Abstract
Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that has been responsible for an epidemic outbreak of unprecedented magnitude in recent years. Since then, significant efforts have been made to better understand the biology of this virus, but we still have poor knowledge of CHIKV interactions with host cell components at the molecular level. Here we describe the extensive use of high-throughput yeast two-hybrid (HT-Y2H) assays to characterize interactions between CHIKV and human proteins. A total of 22 high-confidence interactions, which essentially involved the viral nonstructural protein nsP2, were identified and further validated in protein complementation assay (PCA). These results were integrated to a larger network obtained by extensive mining of the literature for reports on alphavirus-host interactions. To investigate the role of cellular proteins interacting with nsP2, gene silencing experiments were performed in cells infected by a recombinant CHIKV expressing Renilla luciferase as a reporter. Collected data showed that heterogeneous nuclear ribonucleoprotein K (hnRNP-K) and ubiquilin 4 (UBQLN4) participate in CHIKV replication in vitro. In addition, we showed that CHIKV nsP2 induces a cellular shutoff, as previously reported for other Old World alphaviruses, and determined that among binding partners identified by yeast two-hybrid methods, the tetratricopeptide repeat protein 7B (TTC7B) plays a significant role in this activity. Altogether, this report provides the first interaction map between CHIKV and human proteins and describes new host cell proteins involved in the replication cycle of this virus.
INTRODUCTION
Chikungunya virus (CHIKV) is a mosquito-transmitted Togaviridae virus from the Alphavirus genus; it was first reported in 1952 in Tanganyika. It is responsible for an acute infection of abrupt onset, characterized by high fever, polyarthralgia, myalgia, headache, chills, and rash (36, 54). The symptoms are generally of short duration (1 week), and recovery is often complete, although some patients have recurrent episodes for several weeks after infection (36, 54). CHIKV is endemic in Africa, India, and Southeast Asia and is transmitted by Aedes spp. mosquitoes through an urban or sylvatic transmission cycle. In 2006, an outbreak of CHIKV fever occurred in numerous islands of the Indian Ocean (the Comoros, Mauritius, Seychelles, Madagascar, and Reunion island) before jumping to India, where an estimated 1.4 million cases have been reported (9, 32, 41). More recently, imported infections have been described in Europe, and around 200 endemic cases have been reported in Italy (43). Clinically, this CHIKV epidemic was accompanied by more severe symptoms than previous outbreaks, with reports of severe polyarthralgia and myalgia, complications, and deaths.
The CHIKV genome is an 11.8-kb, single-stranded RNA molecule of positive polarity. This virus is closely related to Semliki Forest virus (SFV), Sindbis virus (SINV), and other Old World alphaviruses, and also more distantly related to New World alphaviruses like Venezuelan equine encephalitis virus (23). The genomic RNA is capped and directly translated into a full-length nonstructural polyprotein (nsP) called P1234, which is encoded by the 5′ two-thirds of the genome (27, 29). This precursor cleaves itself to produce P123 and nsP4, which carries the RNA-dependent RNA polymerase activity. These proteins, together with cellular cofactors, assemble into a replication complex that produces antisense genomic RNA molecules. Subsequent cleavage of P123 into nsP1 and P23 gives rise to a polymerase complex making both sense and antisense genomic RNA. Further processing of P23 into nsP2 and nsP3 gives rise to a polymerase complex making only positive-sense genomic RNA molecules. In addition to replicating the viral genome, this viral protein complex transcribes a 26S subgenomic RNA from the 3′ extremity of the viral genome. This mRNA is translated into a polyprotein precursor, which is cleaved by a combination of viral and cellular enzymes to produce a capsid protein (C), two major envelope proteins (E1 and E2), and two smaller accessory peptides, E3 and 6k. Once assembled, CHIKV virions are spherical particles of 65 to 70 nm in diameter, essentially composed of genomic RNA molecules associated with capsid proteins and enveloped in a host-derived lipid membrane decorated by E1-E2 heterodimers organized in an icosahedral lattice (52).
Within the viral replication complex, nsP4 carries the viral RNA-dependent RNA polymerase activity, and nsP3 is an essential cofactor of this enzyme. Both nsP3 and nsP4 have been shown to interact directly with nsP1, a nonstructural protein involved in viral mRNA capping via its guanine-7-methyltransferase and guanylyltransferase enzymatic activities, but also in the synthesis of minus-strand RNA genomes (46). In addition, nsP1 is required for the attachment of the viral replication complex to cytoplasmic membranes and induces filopodium-like structures at the membrane of infected cells (58). The functions of nsP2 are even more complex. It contains an N-terminal RNA triphosphatase domain that performs the first of the viral RNA capping reactions. The same domain is also a nucleotide triphosphatase (NTPase) that fuels the RNA helicase activity carried by the second domain of nsP2. The C-terminal part of the protein exhibits a protease domain responsible for the autocatalytic cleavage of the nonstructural polyprotein and a methyltransferase-like domain of unknown function.
Besides their intrinsic enzymatic activities, nonstructural proteins from SINV and SFV have been shown to interact with numerous host factors. The mapping of interactions between nonstructural viral proteins and cellular factors is essential to understand how the virus hijacks the cellular machinery and blocks the antiviral response to achieve genome replication and production of structural proteins that assemble into new virions. By using a recombinant SINV expressing an enhanced green fluorescent protein (EGFP)-tagged nsP2 protein, Atasheva et al. purified nsP2-associated cellular factors that were subsequently identified by mass spectrometry analysis (3). Viral nsP2 was found to copurify with several RNA-binding proteins, including heterogeneous nuclear ribonucleoproteins (hnRNPs), multiple ribosomal proteins, including ribosomal protein S6 (RpS6), and cellular filament components. Interactions between nsP2 from SINV and hnRNPs, in particular hnRNP-K, were also reported by another group (7). Members of the hnRNP family are involved in RNA processing, trafficking, and translation and have been found associated with alphavirus genomic and/or subgenomic RNA molecules, probably contributing to their synthesis and translation. Interactions between alphavirus nsP2 proteins and ribosomal components, in particular RpS6, were also reported by Montgomery et al. (37) and likely contribute to the hijacking of translational machinery by viral factors. In addition to their different roles as cofactors of the viral polymerase complex, nsP2 proteins from Old World alphaviruses have been described as virulence factors responsible for the transcriptional and translational shutoff observed in infected host cells and the inhibition of interferon (IFN)-mediated antiviral responses (6, 14, 16–19, 21, 33, 49). However, virus-host protein interactions involved are still unknown and remain to be identified.
Cellular interactors of nsP3 and nsP4 have been identified by a similar strategy. Indeed, several groups have engineered recombinant SINV strains expressing either nsP3 or nsP4 with EGFP or 3×-FLAG tags to achieve affinity purification from infected cells. Cellular proteins that copurified with nsP3 or nsP4 were quite similar to those identified with nsP2 as bait, suggesting that all these proteins are bound together into identical virus-induced complexes (10, 11, 15, 22). To further document alphavirus interactions with host cell components, we used for the first time a high-throughput yeast two-hybrid (HT-Y2H) approach to characterize interactions between CHIKV and human proteins. This led to the identification of 22 cellular proteins interacting directly with either nsP2 or nsP4, with most of these interactions being novel with no previous report in the literature. From this list of interactors, we established that both hnRNP-K and ubiquilin 4 (UBQLN4) contribute to CHIKV replication and that tetratricopeptide repeat protein 7B (TTC7B) participates in the shutoff mediated by CHIKV nsP2.
MATERIALS AND METHODS
Plasmid DNA constructs.
Cloning of CHIKV sequences encoding nsP1, nsP2, nsP3, nsP4, C, E3, E2, 6k, and E1 has been described previously (40). Briefly, coding sequences were amplified by standard reverse transcription-PCR (Titan One tube; Roche) from viral RNA samples (CHIKV strain 05115 from La Réunion Island), and cloned by in vitro recombination into pDONR207 using Gateway technology (Invitrogen). PCR primers displayed 20 to 30 specific nucleotides matching open reading frame (ORF) extremities, so that their melting temperature Tm was close to 60°C. To achieve recombinational cloning of PCR products, 5′ ends of forward primers were fused to the attB1.1 recombination sequence, 5′-GGGGACAACTTTGTACAAAAAAGTTGGCATG-3′, while reverse primers were fused to the attB2.1 recombination sequence, 5′-GGGGACAACTTTGTACAAGAAAGTTGGTTA-3′. Recombination of PCR products into pDONR207 was performed following the manufacturer's recommendation (BP cloning reaction). All constructs were transformed and amplified in Escherichia coli DH5α strain. Fragments of nsP2 corresponding to amino acid residues 1 to 605, 1 to 632, 1 to 672, 1 to 767, 469 to 798, 606 to 798, and 633 to 798 were cloned following the same procedure. DNA sequences encoding nsP2 and nsP4 proteins from SFV and SINV were cloned following the same procedure, and the corresponding constructs were previously described (40). SFV clones matched GenBank entry AJ251359. SINV clones corresponded to GenBank entry J02363, but the nsP2 clone contained C-to-T and A-to-G mutations at nucleotide positions 1313 and 1900, respectively. The corresponding nsP2 protein was previously found unable to translocate in nuclei of expressing cells and was defective in shutting off cellular genes (40). Point mutations in the CHIKV nsP2 sequence were introduced using the QuikChange Lightning site-directed mutagenesis kit following the manufacturer's recommendations (Agilent).
Yeast two-hybrid screening procedure.
