Abstract
By screening 74 chordate genomes for endogenous lentiviruses using Pol sequences of exogenous lentiviruses as a reference, we identified a novel endogenous lentivirus in the genome of the ferret (Mustela putorius furo). Phylogenetic analysis suggested that the ferret endogenous lentivirus, denoted ELVmpf, diverged early in the evolution of the mammalian lentiviruses, although with a lack of resolution at key nodes. These data support the notion that lentiviruses have evolved on timescales of millions of years.
TEXT
Lentiviruses (family Retroviridae) cause chronic infections in a broad range of mammalian orders, including primates, artiodactyls, perissodactyls, and carnivores. Although lentiviruses are best known as exogenous infectious agents, endogenous copies were recently discovered in genomes of the European rabbit and christened RELIK (8), in other leporid species (9, 11), and in seven species of lemur (3, 4). Importantly, the existence of these endogenous viral elements indicates that lentiviruses are far older than suggested from many molecular clock-based analyses of their exogenous relatives and may have been present in some host genomes for at least 12 million years (9, 11).
Although endogenous retroviruses are of major importance for our understanding of the pattern and timescale of viral evolution, their known distribution is highly sporadic. In an attempt to expand our knowledge of endogenous lentiviruses, we performed a BLAST analysis of 74 publically available chordate genomes (http://www.ncbi.nlm.nih.gov/sites/genome). We used Pol amino acid sequences of exogenous ovine lentivirus (OLV), simian immunodeficiency virus of mnd2 (host mandrill) (SIVmnd2), human immunodeficiency virus type 1 (HIV-1), HIV-2, feline immunodeficiency virus (FIV), bovine immunodeficiency virus (BIV), and equine infectious anemia virus (EIAV) as queries and employed a cutoff of 35% sequence identity to signify a positive match.
This genomic screening process revealed three positive hits: in rabbit, in mouse lemur, and, for the first time, in ferret (Mustela putorius furo). Ferrets, like feline species, are members of the mammalian order Carnivora. Next, we performed a reverse BLAST analysis using all the endogenous lentiviruses obtained here. This analysis identified a total of five endogenous ferret sequences (with contig lengths shown in parentheses): contigs 098598 (26,030 bp), 023783 (14,373 bp), 006467 (129,423 bp), 111284 (4,088 bp), and 068467 (23,332 bp). These sequences had identifiable Pol protein lengths ranging from 290 to 958 amino acids. These genomic Pol sequences exhibited the closest match to FIV, with E values of 0.0 to 7e−142 and sequence identities of 64 to 41%. Pairwise distance analysis (using uncorrected p-distances) revealed that contigs 023783 and 006467 were genetically identical, while the other endogenous ferret lentivirus sequences differed by 7.6 to 10.6% at the amino acid level. We denote the endogenous lentiviruses in the ferret genome as ELVmpf. Unfortunately, because of the draft nature of the ferret genome, which is currently only available at 2× shotgun coverage, it is not possible to provide precise data on the genomic location of each ELVmpf sequence.
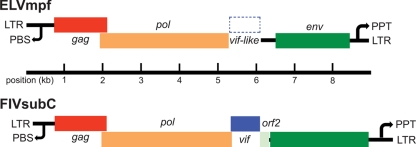
To better characterize the genetic structure of ELVmpf, we retrieved contig 098598 and located the gag, pol, and env genes by aligning them with exogenous lentiviruses. This analysis revealed that ELVmpf has the typical lentivirus genome organization (Fig. 1): two LTRs of approximately 320 bp in length, a primer binding site (PBS) at the 5′ end, and a polypurine tract (PPT) at the 3′ end, a gag gene of 1,375 bp, a pol gene of 3,352 bp, and an env gene of 1,929 bp. The analysis of the retroviral vif gene was more complex. Although there was a 1,216-bp region between the gag and pol genes, we were only able to identify a highly interrupted (i.e., containing multiple insertions and deletions) vif-like element, exhibiting 55.2% nucleotide similarity to FIV subtype C over a 789-bp region and for which no viable amino acid alignment could be obtained. Hence, vif may be present in ELVmpf, as it is in most lentiviruses, with the exception of EIAV, although this requires further verification. We then carried out a third genomic BLAST against the ferret genome using the predicted Gag (457 amino acids) and Pol (835 amino acids) amino acid sequences and a cutoff of 80% sequence identity in contig 098598. The Env protein was not used in this analysis because it is so divergent in sequence that it could not be unambiguously aligned. The analysis using the Gag sequence generated one additional positive hit in the ferret genome, contig 113944, which had a contig length of 1,359 bp, E value of 0.0, and sequence identity of 89%. In sum, our genomic mining identified six copies of endogenous lentiviruses in the ferret genome.
