Abstract
Papillomavirus genomes are thought to be amplified to about 100 copies per cell soon after infection, maintained constant at this level in basal cells, and amplified for viral production upon keratinocyte differentiation. To determine the requirement for E1 in viral DNA replication at different stages, an E1-defective mutant of the human papillomavirus 16 (HPV16) genome featuring a translation termination mutation in the E1 gene was used. The ability of the mutant HPV16 genome to replicate as nuclear episomes was monitored with or without exogenous expression of E1. Unlike the wild-type genome, the E1-defective HPV16 genome became established in human keratinocytes only as episomes in the presence of exogenous E1 expression. Once established, it could replicate with the same efficiency as the wild-type genome, even after the exogenous E1 was removed. However, upon calcium-induced keratinocyte differentiation, once again amplification was dependent on exogenous E1. These results demonstrate that the E1 protein is dispensable for maintenance replication but not for initial and productive replication of HPV16.
INTRODUCTION
Papillomaviruses (PVs) are small, double-stranded DNA viruses that infect stratified squamous epithelium. Human PVs (HPVs) are very important causative agents for various lesions, ranging from verrucas to cancer. Among them, a subset of HPVs, the so-called high-risk types such as type 16 and 18, are associated with more than 90% of all cervical carcinomas as primary etiological factors (45). PVs establish long-term persistent infections in squamous epithelium, and the viral life cycle is tightly linked with the differentiation state of the host keratinocytes (7).
PV genome is replicated and amplified in three different stages: establishment, maintenance, and productive stages of the life cycle. In the establishment stage, soon after infection of the basal layer keratinocytes, a single or a few initial copies of the viral genome amplify and establish residence as multicopy circular extrachromosomal elements (episomes) in the nucleus. In the maintenance stage, the viral genomes in each affected cell replicate approximately once in a cell cycle in proliferating basal layer keratinocytes. Then, in the productive stage, they are exponentially amplified in terminally differentiating keratinocytes and packaged into progeny virions. It is important to understand the molecular mechanisms underlying this triphasic model for development of new therapies against HPV-infected lesions, such as cervical intraepithelial neoplasias, which can progress to cervical cancer.
The regulation of viral DNA replication is thought to differ in these three distinct stages of the viral life cycle. Studies of lesions experimentally induced by rabbit oral papillomavirus (ROPV) infection showed that the genome copy number of ROPV is low in the basal layer and increases up to four orders of magnitude during the terminal differentiation of host keratinocytes (21). This corresponds to more than 13 rounds of continuous replication of the viral genome in the productive stage.
Most of our knowledge of PV replication is derived from short-term replication assays to identify the components required for replication of the viral genome. These transient replication assays suggest that both viral proteins E1, a DNA helicase, and E2, a transcriptional activator and auxiliary replication factor, as well as cis-acting elements, including the E1-binding site (E1BS), several E2-binding sites (E2BS), and an AT-rich region in the long control region (LCR), are all essential for efficient PV DNA replication (4, 6, 20, 23, 30, 35, 37, 38). These studies appear to have analyzed the molecular mechanisms of viral replication in the productive stage, though they did not necessarily examine the three stages separately.
In this context, it is of interest that the HPV genome lacking LCR can replicate in the absence of both E1 and E2 proteins in transient replication assays (1, 16, 29), and a temperature-sensitive (TS) E1 mutant of bovine papillomavirus type 1 (BPV1) can be maintained in mouse C127 cells at a nonpermissive temperature as efficiently as the wild-type BPV1 (17). These reports suggest that E1 might be dispensable for maintenance replication.
To test this hypothesis directly, we here used an E1-defective mutant HPV16 genome featuring a translation termination mutation in E1. We established human dermal keratinocytes (HDKs) containing the E1-defective HPV16 genome with the help of exogenous E1 expression and then removed the exogenous E1 expression cassette with the FLP/FRT system (36). Similar to the wild-type genome, the E1-defective HPV16 genome was maintained for numerous cell generations without exogenous E1 expression. However, unlike the wild-type genomes, the E1-defective HPV16 genome failed to amplify upon differentiation of host cells, with rescue dependent on reexpression of exogenous E1. These results indicate that HPV16 requires E1 protein for the establishment and the productive stages but not for the maintenance stage of viral genome replication. The results have important implications for the development of E1 inhibitors as anti-HPV drugs.
MATERIALS AND METHODS
Cell culture.
Human dermal keratinocytes (HDKs) were purchased from Cell Applications (San Diego, CA). HDKs were immortalized with TERT, a mutant form of CDK4 and cyclin D1 (HDK-K4DT) by lentivirus-mediated gene transfer as described below. The cells were maintained in low-calcium serum-free keratinocyte growth medium (Epilife, Invitrogen, Carlsbad, CA) unless otherwise described. To induce keratinocyte differentiation, cells were exposed to Epilife basal medium supplemented with 1.8 mM CaCl2 for 7 days or more. W12 (20863) cells were obtained from Paul F. Lambert (McArdle Laboratory for Cancer Research, Madison, WI) and cultured on mitomycin-treated Swiss mouse 3T3 cells in F medium as previously described (13).
Plasmid construction.