Yeast culture medium was prepared as previously described (53). ORFs encoding mature CHIKV proteins were transferred by in vitro recombination (LR cloning reaction; Gateway Technology, Invitrogen) from pDONR207 into the Y2H vector pDEST32 (Invitrogen) in order to be expressed in a fusion downstream of the Gal4 DNA-binding domain (Gal4-BD). Bait constructs were transformed into the Saccharomyces cerevisiae AH109 yeast strain (Clontech) by using a standard lithium-acetate procedure. Spontaneous transactivation of the HIS3 reporter gene was not observed in yeast cells transformed with these different constructs, except for GAL4-BD-nsP3, which showed a strong intrinsic transcriptional activity. For this reason, nsP3 was discarded from the screening pipeline. All screens were performed on a synthetic medium lacking histidine (-His medium) and not supplemented with 3-amino-1,2,4-triazole. A mating strategy was used for screening human cDNA libraries established in the Y187 yeast strain (Clontech). Human cDNAs were expressed from the pPC86 vector (Invitrogen), which allows, in yeast cells, the expression of cellular proteins in a fusion downstream of the Gal4 activation domain (Gal4-AD). Human spleen, fetal brain, and HeLa cell cDNA libraries were purchased from Invitrogen. For each screen, 20 to 50 million yeast diploids were produced by mating and grown for 6 days on -His selective medium. In addition, we established a normalized Y2H library from a pool of 12,000 human full-length ORFs cloned in pDONR223 (30) (Open Biosystems) that were transferred by in vitro recombination (LR cloning reaction) into the pPC86 vector (Invitrogen). For each screen performed, only 5 million yeast diploids were generated, since this was sufficient for full coverage of this normalized library. After 6 days of culture on -His selective medium, His+ colonies were picked and purified over 3 weeks by culture on selective medium to eliminate false positives (51). AD cDNAs were amplified by PCR from zymolase-treated yeast colonies using primers that hybridized within the pPC86 regions flanking cDNA inserts. PCR products were sequenced, and cellular interactors were identified by multiparallel BLAST analysis (39).
Cell lines and viruses.
HEK-293T cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-Invitrogen) containing 10% fetal calf serum (FCS), penicillin, and streptomycin at 37°C and 5% CO2. Recombinant alphaviruses with a reporter gene between nsP3 and nsP4 nonstructural proteins have already been produced with success (48). Using this strategy, CHIKV nsP3-Luc chimeric protein was engineered by placing the luciferase insert between the nsP3 and nsP4 coding regions of an infectious clone of CHIKV strain Mauritius-2006 (C. Drosten and B. Kümmerer, unpublished data). The construction of infectious CHIKV expressing luciferase will be described elsewhere (H. H. Gad et al., unpublished data). Briefly, a cassette gene coding for the C-terminal region of nsP3 followed in frame by the nsP1/2 protease recognition site of CHIKV Mauritius, a unique SpeI restriction enzyme site, the Opal codon of nsP3, the nsP3/4 protease recognition site, and the N-terminal region of the nsP4 gene was substituted into the corresponding region of the infectious clone of CHIKV. The resulting construct was designated pCHIKV-SpeI. The Renilla luciferase gene was inserted within the SpeI site, and the resulting plasmid was designated pCHIKV-Luc. Production of capped in vitro transcripts from linearized pCHIKV-Luc was obtained using the mMESSAGE mMACHINE kit (Ambion). Infectious CHIKV-Luc was obtained by electroporation of capped in vitro transcripts into BHK-21 cells. The primary virus stock was collected after 48 h. The first-passage stock (P1) of CHIKV-Luc was used to infect BHK-21 cells and obtain a second-passage stock (P2). Recovered P2 cells were quantified by plaque assay on Vero cells, and the virus titer of CHIKV-Luc was 107 PFU/ml. Luciferase activity from CHIKV-Luc could be detected as soon as 6 h postinfection. The second-passage stock P2 was used to infect Vero cells and obtain a third-passage stock (P3), which was titrated based on the 50% tissue culture infective dose (TCID50) on HEK-293T cells. The virus titer of this P3 stock was 1.4 × 106 PFU/ml and was used in all experiments described in the manuscript.
Sensing protein-protein interactions by protein complementation assay (PCA).
ORF sequences corresponding to cellular partners of CHIKV nsP2 and nsP4 identified by HT-Y2H were cloned by in vitro recombination into pDONR207 as described above (BP cloning reaction). Then, CHIKV nsP2 and nsP4 and cellular ORFs were recombined from pDONR207 into SPICA-N1 and SPICA-N2 vectors so that they could be expressed in fusions with fragments 1 to 93 (Gluc1) and 94 to 169 (Gluc2) of Gaussia princeps luciferase as previously described (8, 42). HEK-293T cells were seeded in 24-well plates at a concentration of 2 × 105 cells per well. After 24 h, cells were transfected with 250 ng of SPICA-N1 and SPICA-N2 plasmid constructs. At 24 h posttransfection, cells were harvested with 30 μl of Renilla lysis buffer (E2820; Promega) for 30 min, and Renilla luciferase enzymatic activity was measured using a Berthold Centro XS LB960 luminometer by injecting 100 μl of Renilla luciferase assay reagent (E2820; Promega) into cell lysates and counting luminescence for 10 s.
Results were expressed as the fold change normalized over the sum of controls, specified herein as the normalized luminescence ratio (NLR). For a given protein pair A-B, the luminescence activity of cells transfected with SPICA-N1-A plus SPICA-N2-B (AB) was divided by the sum of luminescence activity for control wells transfected with SPICA-N1-A plus empty SPICA-N2 vector (CtrlA) and empty SPICA-N1 vector plus SPICA-N2-B (CtlB). Thus, NLR = AB/(CtlA + CtlB). For each interaction, the final result was calculated from the mean NLR of triplicate experiments.
Luciferase reporter gene assays.
ORFs encoding nsP2 from CHIKV or SFV were transferred by in vitro recombination (LR cloning reaction; Gateway Technology, Invitrogen) from pDONR207 into the pCI-neo-3×FLAG vector to be expressed in human cells in a fusion with a 3×FLAG tag (35). HEK-293T cells were plated in 24-well plates (2 × 105 cells per well). One day later, cells were transfected with either cis- or trans-reporter constructs. For the cis-reporter assays, cells were cotransfected with either pISRE-Luc (0.3 μg/well; Stratagene), pNF-κB-Luc (0.3 μg/well; Stratagene), or pCRE-Luc (0.1 μg/well; Stratagene) together with the pCI-neo-3×FLAG vector (0.3 μg/well) (35), either empty or encoding CHIKV nsP2. Cells transfected with pISRE-Luc and pNF-κB-Luc were cultured for 8 h to allow nsP2 expression and then stimulated with recombinant IFN-β (1,000 IU/ml; Biosource) or tumor necrosis factor alpha (TNF-α; 5 ng/ml; R&D Systems), respectively. Expression of a CRE-luciferase reporter gene was induced by cotransfection with the pFC-PKA plasmid expressing the catalytic subunit of protein kinase A (PKA; 0.1 μg/well; Stratagene). For trans-reporter assays, cells were cotransfected with the pGal4-UAS-Luc reporter plasmid (0.3 μg/well), pM vector (0.3 μg/well) expressing Gal4-BD fused to Jun, Fos, or the L-MH2 domain of mothers against decapentaplegic homolog 3 (SMAD3), together with the pCI-neo-3×FLAG vector, either empty or expressing CHIKV nsP2 (0.3 μg/well). Transfections were performed with jetPRIME reagent following the manufacturer's recommendations (Polyplus Transfection). After 24 h of culture, firefly luciferase activity was determined using luciferase assay reagent II following the manufacturer's recommendations (Promega).
Synthesis and transfection of reporter mRNA molecules.
Plasmid DNA containing the gene for firefly luciferase under transcriptional control of the T7 RNA polymerase promoter (luciferase T7 control DNA; Promega) was digested with XmnI to be linearized and purified by using the Qiagen PCR purification kit. Then, both capped and polyadenylated mRNA molecules encoding firefly luciferase were synthesized from this plasmid using the T7 mMessage mMachine T7 Ultra kit from Ambion following the manufacturer's recommendations. Synthesized mRNA molecules were purified with the Qiagen RNeasy minikit. HEK-293T cells were seeded in 48-well plates with 40,000 cells per well. After 24 h, cells were transfected with 250 ng of pCI-neo-3×FLAG vector either empty or encoding CHIKV nsP2 using the jetPRIME reagent. After 24 h of culture to allow nsP2 expression, cells were transfected with 500 ng of luciferase-encoding mRNA molecules by using Lipofectamine 2000 (Invitrogen). Alternatively, cells were transfected with pRL-CMV or pRL-TK reporter vectors (250 ng/well; Promega) using the jetPRIME reagent. With these two vectors, expression of Renilla luciferase gene is induced by viral promoter sequences from cytomegalovirus or herpes simplex virus thymidine kinase, respectively. After 24 h of culture, firefly luciferase activity was determined using the Bright-Glo reagent (Promega) following the manufacturer's recommendations, whereas Renilla luciferase activity was determined using the Renilla-Glo reagent (Promega).
siRNA procedure.
Silencer Select small interfering RNAs (siRNA) were purchased from Invitrogen and transfected into HEK-293T cells following the manufacturer's recommendations. In each well of a 96-well plate, 3 pmol of siRNA was mixed with 20 μl of Opti-MEM (Gibco-Invitrogen) and 0.25 μl of Lipofectamine RNAiMAX transfection reagent (Invitrogen). This mix was incubated for 20 min at room temperature and supplemented with 80 μl of DMEM plus 10% FCS, without penicillin and streptomycin, and 15,000 HEK-293T cells. Cells were incubated for 48 h at 37°C and 5% CO2 and then infected with the recombinant CHIKV expressing the Renilla luciferase at a multiplicity of infection (MOI) of 0.2. After 24 h of culture, Renilla luciferase activity was determined using the Renilla-Glo reagent (Promega) following the manufacturer's recommendations.
