Abstract
Viruses in the genus Bracovirus (BV) (Polydnaviridae) are symbionts of parasitoid wasps that specifically replicate in the ovaries of females. Recent analysis of expressed sequence tags from two wasp species, Cotesia congregata and Chelonus inanitus, identified transcripts related to 24 different nudivirus genes. These results together with other data strongly indicate that BVs evolved from a nudivirus ancestor. However, it remains unclear whether BV-carrying wasps contain other nudivirus-like genes and what types of wasp genes may also be required for BV replication. Microplitis demolitor carries Microplitis demolitor bracovirus (MdBV). Here we characterized MdBV replication and performed massively parallel sequencing of M. demolitor ovary transcripts. Our results indicated that MdBV replication begins in stage 2 pupae and continues in adults. Analysis of prereplication- and active-replication-stage ovary RNAs yielded 22 Gb of sequence that assembled into 66,425 transcripts. This breadth of sampling indicated that a large percentage of genes in the M. demolitor genome were sequenced. A total of 41 nudivirus-like transcripts were identified, of which a majority were highly expressed during MdBV replication. Our results also identified a suite of wasp genes that were highly expressed during MdBV replication. Among these products were several transcripts with conserved roles in regulating locus-specific DNA amplification by eukaryotes. Overall, our data set together with prior results likely identify the majority of nudivirus-related genes that are transcriptionally functional during BV replication. Our results also suggest that amplification of proviral DNAs for packaging into BV virions may depend upon the replication machinery of wasps.
INTRODUCTION
Viruses in the genus Bracovirus (BV) (Polydnaviridae) are distinguished by their large, segmented double-stranded DNA (dsDNA) genomes and symbiotic association with parasitoid wasps (Hymenoptera) in the family Braconidae (summarized in references 10, 44, and 55). Each species of wasp carries a genetically unique BV which is transmitted only vertically to offspring as an integrated provirus. Replication occurs only in pupal- and adult-stage females in a region of the ovary called the calyx. Virions containing multiple circular dsDNAs accumulate to high density in the lumen of the calyx to form “calyx fluid.” Adult wasps then inject calyx fluid along with one or more eggs into each host insect they parasitize. BVs do not replicate in host insects, but infection is vital for wasp survival because viral gene products disable immune defenses and/or cause other alterations required for successful development of offspring (44).
The encapsidated genomes of 5 BVs have been fully sequenced (13, 15, 19, 52), and in all cases many genes of likely wasp origin have been identified that encode products involved in altering host physiology (44). In contrast, no homologs of any viral genes with predicted roles in genome replication, transcription, or virion formation are present. These findings fully explain why BVs do not replicate in the hosts that wasps parasitize. The lack of virus replication genes, however, raised the question of whether BVs evolved from a viral ancestor or are structures wasps evolved that resemble viruses but are not (10, 19, 44). Evidence strongly favoring a viral origin for BVs was recently generated by producing pupal ovary cDNA libraries from the BV-carrying wasps Cotesia congregata and Chelonus inanitus and Sanger sequencing approximately 5,000 expressed sequence tags (ESTs) (9). Among these ESTs were transcripts with homology to 24 known genes from nudiviruses; a genus of large dsDNA viruses that is closely related to the Baculoviridae and which often establishes persistent infections in arthropods (26). That several of the nudivirus-like transcripts identified from C. inanitus encode protein products present in C. inanitus bracovirus (CiBV) virions (54) is also consistent with earlier microscopy studies which noted that BV virions morphologically resemble nudiviruses (43). Since all BV-carrying wasps shared a common ancestor 100 million years ago (33, 55), these data compellingly argue that BVs evolved from an ancient nudivirus, which was associated with the ancestor of the ca. 18,000 wasp species that carry BVs today. They also show that several nudivirus-like genes required for virion formation are part of a proviral genome but are not contained within the episomal DNAs that are packaged into virions.
