Abstract
The adeno-associated virus (AAV) genome encodes four Rep proteins, all of which contain an SF3 helicase domain. The larger Rep proteins, Rep78 and Rep68, are required for viral replication, whereas Rep40 and Rep52 are needed to package AAV genomes into preformed capsids; these smaller proteins are missing the site-specific DNA-binding and endonuclease domain found in Rep68/78. Other viral SF3 helicases, such as the simian virus 40 large T antigen and the papillomavirus E1 protein, are active as hexameric assemblies. However, Rep40 and Rep52 have not been observed to form stable oligomers on their own or with DNA, suggesting that important determinants of helicase multimerization lie outside the helicase domain. Here, we report that when the 23-residue linker that connects the endonuclease and helicase domains is appended to the adeno-associated virus type 5 (AAV5) helicase domain, the resulting protein forms discrete complexes on DNA consistent with single or double hexamers. The formation of these complexes does not require the Rep binding site sequence, nor is it nucleotide dependent. These complexes have stimulated ATPase and helicase activities relative to the helicase domain alone, indicating that they are catalytically relevant, a result supported by negative-stain electron microscopy images of hexameric rings. Similarly, the addition of the linker region to the AAV5 Rep endonuclease domain also confers on it the ability to bind and multimerize on nonspecific double-stranded DNA. We conclude that the linker is likely a key contributor to Rep68/78 DNA-dependent oligomerization and may play an important role in mediating Rep68/78's conversion from site-specific DNA binding to nonspecific DNA unwinding.
INTRODUCTION
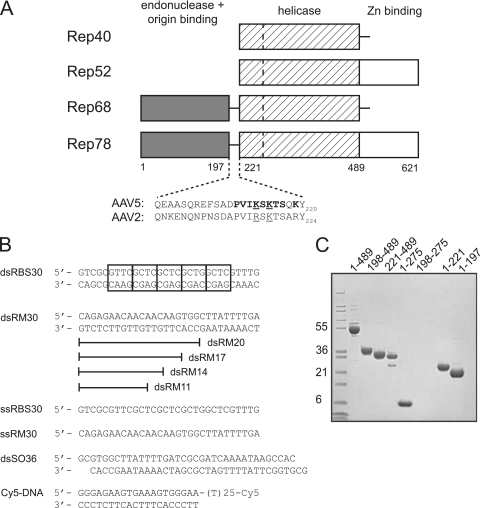
Adeno-associated virus (AAV) is a small virus of the parvovirus family that requires “helper” virus functions from other viruses such as herpesvirus or adenovirus to establish a productive infection (reviewed in reference 18). Of the known serotypes of AAV, AAV type 2 (AAV2) has been studied the most extensively. The single-stranded AAV2 genome is 4.7 kb long, and each end terminates with an inverted terminal repeat (ITR) of ∼145 bases that bears the viral origin of replication. Only two open reading frames are contained in the AAV genome: one encoding the capsid proteins and the other encoding the nonstructural proteins Rep78, Rep68, Rep52, and Rep40 (Fig. 1A), derived by alternative splicing and differential use of viral promoters.
Fig 1.
AAV Rep proteins and oligonucleotides used. (A) Schematic view of the domain organization of the AAV Rep proteins with amino acid numbering for the AAV5 serotype. The dashed line in the helicase domain represents the boundary between a four-helical bundle and the AAA+ domain. The residues comprising the domain linker region are shown for AAV5 and AAV2. Residues mutated in the present study are indicated in boldface: mutant 1 (K213A/K215A/K219A), mutant 2 (P210A/V211A/I212A/K213A), mutant 3 (S214A/K215A/T216A/S217A), and mutant 4 (K213A/S214A/K215A). Underlined residues are those mutated by Urabe et al. (46). (B) Oligonucleotides used for in vitro binding studies and the helicase assay. Cy5 indicates the fluorescent label appended to the 3′ single-stranded DNA end of Cy5-DNA. The boxes in dsRBS30 correspond to tetranucleotide repeats of the AAV5 Rep Binding Site (RBS). (C) SDS-PAGE analysis of AAV5 Rep proteins used. The gel was stained with Coomassie blue, and the molecular mass markers (in kilodaltons) on the left side are Mark12 standards from Invitrogen.
The larger Rep proteins, Rep78 and Rep68, participate in viral replication (7, 23) and site-specific integration (27, 28, 31, 38, 44). It is thought that Rep68/78 orchestrate the completion of DNA replication through the viral ITRs by first binding the so-called “Rep Binding Site” (RBS) located within the ITRs. The viral origin of replication is then melted in the region of a stem-loop structure containing the terminal resolution site (trs), a step that requires Rep's helicase activity (5, 48). A nick is subsequently introduced at the trs using the site-specific endonuclease activity of Rep, thereby providing a free 3-OH group for a polymerase to use to complete replication. Rep52 and Rep40 are identical to Rep78 and Rep68, respectively, except that they lack the N-terminal endonuclease/RBS-binding domain. Rep52 and Rep40 are not needed for DNA replication and instead are required for packaging replicated genomes into preformed capsids (6, 26, 47).
The crystal structure of AAV2 Rep40 (residues 225 to 490 [24]) confirmed that Rep is a member of the SF3 helicase superfamily (17; reviewed in reference 41), whose close relatives—the large T antigen of simian virus 40 (SV40), T-Ag, and the E1 protein of papillomaviruses—readily form hexameric assemblies (see references 12, 14, 16, 30, 35, 40 and references therein). The SF3 helicase superfamily is, in turn, part of a much larger class of ATPases known as AAA+ proteins (ATPases associated with a variety of cellular activities) that form oligomeric assemblies, most often closed hexameric rings (13), in which ATPase active sites are formed and regulated at the subunit interfaces. Consistent with this familial relationship, Rep68/78 has been reported to form large assemblies in the presence of nonspecific DNA (34). However, it is not clear what are the determinants of Rep68/78 multimerization on nonspecific DNA, since there has been little evidence for the assembly of a large multimeric form of the helicase domain itself. In solution on their own, both Rep52 (43) and Rep40 (9, 24, 25, 34) are monomeric, although it has been reported that dimers and trimers can be detected under certain conditions (9).