In pCMV-loxP-HPV16-loxP-puro, the full-length HPV16 genome linearized at the SphI site in the long control region and flanked by loxP recombination sites (18) was inserted between a CMV promoter and a puromycin resistance gene so that the HPV16 genome was located in reverse orientation to the CMV promoter to avoid CMV-driven expression of HPV16 genes. In pCMV-loxP-E1-defective HPV16-loxP-puro, an in-frame stop codon was created at nucleotides (nt) 892 to 894 just downstream of the E1 start codon at nucleotide 865 by site-directed mutagenesis. The segment encoding Cre recombinase with nuclear localization signal in AxCANCre (14) was cloned into pcDNA3 (Invitrogen) to generate pcDNA3-NCre. Detailed methods for the construction of pCMV-loxP-HPV16-loxP-puro, pCMV-loxP-E1-defective-HPV16-loxP-puro, and pcDNA3-NCre are available upon request.
DNA transfection.
HDK-K4DT cells were seeded at a density of 2 × 105 cells onto six-well plates (BD Biosciences, Franklin Lakes, NJ) containing 2 ml of Epilife and incubated overnight and then cotransfected with 1 μg of pcDNA3-NCre and 3 μg of pCMV-loxP-HPV16-loxP-puro (wild-type or E1-defective strains) using FuGENE HD (Roche). One day after transfection, cells were selected by 1 μg/ml of puromycin for 2 days.
Vector construction and retroviral infection.
Construction of lentiviral vectors, CSII-CMV-TERT, CSII-CMV-cyclin D1, and CSII-CMV-CDK4R24C, were described previously (33). CSII-CMV-TetON-ADV contains the TetON-ADV segment from pTet-On Advanced Vector (Clontech, Mountain View, CA). To yield improved E1 gene expression in mammalian cells, the codon-optimized HPV16 E1 gene with an N-terminal hemagglutinin (HA) tag (HA16E1) was synthesized (GenScript, Piscataway, NJ). CSII-TRE-Tight-HA16E1 contains the HA16E1 gene under the control of the tetracycline responsive promoter from pTRE-Tight (Clonetech). pCMSCV-FRT-HA-E1-TKneo consists of the CMV/LTR fusion promoter, the Ψ packaging signal, a mutant (f72) FLP recognition target (5′FRT) (24), the HA16E1 gene, the PKG promoter, and the herpes simplex virus thymidine kinase (HSV-TK) fused to the neomycin-resistant gene (neo), 3′FRT, and 3′LTR, as shown in Fig. 3A. Cells infected with this retrovirus were positively or negatively selected in the presence of 50 μg/ml of G418 or 10 μg/ml of ganciclovir, respectively. The nucleotide sequence of the HA16E1 and the detailed methods for the construction of pCMSCV-FRT-HA16E1-TKneo-FRT, CSII-CMV-TetON-ADV, and CSII-TRE-Tight-HA16E1 are available upon request. The production of recombinant retroviruses and lentiviruses was accomplished as described previously (27, 33).
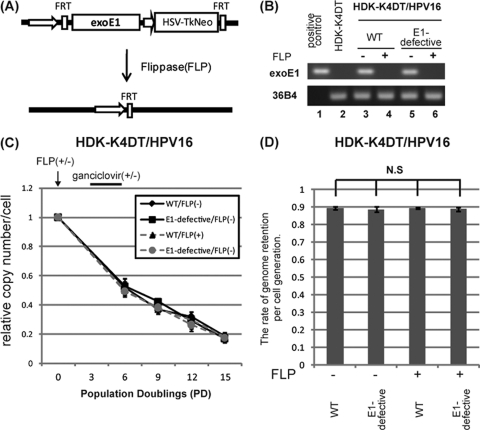
Fig 3.
E1 protein is not essential for maintenance replication of the viral genome. (A) Schematic representation of the viral vector, MSCV-FRT-HA16E1-TKneo-FRT, expressing exogenous E1. The HA-tagged HPV16 E1 gene and the TKneo fusion gene are flanked by FLP recognition target (FRT) sites. When a thermostable FLP-expressing AdV (AxCAFLPe) is infected, the DNA in between the FRT sites is excised so that exogenous E1 as well as TKneo expression is terminated. Cells still expressing E1 and HSV-TK can be negatively selected with ganciclovir. (B) mRNAs containing exogenous E1 message were analyzed to ensure complete removal of E1 expression. Total RNAs isolated from HDK-K4DT/HPV16 cell lines at 21 days postinfection with (lanes 4 and 6) or without (lanes 3 and 5) AxCAFLPe were subjected to RT-PCR with a primer set specific to codon-optimized E1. Representative pictures of an ethidium bromide-stained agarose gel with PCR products indicating exogenous E1 (top) and 36B4 (an internal control; bottom) are shown. pCMSCV-FRT-HA16E1-TKneo-FRT was used as a positive control. (C) HDK-K4DT cells containing the wild type (WT) or the E1-defective HPV16 genome were established in the presence of exogenous E1 expression. Two weeks after transfection, aliquots of cells were infected with AxCAFLPe (FLP +) at a multiplicity of infection of 5, followed by selection with 10 μg/ml of ganciclovir for 1 week. After the selection, cells were cultured for 4 passages at a ratio of 1:8 to examine the retention rate of the wild-type or the E1-defective genome in the presence or the absence of exogenous E1. HDK-K4DT/HPV16 cells which were not infected with AxCAFLPe (FLP−) were also cultured for 5 passages at a ratio of 1:8. Total DNA was extracted just before the infection, at every passage and at the end of the culture. The copy number of HPV16 genomes was measured by real-time PCR and normalized to the total amount of DNA. The graph shows time courses of copy number change during the 5 passages after the infection. The copy number at the each time point is shown as a ratio to the copy number just before the FLP-expressing adenovirus infection (PD0). The end of the ganciclovir selection corresponds to PD6. Means and standard errors of the means are shown. (D) The graph shows the rates of HPV genome retention per cell division. The retention rates were calculated as described in Materials and Methods. Means from three independent experiments and standard deviations are shown as error bars. N.S., not significant, compared with each other.