Since sequence data are limited to a relatively small pool of ESTs from only two wasp species, what remains unclear is whether other nudivirus-like genes exist or whether the genes identified to date are present in all BV-carrying wasps. The absence of any C. congregata or C. inanitus ESTs with homology to genes essential for replication of baculovirus/nudivirus DNA (39) is also notable, because if no such genes exist it would argue that BVs have evolved to rely on the replication machinery of wasps to amplify the DNA that is packaged into virions. To address these questions, we performed massively parallel sequencing of ovary transcripts from Microplitis demolitor, which carries MdBV. Our results likely identify the majority of nudivirus-related transcripts in M. demolitor as well as all wasp genes that are highly expressed during MdBV replication. As such, these data represent the first deep sequencing analysis of both viral and wasp genes during replication of a BV.
MATERIALS AND METHODS
Insects and staging of wasp pupae.
Like most BV-carrying wasps, M. demolitor parasitizes the larval stage of certain species of Lepidoptera, including the moth Pseudoplusia includens (45). Both species were reared as previously described at 27°C with a 16-h-light:8-h-dark photoperiod (47). The different developmental stages of M. demolitor can be precisely monitored under these conditions using time and morphological characteristics (45). In brief, M. demolitor takes a total of 11 days to develop from an egg to an adult. After parasitism of a host larva, the M. demolitor egg hatches and the wasp larva feeds for 6 days. The last-stage wasp larva then emerges from the host's body on the morning of day 7, spins a silken cocoon around itself in 4 h, and pupates between 9 and 12 h postemergence. Newly pupated wasps (stage 1 pupae) are entirely white with light red compound eyes. On day 8, pupae enter stage 2, which is distinguished by wasps having dark red eyes, 3 or 4 black stripes on the dorsal side of the thorax, and a white abdomen. On day 9, stage 3 pupae have an entirely black thorax, black eyes, and a partially darkened abdomen, while on day 10, stage 4 pupae have a fully black thorax and abdomen with brown accents present on each leg. On day 11, pupae emerge from the cocoon as day 1 adults, which in the current study were thereafter fed ad libitum 10% sucrose in water and maintained at 18°C in continuous dark. Females are easily distinguished from males as pupae and adults, because they have much shorter antennae.
PCR-based replication assays and transmission electron microscopy (TEM).
We used quantitative PCR (qPCR) and previously developed methods (6) to determine the abundance of episomal MdBV segment B, integrated (proviral) segment B, and rejoined flanking DNA following segment B excision in ovaries from different wasp stages (see Fig. S1 in the supplemental material). Briefly, ovaries from stage 1 to 4 female pupae and adult female wasps were prepared by homogenizing them in 1× DNase buffer (0.5 mM CaCl2, 2.5 mM MgCl2, 10 mM Tris-HCl, pH 7.5). Following filtration through a 0.45-μm filter, 1 μl of Ambion Turbo DNase was added to some samples for 1 h at 37°C, while others were not DNase treated. After the addition of EDTA (10 mM) to inactivate the DNase, 25 μg of proteinase K (Roche) and 2% sarcosyl were added to samples, followed by incubation at 62°C for 1 h and by phenol-chloroform extraction and ethanol precipitation in the presence of 0.3 M sodium acetate, pH 5.2. DNA pellets were resuspended in 30 μl of H2O and diluted 1:50 with water for use as the template.