To understand the different multimerization properties of the larger and smaller AAV Rep proteins, we investigated the contribution of various Rep domains to multimerization, and we report here an unexpected role for the linker that joins the endonuclease and helicase domains. When the 23-amino-acid linker is appended to the N terminus of the AAV5 helicase domain, the resulting protein readily forms stable oligomeric assemblies on both single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA). These complexes are either single or double hexamers, as judged by elution position on size exclusion chromatography, and hexameric rings can be directly visualized by negative stain electron microscopy. We suggest that this observed DNA-dependent multimerization has implications for the ability of the longer Rep proteins, Rep68/78, to function at different steps of the replication cycle, first as a site-specific DNA-binding protein and subsequently as an origin melting helicase.
MATERIALS AND METHODS
Protein expression and purification.
All deletion versions of AAV5 Rep studied here were cloned into pET-15b, expressed in Escherichia coli BL21(DE3) cells (Novagen), and purified essentially as described for AAV2 Rep40 (24), except that a final preparative gel filtration step on a BioSep SEC-S 3000 column (Phenomenex) was substituted for anion exchange on MonoQ. All purifed proteins were >95% homogeneous, as judged by SDS-PAGE (Fig. 1C; 4 to 12% NuPAGE; Invitrogen). Mutant proteins of Rep198-275 and Rep1-489 were generated using the QuikChange method (Stratagene), and all purified similarly to wild-type except for Rep198-275 mutant 1 (K213A/K215A/K219A), where only a small proportion of the soluble protein was recovered in monomeric form, and Rep1-489 mutant 1, which was not soluble and could not be analyzed further.
DNA-binding assays.
All oligonucleotides were purchased from IDT and used without further purification. The duplex oligonucleotides shown in Fig. 1B were prepared by heating equimolar concentrations of the complementary DNAs in 10 mM Tris (pH 8.0)–1 mM EDTA to 95°C for 5 min, followed by cooling to room temperature over ∼3 h. For binding studies, protein was mixed with DNA in a 6:1.2 or 6:2 protein/DNA ratio (unless otherwise stated) and dialyzed at 4°C for >12 h in binding buffer containing 35 mM Tris (pH 8.0), 50 mM NaCl, 5% (wt/vol) glycerol, and 0.4 mM Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP). The mixtures were then analyzed by size-exclusion chromatography (SEC) at 4°C on either a Pharmacia SmartSystem at a flow rate of 0.05 ml/min using Superose 75, Superdex 200, or Superose 6 columns as indicated (3.2/30; GE Healthcare) or at 10°C on an Agilent 1200 HPLC system using a TSKgel Super SW3000 column (4.6 mm [inner diameter] by 30 cm; Tosoh) at a flow rate of 0.1 ml/min or an SEC-5 column (4.6 by 300 mm; Agilent) at a flow rate of 0.2 ml/min, as indicated. In general, 1-min fractions were collected for subsequent SDS-PAGE analysis.
Nucleotide binding assays.
Complexes were assembled as described above, and MgCl2 and either ADP or the nonhydrolyzable nucleotide analogs AMPPNP and AMPPCP (Jena Bioscience) were added to final concentrations of 1 or 2 mM. For in situ preparation of ADP-AlFx, the order of addition was MgCl2 (to 5 mM), ADP (to 1 mM), NaF (to 12 mM), and finally AlCl3 (to 2 mM). Solutions were then incubated at 25°C for 25 min prior to analysis by SEC.
Colorimetric ATPase assay.
ATP hydrolysis was analyzed by measuring the formation of free phosphate (Pi), as a function of time using procedures adapted from (29). Rep protein alone or a preformed Rep198-489/dsDNA complex was diluted to final concentrations between 0.5 and 4.0 μM in buffer containing 100 mM KCl, 20 mM HEPES (pH 7.5), and 1 mM MgCl2 and then heated to 37°C for 10 min. Reactions were initiated by the addition of ATP (Sigma) to a final concentration of 0.5 mM in a total volume of 180 μl. Samples (20 μl) were removed at various time points and immediately quenched in wells of a 96-well plate, each containing 5 μl of 0.5 M EDTA. An aliquot (150 μl) of a 1 mM malachite green stock solution was added to each well, and the absorbance at 650 nm was measured using a Molecular Devices Spectramax M5 microplate reader. The amount of phosphate released was calculated by comparison to a standard curve generated using KH2PO4.
Helicase assay.
Helicase activity was measured essentially as described by (34). Purified Rep1-489, Rep198-489, and Rep221-489 were diluted into helicase assay buffer (25 mM Tris [pH 8.0], 50 mM NaCl, 5% glycerol, 1 mM TCEP), followed by incubation on ice in the presence of Cy5-labeled DNA (Fig. 1B) for 30 min at a 6:1 ratio of protein to DNA. The complexes were then diluted further in assay buffer to a final concentration of 60 nM. Upon addition of 1 mM MgCl2 and 1 mM ATP, the samples were incubated at 37°C for 30 min. Reactions were quenched by adding 1:1 sample buffer (1× Tris-borate-EDTA [TBE], 0.5% sodium dodecyl sulfate [SDS], 20% glycerol). Samples were analyzed on a 20% TBE gel (Invitrogen) and viewed by a GE Typhoon Trio variable mode imager. Peak areas were quantified using the ImageQuant 5.1 software package.
Electron microscopy.
ADP-AlFx-stabilized Rep198-489 complexes containing dsSO36 were diluted to 0.25 mg/ml and adsorbed to carbon-coated nitrocellulose grids. Grids were washed with buffer (50 mM NaCl, 35 mM Tris [pH 8.0], 5% glycerol, 0.4 mM TCEP), blotted, and stained with 1% uranyl acetate. Samples were visualized using a Technai T12 electron microscope operating at 120 kV, and images were collected at 1.5 to 2.0 μm underfocus with a 2K×2K Gatan charge-coupled device camera at a nominal magnification of ×50,000. Control experiments were performed according to the same protocol.
Image processing.
Individual particles were selected from images of ADP-AlFx-stabilized Rep198-489/dsSO36 complexes using Boxer (33), aligned to reference projections generated from the helicase hexamer of the SV40 large T antigen (EMD-1648) using SPIDER (15), and ranked according to cross-correlation value. Particles with the best correlation (values ranging from 2,900 to 3,200) were used to calculate two-dimensional (2D) averages (see Fig. 6B) in SPIDER. The hexameric nature of the ADP-AlFx-stabilized Rep198-489/dsSO36 complexes was confirmed using the power script in SUPRIM (39), which applied different rotational symmetries to the 289-particle nonsymmetrized average.
