 might be important for LMP1-induced downregulation of miR-203.
Fig 5.
Inhibitors of JNK and NF-κB, but not p38, blocked miR-203 downregulation mediated by EBV-LMP1. (A) Phosphorylation of c-Jun, p38, and NF-κB (p65) was evaluated by Western blotting. (B) The expression of pri-miR-203 after treatment with inhibitors as measured by qRT-PCR. The cells were treated with 5 μM JNK inhibitor (SP600125), p38 inhibitor (SB203580), or NF-κB inhibitor (BAY11-7082). DMSO was used as a vehicle control. After 48 h, the cells were harvested, and expression was analyzed. The experiment was repeated three times. **, P < 0.01.
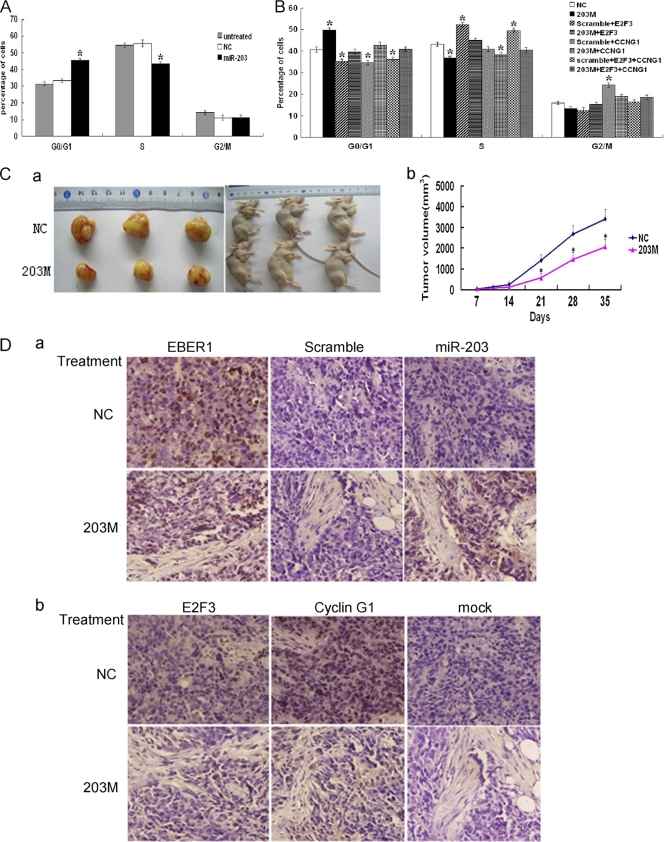
Ectopic expression of miR-203 blocks S-phase entry in EBV-infected cells and suppresses the growth of tumors induced by EBV in vivo.
As previously shown, EBV facilitated entry into S phase of the cell cycle and enhanced the malignant potential of 293-EBV cells (32). To investigate whether downregulation of miR-203 by EBV functionally contributed to the biological changes caused by EBV, miR-203 was ectopically expressed in cell lines and an animal model. As shown in Fig. 6A, at 48 h posttransfection with the 203 M into 293-EBV cells, the number of cells in S phase decreased, and the number in the G1 phase of the cell cycle increased, which demonstrated inhibition of S-phase entry (P < 0.05). This change was overcome by overexpression of the target genes, E2F3 and CCNG1, with miR-203 (Fig. 6B). Intratumoral injection of miR-203 led to a significant decrease in the size of the tumor in nude mice with transplanted tumors (P < 0.05) (Fig. 6C), suggesting that miR-203 inhibits the tumorigenicity of epithelial cells promoted by EBV infection. Expression of E2F3 and cyclin G1 (encoded by the CCNG1 gene) decreased after the introduction of miR-203 (Fig. 6D).
Fig 6.