Fig 1.
Genome organization of an endogenous lentivirus in the ferret genome (ELVmpf, contig 098598). The major genes are shown in comparison to those of exogenous lentiviruses, in this case represented by feline immunodeficiency virus subtype C (FIVsubC). The location of the putative vif-like element in ELVmpf is also shown.
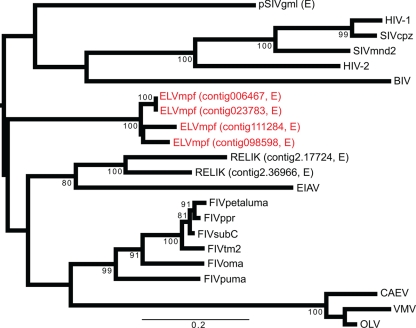
To determine the phylogenetic relationships between ELVmpf and the known endogenous and exogenous lentiviruses, we first aligned Pol amino acid sequences from the ferret endogenous lentiviruses identified here (excluding the very short sequences in contigs 068467 and 113944) with those endogenous lentiviruses from the rabbit (n = 2) and mouse lemur (n = 1), retrieved from Gifford et al. (3), as well as the sequences of representative exogenous lentiviruses (n = 15). Sequence alignment was undertaken using MUSCLE 3.7 (2) and then manually checked using Se-Al (http://tree.bio.ed.ac.uk/software/seal/). This produced a final alignment of 22 sequences, 312 amino acids in length (sequence alignment available from the authors on request). Phylogenetic relationships were then inferred using the maximum likelihood method available in PhyML 3.0 (5), employing the WAG+Γ model of amino acid substitution and with the robustness of each node determined using 1,000 bootstrap replicates. We also used the Gblocks program (10) to remove the divergent and ambiguously aligned regions, resulting in an alignment of 217 amino acids, and performed an equivalent phylogenetic analysis.
Our phylogenetic analysis of the full-length sequence alignment (i.e., 312 amino acid residues) places the ferret endogenous lentivirus in a deep phylogenetic position clearly distinct from those of endogenous lentiviruses previously described in lemurs and leporid species, as well as from the exogenous lentiviruses found in the Carnivora (Fig. 2). However, the tree was also characterized by weak bootstrap support at many of the basal nodes, reflecting the short sequence alignment and its divergent nature, so that the precise phylogenetic position of ELVmpf could not be resolved on these data. Indeed, it is notable that our phylogeny is different from those previously obtained for the lentiviruses (3, 4) in that BIV does not group with caprine arthritis encephalitis virus (CAEV)-OLV-Visna/Maedi virus (VMV). A similarly unresolved phylogeny was generated after using Gblocks to remove divergent positions (tree not shown; available from the authors on request). However, in all phylogenies, the four endogenous lentiviruses in the ferret genome grouped together, suggesting that they arose independently from the endogenous lentiviruses documented in other mammalian species.
Fig 2.