Alternatively, siRNA treatment and culture was performed in a 48-well plates. In each well, 6 pmol of siRNA was mixed with 40 μl of Opti-MEM (Gibco-Invitrogen) and 0.5 μl of Lipofectamine RNAiMAX transfection reagent (Invitrogen). This siRNA mix was incubated for 20 min at room temperature and supplemented with 160 μl of DMEM plus 10% FCS, without penicillin and streptomycin, and 30,000 HEK-293T cells. Cells were incubated for 48 h at 37°C and 5% CO2 and then infected with the recombinant CHIKV expressing the Renilla luciferase at a MOI of 0.2. After 24 h, cultures were harvested, frozen, thawed, and clarified by centrifugation before titration to determine the TCID50 on HEK-293T.
In a parallel experiment, HEK-293T cells were seeded in 48-well plates and siRNA treated as described above. After 48 h of culture, cells were transfected with 60 ng/well of pGal4-UAS-Luc reporter plasmid (provided by Y. Jacob), 60 ng/well of pM vector (Clontech) expressing Gal4-BD fused to Jun, and 80 ng/well of pCI-neo-3×FLAG expressing CHIKV nsP2. Transfection was performed with the jetPRIME reagent following the manufacturer's recommendations. Alternatively, siRNA-treated cells were transfected with 60 ng/well of pISRE-Luc reporter plasmid (Stratagene) and 80 ng/well of pCI-neo-3×FLAG-expressing CHIKV nsP2 and stimulated at 8 h posttransfection with 500 IU/ml of recombinant IFN-β. After 24 h of culture, firefly luciferase activity was determined using the Bright-Glo reagent (Promega) following the manufacturer's recommendations.
Viability of siRNA-treated cells was determined by quantitation of ATP in culture wells in the CellTiter-Glo assay (Promega).
Western blot analysis.
Cells were washed in phosphate-buffered saline (PBS) and then resuspended in lysis buffer (0.5% Nonidet P-40, 20 mM Tris-HCl at pH 8, 120 mM NaCl, and 1 mM EDTA) supplemented with Complete protease inhibitor cocktail (Roche). Cell lysates were incubated on ice for 20 min and then clarified by centrifugation at 14,000 × g for 10 min. Protein extracts were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 4-to-12% NuPAGE bis-Tris gels with morpholinepropanesulfonic acid running buffer (Invitrogen) and transferred to a nitrocellulose membrane. Blocking was performed at room temperature for 1 h in 5% dry milk in TBST (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, and 0.1% Tween 20), and membranes were probed overnight at 4°C with primary antibodies diluted in 5% dry milk in TBST. Rabbit polyclonal anti-hnRNP-K antibody was from Abcam (ab52600) and diluted 1:500 times for blotting. Rabbit polyclonal anti-LCP1 antibody was from Abcam (ab83496) and diluted 1:250 times for blotting. Mouse polyclonal anti-TTC7B antibody was from Sigma-Aldrich (SAB1400809) and diluted 1:100 times for blotting. Anti-rabbit and anti-mouse horseradish peroxidase-conjugated antibodies were obtained from Amersham and diluted 1:7,500 times for blotting.
Immunofluorescence studies.
HeLa cells were infected at a high MOI of 50, which typically results in a synchronized infection in 100% of cells. Infection was performed with CHIKV-98, a virus strain generated from a full-length infectious cDNA clone provided by S. Higgs (50). Three hours postinfection, the inoculum was removed and fresh medium was added. Cells were fixed with 4% paraformaldehyde in PBS for 20 min at 15 h postinfection. Primary antibodies used include rabbit polyclonal antibodies to hnRNP-K (ab52600; Abcam) and UBQLN4 (HPA027920; Sigma-Aldrich). All secondary antibodies used were Alexa Fluor conjugates (Invitrogen). CHIKV nsP2 and capsid immunostaining was performed using mouse monoclonal antibodies developed in Marc Lecuit's laboratory. Cells were fixed with 4% paraformaldehyde in PBS for 20 min, then permeabilized for 30 min in 100× 0.2% Triton (Sigma-Aldrich), and blocked in 5% normal goat serum (Vector Laboratories). Cells were stained with the appropriate primary and secondary antibodies and finally counterstained with Hoechst stain (Vector Lab). Cells were observed with an AxioObserver microscope (Zeiss). Pictures and Z-stacks were obtained using the AxioVision 4.5 software.
RESULTS
Mapping cellular interactors of mature CHIKV proteins.
We recently developed a genome-scale collection of viral ORFs established in a versatile recombination-based cloning system that is dedicated to the characterization of viral protein functions (40). As part of this effort, all ORFs from a CHIKV isolate originating from Reunion Island were cloned in a donor vector of the Gateway system. All nine ORFs, encoding nsP1, nsP2, nsP3, nsP4, C, E3, E2, 6k, and E1, were transferred in a yeast expression vector to be used as bait in our HT-Y2H pipeline. In addition, seven ORF fragments encompassing different domains of nsP2 were cloned and expressed in yeast by using the same strategy. Alphavirus nsP2 is known for its multiple functions, and performing screens with isolated protein domains is well known for increasing HT-Y2H sensitivity, reducing the number of missed interactions. All these viral proteins (except nsP3, which showed an intrinsic transcriptional activity and was discarded) were used to screen four different human libraries, including three cDNA libraries from spleen, fetal brain, and HeLa cells and one normalized library of 12,000 full-length human ORFs.
A total of 966 positive yeast colonies were recovered from the different screens and analyzed by sequencing to identify corresponding interactions. To increase accuracy, interactions supported by only one or two positive yeast colonies were discarded, since they potentially corresponded to false-positive, nonreproducible events (26). In total, 30 distinct interactions that essentially involved not only nsP2 but also nsP4 and E3 were identified (Table 1). Surprisingly, we could not identify any cellular partner for nsP1, C, E2, 6k, or E1 CHIKV proteins. This could reflect some technical limitation of the Y2H system or the fact that these viral proteins are not highly connected to the proteome of infected cells. Unlikely interactions where viral and cellular proteins exhibited discordant localization profiles were also removed in order to generate a high-confidence data set that finally contained 21 interactions supported by nsP2 and only one for nsP4 (Table 1). Since most interactions were identified with nsP2 as bait, this suggests a prominent role of this viral protein in interactions with host cell components. As expected, the screening of multiple libraries and the use of various nsP2 fragments as bait significantly increased numbers of both identified interactions and positive yeast colonies supporting each interaction (Table 1). Among identified interactions, CHIKV nsP2 was found to bind heterogeneous nuclear hnRNP-K and vimentin (VIM), in agreement with previous reports showing that both cellular proteins copurified with SINV nsP2 (3, 7). We also confirmed that nsP2 interacts with 40S ribosomal protein S6 (RpS6), polyadenylate-binding protein (PABPC1), actin (ACTB), and elongation factor 1 (EEF1A1) (3, 37), but these interactions were only supported by single positive yeast colonies and thus discarded from the high-confidence interaction data set. All other identified interactions are novel and were not previously reported in the literature for any alphavirus.
Table 1.