Ectopic expression of miR-203 reversed the phenotype changes of EBV infection. (A) Cell cycle distribution after ectopic expression of miR-203 in 293-EBV cells. Two days after transfection with miR-203 mimics (203 M) and scramble (NC), cells were harvested, fixed with ethanol, and subjected to the measurement by flow cytometry. (B) The overexpression of E2F3 and/or CCNG1 together with miR-203 overcame the effect of miR-203 on the cell cycle distribution. The 293-EBV cells were cotransfected with expression vectors, scramble and the vector pCMV6-XL (for NC), or miR-203 mimics (203 M) (20 nM) and pCMV6-XL (for 203 M), or 203 M/scramble and E2F3/CCNG1 (a mixture of E2F3 and CCNG1) (for the other six groups). Each expression vector contained the entire coding sequence of the gene minus the 3′ UTR. For panels A and B), the values are shown as the mean of three independent experiments (*, P < 0.05 compared with the NC values). (C) The effect of miR-203 on the growth of tumors formed by the 293-EBV cells. Three million cells were injected into mice; three of four mice from each group formed tumors at the site of inoculation. A tumor was first detected at day 7 postinjection. During the intratumoral injection of the 203 M or NC, the xenograft tumor volume was examined. The graph in panel a shows tumor formation in the nude mice. The graph in panel b shows the curve of tumor growth during the treatment with 203 M or NC (*, P < 0.05). The average tumor volume in the 203 M group (n = 3) was statistically significant from the volume in the mice treated with NC. (D) The expression of miR-203 and the two target genes in the tumor tissues at the end of the 203 M intratumoral injection treatment. Serial sectioned specimens were used. The expression of EBER1 and miR-203 (a) was determined by ISH. Scramble was used as a negative control, which was a probe attached to a scrambled sequence. The expression of E2F3 and CCNG1 (b) was determined by immunohistochemical analysis. Mock, no primary antibody.
DISCUSSION
NPC is an EBV-associated malignancy that is common in southern China and Southeast Asia. The mechanism by which EBV causes NPC is unknown. Therefore, it is essential to investigate the interaction between the virus and host cells to further understand the mechanism. miRNAs are thought to have a large impact on cancer. In fact, some are known as onco-miRNAs or tumor-suppressive miRNAs (24, 31). Recent studies have showed that EBV infection deregulates cellular miRNAs in B lymphocytes (7, 29) or EBV-associated B-cell lymphomas (23). Differentiated expression of some miRNAs has also been reported in NPC (11, 41). LMP1 was recently found to activate several miRNAs, including miR-146a and miR-155, in EBV-related B-cell malignancies (8, 20). Shinozaki et al. reported that miR-200 is decreased in EBV-associated gastric carcinoma (44). miR-203 is expressed specifically in the suprabasal layers of stratified epithelia and is thought to function as a tumor suppressor in several epithelial cancers (2, 6, 38, 48). However, whether EBV mediates miR-203 deregulation in NPC and how it functions in EBV-associated carcinogenesis of NPC have not been confirmed. The primary role of miR-203 is to suppress the proliferative capacity of epithelial cells upon differentiation (53). One tumor virus, human papillomavirus (HPV), downregulates the expression of miR-203 (38). However, it has not been confirmed whether miR-203 is involved in the pathogenesis of HPV-associated cervical cancer. The downregulation of miR-203 in 293-EBV cells was 3-fold, as determined by a previous microarray analysis, and 4-fold by qRT-PCR analysis in this study (Fig. 1A). These results suggested that the miR-203 deregulation was an event that occurs during the early stage of EBV latency. Furthermore, analysis of NPC tissues showed that miR-203 is downregulated overall (Fig. 2A). However, it is noteworthy that there were areas in the NPC tissue that were negative for EBV infection but had high levels of miR-203 expression (Fig. 2B). EBV infection is known to be an early event in NPC development; these results suggested that miR-203 downregulation, which is associated with latent EBV infection, might have a role in the development of NPC.
LMP1 is an EBV oncoprotein that transforms human fibroblast cells in vitro and induces many of the characteristic phenotypic changes observed in EBV-transformed LCLs (39, 49, 50). LMP1 expression blocks the differentiation of epithelial cells (14). Our previous results suggested that LMP1 is important for cell proliferation and that tumorigenicity is promoted by EBV (32). In the present study, we demonstrated that LMP1 was essential and necessary to downregulate miR-203 expression in EBV-positive cells and NPC tissues (Fig. 4). As shown in Fig. 4B, LMP1 is sufficient for miR-203 downregulation. Moreover, removal of the EBV genome resulted in the complete restoration of miR-203 expression (in Lm cells, negative for EBV and LMP1) (Fig. 4A), which further confirmed that the miR-203 downregulation was due to EBV infection and LMP1 expression.