AdV.
The thermostable FLP mutant (FLPe)-expressing adenovirus vector (AdV) (AxCAFLPe), a kind gift from Izumi Saito (The Institute of Medical Science, The University of Tokyo), was prepared as described previously (3, 36). Cells were infected with AxCAFLPe at a 5-particle titer multiplicity of infection.
Western analysis.
Western blotting was conducted as described previously (26). Antibodies against HA (16B12; Covance, Princeton, NJ), involucrin (SY5; Sigma-Aldrich, St. Louis, MO), vinculin (Sigma-Aldrich), and loricrin (Covance) were used as probes, and horseradish peroxidase-conjugated anti-mouse, anti-rabbit (Jackson ImmunoResearch Laboratories, West Grove, PA), or anti-goat (sc-2033; Santa Cruz, Santa Cruz, CA) immunoglobulins were employed as secondary antibodies.
PCR and DNA blot hybridization.
Total genomic DNA was isolated by a standard SDS-proteinase K method, and an aliquot (100 ng) was examined by PCR amplification for Cre-mediated HPV DNA excision. Primer sets used for detecting total HPV16 DNA or recombined HPV16 are shown in Table 1. The DNA was amplified by 30 cycles of PCR using Takara Taq DNA polymerase (Takara, Japan) according to the supplier's instructions, with annealing at 60°C and elongation at 72°C for 30 s. PCR products were separated on a 1.5% agarose gel and visualized with ethidium bromide. For Southern blot analyses, digested DNA was separated on a 0.75% agarose gel, soaked in 0.25 M HCl for 15 min, and alkaline transferred onto nylon membranes (Boehringer Mannheim, Mannheim, Germany). The membranes were prehybridized in Hybrisol I (Millipore, Billerica, MA) for 1 h at 42°C. A biotin-labeled probe of the entire HPV16 genome prepared with the NEBlot Phototope kit (New England BioLabs, Ipswich, MA) was applied for hybridization, and the hybridized DNA was visualized with a Phototope-Star detection kit (New England BioLabs) following the protocol provided by the manufacturer. The LAS3000 charge-coupled device (CCD) imaging system (Fujifilm Co. Ltd., Japan) was employed for detection and quantification.
Table 1.
Primers for PCR and RT-PCR
| Name | nt position | Sequence (5′→3′) | Product size |
|---|---|---|---|
| For detecting excision of HPV16 DNAa | |||
| For total HPV DNA, HPV16 E7 | 660 | GGAGGAGGATGAAATAGATGGTC | 136 |
| 795 | AGTACGAATGTCTACGTGTGTGC | ||
| For excised-circular HPV DNA, HPV16 LCR | 7432 | AGGCCCATTTTGTAGCTTC | 271 |
| 7702 | CCTAACAGCGGTATGTAAGG (+loxP 305) | ||
| For detecting mRNAs | |||
| HA-E1 | 7 | CCTTATGACGTGCCAGATTACGC | 144 |
| 150 | GTCATTTTCGTTCTCATCGTCTGAGATG | ||
| E1 ^ E4 | 603 | TTTGCAACCAGAGACAACTGAT | 933 |
| 4014 | AGAGGCTGCTGTTATCCACAAT | ||
| 36B4b | 655 | TCGACAATGGCAGCATCTAC | 223 |
| 877 | GCCTTGACCTTTTCAGCAAG | ||
| For real-time quantitative PCR to quantify HPV16 DNA | |||
| HPV16 L2 | 4474 | CCCACAGCTACAGATACACTTGCT | 143 |
| 4616 | GGAATGGAAGGTACAGATGTTGGTGC |
RNA extraction and RT-PCR analyses.
For detection of mRNAs, total RNA was purified with RNeasy (Qiagen, Valencia, CA) and reverse transcribed to generate cDNAs by the ThermoScript reverse transcription (RT)-PCR system (Invitrogen) using random hexamers according to the supplier's instructions. Primers used for the E1̂E4 spliced transcript, exogenous codon optimized E1, and human acidic ribosomal phosphoprotein P0 (36B4) are shown in Table 1. The thermocycling profile for amplifying E1̂E4, E1, and 36B4 cDNAs was 1 min at 95°C; 40 cycles of 95°C for 30 s, 54°C for 30 s, and 72°C for 30 s (or 1 min for E1̂E4 cDNA); and 4 min of extension at 72°C. PCR products were separated in a 1.5 or 0.9% agarose gel and visualized with ethidium bromide.
Quantitative real-time PCR for genomic DNA.