Cellular interactors of CHIKV proteins identified by HT-Y2H
| Viral bait | Domaina | Ensembl gene ID | No. of positive yeast colonies retrieved from cDNA library |
Gene name | Gene description (HGNC symbol accession no.) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fetal brain | hORFeome | HeLa | HeLab | Spleen | Spleenb | Total | |||||
| nsP2 | 1–605 | ENSG00000005882 | 0 | 0 | 15 | 0 | 131 | 156 | 302 | PDK2 | Pyruvate dehydrogenase kinase, isozyme 2 (8810) |
| nsP2 | Full length | ENSG00000169155 | 11 | 154 | 0 | 0 | 1 | 0 | 166 | ZBTB43 | Zinc finger and BTB domain-containing 43 (17908) |
| nsP2c | 606–798 | ENSG00000030582 | 0 | 0 | 0 | 0 | 0 | 40 | 40 | GRN | Granulin (4601) |
| nsP2 | 469–798 | ENSG00000026025 | 4 | 1 | 17 | 0 | 13 | 2 | 37 | VIM | Vimentin (12692) |
| nsP2 | 469–798 | ENSG00000136436 | 0 | 35 | 0 | 0 | 0 | 2 | 37 | CALCOCO2 | Calcium-binding and coiled-coil domain 2 (29912) |
| nsP2 | 469–798 | ENSG00000100325 | 0 | 0 | 0 | 0 | 14 | 21 | 35 | ASCC2 | Activating signal cointegrator 1 complex subunit 2 (24103) |
| nsP2 | 606–798 | ENSG00000115380 | 0 | 0 | 0 | 0 | 0 | 33 | 33 | EFEMP1 | Epidermal growth factor-containing fibulin-like extracellular matrix protein 1 (3218) |
| nsP2 | 469–798 | ENSG00000183808 | 15 | 0 | 2 | 0 | 6 | 1 | 24 | RBM12B | RNA-binding motif protein 12B (32310) |
| nsP2 | 469–798 | ENSG00000165914 | 14 | 0 | 0 | 0 | 4 | 5 | 23 | TTC7B | Tetratricopeptide repeat domain 7B (19858) |
| nsP2 | Full length | ENSG00000137171 | 0 | 0 | 12 | 0 | 7 | 0 | 19 | KLC4 | Kinesin light chain 4 (21624) |
| nsP2 | 606–798 | ENSG00000165119 | 1 | 0 | 8 | 0 | 0 | 10 | 19 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K (5044) |
| nsP2 | 469–798 | ENSG00000160803 | 4 | 0 | 9 | 2 | 1 | 2 | 18 | UBQLN4 | Ubiquilin 4 (1237) |
| nsP2 | 606–798 | ENSG00000140092 | 0 | 0 | 0 | 0 | 0 | 15 | 15 | FBLN5 | Fibulin 5 (3602) |
| nsP2 | 606–798 | ENSG00000163743 | 0 | 0 | 0 | 0 | 0 | 14 | 14 | RCHY1 | Ring finger and CHY zinc finger domain-containing 1 (17479) |
| nsP2 | Full length | ENSG00000112081 | 0 | 0 | 10 | 0 | 0 | 0 | 10 | SRSF3 | Serine/arginine-rich splicing factor 3 (10785) |
| nsP2 | 469–798 | ENSG00000182944 | 0 | 0 | 3 | 0 | 5 | 1 | 9 | EWSR1 | Ewing sarcoma breakpoint region 1 (3508) |
| nsP2 | Full length | ENSG00000123124 | 0 | 0 | 6 | 0 | 2 | 0 | 8 | WWP1 | WW domain-containing E3 ubiquitin protein ligase 1 (17004) |
| nsP2 | 469–798 | ENSG00000204713 | 0 | 2 | 1 | 0 | 0 | 5 | 8 | TRIM27 | Tripartite motif-containing 27 (9975) |
| nsP2 | 606–798 | ENSG00000172638 | 0 | 0 | 0 | 0 | 0 | 7 | 7 | EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 (3219) |
| nsP2 | 469–798 | ENSG00000110799 | 0 | 0 | 0 | 0 | 1 | 5 | 6 | VWF | von Willebrand factor (12726) |
| nsP2 | 606–798 | ENSG00000178988 | 0 | 0 | 0 | 0 | 0 | 6 | 6 | MRFAP1L1 | Morf4 family-associated protein 1-like 1 (28796) |
| nsP2 | Full length | ENSG00000185811 | 0 | 6 | 0 | 0 | 0 | 0 | 6 | IKZF1 | IKAROS family zinc finger 1 (Ikaros) (13176) |
| nsP2 | Full length | ENSG00000013810 | 0 | 0 | 5 | 0 | 0 | 0 | 5 | TACC3 | Transforming, acidic coiled-coil-containing protein 3 (11524) |
| nsP2 | Full length | ENSG00000131095 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | GFAP | Glial fibrillary acidic protein (4235) |
| nsP2 | 469–798 | ENSG00000047410 | 0 | 0 | 0 | 0 | 1 | 2 | 3 | TPR | Translocated promoter region (to activated MET oncogene) (12017) |
| nsP2 | Full length | ENSG00000138180 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | CEP55 | Centrosomal protein 55 kDa (1161) |
| nsP2 | 606–798 | ENSG00000145147 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | SLIT2 | Slit homolog 2 (Drosophila) (11086) |
| nsP4 | Full length | ENSG00000136167 | 0 | 0 | 0 | 0 | 8 | 0 | 8 | LCP1 | Lymphocyte cytosolic protein 1 (L-plastin) (6528) |
| E3 | Full length | ENSG00000157110 | 0 | 8 | 0 | 0 | 0 | 0 | 8 | RBPMS | RNA-binding protein with multiple splicing (19097) |
| E3 | Full length | ENSG00000124788 | 4 | 0 | 0 | 0 | 0 | 0 | 4 | ATXN1 | Ataxin 1 (10548) |
Indicates for each viral protein the shortest fragment (amino acid residues) that was found to capture the corresponding cellular protein.
Indicates results obtained when the indicated libraries were screened with nsP2 fragments.
Shaded rows correspond to interactions with inconsistent localization profiles.
Identified virus-host interactions are mostly conserved among different alphaviruses.
To further evaluate nsP2 and nsP4 binding to cellular partners identified by HT-Y2H, we used a protein-binding assay based on luciferase complementation (8, 42). In this PCA, bait and prey proteins are fused to two inactive fragments of Gaussia princeps luciferase that restore a significant enzymatic activity in mammalian cells when brought in close proximity by interacting proteins. Results are expressed as NLRs (see Material and Methods). In a recent publication, we benchmarked this protein-binding assay by using both a positive reference set (PRS) of 143 protein pairs that are known to interact and a random reference set (RRS) of 100 a priori noninteracting protein pairs (8, 42). We observed a clear segregation of NLR values for these two reference sets. When selecting an NLR threshold of 3.5, more than 70% of PRS interactions scored positive, whereas only 2.5% of random protein pairs from the RRS showed NLR values above a 3.5 cutoff. These results demonstrated the high sensitivity and low background of this protein-binding assay. CHIK-host interaction data identified by HT-Y2H were thus evaluated for their overall quality using this system. Of the 22 interactions tested in the present study, 15 showed NLR values higher than 3.5 and thus were considered validated (Table 2). Since only 70% of PRS interactions scored positive in this assay when using 3.5 as a threshold for NLR values, the 7 interactions scoring negative with CHIKV nsP2 could correspond to false negatives of the PCA system rather than false positives of the HT-Y2H screen. Altogether, these results confirm that our HT-Y2H data set is of high quality.
Table 2.
Biochemical validation of nsP2 interactions from the Gaussia princeps luciferase-based protein complementation assay
| Ensembl ID | Gene name | Fold change in NLR compared to controla |
|||||
|---|---|---|---|---|---|---|---|
| CHIKV |
SFV |
SINV |
|||||
| Gluc1-nsP2 + Gluc2-host factor | Gluc2-nsP2 + Gluc1-host factor | Gluc1-nsP2 + Gluc2-host factor | Gluc2-nsP2 + Gluc1-host factor | Gluc1-nsP2 + Gluc2-host factor | Gluc2-nsP2 + Gluc1-host factor | ||
| ENSG00000169155 | ZBTB43 | 25.2 | 116.1 | ND | 15.2 | ND | 40.8 |
| ENSG00000112081 | SFRS3 | 21.8 | 35.3 | ND | 19.6 | ND | 105.8 |
| ENSG00000026025 | VIM | 6.1 | 34.4 | ND | 15.2 | ND | 46.7 |
| ENSG00000185811 | IKZF1 | 3.8 | 27.2 | ND | 6.4 | ND | 10.6 |
| ENSG00000178988 | MRF4P1L1 | 5.6 | 20.8 | ND | 6.8 | ND | 24.0 |
| ENSG00000005882 | PDK2 | 3.9 | 22.1 | ND | 6.6 | ND | 11.4 |
| ENSG00000123124 | WWP1 | 7.2 | 9.3 | ND | 4.2 | ND | 14.6 |
| ENSG00000131095 | GFAP | 1.0 | 9.3 | ND | 18.4 | ND | 101.7 |
| ENSG00000136436 | CALCOCO2 | 1.0 | 5.2 | ND | 1.9 | ND | 389.9 |
| ENSG00000138180 | CEP55 | 20.9 | 5.3 | 5.8 | ND | 4.9 | ND |
| ENSG00000137171 | KLC4 | 10.8 | 5.1 | 2.1 | ND | 7.6 | ND |
| ENSG00000165119 | HNRNPK | 0.4 | 4.9 | ND | 4.4 | ND | 28.7 |
| ENSG00000183808 | RBM12B | 0.2 | 4.5 | ND | 2.2 | ND | 6.3 |
| ENSG00000204713 | TRIM27 | 1.3 | 5.5 | ND | 3.1 | ND | 50.9 |
| ENSG00000165914 | TTC7B | 6.9 | 1.4 | 1.4 | ND | 3.6 | ND |
| ENSG00000013810 | TACC3 | 0.1 | 0.7 | ND | 1.2 | ND | 5.4 |
| ENSG00000182944 | EWSR1 | 0.6 | 0.2 | ND | 0.9 | ND | 2.8 |
| ENSG00000160803 | UBQLN4 | 0.1 | 0.4 | ND | 0.6 | ND | 5.1 |
| ENSG00000047410 | TPR | 0.3 | 0.3 | ND | 0.2 | ND | 2.3 |
| ENSG00000100325 | ASCC2 | 0.3 | 0.2 | ND | 0.1 | ND | 0.1 |
| ENSG00000163743 | RCHY1 | 0.1 | 0.3 | ND | 0.5 | ND | 1.6 |
| ENSG00000136167b | LCP1 | 1.5 | 2.6 | 3.0 | 4.7 | 3.0 | 5.0 |
Gluc1 and Gluc2 correspond to complementary fragments of Gaussia princeps luciferase encoded by SPICA-N1 and SPICA-N2, respectively. Values above the 3.5-fold cutoff are shown in bold. ND, not determined.
ENSG00000136167 (LCP1) was the only gene retrieved with nsP4 as bait. Thus, results shown are for nsP4 and not nsP2.
The same assay was used to determine if nsP2 and nsP4 proteins from SFV or SINV could also interact with the same cellular partners. In total, 10 interactions were validated with all three viruses, and 6 were validated with two out of three viruses. Interestingly, most interactions showed higher NLR values when performed with nsP2 from SINV rather than CHIKV or SFV. This could be explained by the fact that the protein-binding assay was performed with a mutant of SINV nsP2, which was previously found unable to translocate in nuclei of expressing cells and defective for shutting off cellular genes (40). Despite some technical limitations potentially related to the toxicity of nsP2 proteins, we can conclude from these experiments that several members of the Alphavirus genus share a large fraction of reported interactions.
Building an interaction map between alphaviral and host proteins.