These results indicated that the EBV protein LMP1 mediated downregulation of miR-203 expression at the primary transcript level. LMP1 functions through its CTAR effector domains via multiple pathways. To investigate whether these pathways are involved in LMP1-mediated downregulation of miR-203, we used inhibitors of these pathways. Inhibition of the JNK and NF-κB pathways largely restored miR-203 expression (Fig. 5). Additionally, LMP1-mediated miR-203 downregulation depended on activation of the JNK and NF-κB pathways; the mechanism is currently being studied.
Studies have shown that miR-203 decreases cell growth and inhibits tumor invasion by regulating the expression of its target genes (13, 18, 35). EBV and LMP1 facilitate the G1/S transition in cell cycle progression and transformation (32, 33, 51). In the present study, overexpression of miR-203 blocks the G1/S transition in EBV-infected cells and suppresses tumor growth in vivo (Fig. 6). Thus, miR-203 and EBV-LMP1 play opposite roles in cell growth and transformation. On the other hand, E2F3 and CCNG1 (encoding cyclin G1) were verified to be novel targets of miR-203 (Fig. 3). Furthermore, E2F3 and CCNG1 levels were reduced when miR-203 was ectopically expressed in epithelial cells and tumor models (Fig. 3D and 6D). The activity of E2F family members plays an important role in the cell cycle of proliferating cells (52). The release of E2F factors from retinoblastoma protein (Rb) is critical for entry into S phase, and E2F3 is sufficient to support G1/S-specific gene expression (12). E2F factors are also involved in oncogene-mediated transformation of mouse embryonic fibroblasts (43). Additionally, cyclin G1 is a regulator of the cell cycle. It is one of the first target genes of the suppressor p53 identified. Overexpression of cyclin G1 reduces the levels of p53 (40), and the loss of cyclin G1 decreases tumor susceptibility and significantly lowers tumor incidence in experimental hepatocarcinogenesis models (25). Suppression of cyclin G1 results in the inhibition of tumor growth (10, 22). As shown in Fig. 6B, overexpression of E2F3 and cyclin G1 reversed the cell cycle changes induced by miR-203. Moreover, E2F3 and cyclin G1 are upregulated in NPC tissues, as demonstrated by microarray analysis (4). This study indicates that E2F3 and cyclin G1 might contribute to the progression of EBV-associated NPC by downregulating miR-203.
In conclusion, miR-203 is downregulated in EBV-infected epithelial cells and EBV-associated NPC, and this occurs during the early stages of EBV latency. The EBV-encoded oncoprotein LMP1 was the essential viral factor responsible for miR-203 downregulation. Removal of EBV infection and suppression of LMP1 resulted in the restoration of miR-203 expression. Ectopic expression of miR-203 inhibited EBV-induced S-phase entry and transformation. EBV-LMP1 mediated miR-203 downregulation at the primary transcript level. E2F3 and cyclin G1, which are important for cell cycle regulation and tumor incidence, were identified as novel targets of miR-203. Furthermore, they contributed to the phenotypic changes caused by miR-203 downregulation. Additionally, the LMP1-mediated JNK and NF-κB pathways were involved in miR-203 downregulation. These results implied that EBV contributed to epithelial cancer etiology through LMP1, which downregulates cellular miR-203. The results of this study reflect the interaction between the virus and host during cancer development. Furthermore, miR-203 is a potential biomarker for the diagnosis and therapy of EBV-associated NPC.
ACKNOWLEDGMENTS
We thank the GSF-National Research Center for Environment and Health (Germany) and Wolfgang Hammerschmidt for allowing us to use the Maxi-EBV system. We also thank James McCarthy (Tumor Biology and Progression Program in Masonic Cancer Center, University of Minnesota) for his suggestions. We are grateful for the help of Yanhong Zhou in the analysis of flow cytometry, Lei Shi in collecting clinical specimens, and Shuping Peng, Minghua Wu, Wei Xiong, Ming Zhou, Xiaoling Li, Jian Ma, and Juanjuan Xiang in sharing their ideas.