Reactions were prepared in a volume of 10 μl containing 1× quantitative PCR (qPCR) master mix of KAPA SYBR FAST qPCR kits (Kapa Biosystems, Woburn, MA) and 300 nM each primer. PCR was performed using StepOnePlus (Applied Biosystems) with 10 s of denaturation at 95°C followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Serial dilutions of linearized HPV16 genome from pUC-HPV16 plasmid DNA by BamHI digestion were used as controls to measure the amounts of HPV16 genomic DNA. All real-time PCRs were run in triplicate. Total DNA was digested with DpnI before PCR amplification of the HPV16 genome with a primer set amplifying a product containing two DpnI sites (Table 1). HPV16 DNA copy number was expressed as copies per cell assuming that the total human genomic DNA is 6.6 pg/diploid cell. The rate of HPV16 genome retention (RR) was calculated as follows: RR = (copy number of viral genomes at the end/copy number of viral genomes at the beginning) 1/PD, where population doubling (PD) of cells was calculated as follows: PD = log(number of cells obtained/initial number of cells)/log2.
RESULTS
Establishment of keratinocytes containing the HPV16 genome.
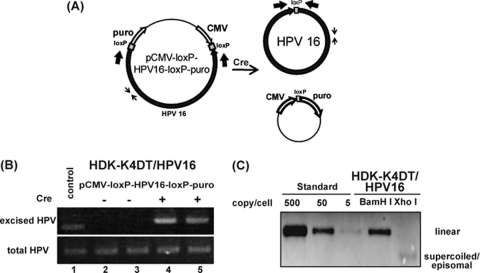
Primary human keratinocytes are often used for establishment of HPV-containing cell lines. However, they have a finite life span, and cells harboring HPV genomes tend to preferentially grow in culture. To minimize this effect and to obtain reproducible results, we first immortalized human dermal keratinocytes (HDKs) with TERT, a mutant form of CDK4 and cyclin D1 (HDK-K4DT). HDK-K4DT formed fully stratified squamous epithelium in an organotypic raft culture (data not shown). Then, we established keratinocytes harboring HPV genomes as episomes. HDK-K4DT cells were transfected with pCMV-loxP-HPV16-loxP-puro, designed to generate a circular 7.9-Kb HPV16 genome and a circular pCMV-puro expression cassette (Fig. 1A). With the Cre expression plasmid, about 5 to 10% of the transfected cells survived after short-term puromycin selection, whereas only a few cells survived without Cre recombinase (data not shown). At 2 days posttransfection without puromycin selection, total DNA was analyzed by PCR using two sets of primers, one for total HPV16 DNA and the other specific for the circularized HPV16 genome (Fig. 1A). In cells cotransfected with Cre, PCR products corresponding to the recombined circular HPV16, whose size should be bigger by 34 bp due to an inserted loxP sequence, were observed (Fig. 1B, lanes 4 and 5), whereas no circularized HPV16 genome was detected in cells without Cre expression (Fig. 1B, lanes 2 and 3). Southern blot analysis of total DNA extracted from cells at 21 days posttransfection suggested that the established HDK-K4DT cells harbored more than 50 viral genome copies per cell (Fig. 1C). Repeated transfection experiments confirmed the reproducibility of this technique. At 60 days posttransfection, after nine serial passages at a ratio of 1:8, we still detected 10 to 20 copies of the viral genomes per cell (data not shown). The rate of HPV16 genome retention was calculated as 90% per cell division.
Fig 1.
HPV genome excision and establishment of the cell line containing the HPV16 genome. (A) Schematic representation describing the parental pCMV-loxP-HPV16-loxP-puro, the Cre recombinase-excised HPV16 genome, and the pCMV-puro plasmid. PCR primers (thin and thick arrows) to detect total and excised HPV DNA, respectively, are indicated. (B) HDK-K4DT cells were cotransfected with the pCMV-loxP-HPV16-loxP-puro plasmid with (lanes 4 and 5) or without (lanes 2 and 3) the NCre expression plasmid. Total DNA was extracted from the cells 2 days after the transfection without puromycin selection. Representative pictures of an ethidium bromide-stained agarose gel with PCR products indicating total HPV16 DNA (bottom) and excised circular HPV16 DNA (top), containing a surplus of 34 bp of loxP DNA, are shown. (C) Southern blot hybridization for the HPV genome in HDK-K4DT cultures. DpnI and BamHI- or XhoI-digested total DNA isolated from HDK-K4DT at 3 weeks after transfection with pCMV-loxP-HPV16-loxP-puro plasmid and the NCre expression plasmid is shown. Digestion with BamHI, which cuts the HPV16 genome once, produced results of the expected size for the HPV16 genome. Digestion with XhoI, which does not cut the HPV16 genome, showed supercoiled plasmid of HPV16 genome. The BamHI-linearized HPV16 plasmid was used for length and copy number standards.
E1 protein is required for establishment of HPV16 genomes as episomes in HDK-K4DT cells.