Fig 6.
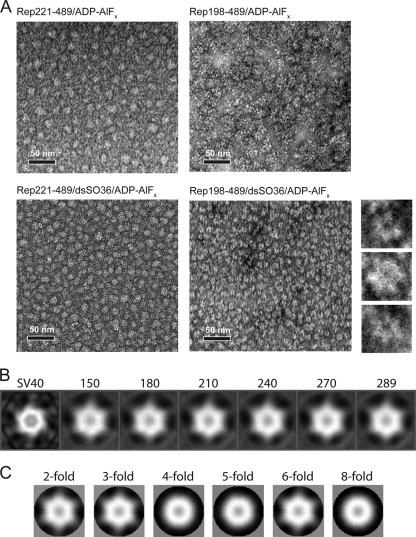
Rep198-489 bound to nonspecific dsDNA forms hexameric assemblies in the presence of ADP-AlFx. (A) Negatively stained electron micrographs of Rep complexes. Images show Rep221-489 in the presence of ADP-AlFx (top left), Rep198-489 in the presence of ADP-AlFx (top right), Rep221-489 in the presence of dsSO36 DNA and ADP-AlFx (bottom left), and Rep bound to dsSO36 DNA in the presence of ADP-AlFx (bottom right). The smaller images to the right show three representative close-up images of Rep198-489/dsSO36/ADP-AlFx complexes that illustrate the apparent 6-fold symmetry. Scale bars, 50 nm. (B) Reference-based classification and 2D averaging of individual Rep198-489/dsSO36/ADP-AlFx complexes. Individual particles were compared to reference projections derived from the helicase hexamer of the SV40 large T antigen (EMD-1648) (labeled SV40 on right) and ranked according to cross-correlation values. Averages calculated from the top 150, 180, 210, 240, 270, and 289 particles in the data set with the highest cross-correlation values are shown. Note the hexameric appearance in the absence of externally applied symmetry constraints. (C) 2D averages retain structural features only when 2-, 3-, or 6-fold symmetry is applied.
RESULTS
Rep221-489 binds and multimerizes on DNA when the 23-residue domain linker is appended.
To investigate the contribution of various Rep protein domains to DNA binding, we cloned, expressed, and purified several truncated forms of AAV5 Rep (Fig. 1C), a protein whose domains have previously been shown to be highly expressed, soluble, and stable when expressed recombinantly in E. coli (20, 21). All forms were expressed as N-terminal His-tagged fusion proteins and purified by Ni-affinity chromatography, followed by thrombin cleavage to remove the His tag and then preparative-scale SEC.
Binding was initially evaluated using four different oligonucleotides (Fig. 1B): a double-stranded 30-mer containing the AAV5 RBS sequence (dsRBS30), a 30-mer of unrelated (random; RM) sequence as a control (dsRM30), a single-stranded 30-mer consisting of the “top” strand of dsRBS30 (ssRBS30), and a single-stranded 30-mer of the control sequence (ssRM30). We initally used 30-mers since preliminary studies indicated that this is a critical length for the assembly of Rep198-489 complexes (data not shown).
Binding was assessed based on comigration of protein and DNA during SEC as a measure of complex formation. This approach has the advantages that binding can be assessed under controlled buffer and temperature conditions, and the sizes of any resulting complexes estimated from migration times relative to known molecular weight standards. However, it is not an equilibrium method and does require that complexes remain stable for the duration of the elution (typically 40 min at 4 to 10°C) to be detected. This method has been previously used to study DNA binding by AAV5 (21) and AAV2 Rep proteins (34, 42).
When the Rep helicase domain, Rep221-489, was incubated with each of the oligonucleotides in a 6:1.2 protein/DNA ratio, no binding was detected (Fig. 2B to E, left), and each chromatogram was essentially a superposition of the elution positions of the oligonucleotide (data not shown) and Rep221-489 alone. Rep221-489 eluted at a position (31.8 min) consistent with a monomer (Fig. 2A), as has been previously reported (9, 24, 25, 34).
Fig 2.
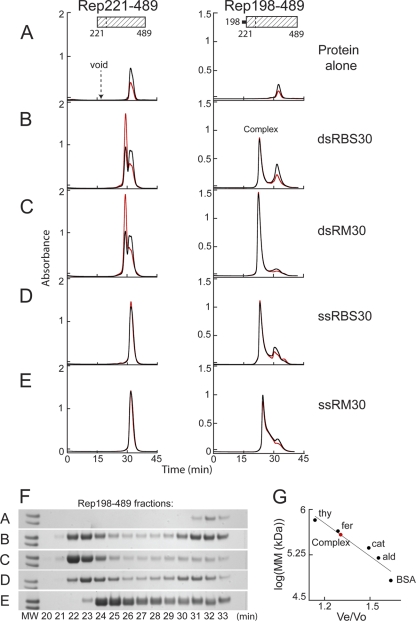
The Rep helicase domain binds DNA when the linker region is appended. Binding was assessed on a Superdex 200 column based on comigration of protein and DNA during SEC after Rep221-489 (left) or Rep198-489 (right) at 82 μM (2.5 and 2.8 mg/ml, respectively) was mixed with a 6:1.2 molar ratio of the indicated DNA oligonucleotides and then dialyzed overnight in binding buffer (see Materials and Methods). The resulting chromatograms are shown in panels B to E. Protein alone similarly dialyzed is shown in panel A. Note that Rep198-489 is markedly less soluble than Rep221-489 in binding buffer. The A260 is shown in red, the A280 is shown in black, and the elution position of blue dextran (corresponding to the column void) is marked. The complex at ∼22.5 min that is formed with Rep198-489 is indicated. (F) SDS-PAGE of Rep198-489 fractions collected during chromatography; the indicated fraction number corresponds to the elution time in minutes. The two molecular weight (MW) markers on the left of each gel (Mark12; Invitrogen) represent Mr values of 36,000 (top) and 31,000 (bottom). (G) Standard curve used to determine the apparent molecular masses (MM) of Rep198-489/DNA complexes. The elution positions on a Superdex 200 column of the molecular mass standards (•) thyroglobulin (thy; 669 kDa), ferritin (fer; 443 kDa), catalase (cat; 232 kDa), aldolase (ald; 158 kDa), and bovine serum albumin (BSA; 67 kDa) are plotted on a semilog plot as log(MM) versus Ve/Vo, where Ve is the elution volume and Vo is the void volume elution time. The curve yields an apparent molecular mass of ∼385 kDa for the Rep198-489 complexes eluting at ∼22.5 min (red circle).