After quantifying DNA concentrations, PCRs were run using a Bio-Rad thermocycler and 25-μl reaction volumes containing DNA (10 ng) from day 5 adults as the template, DNA primers (6.25 pmol), and 1.25 units of Hotmaster Taq polymerase (5 Prime) under the following cycling conditions: initial denaturation at 94° for 2 min, followed by 35 cycles of denaturation at 94°C for 20 s, annealing for 20 s at the specified temperature for the primers used, and extension at 65°C for 30 s with a final extension at 65°C for 7 min. Primer sequences can be found in Table S1 in the supplemental material. Products were cloned into pSC-A-amp/kan (Stratagene) and propagated in Escherichia coli. Following overnight culture, plasmid DNA was then isolated using the Fermentas GeneJet plasmid miniprep kit and quantified, and their identity confirmed by sequencing. As a control, primers were also designed for M. demolitor elongation factor 1α (EF1α) (see Table S1 in the supplemental material), which was similarly amplified and cloned, and its corresponding plasmid purified. Each of these plasmids was then used to make absolute standard curves to determine the abundance of integrated genomic DNA B, episomal genomic B, the empty B locus, and EF1α in ovary DNA. Ten-microliter qPCR mixtures containing 1 μl of DNA template (wasp DNA or serially diluted amounts [102 to 107 copies] of each plasmid standard) and specific primers were then run using a Rotor-Gene 3000 cycler (Corbett) and previously described reaction conditions (6). Melting curves of products were checked for amplification specificity, and threshold cycle (CT) values for each sample were fit to the standard curve generated from the plasmid DNA template dilutions. Per-ovary copy numbers were calculated by multiplying the qPCR estimate of copy number by the dilution factor and elution volume and dividing by the number of ovaries used in each sample. Two or 3 independently acquired biological replicates were performed for each wasp developmental stage with each sample internally replicated 4 times. For transmission electron microscopy (TEM), ovaries were collected, processed, and examined as previously described (46).
RNA isolation, library preparation, and transcriptome sequencing.
Total RNA was isolated from 6 ovary samples collected from M. demolitor stage 1 pupae and day 1 adult females. Each ovary sample consisted of 15 to 20 ovary pairs with total RNA extracted using the Roche high pure RNA isolation kit followed by a second DNase treatment using Ambion Turbo DNA-free reagents. Sequencing libraries were prepared by the University of Georgia Genomics Facility using the Illumina TruSeq DNA sample preparation kit and the standard low-throughput protocol. RNA was fragmented thermally by ramping up to 94°C followed by immediate cooling to 4°C. Each biological replicate was labeled with an individual indexed adapter. Libraries were then pooled to equal concentrations using quantitative PCR data and size selected by gel extraction for an average library size of 441 bp. Libraries were clustered on one lane at 6 pM followed by 100 cycles of paired-end sequencing on the Illumina HiSeq system housed at the HudsonAlpha Institute for Biotechnology (Huntsville, AL).
Sequence read processing, assembly, and analyses.
Sequence data were demultiplexed using CASAVA v1.8 software (Illumina). Reads were filtered for quality by removing those that had <90% of bases with >Q10 quality scores (based upon the Illumina TruSeq quality scoring system). High-quality reads were assembled using velvet v1.1.04 and Oases v0.2.21 (60), with k-mer length 51, and parameters cov_cutoff = 3, min_trans_length = 200, ins_length = 441. Sequence reads were mapped to transcripts using the Burrows-Wheeler Aligner bwa-sw algorithm and samtools (27, 28). Use of the longer-read algorithm improved the percentage of mapped reads over short-read algorithms (data not shown). If multiple matches were found for a single read, bwa-sw chose the reported mapping location at random. Counts of aligned reads could therefore be summed for all possible alternative transcripts of a single locus to give the raw number of reads mapping to a locus for each sample. Raw counts were converted to reads per kilobase per million reads mapped (RPKM) to normalize for average transcript length and the total number of reads for a sample (32). Many loci had low RPKM values that did not allow meaningful statistical analyses for differences in expression. Loci with a cumulative RPKM value for all samples of <10 were considered unsuitable for further analyses due to their low transcriptional abundance and were removed from the data set. The difference between RPKM values for high-abundance loci in adult and pupal samples was tested by the t test function in R.