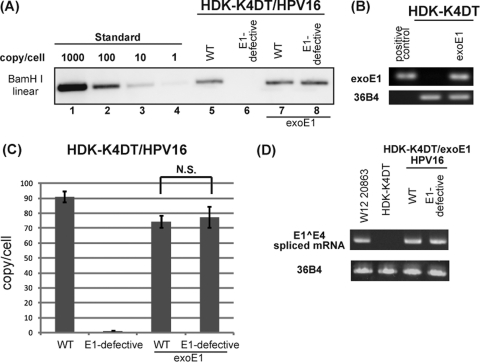
To study the role of E1 in each stage of the viral life cycle, we prepared an E1-defective mutant HPV16 genome containing an E1 translation termination mutation at nt 892 to 894 to abrogate E1 protein expression. Unlike the wild-type HPV16 genome, the HPV16 E1-defective genome failed to establish in HDK-K4DT cells as episomes (Fig. 2A, lanes 5 and 6). However, in HDK-K4DT cells expressing exogenous E1 from the retrovirus, MSCV-FRT-HA16E1-TKneo-FRT, the E1-defective HPV16 genome could establish as episomes as efficiently as the wild type HPV16 genome (Fig. 2A, lanes 7 and 8). We confirmed the expression of exogenous E1 driven by the LTR promoter by RT-PCR using primers specific for exogenous E1 (Fig. 2B), though the E1 protein was undetectable by Western blot analysis. The copy numbers of the E1-defective HPV16 genome were comparable to that of the wild-type HPV16 genome in the presence of exogenous E1 expression (Fig. 2C). The translation termination mutation inserted in the downstream of the splice donor site for E1̂E4 (nt 880) did not disrupt normal E1̂E4 splicing (Fig. 2D). These data indicate that the E1 protein is required for initial replication and/or maintenance of the viral genome.
Fig 2.
Requirement for E1 protein in the establishment stage. (A) Southern blot hybridization of the HPV genome in HDK-K4DT cultures. Parental (lanes 5 and 6) and exogenous E1 expressing HDK-K4DT cells (lanes 7 and 8) were transfected with the wild-type (WT) or the E1-defective pCMV-loxP-HPV16-loxP-puro plasmid and the NCre expression plasmid. Total DNA was extracted at 21 days posttransfection. DpnI- and BamHI-digested total DNA from parental or exogenous E1 expressing HDK-K4DT cultures was analyzed. The BamHI-linearized HPV16 plasmid was used for length and copy number standards (lanes 1 to 4). (B) mRNAs for exogenous E1 of HDK1-K4DT cells transduced with the retroviral vector expressing HA16E1 were analyzed. Total RNAs isolated from cells with or without the retroviral transduction were subjected to reverse transcription (RT)-PCR with a primer set specific to codon-optimized E1. 36B4 mRNA was also detected as an internal control. The PCR products of exogenous E1 (top) and 36B4 (bottom) visualized in ethidium bromide-stained agarose gels are shown. pCMSCV-FRT-HA16E1-TKneo-FRT was used as a positive control. (C) The copy number of the HPV16 genomes at 14 days posttransfection in each HDK-K4DT cell lines was determined by real-time PCR. The deviations of three independent sets of transfectants are shown as error bars. N.S., not significant. (D) Expression of the E1̂E4 spliced mRNAs in E1-expressing HDK-K4DT cells harboring the wild type or the E1-defective HPV16 genomes at 35 days posttransfection. Total RNAs were subjected to RT-PCR with an E1̂E4-specific primer set (Table 1). RNAs from parental HDK-K4DT cells and W12 cells were used as controls. PCR products of E1̂E4 (top) and 36B4 (bottom) are shown, as described for panel B.
E1 protein is dispensable for maintenance replication of the viral genome.
In order to assess the requirement of E1 protein for maintenance replication of the viral genome, we used the HDK-K4DT cells harboring the E1-defective HPV16 genomes established with exogenous E1 expression from the integrated MSCV-FRT-HA16E1-TKneo-FRT retrovirus. Upon infection of FLPe-expressing AdV, the exogenous E1 expression cassette as well as the TKneo gene was excised by FLP at an efficiency of around 50% (Fig. 3A). Then we isolated HDK-K4DT cells which no longer expressed exogenous E1 by ganciclovir selection (Fig. 3B). After several passages of cells at a ratio of 1:8, the copy number of HPV16 genomes was determined by real-time PCR. We detected 70 to 100 copies of the wild-type or the E1-defective HPV16 genomes per cell just before the AdV infection. Copy numbers of both the wild-type and the E1-defective HPV16 genomes gradually decreased during the passages (Fig. 3C). However, about 10 to 20 copies of the E1-defective HPV16 genomes per cell still remained after several passages of cells, even in the absence of exogenous E1 expression. No difference in the rate of genome retention was observed between the E1-defective and the wild-type HPV16 genomes. The rates were calculated as approximately 90% per cell division, irrespective of exogenous E1 expression, and proved quite constant in three independent experiments (Fig. 3D). These data indicate that the E1 protein is dispensable for maintenance replication of the viral genome.
E1 protein is required for viral genome amplification upon differentiation.