In marked contrast, we observed the appearance of a new peak when the experiment was repeated (Fig. 2B to E, right) with the protein corresponding to Rep residues 198 to 489, which is the helicase domain plus the 23-amino-acid linker between the endonuclease and helicase domains that precedes it. This peak was distinct from the position corresponding to the column void volume (∼17.3 min), indicating that it does not correspond to aggregated material, and also from the position of Rep198-489 alone (Fig. 2A, 32.4 min), which was poorly soluble in the absence of DNA at a lower ionic strength. The elution times for the Rep198-489 complexes formed with dsDNA (Fig. 2B and C) or with ssRBS30 (Fig. 2D) were all ∼22.5 min. The Rep198-489 complex formed with ssRM30 eluted slightly later (24.5 min), and the peak was more asymmetric with a trailing edge (Fig. 2E). SDS-PAGE analysis of the eluted fractions confirmed that Rep198-489 is present in all of the complexes (Fig. 2F); DNA is also present as demonstrated by the increase in the 260-nm/280-nm absorbance ratio relative to protein alone and the near-quantitative shift of DNA absorbance into the complex peaks. Shorter oligonucleotides (10- to 15-mers) did not show binding, whereas slightly longer oligonucleotides (20- to 25-mers) gave rise to a broad envelope of unresolved peaks eluting between 25 and 31 min, a finding suggestive of multimeric species such as dimers, trimers, and tetramers (data not shown). This could indicate either incomplete assembly or disassociation during SEC.
The molecular masses of the discrete complexes eluting during SEC were estimated using a standard curve (Fig. 2G) generated from gel filtration standards (Amersham Biosciences). The ∼22.5-min elution time (Fig. 2B to D) is very similar to that of apoferritin (443 kDa; 22.2 min) and corresponds to a molecular mass of ∼385 kDa. Since Rep198-489 has a molecular mass of 33.4 kDa, it appears that the assembled complexes are not single hexamers, but rather most likely dodecamers or double hexamers (12 × 33.4 kDa = 401 kDa; ssRBS30 = 9.2 kDa; dsRM30 = 18.4 kDa; dsRBS30 = 18.4 kDa). Since SEC analysis of Rep198-489 complexes formed at a 6:1 protein/DNA ratio generally showed quantitative DNA binding, whereas those formed at a 6:2 ratio invariably showed excess DNA (data not shown), the stoichiometry of Rep198-489/DNA complexes is most likely 6:1 and, under the assembly conditions used here, Rep198-489 forms double hexamer complexes that contain two DNA molecules.
It is intriguing that the Rep198-489 complex formed with ssRM30 eluted at a position consistent with a complex of apparent molecular mass of ∼220 kDa (using the standard curve in Fig. 2G), most likely corresponding to a single hexamer (6 × 33.4 kDa = 200 kDa; molecular mass [MM] of ssRM30 = 9.2 kDa). Other random single-stranded 30-mer oligonucleotides of different sequence yielded various results, some eluting at the single hexamer position and others at the double hexamer position (data not shown). We have not observed any sequence specificity to this varied behavior and do not yet understand its basis, although we cannot rule out that different single-stranded oligonucleotides might have different secondary structure features that affect assembly. Under our binding buffer conditions, the smaller Rep198-489 complexes appear less stable than those formed with the other 30-mer oligonucleotides shown in Fig. 2, as judged by the less symmetric shape of the eluted peak during SEC and a long protein tail following the complex peak (in Fig. 2F, compare fractions 25 to 29).
Effect of the domain linker on the small α-helical bundle of the Rep helicase.
In the AAV2 Rep40 crystal structure, the AAA+ domain is preceded by a small four-helix domain spanning residues 225 to 279 (24). If the 23-amino-acid linker (residues 198 to 220) is the sole determinant of the observed AAV5 Rep198-489 multimerization, we reasoned that it should also confer on the isolated helical bundle the ability to oligomerize in the presence of DNA. Therefore, we assessed the DNA-binding properties of AAV5 Rep198-275 using our SEC comigration assay. As shown in Fig. 3A, the ability of Rep198-489 to bind to non-RBS DNA was recapitulated with Rep198-275. In contrast to Rep198-489, however, the sizes of the resulting complexes (as judged by elution position) were dependent on DNA length and, at shorter DNA lengths, Rep198-275 bound more readily to dsDNA than to ssDNA (for example, compare ssRM17 to dsRM17). Thus, the AAA+ domain does not appreciably contribute to DNA binding by Rep198-489 (as expected, given the lack of detectable binding by Rep221-489 itself) but perhaps plays an architectural role in either limiting the size of the resulting protein/DNA complex or organizing it.
Fig 3.
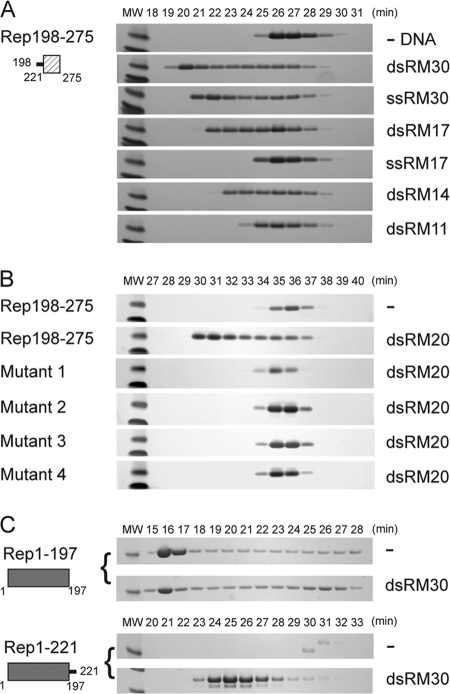
The linker region confers DNA binding to the small helical bundle of the Rep helicase. (A) Rep198-275 (2.2 mg/ml; 235 μM) was mixed with the indicated oligonucleotides at a 6:2 molar ratio and dialyzed into binding buffer as described in the text. Fractions eluting during SEC on a Superdex 75 column were analyzed by SDS-PAGE, and the numbers correspond to elution times in minutes. The two molecular weight (MW) markers at the left of each gel are 6,000 (top) and 3,500 (bottom). (B) Similarly, the binding of dsRM20 by wild-type Rep198-275, mutant 1 (K213A/K215A/K219A), mutant 2 (P210A/V211A/I212A/K213A), mutant 3 (S214A/K215A/T216A/S217A), and mutant 4 (K213A/S214A/K215A) was assessed on a Superdex 200 column. The protein concentrations used were 300 to 400 μM (between 3 to 4 mg/ml), except for mutant 1, where the concentration was only 110 μM, due to limited recovery of the monomeric form. (C) Binding of dsRM30 by Rep1-197 and Rep1-221. Rep1-197 (205 μM) and Rep1-221 (215 μM) were mixed with dsRM30 at 6:2 molar ratio and dialyzed in binding buffer prior to SEC on a Superdex 200 column. The molecular weight (MW) marker on the left of each SDS-PAGE gel corresponds to Mr 21,000. The numbers above the lanes correspond to elution times in minutes.