Some transcripts with homology to baculovirus or nudivirus genes (pif-1, p74, lef-8, HzNVorf9-1 and -2, and HzNVorf64) were fragmented in assembly into two or more different contigs, while others had frameshift mutations which suggested that they were inactivated. To assess whether these alterations reflected sequencing or assembly errors, we synthesized gene-specific primers that spanned an assembly gap or potential mutation site (see Table S1 in the supplemental material). Standard 25-μl reaction volumes containing DNA (10 ng) from stage 1 pupae and the appropriate primer pair were then run using a Bio-Rad thermocycler. Products diluted 1:2 in H2O were sequenced by the Sanger method followed by assembly of the resulting sequence reads with DNASTAR SeqMan software. Resequencing showed that each of these genes was intact in the M. demolitor genome. Thus, the fragmented transcript sequences were merged into one locus. Two ultra-high-abundance genes (vp39 and 17a) were fragmented during assembly due to excessive read coverage and were identified by reassembling 8.3% of all reads using the same assembly parameters described above.
Orthologous genes among the M. demolitor transcripts were identified by tblastn (orthologous protein candidates against translated transcripts) followed by reciprocal BLAST, using blastx to identify orthologs of translated transcripts in the NCBI nonredundant database of 3 June 2011 (3). Percent identity was calculated by tblastn alignment of the best query protein to the M. demolitor transcript. Coordinates for alignment boundaries of M. demolitor transcripts to protein orthologs to identify gene duplicates were gleaned from tblastn alignment results. The longest translated open reading frames (ORFs) were then aligned with orthologous protein-coding sequences from the NCBI protein database using MUSCLE (18). Phylogenetic reconstruction of the evolution of these sequences was performed by maximum likelihood analysis using RAxML PROTCATLG (with the LG substitution matrix) and 1,000 bootstrap replicates, with the CIPRES online resource (42).
HMM searches for conserved protein domains.
To identify matches to a specific Hidden Markov model (HMM) in the M. demolitor transcripts, HMM profiles served as queries searched against a database of translated transcript sequences using HMMER 3.0 hmmsearch (http://hmmer.janelia.org). Translations were performed using EMBOSS getorf to find the longest open reading frames of >150 nucleotides in length between stop codons (38). DNA polymerase genes were identified using HMMER 3.0 hmmsearch with viral and eukaryotic DNA polymerase HMM queries (DNAPolymera_Pol, DNAP_B_exo_N, DNA_polA, DNA_pol_B, DNA_pol_B2, DNA_pol_B3, DNA_pol_B_exo1, DNA_pol_B_exo2, DNA_pol_lamd_f, DNA_pol_viral_C, DNA_pol_viral_N) against M. demolitor transcripts (20). To provide further evidence for the presence or absence of homologs to core nudivirus genes in the M. demolitor data set, the transcripts were searched with HMMs built for all nudivirus core genes using the nudivirus orthologs from all currently sequenced nudivirus genomes. Finally, conserved amino acid residues between nudivirus, baculovirus, and eukaryotic DNA polymerases were manually identified within regions of broad conservation (1). One region (III) was highly conserved and was characterized by the motif KXXXXSXYGXXGX[6–11]AXXXTXXGR in nudiviruses and baculovirus polymerases. Using perl programming, pattern matching searches were performed against all translated transcripts and also raw contigs from the velvet assembly that were used to build transcript sequences. Due to the short length of this motif, proteins should be identifiable using this method even if highly fragmented during assembly. To identify any possible conserved protein domains in a particular transcript of interest, the translated transcript sequence was searched against the Pfam-A database of HMMs using HMMER 3.0 hmmscan (20). Only hits with scores above the trusted cutoff were used.
Reverse transcription and qPCR measurement of transcript abundance.