Cellular interactors of CHIKV identified by HT-Y2H were analyzed in the context of interaction data already available in the literature for other Old World alphaviruses. Several groups have already identified cellular interactors of nsP2, nsP3, and nsP4 for SINV, in particular, by using recombinant viruses expressing tagged viral proteins that allowed the affinity purification and mass spectrometry-based identification of cellular partners (3, 7, 10, 11, 15, 22, 37). Interaction data reported in these manuscripts were gathered and reformatted using identical protein ID numbers (see Table S1 in the supplemental material) and collapsed with our HT-Y2H data to build a map of cellular factors targeted by nonstructural proteins of Old World alphaviruses (Fig. 1). Numerous interactors identified by co-affinity purification and mass spectrometry analysis are shared between nsP2, nsP3, and nsP4, suggesting that all these proteins are bound together into identical virus-induced complexes, such as viral factories. Nevertheless, it is impossible to determine from these data if reported virus-host interactions are direct or not. In contrast, HT-Y2H data obtained with CHIKV nsP2 and nsP4 as bait are most likely direct interactions, since they were detected in the yeast system, which is a highly heterologous system.
Fig 1.
Combined virus-host interaction data for CHIKV and other Old World alphaviruses. Virus-host interactions identified in this study by HT-Y2H using CHIKV proteins as bait (blue lines) were combined with interaction data obtained by literature mining for alphavirus-host interactions (black lines). Several groups have already identified cellular interactors for nsP2, nsP3, and nsP4 of SINV, in particular, by using recombinant viruses expressing tagged viral proteins that allow the co-affinity purification and mass spectrometry-based identification of cellular partners (3, 7, 10, 11, 15, 22, 37). Retrieved interactions were displayed using the Cytoscape 2.8 network analysis tool. Thick lines correspond to interactions reported by more than one laboratory. Statistical enrichment for specific GO terms was determined using the ClueGO plug-in and is displayed on the map by coloring of the corresponding nodes.
In this virus-host interaction map, we searched for possible functional enrichment by interrogating the Gene Ontology database using Cytoscape 2.8 and ClueGO plugin (2, 4, 47). We found this virus-host protein interaction map particularly enriched for components of translational machinery and RNA splicing factors, in agreement with previous reports showing that such cellular factors are hijacked by alphaviruses to replicate (7, 24, 37). In addition to hnRNP-K, we identified SRSF3 (splicing factor, arginine/serine-rich 3) as a novel component of the RNA splicing machinery targeted by CHIKV nsP2. We also found that nsP2 interacts with several cytoskeleton components, like VIM, TACC3 (transforming, acidic coiled-coil containing protein 3), CEP55 (centrosomal protein 55 kDa), and KLC4 (kinesin light chain 4), suggesting that nsP2 hijacks these proteins to achieve transport inside infected cells. However, most cellular proteins identified by HT-Y2H with nsP2 as bait do not cluster with these functional categories but correspond to regulators of gene transcription, like ASCC2 (activating signal cointegrator 1 complex subunit 2), TRIM27 (tripartite motif 27), MRFAP1L1 (Morf4 family-associated protein 1-like 1), EWSR1 (Ewing sarcoma breakpoint region 1), IKZF1 (IKAROS family zinc finger 1), and ZBTB43 (zinc finger and BTB domain containing 43), and host factors involved in protein degradation and/or autophagy, like CALCOCO2/NDP52 (calcium-binding and coiled-coil domain 2/nuclear dot protein 52 kDa), UBQLN4 (ubiquilin 4), RCHY1 (ring finger and CHY zinc finger domain-containing 1), and WWP1 (WW domain-containing E3 ubiquitin protein ligase 1). This suggests that CHIKV interacts through nsP2 with these cellular functions. We thus evaluated the role of these cellular proteins in the in vitro replication of CHIKV and in the cellular shutoff induced by nsP2.
hnRNP-K and UBQLN4 contribute to CHIKV replication in HEK-293T cells.
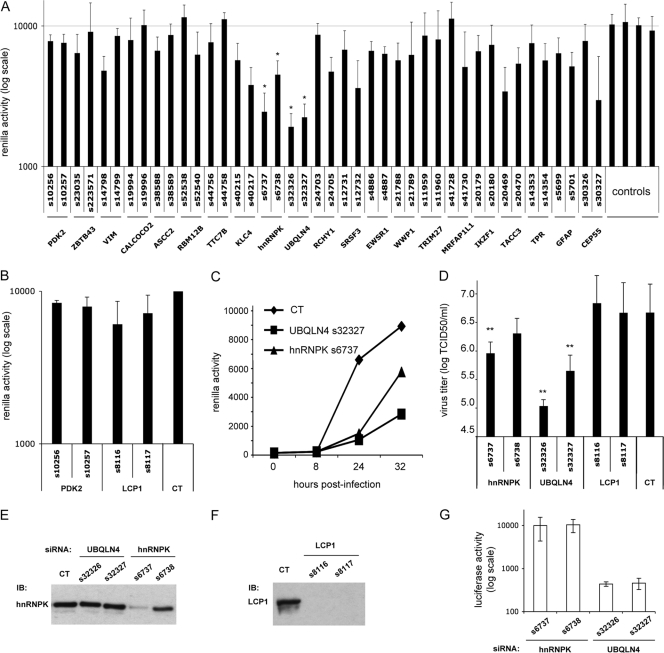
For all nsP2 and nsP4 interactors identified by HT-Y2H, we evaluated the impact of gene knockdown on CHIKV replication (Fig. 2A and B). HEK-293T cells were transfected with two distinct siRNAs for each target gene and cultivated for 48 h. We verified from a sister plate that cell viability, as determined by quantitation of cellular ATP, was virtually not affected by the siRNA treatment (cell viability of ≥70% [data not shown]). We also verified our silencing protocol by measuring expression levels of two targeted proteins: hnRNP-K and LCP1. As shown in Fig. 2E, the expression level of hnRNP-K protein was identical in cells either untreated or treated with off-target siRNA directed against UBQLN4. In contrast, specific siRNA either strongly (s6737) or partially (s6738) reduced hnRNP-K expression. Similarly, LCP1 protein was undetectable when treating cells with specific siRNA (Fig. 2F). Since we could not find an appropriate antibody for the Western blot analysis of UBQLN4 expression, we used a surrogate approach to validate corresponding siRNA molecules. Cells were transfected with siRNA molecules either off-target or specific for UBQLN4 sequence. Six hours later, cells were transfected again with an expression plasmid encoding firefly luciferase fused to UBQLN4 (Luc-UBQLN4). As shown in Fig. 2G, the efficacy of siRNA molecules targeting the UBQLN4 sequence was confirmed by measurement of luciferase activity. Although we could not confirm that each 1 of the 22 targeted proteins was properly suppressed by the siRNA we used, observations reported above at least validate our silencing protocol. Cells treated with siRNA were subsequently infected with a recombinant CHIKV expressing the Renilla luciferase (CHIKV-Luc; MOI, 0.2). At 24 h postinfection, Renilla luciferase activity was measured to evaluate CHIKV replication in siRNA-treated cells. Although several siRNAs somewhat affected CHIKV replication, a substantial inhibition was observed only with the two siRNAs targeting hnRNP-K or UBQLN4 (Fig. 2A). SINV replication was previously shown to be impaired when knocking down hnRNP-K expression (7), and we now report consistent results with CHIKV. Our observation that UBQLN4, a member of the ubiquilin family, participates in CHIKV replication is new and more surprising. We also analyzed CHIKV replication in a time course experiment, and similar inhibitions were observed when silencing hnRNP-K or UBQLN4 (Fig. 2C). We thus measured the production of infectious particles in HEK-293T cells treated or not with siRNA targeting either hnRNP-K, UBQLN4, or LCP1, which we used as a negative control. As shown in Fig. 2D, knocking down hnRNP-K reduced the production of infectious particles, but this was even more pronounced when targeting UBQLN4. This observation further supports the role of hnRNP-K and UBQLN4 in the replication cycle of CHIKV.
Fig 2.
Some cellular interactors of nsP2 contribute to CHIKV replication in vitro. (A) Human HEK-293T cells were transfected with targeted siRNA to knock down cellular partners of nsP2 and infected 48 h later with a recombinant CHIKV expressing Renilla luciferase as a reporter (CHIKV-Luc) at an MOI of 0.2. At 24 h postinfection, CHIKV infection was determined by measuring Renilla luciferase activity. Results presented are the means of 4 independent experiments, each performed in triplicate. *, P < 0.02 compared to all four control wells. (B) The same experiment was performed to target the nsP4 cellular partner LCP1. Results presented are the means of 2 independent experiments, each performed in triplicate. (C) The same experiment was performed without or with siRNA targeting hnRNPK or UBQLN4, but CHIKV replication, as assessed by Renilla luciferase activity, was measured at 0, 8, 24, and 32 h postinfection. (D) HEK-293T cells were transfected with siRNA against targeted genes and infected 48 h later with CHIKV-Luc at an MOI of 0.2. Cell cultures were harvested 24 h postinfection, and virus titers were determined based on the TCID50. **, P < 0.01. (E) hnRNP-K silencing was assessed by Western blot analysis of protein extracts from HEK-293T cells either untreated (CT) or treated with siRNA targeting hnRNP-K or UBQLN4, which was used as a control. (F) LCP1 silencing was assessed by Western blot analysis of protein extracts from HEK-293T cells either untreated (CT) or treated with siRNA targeting LCP1. (G) To determine the efficiency of siRNA targeting UBQLN4, cells were transfected with either specific siRNAs (s32326 or s32327) or off-target siRNAs (s6737 or s6738). Six hours later, cells were transfected with a pCi-neo plasmid carrying the luciferase enzyme fused to UBQLN4 (Luc-UBQLN4). After 24 h of culture, Luc-UBQLN4 expression was assessed based on luciferase activity.
hnRNP-K and UBQLN4 subcellular localization patterns are affected by CHIKV infection.