Mutations in the domain linker inhibit DNA binding by Rep198-275 and Rep1-489.
To confirm that DNA binding by Rep198-275 is mediated only by the linker, we created a series of mutants (indicated in boldface in Fig. 1A) in which clusters of linker residues were mutated to alanine. Each cluster contained at least one strictly or highly conserved basic residue. Rep198-275 mutant 1 (K213A/K215A/K219A), mutant 2 (P210A/V211A/I212A/K213A), mutant 3 (S214A/K215A/T216A/S217A), and mutant 4 (K213A/S214A/K215A) were expressed and purified as for the other Rep proteins. Binding studies were performed using a double-stranded random 20-mer (dsRM20) and, in all cases, when the resulting solutions were dialyzed into low ionic strength and analyzed by SEC, no binding was observed. Most of the mutants showed no differences in expression or purification properties relative to wild-type Rep198-275, except for mutant 1, which was largely aggregated (>80%) when subjected to the final SEC purification step (see Materials and Methods). For the binding studies for all of the mutants, only material eluting at the monomer position was used, and the results suggest that the inability to bind DNA is due to the mutation of residues crucial for DNA binding rather than protein folding or stability problems.
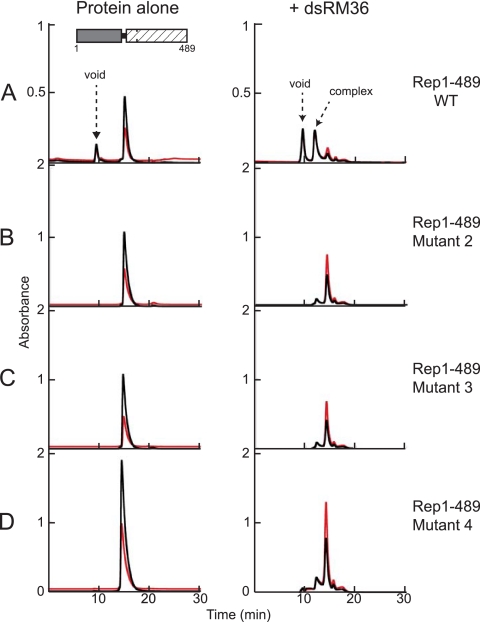
To determine whether the mutations in the domain linker have a similar effect in the context of the larger Rep proteins, we introduced the mutations corresponding to mutants 1 to 4 into AAV5 Rep1-489 (21). We were unable to purify mutant 1 due to poor solubility but were able to repeat the binding studies with mutants 2 to 4 using dsSO36. In control experiments, mutants 2 to 4 all bind dsRBS30 similarly to Rep1-489 (data not shown), indicating that some elements of correct protein folding have been retained. Overall, the DNA binding experiments were plagued by problems of precipitation at low ionic strength, not only of the mutant proteins and Rep1-489 but also of complexes of Rep1-489 with nonspecific dsDNA. Nevertheless, as shown in Fig. 4A (right), SEC analysis indicates that in the presence of dsSO36, Rep1-489 forms two distinct soluble species that contain DNA, one that elutes in the column void volume and a smaller complex that elutes at a position (11.9 min) between those of the molecular mass standards thyroglobulin (667 kDa, 11.7 min) and ferritin (440 kDa, 12.8 min). The estimated molecular mass of this complex is ∼600 kDa, which is consistent with a double hexamer (Rep1-489 = 55.8 kDa), although we cannot rule out other possibilities. Rep1-489 mutants 2 to 4 do not form the larger soluble species, and the height of the peak corresponding to the smaller complex is substantially reduced (Fig. 4B to D), suggesting that mutations in the linker affect the ability of Rep1-489 to bind nonspecific dsDNA.
Fig 4.
Mutations in the linker region prevent nonspecific dsDNA binding by Rep1-489. Proteins were mixed with dsSO36 at a 6:2 molar ratio and dialyzed in binding buffer as described in the text. Complexes were subsequently analyzed using an Agilent SEC-5 column at a flow rate of 0.2 ml/min. Samples without DNA (left) were run in 0.5 M NaCl-containing buffer, and the samples with DNA (right) in 50 mM NaCl binding buffer. Prior to dialysis, the concentration of the wild-type Rep1-489 was 2.5 mg/ml, and the concentrations of the mutants were all 4 to 5 mg/ml. The A260 is indicated in red; the A280 is indicated in black. (A) Rep1-489. (B) Rep1-489 mutant 2 (P210A/V211A/I212A/K213A). (C) Rep1-489 mutant 3 (S214A/K215A/T216A/S217A). (D) Rep1-489 mutant 4 (K213A/S214A/K215A). After dialysis, light precipitate was observed in the mixture of Rep1-489 and dsSO36, and very heavy precipitate was observed with the mutants, a finding similar to that observed for the mutant proteins in binding buffer in the absence of DNA.
Effect of the domain linker on the Rep endonuclease domain.