Selected transcripts were analyzed by qPCR in time course studies. Total RNA was isolated per replicate from a single last-instar M. demolitor larva, a female pupa from stage 1 to 4, or an adult female wasp as described above. RNA concentrations were then quantified using a Nanodrop spectrophotometer followed by first-strand cDNA synthesis, using 100 or 500 ng of total RNA, poly(dT) primers, and Superscript III (Invitrogen) as outlined by the manufacturer. The corresponding cDNA for each gene of interest was PCR amplified using specific primers (see Table S1 in the supplemental material) and cloned into pSC-A-amp/kan, and its identity was confirmed by sequencing as outlined above. After propagating and isolating each plasmid from minipreps, standard curves were generated followed by determination of copy numbers for each transcript in ovary and whole-body samples with values standardized per 1 μg of total RNA. Three independently acquired biological replicates were analyzed per stage for each transcript, with each sample internally replicated 4 times.
Nucleotide sequence accession numbers.
A total of 66,425 transcripts were deposited in GenBank with accession numbers JO913492 through JO979916 and JR139425 through JR139430 (see below).
RESULTS
MdBV replication begins in stage 2 pupae.
BV replication in calyx cells of the wasp ovary consists of three major events: (i) amplification of viral DNA, (ii) virion formation and packaging of DNAs that have been excised and circularized from amplified segments, and (iii) lysis of calyx cells and accumulation of virions in the calyx lumen to form calyx fluid (2, 4, 30, 40, 41, 58). The encapsidated genome of MdBV consists of 15 segments (A to O), which are individually packaged into virions that consist of a single nucleocapsid enveloped by a unit membrane (6, 46, 52). Prior results also report the complete sequence of each packaged segment and describe the wasp-viral boundary sequences for selected proviral segments including B (6, 8). We thus characterized the timing of MdBV proviral DNA amplification, segment excision/circulation, and packaging by conducting qPCR assays using genomic segment B as a marker (see Fig. S1 in the supplemental material). The copy number of each product was determined in ovaries from stage 1 to 4 pupae and adults, while the copy number of EF1α served as a genetically unlinked control gene in the wasp genome.
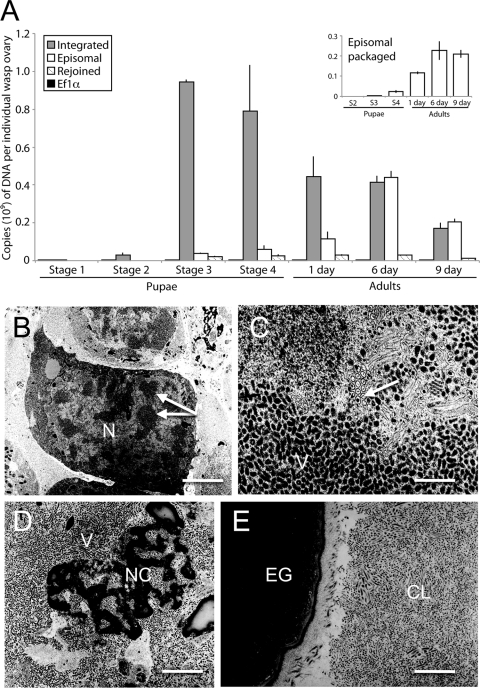
In stage 1 pupae, we detected approximately equal copy numbers of integrated segment B and EF1α but no circularized segment B or empty B locus (Fig. 1A). We also detected no circularized segment B or empty B locus in stage 2 pupae, but the copy number of integrated segment B was nearly an order of magnitude higher than that of EF1α, indicating specific amplification of the number of DNA copies of the region of the genome containing segment B (Fig. 1A). The copy number of integrated segment B then dramatically increased in stage 3 pupae, which was followed by a progressive decline during subsequent stages of wasp development (Fig. 1A). Reciprocally, the abundance of episomal segment B began to rise in stage 3 pupae but did not reach a maximum until adult wasps were 6 days old (Fig. 1A). We also first detected products for the segment B “empty locus” (Fig. 1A, Rejoined) in stage 3 pupae, but its abundance did not change thereafter (Fig. 1A). Including a DNase step before isolating DNA from ovary homogenates (6) further allowed us to determine the copy number of episomal segment B in virions, which protect packaged DNAs from DNase degradation. Packaged episomal segment B was first detectable in stage 3 pupae and reached a maximum in 6-day-old adult wasps (Fig. 1A, inset). Comparison of these data to the total amount of episomal segment B indicated that less than half was packaged in ovaries from stage 3 and 4 pupae and day 6 adults. However, in day 9 adults, most copies of episomal segment B were packaged into virions.