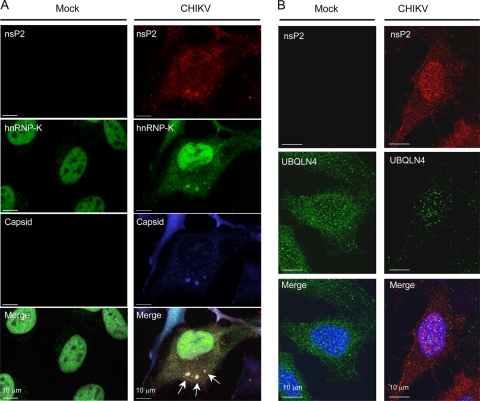
To determine consequences of CHIKV infection on the localization of hnRNP-K and UBQLN4, HeLa cells were infected with CHIKV and costained for nsP2 and hnRNP-K or UBQLN4. As shown in Fig. 3A, hnRNP-K localized in nuclei of mock-infected cells. Upon CHIKV infection, a fraction of hnRNP-K was found in the cytoplasm of infected cells. Furthermore, hnRNP-K colocalized with nsP2 into cytoplasmic aggregates that also stained positive for viral capsid protein C. Since they contain both nsP2 and C, these structures likely correspond to viral factories where CHIKV replicates. Therefore, our observations show that a fraction of hnRNP-K is recruited from the nucleus into these structures. In mock-infected cells, UBQLN4 exhibited a punctate pattern in both cytoplasmic and nuclear regions (Fig. 3B). Upon CHIKV infection, UBQLN4 staining essentially disappeared from cytoplasmic regions but was maintained in nuclei of infected cells. In addition, we could not detect a significant colocalization between nsP2 and UBQLN4. This could be explained if, upon nsP2 binding, UBQLN4 were degraded and eliminated from the cytoplasmic compartment, as previously shown in another experimental system that involved autophagy (44).
Fig 3.
hnRNP-K and UBQLN4 localization in CHIKV-infected cells. (A) HeLa cells were infected with CHIKV or treated with mock supernatant, incubated for 15 h, and then labeled for hnRNP-K, CHIKV nsP2, and capsid. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Arrows indicate protein aggregates where hnRNP-K, CHIKV nsP2, and capsid colocalized. Bars, 10 μm. (B) HeLa cells were infected with CHIKV or treated with mock supernatant, incubated for 15 h, and then labeled for detection of UBQLN4 and CHIKV nsP2. Cell nuclei were stained with DAPI. Bars, 10 μm.
CHIKV nsP2 is a potent inhibitor of gene expression.
Alphavirus nsP2 proteins have been identified as major virulence factors involved in the transcriptional and translational shutoff of cellular genes (6, 14, 16–19, 21, 33, 49). More recently, CHIKV nsP2 was also shown to block the host antiviral response by inhibiting the cellular response to type I interferons (IFN-α/β [17]) and NF-κB-dependent transactivation (40). However, there is a lack of data clearly demonstrating that CHIKV nsP2 induces a cellular shutoff. We thus tested the effect of CHIKV nsP2 on 8 different reporter genes, including both cis-reporter and trans-reporter systems. Most interestingly, we found that all reporter genes were strongly inhibited by nsP2 expression (Fig. 4A, B, C, and D). Activation of interferon-stimulated response elements (ISRE), NF-κB response elements, and cyclic AMP response elements (when induced by IFN-β, TNF-α, and PKA, respectively) was deeply affected by nsP2 expression. Similarly, gene expression induced by transcription factors like Jun, Fos, or SMAD3 was strongly inhibited by nsP2 expression. Reporter genes driven by viral promoter sequences from CMV or HSV thymidine kinase were also affected, provided that transfection of reporter plasmids was performed when nsP2 expression was already established (Fig. 4D). This could be explained by the induction of a transcriptional shutoff, a translational shutoff, or both. We thus determined if CHIKV nsP2 could block cellular mRNA translation. HEK-293T cells were transfected with a mammalian expression plasmid that was either empty or carried CHIKV nsP2. After 24 h of culture to allow nsP2 expression, cells were transfected with in vitro-transcribed and purified mRNA molecules encoding the luciferase protein. Most surprisingly, CHIKV nsP2 expression did not block but rather enhanced mRNA translation as assessed by luciferase activity measures (Fig. 4D). This demonstrated that the inhibitory effects of CHIKV nsP2 on a large panel of promoter sequences and transcription factors essentially rely on the induction of a transcriptional shutoff.
Fig 4.
CHIKV nsP2 alone is sufficient to induce gene expression shutoff. (A) Human HEK-293T cells were transfected with pISRE-Luc, a cis-reporter plasmid for the IFN-β signaling pathway, together with pCI-neo-3×FLAG vector expressing either wild-type or mutant nsP2 proteins. Eight hours later, cells were left unstimulated or stimulated with recombinant IFN-β at 1,000 IU/ml. (B) Human HEK-293T cells were cotransfected with the pGal4-UAS-Luc reporter plasmid and pM vector expressing Gal4-BD fused to Jun, together with the pCI-neo-3×FLAG vector expressing either wild-type or mutant nsP2 proteins. (C) Human HEK-293T cells were transfected with pNF-κB-Luc, a cis-reporter plasmid for the TNF-α signaling pathway. Eight hours later, cells were left unstimulated or stimulated with recombinant TNF-α at 5 ng/ml (upper left panel). Alternatively, cells were transfected with pCRE-Luc, a cis-reporter plasmid for the cyclic AMP signaling pathway, together with the pFC-PKA plasmid expressing the catalytic subunit of PKA to activate this pathway (upper right panel). For trans-reporter assays, cells were cotransfected with the pGal4-UAS-Luc reporter plasmid together with a pM vector expressing Gal4-BD fused to Fos or the SMAD3 L-MH2 domain (middle panels). In all these experiments, cells were cotransfected with the pCI-neo-3×FLAG vector, either empty or expressing CHIKV nsP2. (A to C) Luciferase expression was determined on cell lysates 24 h posttransfection. The data presented correspond to one representative experiment of three, each performed in duplicate. (D) Human HEK-293T cells were transfected with the pCI-neo-3×FLAG vector, either empty or expressing CHIKV nsP2. After 24 h of culture to allow nsP2 expression, cells were transfected with pRL-CMV or pRL-TK reporter plasmids, in which CMV or HSV thymidine kinase promoter sequences constitutively activate Renilla luciferase expression. Alternatively, cells were transfected with in vitro-synthesized mRNA molecules encoding firefly luciferase. Luciferase expression was determined 24 h later. Data presented correspond to one representative experiment of three, each performed in triplicate. (E) Diagram of CHIKV nsP2 domains based on data reported by Mayuri et al. (33) and InterPro annotation and the point mutations that were tested in panels A and B.
We then searched for mutations affecting the cellular shutoff induced by this viral protein. We thus generated mutants specifically targeting the NTPase (Fig. 4E, K192N) or the protease catalytic site of CHIKV nsP2 (Fig. 4E, C478A, H548A, and W549A) (20, 45). We also introduced mutations in conserved residues in the C-terminal region of CHIKV nsP2, which correspond to amino acid positions essential to the cellular shutoff induced by SINV nsP2 (Fig. 4E, R606A, P718G, and P718L) (18, 21, 33). In agreement with a previous report on SINV, the cellular shutoff induced by CHIKV nsP2 was not affected by mutations in the protease catalytic site (Fig. 4A and B) (18). We also found that CHIKV nsP2 mutated at the NTPase site mostly retained its ability to block gene expression. In contrast, its activity was significantly affected by mutations localized in the C-terminal region of the protein (R606A, P718G, and to some extent P718L), thus confirming the critical role of nsP2 methyltransferase-like domain in the cellular shutoff induced by alphaviruses (Fig. 4A and B).
TTC7B is an important cofactor of nsP2 in the induction of a cellular shutoff.
We tested if knocking down CHIKV nsP2 partners by siRNA altered the cellular shutoff induced by this viral protein. HEK-293T cells were transfected with two distinct siRNAs for each target gene and cultivated for 48 h. Then, cells were cotransfected with CHIKV nsP2 and the pISRE-Luc reporter plasmid, for which expression was induced by IFN-β, as illustrated in Fig. 4A. Alternatively, cells were cotransfected with CHIKV nsP2 and elements of the luciferase-based Jun-dependent reporter system presented in Fig. 4B. After 24 h, reporter gene expression was determined by measuring luciferase activity in culture wells. Although most siRNAs had virtually no or limited effects, a substantial alteration of nsP2-mediated shutoff was observed when the target was TTC7B (Fig. 5). Indeed, siRNA molecules directed against TTC7B alleviated by 9 to 16% the inhibition of Jun-mediated transcription and by 16 to 27% the inhibition of an ISRE-luciferase reporter gene by CHIKV nsP2. This is the only gene for which consistent results were obtained with both siRNAs and when considering the two reporter systems presented in Fig. 5. Furthermore, we found that TTC7B silencing was only observed with gene-specific siRNA, as assessed by Western blot analysis (Fig. 5, upper right panel). This demonstrated that TTC7B, a protein of unknown function exhibiting multiple tetratricopeptide repeats, is somehow involved in the induction of a cellular shutoff by CHIKV nsP2. Although the presented data suggest that other nsP2 partners, like ZBTB43, could be involved in the cellular shutoff induced by alphavirus nsP2, this will require further investigations.
Fig 5.