If the 23-amino-acid linker between the Rep endonuclease and helicase domains is both necessary and sufficient for DNA binding, then the addition of the linker to the endonuclease domain should similarly result in a protein capable of binding nonspecific DNA. Since the endonuclease domain alone, AAV5 Rep1-197, binds not only to dsDNA containing the RBS sequence (21) but also to nonspecific ssDNA (data not shown and also reported for the AAV2 endonuclease domain [34]), this limited binding studies with our standard oligonucleotides (Fig. 1B) to dsRM30: the negative control is not possible for the others. As shown by combined SEC and subsequent SDS-PAGE analysis (Fig. 3C), both Rep1-197 and Rep1-221 are poorly behaved when dialyzed into low-ionic-strength buffer in the absence of DNA: Rep1-197 aggregates (the protein elutes in the column void volume, corresponding to fractions 16 and 17), whereas Rep1-221 precipitates and only tiny amounts remain soluble (fraction 31). When Rep1-197 was incubated with dsRM30 DNA, the protein remained largely aggregated, suggesting that it does not recognize or bind nonspecific dsDNA. In contrast, incubation of Rep1-221 with dsRM30 resulted in the formation of a discrete complex eluting at ∼24.6 min, corresponding to an apparent MM of ∼227 kDa. A complex of this size is consistent with approximately eight Rep1-221 monomers (MM = 25.6 kDa) bound to dsRM30, although other combinations are possible. Collectively, these results demonstrate that the linker region confers the ability to bind to nonspecific dsDNA when it is added to either the Rep domain that precedes it or that which follows it.
Complexes of Rep198-489 bound to DNA also bind nucleotides and have stimulated ATPase and helicase activities.
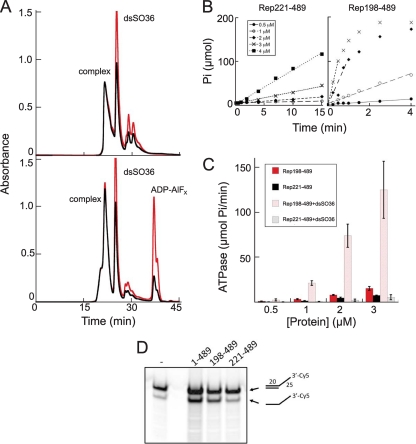
Although Rep198-489/DNA complexes form readily in the absence of nucleotides, we wondered whether nucleotides or nucleotide analogs could bind to these complexes. When preformed complexes were incubated with various nucleotides, all of the nucleotides we tested (ADP, AMP-PNP, AMP-PCP, and ADP-AlFx) showed evidence for binding. For example, as shown in Fig. 5A for ADP-AlFx, addition of the transition state analog is associated with an increase in the 260-nm/280-nm ratio in the complex and a concomitant narrowing of the eluted complex peak.
Fig 5.
Properties of Rep198-489/dsDNA complexes. (A) Size exclusion chromatographic analysis (on TSKgel Super SW3000) of Rep198-489/dsSO36 complexes formed at 3.9 mg/ml (120 μM) and a 6:2 protein/DNA molar ratio with (bottom) or without (top) ADP-AlFx. Peaks corresponding to the formed complex, unbound DNA (dsSO36), and excess ADP-AlFx are marked. The A260 is indicated in red; the A280 is indicated in black. (B) Representative ATPase data for Rep221-489 (left) and Rep198-489 (right), both in the presence of dsSO36 DNA, plotted as μmol of free phosphate (Pi) released as a function of time. (C) The initial rate of ATP hydrolysis as a function of time was determined for Rep221-489 and Rep198-489 with or without DNA. The error bars indicate the standard deviations calculated for four separate experiments. (D) The helicase activities of Rep1-489, Rep198-489, and Rep221-489 were compared using a 3′-Cy5-labeled oligonucleotide with a 3′-single stranded T25 extension. The first lane corresponds to the annealed substrate (shown schematically on the right, top) in the absence of protein. The percent substrate converted to product (after corrections for background and contaminating product in the starting substrate) are 25% (Rep1-489), 21% (Rep198-489), and 3.5% (Rep221-489).
The ability of Rep198-489 to bind both DNA and nucleotides allows us to begin probing the biochemical properties of Rep198-489/DNA complexes. We first used a malachite green-based ATPase assay (29) to measure the activity of Rep221-489 and Rep198-489 as a function of protein concentration and nonspecific dsDNA (Fig. 5B and C). Typical results are shown in Fig. 5B. As shown in Fig. 5C, in the absence of DNA, both Rep221-489 and Rep198-489 have similar (within 2-fold) activities. In the presence of dsSO36 DNA, an oligonucleotide that does not contain the RBS sequence, Rep221-489 shows no increase in activity consistent with our observation that it does not measurably bind DNA; in contrast, the ATPase activity of Rep198-489 is stimulated ∼8-fold.
We also assayed the helicase activities of Rep1-489, Rep198-489, and Rep221-489 using a Cy5-labeled short double-stranded nonspecific oligonucleotide with a poly(T) 3′ overhang (34). As shown in Fig. 5D, Rep198-489 demonstrates robust helicase activity comparable to that of Rep1-489, and which is stimulated ∼6-fold relative to that of Rep221-489. Thus, the linker region not only confers DNA-binding ability to the Rep helicase domain but also markedly stimulates both its ATPase and helicase activities. This activation strongly suggests that the complex represents a catalytically and multimerically relevant state of AAV Rep, most likely akin to that seen for other AAA+ proteins, in which ATP is bound between monomers with Walker A and B motifs from one subunit and an arginine finger of an adjacent subunit contributing to active site formation (49), rather than protein nonspecifically bound to DNA.
Rep198-489/DNA complexes form hexameric rings.
To visualize complexes of Rep198-489 directly, we obtained negatively stained electron microscopy images of complexes assembled with dsSO36 in the presence of ADP-AlFx (Fig. 6A, lower right panel). The dense lawn of complexes reveals apparently homogenous particles ∼12 nm in diameter, which consistent with the 12- to 14-nm diameters of other hexameric helicases (37) and also with the hexameric model based on the AAV2 Rep40 crystal structure (24). Inspection of individual particles suggests that, as expected for a SF3 helicase, they are ring-like structures with a channel or hole in the middle and six distinct lobes (Fig. 6A, zoomed inserts on right). The hexameric nature of these complexes was confirmed by reference-based classification and 2D averaging (Fig. 6B). It is important to note that the 6-fold symmetry emerges without applying any external symmetry constraints. Furthermore, the structural features of the averaged images are only preserved when 2-, 3-, or 6-fold symmetry is applied (Fig. 6C). In the absence of dsSO36 or when Rep221-489 was substituted for Rep198-489, no discrete hexameric assemblies were observed, and the mixtures appeared to form amorphous clumps of various sizes and no discernible symmetry (Fig. 6A).