Fig 1.
Timing of proviral segment B amplification, excision/circularization of episomal segment B, and MdBV virion formation in M. demolitor. (A) Mean copy number ± standard error (SE) of qPCR products corresponding to integrated segment B, episomal segment B, the empty segment B locus, and EF1α. The total number of copies of each DNA in an individual wasp ovary is presented along the y axis, while the stages of wasp development are presented along the x axis. The inset shows mean copy number ± SE of episomal segment B at each wasp stage that is packaged into virions. The data for each wasp stage derives from two or three independently collected ovary samples. (B to E) TEM micrographs of MdBV nucleocapsids and virions during particular phases of replication. (B) Low-magnification image of calyx cells from a stage 3 pupa. Note the enlarged nucleus (N) for the calyx cell in the center of the image. Arrows indicate regions in the nucleus where MdBV particles are being assembled in proximity to virogenic stroma. Bar = 1.5 μm (C) High-magnification image of a calyx cell nucleus from a stage 3 pupa. The arrow indicates a region where both empty viral envelopes and envelopes containing a capsid are visible, while below this region are an abundance of mature virions (V). Bar = 100 nm. (D) High-magnification image of a lysed calyx cell from a stage 4 pupa. The nuclear membrane of the cell has deteriorated, resulting in release of mature virions (V) and dense nuclear chromatin (NC). Bar = 500 nm. (E) Low-magnification image from a day 1 adult showing densely packed MdBV virions in the calyx lumen (CL) that is adjacent to an M. demolitor egg (EG). Bar = 1 μm.
We linked our qPCR data to the timing of virion formation by conducting TEM studies. These observations showed that no capsids were visible in calyx cell nuclei from stage 1 and 2 pupae (data not shown), whereas calyx cells from stage 3 pupae had enlarged nuclei that contained MdBV virions (Fig. 1B). At higher magnification, MdBV particles at different stages of assembly were readily visible within these calyx cells (Fig. 1C). In stage 4 pupae and day 1 adults, we observed calyx cells where MdBV particles were in the process of being assembled and also calyx cells that were in the process of lysing (Fig. 1D). Calyx cell lysis in turn resulted in the accumulation of a high density of virions in the calyx lumen (Fig. 1E). Overall, these data showed that amplification of DNA containing proviral segment B began in stage 2 pupae, while virion formation and packaging of episomal segment B began in stage 3 pupae. Replication then continued into adulthood.
Illumina sequencing generated a rich catalog of transcripts for M. demolitor ovaries.
We used the preceding results to select stage 1 pupae as a source of prereplication ovary RNAs and day 1 adults as a source of ovary RNAs during active MdBV replication. While replication in stage 3 and 4 pupae was higher than that in day 1 adults, we selected the latter as a source of RNAs during active replication because replication remained high at this period and adults are easier to collect than pupae. We produced 6 libraries for each stage and sequenced them in a single reaction using the Illumina HiSeq system. Our results yielded a total 371 million paired 100-bp reads, of which 330 million had an identifiable index tag. The number of reads in each index group ranged from 19 to 41 million. After quality filtering, 94 million read pairs and 33 million single-end reads remained, for a total of 222 million reads representing 22 Gb of sequence. From this data set, 197 million reads assembled into 32,711 loci that were 200 bp in length or greater. Alternative splicing was also predicted for 9,973 loci for a total of 66,425 transcripts. These were deposited in GenBank with accession numbers JO913492 through JO979916 and JR139425 through JR139430. BLASTN identified 66 loci (0.2% of our total) that corresponded to known intergenic regions or ORFs in MdBV genomic DNAs that are packaged into virions, many of which were misassembled (see Table S2 in the supplemental material). Since mean RPKM values for these loci were much higher in our adult than in our pupal libraries, we reasoned most of these reads arose from low-level contamination by MdBV genomic segments that were massively amplified during replication and removed them from further analyses. However, recent results do indicate that some genes in the MdBV encapsidated genome are expressed in M. demolitor adults (11). While we could not distinguish DNA contamination from transcripts in our data set, we indicate in Table S2 in the supplemental material the viral transcripts identified by Bitra et al. (11) that potentially were not contaminants in this data set. In contrast, the low RPKM values for these loci in our stage 1 pupal samples argued that little or no contamination of our libraries by cellular genomic DNA occurred generally.