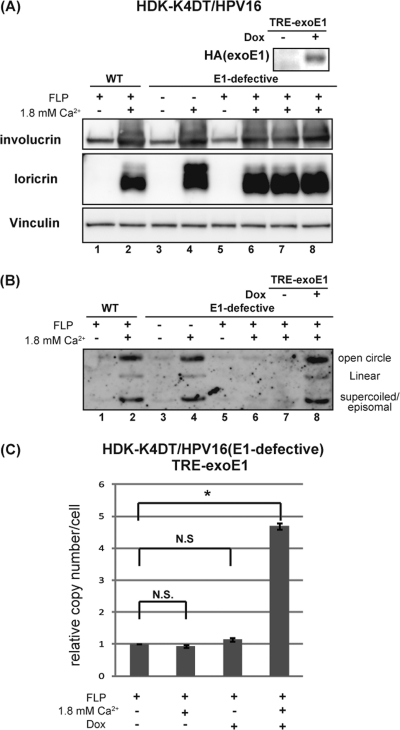
To confirm that E1 protein is required for the productive stage of viral replication, differentiation-dependent viral genome amplification was examined with the same series of the cells established for the previous section (Fig. 3). HDK-K4DT cells harboring the wild-type or the E1-defective HPV16 genomes in the presence or the absence of exogenous E1 expression were exposed to a high calcium concentration. Induction of differentiation was confirmed by expression of keratinocyte differentiation markers, involucrin and loricrin (Fig. 4A). Southern blot analyses of DNA extracted from sister cultures showed the wild-type HPV16 genomes to be amplified episomally upon differentiation in the absence of exogenous E1 expression (Fig. 4B, lanes 1 to 2). However, the E1-defective HPV16 genomes were amplified only in the presence of exogenous E1 expression (Fig. 4B, lanes 3 to 6). Reintroduction of E1 with lentiviruses, CSII-CMV-tetON and CSII-TRE-Tight-HA16E1, to the cells whose exogenous E1 cassette had been excised by FLP rescued the E1-defective HPV16 genome amplification upon differentiation only when the E1 expression was induced by doxycycline (Fig. 4B, lanes 7 and 8), whereas induction of E1 alone without high calcium failed to rescue the genome amplification (Fig. 4C). These data indicate that E1 protein is required for viral genome amplification upon differentiation. Since the same series of cells used for Fig. 3 were used in these experiments, the results confirmed that the E1-defective HPV16 genomes were episomally maintained in the absence of E1 expression.
Fig 4.
E1 protein is required for viral genome amplification upon differentiation. (A) HDK-K4DT cells harboring the wild-type (lanes 1 and 2) or the E1-defective (lanes 3 to 8) HPV16 genomes in the presence (FLP−, lanes 3 and 4) or the absence (FLP+, lanes 1 and 2, 5 to 8) of exogenous E1 expression were seeded at 2 × 105 cells per well (6-well plate) in Epilife complete growth medium, and then cells were exposed to 1.8 mM calcium (lanes 2, 4, 6, 7, 8) to induce keratinocyte differentiation. Total cell lysates were made, and total DNAs were extracted from the sister cultures just before and 10 days after calcium exposure. The expression of involucrin and loricrin, keratinocyte differentiation markers, was analyzed by Western blotting. Expression of reintroduced exogenous E1 controlled by doxycycline was detected by anti-HA antibody (lanes 7 and 8). Vinculin was detected as a loading control. (B) Southern blot hybridization for episomal HPV16 genomes in HDK-K4DT cells. XhoI-digested total DNA from each HDK-K4DT culture was loaded. A representative image of three independent experiments is shown. (C) The copy number of the E1-defective HPV16 genomes in HDK-K4DT cells in the indicated condition was determined by real-time PCR. Three replicates are shown and standard deviations are shown as error bars. N.S., not significant. The single asterisk indicates P values of <0.05.
DISCUSSION
In this study, we demonstrated that E1 is dispensable for maintenance replication of the HPV16 genome but required for replication in the establishment and productive stages. Taking the previous study using a TS E1 mutant of BPV1 (17) into account, it is likely that E1 is dispensable for the maintenance replication of other PVs, too.
Thus, HPVs have at least two replication modes and control the copy number of the viral genome depending on the situation. Such a strategy of the virus would clearly be beneficial for persistent infection and continuous virus production. In the maintenance phase, minimal expression of viral proteins in host cells with low copy numbers of viral genomes would allow HPV to evade cellular immune surveillance. Moreover, as recent studies indicate, a high level of E1 expression in basal-layer cells could activate an ATM-dependent damage response and cause growth suppression (10, 32).
It is reasonable to speculate that the E1-independent maintenance replication employs the cellular replication machinery to support viral genome maintenance under S phase control. Mechanisms of viral genome DNA replication control by host cell factors have been well studied for the Epstein-Barr virus (EBV). The EBV genome DNA is replicated once per S phase in the latent phase of infection (19, 44). In this phase, EBV employs replication licensing proteins, MCMs and ORC (19, 22), which assemble on the latent origin of replication of EBV, OriP. A low copy number for the viral genome may be an appropriate common strategy for episomal viruses to sustain latent infection. Although it is not known whether and how MCMs and ORC are involved in HPV DNA replication, E1- and E2-independent cis-replicating elements may reside outside the LCR and possibly in the late region (L2-L1 open reading frames [ORFs]) of the HPV16 genome (28, 29).
The observed rate of HPV16 genome retention was about 90% per cell generation, which is comparable to the reported rate for the EBV genome (25). With this retention rate, the viral genomes of 100 copies per cell can be maintained for 2 to 3 months, corresponding to approximately 40 cell divisions under our culture conditions. In the stratified squamous epithelium, stem cells self-renew by dividing infrequently and generate a population of cells that undergo limited but more frequent divisions before giving rise to nonproliferative, terminally differentiating cells. In the natural life cycle of HPVs, the virus must infect epithelial stem cells (8, 34), which would divide much less frequently than cultured cells do. Therefore, the viral genome in the stem cells in vivo would be able to persist for much longer period than in cultured cells, even if the genome retention rate is the same as that found in our study.