DISCUSSION
Accumulating evidence indicates that there are at least two distinct modes of DNA binding by Rep68/78, corresponding to the multiple roles that these proteins play during viral replication. One of the initial functions of Rep68/78 is to recognize and bind specifically to the viral origin of replication. This is mediated by the N-terminal endonuclease domain, which binds specifically to the double-stranded form of the RBS, most likely forming a spiral of Rep molecules along the DNA, as seen for the isolated endonuclease domain (21). This type of spiral assembly presumably accounts for the observation of AAV2 Rep68/78 hexamers in the presence of AAV origin sequences (10, 42), although it has recently been suggested that this complex might be only pentameric (34). The second mode of binding occurs in the presence of random ssDNA or dsDNA and relates to the helicase function of Rep68/78. These assemblies seem likely to be hexameric or dodecameric, although double octamers have been reported (34).
In vitro studies to understand the apparent ability of Rep68/78 to switch conformations from a spiral assembly to a planar ring expected for an SF3 helicase—or to be capable of adopting both conformations—have been hampered by the poor biophysical properties of the AAV2 and AAV5 Rep 68/78 proteins, particularly in the low-ionic-strength buffer conditions required to detect DNA binding (10, 34; this study). Here, we circumvented this problem by creating a series of deletion versions of Rep68/78 in an effort to understand the contributions of various domains to DNA binding and helicase assembly. Our results establish that the 23-amino-acid linker sequence located between the endonuclease and helicase domains of AAV5 Rep68/78 proteins is a crucial contributor to the ability of the AAV5 Rep helicase domain to form discrete oligomeric complexes on both ssDNA and dsDNA. These complexes are functionally active, demonstrating both stimulated ATPase and helicase activity relative to the isolated helicase domain.
The linker region appears to be a strong driver of DNA binding. For example, it confers nonspecific dsDNA binding to the isolated helicase domain (Fig. 2), the endonuclease domain (Fig. 3C), and even the small subdomain comprised of only the four-helix bundle that precedes the Rep AAA+ domain (Fig. 3A). However, it is possible that the architecture of these protein-DNA complexes may be fundamentally different. For example, in the case of Rep198-275, which does not have the AAA+ domain, DNA binding is evident on short oligonucleotides, and it appears that the number of bound protein molecules depends on the DNA length. In contrast, Rep198-489 forms discrete hexameric complexes (Fig. 6) only on DNA longer than ∼30 nucleotides. Thus, although the AAA+ domain does not detectably contribute to DNA binding, it is clearly important in establishing the architecture of the assembled proteins on DNA. Stable assembly on DNA appears to require sufficient DNA to fully pass through the central cavity of the assembled hexamer, since 30 nucleotides is on the same order as the number of base pairs of dsDNA that might be expected to be accommodated in the observed central channels of the SF3 helicase domains that have been structurally characterized. In the case of the SV40 helicase domain, the central channel is ∼80 Å long (30), whereas that of the E1 helicase domain is ∼60 Å (12).
Our data strongly suggest that the linker region plays an important role in the oligomerization of the larger Rep proteins on nonspecific ssDNA and dsDNA. It is tempting to speculate that the linker might serve as a hook that helps to hold Rep68/78 in place on DNA as it transitions between the first mode and second mode of DNA binding. During the transition, the linker region might be engaged as we observed for Rep198-275, perhaps forming a “spiral coat” along the DNA molecule limited only by the availability of accessible DNA or the number of Rep protomers and recapitulating the spiral observed for endonuclease binding to the RBS (21). In the second mode of binding, the presence of the linker region stimulates both the ATPase and helicase activities, suggesting that it actively contributes to the organization of the assembly.
The importance of the linker region has been previously demonstrated for AAV2 since mutation of either R217 or K219 (corresponding to K213 and K215 of AAV5; underlined in Fig. 1A) to alanine in Rep78 results in a protein unable to nick at the trs or mediate site-specific integration into AAVS1, whereas RBS binding is maintained (46). The lost activities are consistent with a model in which mutations in the linker region prevent the formation of an active helicase-competent assembly, shown to be necessary for trs nicking (5, 48). Each of our cluster mutants 1 to 4 contained a mutation of either K213 or K215, and the associated loss of DNA binding (Fig. 2 and 4) provides a potential mechanistic explanation for the observations of Urabe et al. (46).
For other members of the SF3 helicase superfamily, such as T-Ag and E1, combined biochemical and structural studies have shown that the N-terminal origin binding domain and the helicase AAA+ domain are linked by an intervening domain that mediates oligomerization (12, 30, 32, 45; reviewed in reference 22). In the case of T-Ag, the intervening domain is a Zn2+-binding domain; for E1, the intervening domain forms a four-helix bundle that is structurally unrelated to the T-Ag Zn2+-binding domain but is in turn structurally homologous to the hexamerization domain of the RCR replication initiator protein of pMV158 (3). AAV Rep appears to have dealt with the intervening region somewhat differently by dividing it into two parts depending on protein context. For Rep40 and Rep52, the intervening region consists of a small four-helix bundle that alone does not have potent DNA binding or oligomerization properties. For the larger Rep proteins, the intervening domain appears to be functionally comprised of the four-helix bundle supplemented by the linker.
Since Rep40 and Rep52 lack the linker sequence, our results here do not shed direct light onto the functions of these smaller Rep proteins during genome packaging, which might be reasonably expected to be mediated by a multimeric molecular motor that pumps DNA into preformed capsids. It would be interesting to establish whether the ability of Rep40 to oligomerize when extended by a short peptide sequence mimics the properties of Rep40 and Rep52 when bound to other proteins or protein complexes such as the viral capsid proteins (1, 11). The need for accessory proteins to aid the assembly of an active motor protein is not unprecedented: for example, the eukaryotic MCM2-7 replicative helicase requires the assistance of accessory proteins ORC1-6 and Cdc6 to load onto DNA (2), and deposition of the E. coli DnaB helicase at replication forks requires direct interactions with two other proteins, DnaA and DnaC (25).
Hexameric helicases continue to intrigue, and the relevance of double hexamers encircling dsDNA continues to be discussed (4). We do not yet know whether the double hexamers we observed represent an authentic assembly along the AAV replication pathway. It is nonetheless clear that the linker region is not a passive tether joining two independent protein domains but rather contributes to the oligomeric properties of AAV Rep in the presence of DNA.
ACKNOWLEDGMENTS
This study was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institutes of Health, Bethesda, MD. J.S.C. was supported by a Nancy Nossal Fellowship Award from the NIDDK.