As the M. demolitor genome is not sequenced, we evaluated the breadth of gene sampling in our transcriptional data set by comparing it to the sequenced genomes of two other hymenopterans: Nasonia vitripennis and Apis mellifera (24, 53). Orthologs between genome/transcriptome pairs were determined using BLAST, and we counted shared genes as those with reciprocal hits between the data sets. Using this method, the N. vitripennis and A. mellifera genomes shared 3531 of 9252 and 10560 genes respectively, while the M. demolitor data set shared 2428 orthologs with N. vitripennis and 2697 with A. mellifera. Assuming each species pairing shares the same number of genes (approximately 3,500), these data suggested that 69–77% of genes shared among species were present in the data set we generated, and that a large percentage of genes in the M. demolitor genome were sequenced. This analysis also suggested that all of the highly expressed genes in ovaries were sampled, because random transcript sequencing results in highly expressed genes being sequenced more often by chance than genes expressed at low levels (32).
M. demolitor ovaries express several nudivirus/baculovirus-like genes.
Given prior results with C. congregata and C. inanitus (9), our first priority was to screen the M. demolitor transcriptome for genes with similarity to known nudivirus and/or baculovirus genes. We identified a total of 41 such transcripts (Table 1; also see Table S3 in the supplemental material), whose known functions from studies of baculoviruses include RNA transcription, oral infectivity, and virion formation (39). Of particular note was the presence of all 4 genes (p47, lef-4, lef-8, and lef-9) for the subunits of the unique RNA polymerase that baculoviruses and nudiviruses contain, and two genes similar to lef-5 and vlf-1, which regulate the hyperexpression of baculovirus very-late genes. We also identified homologs of all six per os infectivity factors (i.e., pif genes p74, pif-1, pif-2, pif-3, 19K, odv-e56) which are envelope proteins, and odv-e66, which functions as both an envelope protein and hyaluronidase in baculoviruses that infect Lepidoptera (39). Last, we detected three genes similar to vp39, vp91, and 38K, which together with vlf-1 encode capsid proteins conserved between nudiviruses and baculoviruses. Four other nucleocapsid proteins present in all baculovirus genomes (gp41, vp1054, p6.9, and odv-ec27) but unknown in nudiviruses were absent from the M. demolitor ovary transcriptome. However, we did identify homologs of the predicted structural genes HzNVorf9, HzNVorf106, HzNVorf140, and PmV hypothetical protein, identified from Helicoverpa zea nudivirus 1 (Hz-1) (14) and Penaeus monodon baculovirus (MBV), which is in actuality a nudivirus recently renamed P. monodon nudivirus (PmNV) (26, 51). Homologs of these genes were also identified by proteomic analysis of CiBV particles from the wasp Chelonus inanitus (54).
Table 1.