This work was supported by Projects of Hunan Provincial Natural Science Foundation of China (11JJ2043), the National Natural Science Foundation of China (81171931), the Research and Development Project of Hunan Development and Reform Commission (2009-13910), Graduate Degree Thesis Innovation Foundation of Central South University (2010ybfz102), and the Doctor Innovation Research Project of Hunan Province (CX2010B040).
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 2. Bo J, et al. 2011. microRNA-203 suppresses bladder cancer development by repressing bcl-w expression. FEBS J. 278:786–792 [DOI] [PubMed] [Google Scholar]
- 3. Bornkamm GW, Hammerschmidt W. 2001. Molecular virology of Epstein-Barr virus. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 356:437–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bose S, et al. 2009. The ATM tumour suppressor gene is down-regulated in EBV-associated nasopharyngeal carcinoma. J. Pathol. 217:345–352 [DOI] [PubMed] [Google Scholar]
- 5. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113:25–36 [DOI] [PubMed] [Google Scholar]
- 6. Bueno MJ, et al. 2008. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13:496–506 [DOI] [PubMed] [Google Scholar]
- 7. Cameron JE, et al. 2008. Epstein-Barr virus growth/latency III program alters cellular microRNA expression. Virology 382:257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cameron JE, et al. 2008. Epstein-Barr virus latent membrane protein 1 induces cellular MicroRNA miR-146a, a modulator of lymphocyte signaling pathways. J. Virol. 82:1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen CZ, Li L, Lodish HF, Bartel DP. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86 [DOI] [PubMed] [Google Scholar]
- 10. Chen DS, et al. 1997. Retroviral vector-mediated transfer of an antisense cyclin G1 construct inhibits osteosarcoma tumor growth in nude mice. Hum. Gene Ther. 8:1667–1674 [DOI] [PubMed] [Google Scholar]
- 11. Chen HC, et al. 2009. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br. J. Cancer 100:1002–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chong JL, et al. 2009. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol. Cell. Biol. 29:414–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Craig VJ, Cogliatti SB, Rehrauer H, Wundisch T, Muller A. 2011. Epigenetic silencing of microRNA-203 dysregulates ABL1 expression and drives Helicobacter-associated gastric lymphomagenesis. Cancer Res. 71:3616–3624 [DOI] [PubMed] [Google Scholar]
- 14. Dawson CW, Rickinson AB, Young LS. 1990. Epstein-Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature 344:777–780 [DOI] [PubMed] [Google Scholar]
- 15. Delecluse HJ, Hammerschmidt WW. 2000. The genetic approach to the Epstein-Barr virus: from basic virology to gene therapy. Mol. Pathol. 53:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eliopoulos AG, Young LS. 2001. LMP1 structure and signal transduction. Semin. Cancer Biol. 11:435–444 [DOI] [PubMed] [Google Scholar]
- 18. Faber J, Gregory RI, Armstrong SA. 2008. Linking miRNA regulation to BCR-ABL expression: the next dimension. Cancer Cell 13:467–469 [DOI] [PubMed] [Google Scholar]
- 19. Feber A, et al. 2008. MicroRNA expression profiles of esophageal cancer. J. Thorac. Cardiovasc. Surg. 135:255–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gatto G, et al. 2008. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway. Nucleic Acids Res. 36:6608–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glaser R, et al. 1989. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc. Natl. Acad. Sci. U. S. A. 86:9524–9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gordon EM, et al. 2000. Inhibition of metastatic tumor growth in nude mice by portal vein infusions of matrix-targeted retroviral vectors bearing a cytocidal cyclin G1 construct. Cancer Res. 60:3343–3347 [PubMed] [Google Scholar]
- 23. Imig J, et al. 2011. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 39:1880–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iorio MV, Croce CM. 2009. MicroRNAs in Cancer: Small molecules with a huge impact. J. Clin. Oncol. 27:5848–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jensen MR, et al. 2003. Reduced hepatic tumor incidence in cyclin G1-deficient mice. Hepatology 37:862–870 [DOI] [PubMed] [Google Scholar]
- 26. Kaye KM, Izumi KM, Kieff E. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. U. S. A. 90:9150–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li HM, et al. 2004. Epstein-Barr virus latent membrane protein 1 (LMP1) upregulates Id1 expression in nasopharyngeal epithelial cells. Oncogene 23:4488–4494 [DOI] [PubMed] [Google Scholar]