In maintenance replication, HPV may still employ two different modes of replication. Hoffmann et al. showed that HPV16 DNA replicates once per S phase in W12 cells, while HPV31 DNA replicates via a random-choice mechanism with some multiple rounds of the viral genome replication per S phase in CIN612-9E cells, and that forced expression of E1 in W12 cells converted HPV16 DNA replication to random-choice replication (12). Interestingly, when HPV16 or HPV31 DNAs are separately introduced into NIKS cells, they both replicate randomly (12). Thus, it is likely that the difference between W12 and CIN612-9E cells depends on expression levels of E1. It is possible that occasional or low-level expression of auxiliary E1 hinders the copy number loss for an even longer period of maintenance, as indicated by previous studies (18, 39). Theoretically, E1 protein could be supplied from the infected virion and/or by de novo synthesis from the infected viral genome in the establishment stage. In this regard, it is not clear whether our experimental system recapitulates the actual establishment stage of the HPV life cycle, since E1 can be supplied only by de novo synthesis. At present, little is known about the underlying mechanisms of the establishment stage and the mechanism(s) of switching to the subsequent maintenance stage. Clearly, it needs to be examined whether the E1 protein is included in infectious virions and how the E1 expression is regulated in the three different stages.
PV E1 protein forms double hexamers at the replication origin in the LCR with the help of E2 (2, 11, 43) and unwinds DNA through helicase activity ahead of replication forks powered by the hydrolysis of ATP (31). Since E1 is the only viral protein with enzymatic activities, it is an attractive target for development of novel therapeutic agents to treat HPV-associated benign lesions where the whole viral life cycle is completed. Indeed, some candidate small molecules have been reported to inhibit the E1 function (5, 9, 15, 40–42). However, their identification and evaluation was done using biochemical assays or surrogate cell-based assays, and a true antiviral activity has yet to be tested. Based on our present study, the effectiveness of E1 inhibitors as antiviral drugs may be restricted, since they cannot inhibit E1-independent HPV replication in long-living basal cells. Inhibition of E1 protein function could prevent amplification of the viral genome in the establishment and productive stages. Thus, it may prevent HPV infection and reduce pathogenesis, including papilloma formation and virion production. In the case of cervical intraepithelial neoplasias (CINs), continuous inhibition of E1 might reverse low-grade lesions to apparently healthy mucosa, but interruption of the inhibition might lead to recurrence of the lesions. More importantly, it may not be able to eliminate the HPV genomes replicating in undifferentiated basal cells, which are thought to be the histogenetic origin of cervical cancer. Thus, E1 inhibition might not be able to prevent CIN lesions from progressing into cancer.
In summary, we have established an experimental system which can evaluate the requirement of any viral gene of interest in the viral life cycle by supplying and deleting exogenous expression of the gene and demonstrated that E1 is entirely dispensable for maintenance replication of the HPV16 genome in human keratinocytes. Thus, inhibition of E1 may not be able to eliminate the viral genome from the basal cell layer. The rationale for development of E1 inhibitors as anti-HPV drugs may be more restricted than formerly envisaged. Further studies will be required to elucidate the roles of cellular replication factors and the cis elements of the HPV genome in E1-independent maintenance replication.
ACKNOWLEDGMENTS
We express our appreciation to Takako Ishiyama for expert technical assistance. We are grateful to John H. Lee (University of Iowa Hospitals and Clinics) for the loxP-HPV16-loxP construct, Izumu Saito (University of Tokyo and RIKEN RDB) for adenovirus vectors, and Hiroyuki Miyoshi (RIKEN BRC) for lentivirus vectors and packaging constructs.
This work was supported in part by Grants-in-Aid for Cancer Research from the Ministry of Health Labor and Welfare to T.K., and for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to N.E., T.N., and T.K.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Angeletti PC, Kim K, Fernandes FJ, Lambert PF. 2002. Stable replication of papillomavirus genomes in Saccharomyces cerevisiae. J. Virol. 76:3350–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berg M, Stenlund A. 1997. Functional interactions between papillomavirus E1 and E2 proteins. J. Virol. 71:3853–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buchholz F, Angrand PO, Stewart AF. 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16:657–662 [DOI] [PubMed] [Google Scholar]
- 4. Chiang CM, et al. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. U. S. A. 89:5799–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. D'Abramo CM, Archambault J. 2011. Small molecule inhibitors of human papillomavirus protein-protein interactions. Open Virol. J. 5:80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Del Vecchio AM, Romanczuk H, Howley PM, Baker CC. 1992. Transient replication of human papillomavirus DNAs. J. Virol. 66:5949–5958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doorbar J. 2006. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. (Lond.) 110:525–541 [DOI] [PubMed] [Google Scholar]
- 8. Egawa K. 2003. Do human papillomaviruses target epidermal stem cells? Dermatology 207:251–254 [DOI] [PubMed] [Google Scholar]
- 9. Faucher AM, et al. 2004. Discovery of small-molecule inhibitors of the ATPase activity of human papillomavirus E1 helicase. J. Med. Chem. 47:18–21 [DOI] [PubMed] [Google Scholar]