TTC7B contributes to nsP2-mediated gene expression shutoff. Human HEK-293T cells were transfected with targeted siRNA and transfected 48 h later with the pGal4-UAS-Luc reporter plasmid and pM vector carrying Gal4-BD fused to Jun for transcriptional activation (black bars), together with the pCI-neo-3×FLAG vector, either empty or expressing nsP2 to blunt gene expression. CT1 control corresponds to the pGal4-UAS-Luc reporter plasmid in the absence of Gal4-BD-Jun for transcriptional activation. CT2 and CT3 controls correspond to the pGal4-UAS-Luc reporter plasmid in the presence of Gal4-BD-Jun without or with CHIKV nsP2, respectively. Alternatively, siRNA-treated cells were transfected with the pISRE-Luc reporter plasmid together with pCI-neo-3×FLAG expressing or not CHIKV nsP2 and then stimulated 8 h later with recombinant IFN-β (open bars). The CT1 control corresponds to the pISRE-Luc reporter plasmid in the absence of IFN-β. CT2 and CT3 controls correspond to pISRE-Luc reporter plasmid in the presence of IFN-β without or with CHIKV nsP2, respectively. Results presented are the means of 2 independent experiments, each performed in triplicate. *, P < 0.05 compared to control wells. TTC7B silencing was assessed by Western blot analysis of protein extracts from HEK-293T cells either untreated (CT) or treated with siRNA targeting TTC7B or UBQLN4, which was used as a control.
DISCUSSION
From previous reports, a list of cellular proteins associated with the viral replication complex of alphaviruses could be established, providing a framework to investigate alphavirus relationships with host cell components. However, all these reports were based on the expression of tagged viral proteins in infected cells, and the identification of associated cellular proteins by affinity purification and mass spectrometry analysis. Cellular proteins that are stably trapped inside virus-induced protein complexes can be efficiently identified with this kind of approach, but more transient protein interactions are frequently lost (1). In addition, and despite its undisputable interest, this approach doesn't discriminate between direct and indirect protein-protein interactions. Finally, it is well known that using only one method is insufficient to map all the binding partners of a given protein, and several techniques must be combined to increase coverage and accuracy (34). In this report, we used for the first time an HT-Y2H approach to characterize interactions between CHIKV and human proteins. This led to the identification of 22 cellular proteins interacting directly with either nsP2 or nsP4, most of these interactions being novel with no previous reports in the literature. From this list of interactors, we established that both hnRNP-K and UBQLN4 contribute to CHIKV replication and that TTC7B participates in the shutoff mediated by CHIKV nsP2. Altogether, our results provide a more complete picture of virus-host interactions that can be developed by Old World alphaviruses.
In our HT-Y2H assay, nsP2 was much more prone to interact with host cell factors than other CHIKV mature proteins. This could reflect some technical limitation of the Y2H system. For example, conventional Y2H is not a suitable system to efficiently identify binding partners of extracellular or membrane proteins like E1 or E2, because interactions need to occur in yeast nuclei. Nevertheless, it is somehow expected that nsP2 interacts with more host factors, considering its dual role both as a key component of viral replication complex and as an important virulence factor inhibiting the host antiviral response. In contrast, structural proteins do not necessarily need to interact with multiple cellular proteins to properly play their role and self-assemble into viral particles. Besides, some CHIKV proteins like nsP1 have evolved to preferentially interact with host lipids rather than cellular proteins, and such interactions cannot be detected by Y2H. This argues for a critical role of nsP2 in mediating CHIKV interactions with host cell proteins. Unfortunately, we could not screen CHIKV nsP3, since it exhibited a strong intrinsic transcriptional activity when fused to Gal4-BD. The acidic and intrinsically disordered C-terminal region of CHIKV nsP3 (amino acids 321 to 523) was found responsible for this property (data not shown). Consequently, the N-terminal region of nsP3 could be used as bait in the Y2H system. However, numerous nsP3 interactors would be missed, since intrinsically disordered regions of proteins are usually prone to interact with multiple partners (12). Although CHIKV nsP3 is likely to interact with several cellular partners, as suggested by previous reports on SINV, this should be investigated with other methods than Y2H in order to capture most of its partners.
Interestingly, we showed that cellular hnRNP-K interacts with nsP2, colocalizes with nsP2 and capsid, and contributes to CHIKV replication, as reported for SINV (3, 7). In addition, CHIKV nsP2 was found to bind SRSF3, another RNA splicing machinery component, in agreement with previous reports showing that SINV nsP2 targets numerous factors involved in this cellular process. Our results provide further evidence that alphaviruses subvert host cell factors involved in RNA splicing to the benefit of their own replication. Most interestingly, nsP2 was also found to bind cellular proteins involved in protein degradation and/or autophagy. Whereas RCHY1 and WWP1 are E3 ubiquitin ligases that can target proteins for degradation, UBQLN4 and CALCOCO2/NDP52 display specific domains to interact simultaneously with ubiquitinated cargo and either autophagy machinery or proteasome subunits. Members of the UBQLN family exhibit a ubiquitin-like domain (UBL) and a ubiquitin-associated domain (UBA) at their N and C termini, respectively. They function as regulators of the ubiquitination complex and protein degradation machinery. More recently, UBQLN1 has also been involved in autophagy, mediating protein targeting to autophagosomes by its interaction with LC3 (44). Thus, the nsP2 interaction with UBQLN4 could be either involved in the negative regulation of antiviral mechanisms through the degradation of cellular factors like p53 or participate in the hijacking of cellular process like autophagy to the benefit of CHIKV replication, as previously shown for several positive-strand RNA viruses, including CHIKV (28, 31). Indeed, although autophagy was recently shown to protect mice from SINV infection of the central nervous system (38), this cellular process is hijacked by numerous positive-strand RNA viruses to produce multimembrane structures involved in viral replication and assembly (31). While the manuscript was under revision, P. Krejbich-Trotot et al. reported that CHIKV triggers autophagy and that this promotes viral replication (28). Moreover, our collaborators recently showed that the nsP2 interaction with CALCOCO2/NDP52, which is described in here, is involved in the formation of viral factories and participates in the hijacking of autophagy components by CHIKV (D. Judith, submitted for publication). Besides autophagy, members of the ubiquilin family, like UBQLN1, have been involved in the formation of high-molecular-mass complexes and their delivery to aggresomes or autophagosomes for degradation (25, 44). Thus, the nsP2 interaction with UBQLN4 could somehow contribute to the subversion of aggresome structures and/or autophagosomes by the CHIKV replication complex (31, 56). Alternatively, the UBQLN4 interaction with nsP2 could be involved in the inhibition of host cellular defenses by CHIKV, as suggested by a recent report showing that UBQLN1 inhibits Toll-like receptor 3 (TLR3) signaling (5). A small hydrophobic protein encoded by Kaposi's sarcoma-associated virus (HHV8) named K7 has also been shown to interact with UBQLN1 (13). This interaction was found to promote the degradation of both p53 and the NF-κB inhibitor IκB and to protect cells from apoptosis. More recently, another small hydrophobic protein from mumps virus named SH was also reported to interact with both UBQLN1 and UBQLN4 (57). This interaction relocalized SH to punctate structures positive for proteosomal markers, but the functional role of this interaction remains to be determined. In this report, we showed that silencing of UBQLN4 expression in human cells inhibits CHIKV replication and that UBQLN4 disappears from the cytoplasmic compartment of CHIKV-infected cells. Altogether, these findings suggest that UBQLN4 is hijacked by nsP2 to the benefit of viral replication, and this could lead to UBQLN4 consumption and degradation. This could possibly involve autophagy, since both CHIKV and UBQLN4 interact with this pathway. However, further studies will be required to determine the exact role and contribution of the nsP2 interaction with this cellular factor.
Numerous nsP2 partners identified in the HT-Y2H assay are cellular factors involved in the control of gene expression, like ASCC2, TRIM27, MRFAP1L1, EWSR1, IKZF1, and ZBTB43. This is reminiscent of nsP2 function as a virulence factor for Old World alphaviruses (6, 14, 16–19, 21, 33, 49). Indeed, SINV, SFV, and more recently CHIKV (55) have been shown to induce a transcriptional and translational shutoff that blunts the host antiviral response and finally kills infected cells. Mutations within the methyltransferase-like domain of nsP2 abrogate this activity. However, all these experiments measuring nsP2-induced transcriptional and translational shutoff have been performed with either infectious viruses or viral replicons expressing nsP2. Consequently, it remains unclear if nsP2 alone induces both a transcriptional and a translational shutoff or if it only induces a transcriptional shutoff that favors the replication of viruses or viral replicons and thus mediates a translational shutoff indirectly through the activation of PKR or other pathways. Here, nsP2 from CHIKV was shown to inhibit the expression of various reporter genes when expressed in human cells, and it concomitantly enhanced the translation of transfected mRNA molecules. This demonstrated that when expressed alone, CHIKV nsP2 both induces a transcriptional shutoff and promotes mRNA translation. In a recent report, it was shown that CHIKV nsP2 inhibits type I/II signaling in human cells by blocking STAT1 (signal transducer and activator of transcription 1) phosphorylation and/or nuclear translocation, and this was clearly unrelated to any kind of translational shutoff (17). We fully agree with the results of Fros et al., but those authors concluded that type I/II signaling is specifically targeted by CHIKV nsP2. In contrast, our data show that other signaling pathways are also deeply impaired by nsP2 expression, thus demonstrating that the effects of this virulence factor on gene transcription extend far beyond the type I/II signaling pathway. Furthermore, our data show that CHIKV nsP2 did not induce a translational shutoff, but rather enhanced mRNA translation when expressed alone. Interactions between nsP2 and ribosomal factors or RNA-binding proteins like hnRNP-K could be involved in this process. Whether nsP2 from SINV and SFV exhibit a similar property when expressed alone in human cells will require further investigations. In conclusion, this dual activity of CHIKV nsP2 is probably essential to the production of a large number of progeny in vivo, since inhibiting cellular transcription could block antiviral mechanisms, while promoting translation could increase viral protein synthesis.