- 28. Li QX, et al. 1992. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature 356:347–350 [DOI] [PubMed] [Google Scholar]
- 29. Linnstaedt SD, Gottwein E, Skalsky RL, Luftig MA, Cullen BR. 2010. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J. Virol. 84:11670–11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lo KW, To KF, Huang DP. 2004. Focus on nasopharyngeal carcinoma. Cancer Cell 5:423–428 [DOI] [PubMed] [Google Scholar]
- 31. Lu J, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838 [DOI] [PubMed] [Google Scholar]
- 32. Lu JH, et al. 2010. Epstein-Barr virus facilitates the malignant potential of immortalized epithelial cells: from latent genome to viral production and maintenance. Lab. Invest. 90:196–209 [DOI] [PubMed] [Google Scholar]
- 33. Mainou BA, Everly DN, Jr, Raab-Traub N. 2007. Unique signaling properties of CTAR1 in LMP1-mediated transformation. J. Virol. 81:9680–9692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mathe EA, et al. 2009. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin. Cancer Res. 15:6192–6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McKenna DJ, McDade SS, Patel D, McCance DJ. 2010. MicroRNA 203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J. Virol. 84:10644–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mei YP, et al. 2007. Silencing of LMP1 induces cell cycle arrest and enhances chemosensitivity through inhibition of AKT signaling pathway in EBV-positive nasopharyngeal carcinoma cells. Cell Cycle 6:1379–1385 [DOI] [PubMed] [Google Scholar]
- 37. Meisler AI. 1996. Epstein-Barr virus and nasopharyngeal cancer. N. Engl. J. Med. 334:122–123 [DOI] [PubMed] [Google Scholar]
- 38. Melar-New M, Laimins LA. 2010. Human papillomaviruses modulate expression of microRNA 203 upon epithelial differentiation to control levels of p63 proteins. J. Virol. 84:5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moorthy RK, Thorley-Lawson DA. 1993. All three domains of the Epstein-Barr virus-encoded latent membrane protein LMP-1 are required for transformation of rat-1 fibroblasts. J. Virol. 67:1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ohtsuka T, Jensen MR, Kim HG, Kim KT, Lee SW. 2004. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene 23:5405–5408 [DOI] [PubMed] [Google Scholar]
- 41. Sengupta S, et al. 2008. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc. Natl. Acad. Sci. U. S. A. 105:5874–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shannon-Lowe CD, Neuhierl B, Baldwin G, Rickinson AB, Delecluse HJ. 2006. Resting B cells as a transfer vehicle for Epstein–Barr virus infection of epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 103:7065–7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma N, et al. 2006. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J. Biol. Chem. 281:36124–36131 [DOI] [PubMed] [Google Scholar]
- 44. Shinozaki A, et al. 2010. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 70:4719–4727 [DOI] [PubMed] [Google Scholar]
- 45. Sonkoly E, et al. 2007. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2:e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sugden B, Mark W. 1977. Clonal transformation of adult human leukocytes by Epstein-Barr virus. J. Virol. 23:503–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang YL, et al. 2007. Lytic replication and inductive production of recombinant Epstein-Barr virus visualized. Prog. Biochem. Biophys. 34:418–424 [Google Scholar]
- 48. Viticchie G, et al. 2011. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle 10:1121–1131 [DOI] [PubMed] [Google Scholar]
- 49. Wang D, Liebowitz D, Kieff E. 1985. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell 43:831–840 [DOI] [PubMed] [Google Scholar]
- 50. Wang D, et al. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang LT, et al. 2011. Functional interaction of Ugene and EBV infection mediates tumorigenic effects. Oncogene 30:2921–2932 [DOI] [PubMed] [Google Scholar]
- 52. Wu L, et al. 2001. The E2F1-3 transcription factors are essential for cellular proliferation. Nature 414:457–462 [DOI] [PubMed] [Google Scholar]
- 53. Yi R, Poy MN, Stoffel M, Fuchs E. 2008. A skin microRNA promotes differentiation by repressing “stemness.” Nature 452:225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]
- 55. Yu Z, et al. 2011. A precise excision of the complete Epstein-Barr virus genome in a plasmid based on a bacterial artificial chromosome. J. Virol. Methods 176:103–107 [DOI] [PubMed] [Google Scholar]