- 10. Fradet-Turcotte A, et al. 2011. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J. Virol. 85:8996–9012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frattini MG, Laimins LA. 1994. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. U. S. A. 91:12398–12402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffmann R, Hirt B, Bechtold V, Beard P, Raj K. 2006. Different modes of human papillomavirus DNA replication during maintenance. J. Virol. 80:4431–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeon S, Lambert PF. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 92:1654–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanegae Y, et al. 1996. Efficient gene activation system on mammalian cell chromosomes using recombinant adenovirus producing Cre recombinase. Gene 181:207–212 [DOI] [PubMed] [Google Scholar]
- 15. Kasukawa H, Howley PM, Benson JD. 1998. A fifteen-amino-acid peptide inhibits human papillomavirus E1–E2 interaction and human papillomavirus DNA replication in vitro. J. Virol. 72:8166–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim K, Angeletti PC, Hassebroek EC, Lambert PF. 2005. Identification of cis-acting elements that mediate the replication and maintenance of human papillomavirus type 16 genomes in Saccharomyces cerevisiae. J. Virol. 79:5933–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim K, Lambert PF. 2002. E1 protein of bovine papillomavirus 1 is not required for the maintenance of viral plasmid DNA replication. Virology 293:10–14 [DOI] [PubMed] [Google Scholar]
- 18. Lee JH, et al. 2004. Propagation of infectious human papillomavirus type 16 by using an adenovirus and Cre/LoxP mechanism. Proc. Natl. Acad. Sci. U. S. A. 101:2094–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindner SE, Sugden B. 2007. The plasmid replicon of Epstein-Barr virus: mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid 58:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu JZ, Sun YN, Rose RC, Bonnez W, McCance DJ. 1993. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J. Virol. 67:7131–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maglennon GA, McIntosh P, Doorbar J. 2011. Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression. Virology 414:153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maiorano D, Lemaitre JM, Mechali M. 2000. Stepwise regulated chromatin assembly of MCM2-7 proteins. J. Biol. Chem. 275:8426–8431 [DOI] [PubMed] [Google Scholar]
- 23. Mungal S, Steinberg BM, Taichman LB. 1992. Replication of plasmid-derived human papillomavirus type 11 DNA in cultured keratinocytes. J. Virol. 66:3220–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakano M, Ishimura M, Chiba J, Kanegae Y, Saito I. 2001. DNA substrates influence the recombination efficiency mediated by FLP recombinase expressed in mammalian cells. Microbiol. Immunol. 45:657–665 [DOI] [PubMed] [Google Scholar]
- 25. Nanbo A, Sugden A, Sugden B. 2007. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 26:4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Narisawa-Saito M, et al. 2007. HPV16 E6-mediated stabilization of ErbB2 in neoplastic transformation of human cervical keratinocytes. Oncogene 26:2988–2996 [DOI] [PubMed] [Google Scholar]
- 27. Naviaux RK, Costanzi E, Haas M, Verma IM. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pittayakhajonwut D, Angeletti PC. 2008. Analysis of cis-elements that facilitate extrachromosomal persistence of human papillomavirus genomes. Virology 374:304–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pittayakhajonwut D, Angeletti PC. 2010. Viral trans-factor independent replication of human papillomavirus genomes. Virol. J. 7:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rabson MS, Yee C, Yang YC, Howley PM. 1986. Bovine papillomavirus type 1 3′ early region transformation and plasmid maintenance functions. J. Virol. 60:626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rocque WJ, et al. 2000. Replication-associated activities of purified human papillomavirus type 11 E1 helicase. Protein Expr. Purif. 18:148–159 [DOI] [PubMed] [Google Scholar]
- 32. Sakakibara N, Mitra R, McBride A. 2011. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 85:8981–8995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sasaki R, et al. 2009. Oncogenic transformation of human ovarian surface epithelial cells with defined cellular oncogenes. Carcinogenesis 30:423–431 [DOI] [PubMed] [Google Scholar]
- 34. Schmitt A, et al. 1996. The primary target cells of the high-risk cottontail rabbit papillomavirus colocalize with hair follicle stem cells. J. Virol. 70:1912–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sverdrup F, Khan SA. 1994. Replication of human papillomavirus (HPV) DNAs supported by the HPV type 18 E1 and E2 proteins. J. Virol. 68:505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takata Y, Kondo S, Goda N, Kanegae Y, Saito I. 2011. Comparison of efficiency between FLPe and Cre for recombinase-mediated cassette exchange in vitro and in adenovirus vector production. Genes Cells 16:765–777 [DOI] [PubMed] [Google Scholar]
- 37. Ustav M, Stenlund A. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ustav M, Ustav E, Szymanski P, Stenlund A. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang HK, Duffy AA, Broker TR, Chow LT. 2009. Robust production and passaging of infectious HPV in squamous epithelium of primary human keratinocytes. Genes Dev. 23:181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White PW, Faucher AM, Goudreau N. 2011. Small molecule inhibitors of the human papillomavirus E1-E2 interaction. Curr. Top. Microbiol. Immunol. 348:61–88 [DOI] [PubMed] [Google Scholar]
- 41. White PW, et al. 2005. Biphenylsulfonacetic acid inhibitors of the human papillomavirus type 6 E1 helicase inhibit ATP hydrolysis by an allosteric mechanism involving tyrosine 486. Antimicrob. Agents Chemother. 49:4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. White PW, et al. 2003. Inhibition of human papillomavirus DNA replication by small molecule antagonists of the E1-E2 protein interaction. J. Biol. Chem. 278:26765–26772 [DOI] [PubMed] [Google Scholar]
- 43. Yang L, Li R, Mohr IJ, Clark R, Botchan MR. 1991. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature 353:628–632 [DOI] [PubMed] [Google Scholar]
- 44. Yates JL, Guan N. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. zur Hausen H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342–350 [DOI] [PubMed] [Google Scholar]