Deciphering mechanisms involved in nsP2-mediated shutoff is a most challenging puzzle to be solved. We found that in cells where TTC7B expression has been silenced, nsP2-mediated inhibition of Jun and ISRE-dependent reporter genes is less pronounced. This suggests that the nsP2 interaction with TTC7B is somehow involved in the cellular shutoff induced by this viral protein. Because TTC7B knockdown had significant but limited effects on nsP2-mediated shutoff, we propose that TTC7B is only one component of a larger heterocomplex recruited by CHIKV nsP2 to blunt transcription. Unfortunately, TTC7B is a protein of unknown function that has only been characterized by the presence of tetratricopeptide peptide repeats in its sequence. Altogether, this provides a first lead in better understanding the mode of action of alphavirus nsP2 and its critical role as a virulence factor.
Besides hnRNP-K, UBQLN4, and TTC7B, knocking down nsP2 or nsP4 partners identified by HT-Y2H only had limited or no effects on CHIKV replication or nsP2-induced shutoff in tested functional assays. However, biological and technical aspects could account for this observation. First of all, we could not verify if all tested siRNAs were truly active and efficiently blocked the expression of targeted proteins. Furthermore, our assay may not have been suitable or sensitive enough to detect the impact of gene silencing on the CHIKV replication cycle. In addition, some level of gene redundancy exists in host cells. Finally, several identified interactions could play a role only in specific cell types and/or under specific environmental constraints. Future studies will aim at deciphering the role of all other interactions identified by HT-Y2H in the CHIKV replication cycle.
Supplementary Material
ACKNOWLEDGMENTS
M.B. was supported by grants from Région Ile-de-France (DIM-Malinf). This work was supported by the “Programme Interdisciplinaire—CNRS—Maladies Infectieuses Emergentes” and the Institut Pasteur “Programmes Traversaux de Recherche.”
We thank Marc Le Breton for his technical input with the HT-Y2H screens.
Footnotes
Published ahead of print 18 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Aloy P, Russell RB. 2002. The third dimension for protein interactions and complexes. Trends Biochem. Sci. 27:633–638 [DOI] [PubMed] [Google Scholar]
- 2. Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J. Virol. 81:5046–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bindea G, et al. 2009. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biswas N, et al. 2011. The ubiquitin-like protein PLIC-1 or ubiquilin 1 inhibits TLR3-Trif signaling. PLoS One 6:e21153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breakwell L, et al. 2007. Semliki Forest virus nonstructural protein 2 is involved in suppression of the type I interferon response. J. Virol. 81:8677–8684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burnham AJ, Gong L, Hardy RW. 2007. Heterogeneous nuclear ribonuclear protein K interacts with Sindbis virus nonstructural proteins and viral subgenomic mRNA. Virology 367:212–221 [DOI] [PubMed] [Google Scholar]
- 8. Cassonnet P, et al. 2011. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat. Methods 8:990–992 [DOI] [PubMed] [Google Scholar]
- 9. Charrel RN, de Lamballerie X, Raoult D. 2007. Chikungunya outbreaks: the globalization of vector-borne diseases. N. Engl. J. Med. 356:769–771 [DOI] [PubMed] [Google Scholar]
- 10. Cristea IM, et al. 2006. Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem. 281:30269–30278 [DOI] [PubMed] [Google Scholar]
- 11. Cristea IM, et al. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol. 84:6720–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunker AK, et al. 2008. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics 9(Suppl. 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng P, et al. 2004. Kaposi's sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domain-containing protein to promote protein degradation. Mol. Cell. Biol. 24:3938–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frolov I, Garmashova N, Atasheva S, Frolova EI. 2009. Random insertion mutagenesis of Sindbis virus nonstructural protein 2 and selection of variants incapable of downregulating cellular transcription. J. Virol. 83:9031–9044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frolova E, et al. 2006. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 80:4122–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frolova EI, et al. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fros JJ, et al. 2010. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J. Virol. 84:10877–10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garmashova N, Gorchakov R, Frolova E, Frolov I. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garmashova N, et al. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 81:2472–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gomez de Cedron M, Ehsani N, Mikkola ML, Garcia JA, Kaariainen L. 1999. RNA helicase activity of Semliki Forest virus replicase protein NSP2. FEBS Lett. 448:19–22 [DOI] [PubMed] [Google Scholar]
- 21. Gorchakov R, Frolova E, Frolov I. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397–9409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gorchakov R, Garmashova N, Frolova E, Frolov I. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 82:10088–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Griffin DE. 2007. Alphaviruses, p. 1023–1067 In Howley PM, Knipe DH. (ed.), Fields virology, 5th ed, vol. 1 Wolters Kluwer, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 24. Gui H, Lu CW, Adams S, Stollar V, Li ML. 2010. hnRNP A1 interacts with the genomic and subgenomic RNA promoters of Sindbis virus and is required for the synthesis of G and SG RNA. J. Biomed. Sci. 17:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haapasalo A, et al. 2010. Emerging role of Alzheimer's disease-associated ubiquilin-1 in protein aggregation. Biochem. Soc. Trans. 38:150–155 [DOI] [PubMed] [Google Scholar]
- 26. Ito T, et al. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 98:4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jose J, Snyder JE, Kuhn RJ. 2009. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 4:837–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krejbich-Trotot P, et al. 2011. Chikungunya triggers an autophagic process which promotes viral replication. Virol. J. 8:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhn RJ. 2007. Togaviridae: the viruses and their replication, p. 1001–1022 In Howley PM, Knipe DM. (ed.), Fields virology, 5th ed, vol. 1 Wolters Kluwer, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 30. Lamesch P, et al. 2007. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89:307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin LT, Dawson PW, Richardson CD. 2010. Viral interactions with macroautophagy: a double-edged sword. Virology 402:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mavalankar D, Shastri P, Raman P. 2007. Chikungunya epidemic in India: a major public-health disaster. Lancet Infect. Dis. 7:306–307 [DOI] [PubMed] [Google Scholar]
- 33. Mayuri Geders TW, Smith JL, Kuhn RJ. 2008. Role for conserved residues of Sindbis virus nonstructural protein 2 methyltransferase-like domain in regulation of minus-strand synthesis and development of cytopathic infection. J. Virol. 82:7284–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mendez-Rios J, Uetz P. 2010. Global approaches to study protein-protein interactions among viruses and hosts. Future Microbiol. 5:289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mendoza JA, Jacob Y, Cassonnet P, Favre M. 2006. Human papillomavirus type 5 E6 oncoprotein represses the transforming growth factor beta signaling pathway by binding to SMAD3. J. Virol. 80:12420–12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mohan A. 2006. Chikungunya fever: clinical manifestations and management. Indian J. Med. Res. 124:471–474 [PubMed] [Google Scholar]
- 37. Montgomery SA, Berglund P, Beard CW, Johnston RE. 2006. Ribosomal protein S6 associates with alphavirus nonstructural protein 2 and mediates expression from alphavirus messages. J. Virol. 80:7729–7739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Orvedahl A, et al. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pellet J, et al. 2009. pISTil: a pipeline for yeast two-hybrid Interaction sequence tags identification and analysis. BMC Res. Notes 2:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pellet J, et al. 2010. ViralORFeome: an integrated database to generate a versatile collection of viral ORFs. Nucleic Acids Res. 38:D371–D378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. 2007. Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 7:319–327 [DOI] [PubMed] [Google Scholar]
- 42. Remy I, Michnick SW. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3:977–979 [DOI] [PubMed] [Google Scholar]
- 43. Rezza G, et al. 2007. Infection with Chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370:1840–1846 [DOI] [PubMed] [Google Scholar]
- 44. Rothenberg C, et al. 2010. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum. Mol. Genet. 19:3219–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Russo AT, White MA, Watowich SJ. 2006. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 14:1449–1458 [DOI] [PubMed] [Google Scholar]
- 46. Salonen A, et al. 2003. Properly folded nonstructural polyprotein directs the Semliki Forest virus replication complex to the endosomal compartment. J. Virol. 77:1691–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. 2011. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tamberg N, et al. 2007. Insertion of EGFP into the replicase gene of Semliki Forest virus results in a novel, genetically stable marker virus. J. Gen. Virol. 88:1225–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tamm K, Merits A, Sarand I. 2008. Mutations in the nuclear localization signal of nsP2 influencing RNA synthesis, protein expression and cytotoxicity of Semliki Forest virus. J. Gen. Virol. 89:676–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vanlandingham DL, et al. 2005. Development and characterization of a double subgenomic Chikungunya virus infectious clone to express heterologous genes in Aedes aegypti mosquitoes. Insect Biochem. Mol. Biol. 35:1162–1170 [DOI] [PubMed] [Google Scholar]
- 51. Vidalain PO, Boxem M, Ge H, Li S, Vidal M. 2004. Increasing specificity in high-throughput yeast two-hybrid experiments. Methods 32:363–370 [DOI] [PubMed] [Google Scholar]
- 52. Voss JE, et al. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712 [DOI] [PubMed] [Google Scholar]
- 53. Walhout AJ, Vidal M. 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24:297–306 [DOI] [PubMed] [Google Scholar]
- 54. Weaver SC, Frolov IV. 2005. Togaviruses, p. 1010–1024 In Mahy B, ter Meulen V. (ed.), Topley and Wilson's microbiology and microbial infections: virology, 10th ed, vol. 2 Hodder Arnold, London, United Kingdom [Google Scholar]
- 55. White LK, et al. 2011. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J. Virol. 85:606–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wileman T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61:149–167 [DOI] [PubMed] [Google Scholar]
- 57. Woznik M, et al. 2010. Mumps virus small hydrophobic protein targets ataxin-1 ubiquitin-like interacting protein (ubiquilin 4). J. Gen. Virol. 91:2773–2781 [DOI] [PubMed] [Google Scholar]
- 58. Zusinaite E, et al. 2007. Mutations at the palmitoylation site of non-structural protein nsP1 of Semliki Forest virus attenuate virus replication and cause accumulation of compensatory mutations. J. Gen. Virol. 88:1977–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.