We thank Bob Craigie and Andrea Regier Voth for helpful comments on the manuscript, Wei Yang for access to the microplate reader, Jenny Hinshaw for the use of the electron microscope, and Shunming Fang for assistance with single particle image processing.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Bleker S, Pawlita M, Kleinschmidt JA. 2006. Impact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. J. Virol. 80:810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bochman ML, Schwacha A. 2009. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 73:652–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boer DR, et al. 2009. Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains. EMBO J. 28:1666–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Botchan M, Berger J. 2010. DNA replication: making two forks from one prereplication complex. Mol. Cell 40:860–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brister JR, Muzyczka N. 1999. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J. Virol. 73:9325–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chejanovsky N, Carter BJ. 1989. Mutation of an AUG codon in the adeno-asociated virus rep gene: effects on viral DNA replication. Virol. 173:120–128 [DOI] [PubMed] [Google Scholar]
- 7. Chejanovsky N, Carter BJ. 1990. Mutation of a consensus purine nucleotide binding site in the adeno-associated virus rep gene generates a dominant-negative phenotype for DNA replication. J. Virol. 64:1764–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reference deleted.
- 9. Dignam SS, et al. 2007. Coupled ATP and DNA binding of adeno-associated virus Rep40 helicase. Biochem. 46:568–576 [DOI] [PubMed] [Google Scholar]
- 10. Dignam SS, Correia JJ, Nada SE, Trempe JP, Dignam JD. 2007. Activation of the ATPase activity of adeno-associated virus Rep68 and Rep78. Biochem. 46:6364–6374 [DOI] [PubMed] [Google Scholar]
- 11. Dubielzig R, King JA, Weger S, Kern A, Kleinschmidt JA. 1999. Adeno-associated virus type 2 protein interactions: formation of pre-encapsidation complexes. J. Virol. 73:8989–8998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Enemark EJ, Joshua-Tor L. 2006. Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442:270–275 [DOI] [PubMed] [Google Scholar]
- 13. Erzberger JP, Berger JM. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35:93–114 [DOI] [PubMed] [Google Scholar]
- 14. Fouts ET, Yu X, Egelman EH, Botchan MR. 1999. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J. Biol. Chem. 274:4447–4458 [DOI] [PubMed] [Google Scholar]
- 15. Frank J, Shimkin B, Dowse H. 1981. SPIDER: a modular software system for electron image processing. Ultramicroscopy 6:343–358 [Google Scholar]
- 16. Gomez-Lorenzo MG, et al. 2003. Large T antigen on the simian virus 40 origin of replication: a 3D snapshot prior to DNA replication. EMBO J. 22:6205–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gorbalenya AE, Koonin EV, Wolf YI. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145–148 [DOI] [PubMed] [Google Scholar]
- 18. Henckaerts E, Linden RM. 2010. Adeno-associated virus: a key to the human genome? Future Virol. 5:555–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reference deleted.
- 20. Hickman AB, Ronning DR, Kotin RM, Dyda F. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell 10:327–337 [DOI] [PubMed] [Google Scholar]
- 21. Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. 2004. The nuclease domain of adeno-associated virus Rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell 13:403–414 [DOI] [PubMed] [Google Scholar]
- 22. Hickman AB, Dyda F. 2005. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15:77–85 [DOI] [PubMed] [Google Scholar]
- 23. Im DS, Muzyczka N. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447–457 [DOI] [PubMed] [Google Scholar]
- 24. James JA, et al. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025–1035 [DOI] [PubMed] [Google Scholar]
- 25. James JA, Aggarwal AK, Linden RM, Escalante CR. 2004. Structure of adeno-associated virus type 2 Rep40-ADP complex: insight into nucleotide recognition and catalysis by superfamily 3 helicases. Proc. Natl. Acad. Sci. U. S. A. 101:12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. King JA, Dubielzig R, Grimm D, Kleinschmidt JA. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kotin RM, et al. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 87:2211–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kotin RM, Linden RM, Berns KI. 1992. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 11:5071–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leonard M, Song BD, Ramachandran R, Schmid SL. 2005. Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol. 404:490–503 [DOI] [PubMed] [Google Scholar]
- 30. Li D, et al. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512–518 [DOI] [PubMed] [Google Scholar]
- 31. Linden RM, Winocour E, Berns KI. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. U. S. A. 93:7966–7972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loeber G, et al. 1991. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J. Virol. 65:3167–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 34. Mansilla-Soto J, et al. 2009. DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68. PLoS Pathog. 5:e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mastrangelo IA, et al. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658–662 [DOI] [PubMed] [Google Scholar]
- 36. Reference deleted.
- 37. Patel SS, Picha KM. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651–697 [DOI] [PubMed] [Google Scholar]
- 38. Samulski RJ, et al. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10:3941–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schroeter JP, Bretaudiere JP. 1996. SUPRIM: easily modified image processing software. J. Struct. Biol. 116:131–137 [DOI] [PubMed] [Google Scholar]
- 40. Sedman J, Stenlund A. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singleton MR, Dillingham MS, Wigley DB. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76:23–50 [DOI] [PubMed] [Google Scholar]
- 42. Smith RH, Spano AJ, Kotin RM. 1997. The Rep78 gene product of adeno-associated virus (AAV) self-associates to form a hexameric complex in the presence of AAV ori sequences. J. Virol. 71:4461–4471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith RH, Kotin RM. 1998. The Rep52 gene product of adeno-associated virus is a DNA helicase with 3′-to-5′ polarity. J. Virol. 72:4874–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Surosky RT, et al. 1997. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J. Virol. 71:7951–7959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Titolo S, et al. 2000. Identification of domains of the human papillomavirus type 11 E1 helicase involved in oligomerization and binding to the viral origin. J. Virol. 74:7349–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Urabe M, et al. 1999. Charged-to-alanine scanning mutagenesis of the N-terminal half of adeno-associated virus type 2 Rep78 protein. J. Virol. 73:2682–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoon-Robarts M, et al. 2004. Residues within the B' motif are critical for DNA binding by the superfamily 3 helicase Rep40 of adeno-associated virus type 2. J. Biol. Chem. 279:50472–50481 [DOI] [PubMed] [Google Scholar]
- 48. Walker SL, Wonderling RS, Owens RA. 1997. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J. Virol. 71:6996–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang X, Wigley DB. 2008. The ‘glutamate switch’ provides a link between ATPase activity and ligand binding in AAA+ proteins. Nature Struct. Mol. Biol. 15:1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]