Phylogenetic relationships of exogenous and endogenous (E) lentiviruses. ELVmpf sequences are shown in red. Bootstrap values greater than 70% are shown, and branch lengths are drawn to a scale of amino acid substitutions per site. The tree is midpoint rooted for purposes of clarity only. Abbreviations (with GenBank accession numbers shown in parentheses) of exogenous lentiviruses are as follows: OLV, ovine lentivirus (YP_536867); BIV, bovine immunodeficiency virus (NP_040563); SIVcpz, simian immunodeficiency virus cpz, host chimpanzee (AAF18575); SIVmnd2, simian immunodeficiency virus mnd2, host mandrill (NP_758887); HIV-1, human immunodeficiency virus 1 (NP_057849); HIV-2, human immunodeficiency virus 2 (NP_663784); VMV, Visna/Maedi virus (NP_040840); CAEV, caprine arthritis encephalitis virus (NP_040939); FIVoma, feline immunodeficiency virus strain Oma (AAB49923); FIVpuma, feline immunodeficiency virus, host Puma concolor (ABO69501); FIVppr, feline immunodeficiency virus PPR isolate (AAA43076), FIVsubC, feline immunodeficiency virus subtype C (AAM13444); FIVpetaluma, feline immunodeficiency virus isolate Petaluma (P16088); FIVtm2, feline immunodeficiency virus isolate TM2 (P31822); and EIAV, equine infectious anemia virus (NP_056902). For endogenous lentiviruses, we used the names provided by Gifford et al. (3) and Katzourakis et al. (8).
To estimate the insertion time of ELVmpf into the ferret genome, we used the percent divergence among LTRs (7), assuming that all LTR differences appeared postintegration and are evolving neutrally. Accordingly, we compared the LTRs of ELVmpf in contig 098598 that exhibit 9.2% nucleotide sequence divergence (5′-LTR length of 322 nucleotides [nt] and 3′-LTR of 326 nt). By using a synonymous nucleotide substitution rate for carnivore genes of 0.38% substitutions per site per million years (1), we tentatively estimate that this particular ELVmpf element inserted into the ferret genome approximately 12 million years ago (mya). However, the lack of a recent and comprehensive estimate of the nucleotide substitution rate in carnivores, as well as the short sequence analyzed, means that this proposed integration date should be treated with caution. This uncertainty notwithstanding, a time scale of ∼12 mya is younger than the divergence of the Mustelidae (commonly referred to as the weasel family and which includes ferrets) from their closest relatives, the Procyonidae (raccoons, coatis, etc.), at about 29 mya (6), and hence again suggests that the insertion of this ELVmpf element was an independent event in the Mustelidae. Studies of endogenous lentiviruses in the mouse lemur and leporid species similarly suggest that they entered their respective host genomes independently: for leporid species, the lentiviral integration event occurred more than 12 mya (9, 11), while for the mouse lemur species the equivalent invasion took place around 4.2 mya (4). Together with our finding of endogenous lentiviruses in ferrets, these studies clearly indicate that the invasion of lentiviruses into mammalian genomes has occurred on multiple occasions and on a timescale of millions of years.
The eventual release of a high-quality ferret genome will enable a more refined analysis of the endogenous lentiviruses present in this species. However, irrespective of its exact phylogenetic placement and time of insertion, the discovery of ELVmpf in the ferret genome confirms that lentiviruses are a broadly distributed group of retroviruses and one that likely has an ancient origin.
ACKNOWLEDGMENTS
This work was in part funded by grant R01 GM080533-05 from the National Institute of General Medical Sciences, National Institutes of Health.
We thank two anonymous reviewers for valuable comments.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Bulmer M, Wolfe KH, Sharp PM. 1991. Synonymous nucleotide substitution rates in mammalian genes: implications for the molecular clock and the relationship of mammalian orders. Proc. Natl. Acad. Sci. U. S. A. 88:5974–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gifford RJ, et al. 2008. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U. S. A. 105:20362–20367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gilbert C, Maxfield DG, Goodman SM, Feschotte C. 2009. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 5:e1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 6. Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22:2971–2972 [DOI] [PubMed] [Google Scholar]
- 7. Johnson WE, Coffin JM. 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc. Natl. Acad. Sci. U. S. A. 96:10254–10260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Katzourakis A, Tristem M, Pybus OG, Gifford RJ. 2007. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U. S. A. 104:6261–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. 2009. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology 384:7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564–577 [DOI] [PubMed] [Google Scholar]
- 11. van der Loo W, Abrantes J, Esteves PJ. 2009. Sharing of endogenous lentiviral gene fragments among leporid lineages separated for more than 12 million years. J. Virol. 83:2386–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]