Nudivirus-like transcripts from female stage 1 pupae and day 1 adult M. demolitora
| Gene name | Locus | Adult mean RPKM (n = 6) | Pupal mean RPKM (n = 6) | P valueb | Putative functional role |
|---|---|---|---|---|---|
| helicase | 3891 | 29 | 0 | 1.1e−05 | DNA replication |
| integrase | 3978 | 53 | 1 | 1.9e−07 | DNA replication |
| 1667 | 48 | 0 | 6.7e−07 | DNA replication | |
| p47 | 703 | 128 | 233 | 0.03 | RNA polymerase subunit |
| lef-4 | 2025 | 36 | 14 | 0.0002 | RNA polymerase subunit |
| lef-8 | 283 | 176 | NS | RNA polymerase subunit | |
| lef-9 | 3218 | 72 | 54 | NS | RNA polymerase subunit |
| lef-5 | 539: transcripts 2, 7, 8, 9 | 159 | 231 | NS | Late gene expression |
| vlf-1 | 539: transcripts 1, 3,4, 5, 6, 10 | 144 | 98 | NS | Late gene expression, capsid protein, DNA packaging |
| HzNVorf140 (vlf-1b) | 3488 | 88 | 0 | 6.4e−08 | Nudcleocapsid protein, DNA packaging |
| 2966 | 197 | 0 | 1.9e−05 | Nudcleocapsid protein, DNA packaging | |
| 2965 | 519 | 0 | 2.2e−08 | Nudcleocapsid protein, DNA packaging | |
| 38K | 318 | 511 | 2 | 4.0e−05 | Nucleocapsid protein |
| vp91 | 4098 | 210 | 0 | 7.5e−08 | Nucleocapsid protein |
| vp39 | 4,109 | 3 | 6.2e−06 | Nucleocapsid protein | |
| p74 | 1,320 | 2 | 1.1e−07 | Per os infectivity | |
| pif-1 | 382 | 0 | 6.4e−09 | Per os infectivity | |
| pif-2 | 226 | 448 | 0 | 9.4e−09 | Per os infectivity |
| pif-3 | 767 | 150 | 0 | 9.4e−08 | Per os infectivity |
| 19 kDa | 1791 | 597 | 2 | 1.7e−07 | Per os infectivity |
| odv-e56 | 13843 | 5 | 0 | 1.2e−08 | Per os infectivity |
| 1447 | 329 | 1 | 7.3e−06 | Per os infectivity | |
| 1083 | 217 | 1 | 4.6e−07 | Per os infectivity | |
| odv-e66 | 175 | 405 | 0 | 2.0e−09 | ODV envelope protein |
| 1139 | 91 | 0 | 4.7e−13 | ||
| 2331 | 197 | 0 | 3.1e−09 | ||
| 4186 | 207 | 0 | 1.0e−08 | ||
| 2730 | 244 | 0 | 1.0e−08 | ||
| HzNVorf9-1 | 864 | 2 | 1.0e−06 | Structural protein | |
| HzNVorf9-2 | 761 | 0 | 1.7e−05 | Structural protein | |
| HzNVorf106 | 332 | 508 | 1 | 8.2e−08 | Structural protein |
| PmV hypothetical protein | 756 | 546 | 0 | 1.5e−06 | Structural protein |
| Structural protein | |||||
| ac92 | 5801 | 95 | 0 | 6.9e−08 | Sulfhydryl oxidase |
| 2756 | 162 | 0 | 1.30E−08 | ||
| HzNVorf64 | 1,043 | 12 | 7.9e−08 | Unknown | |
| HzNVorf94 | 318 | 511 | 2 | 4.0e−05 | Unknown |
| HzNVorf128 | 1167 | 107 | 85 | NS | Unknown |
Expression levels in pupae and adults are indicated by RPKM values (see Materials and Methods).
NS, not significant.
We noted two other important features about our data set in relation to nudiviruses and baculoviruses. First, with the exception of a nudivirus/baculovirus-like helicase and integrase (also known as HzNVorf144), other genes considered essential for DNA replication (39) including a DNA polymerase (dnapol), DNA primases (lef-1, lef-2), DNA ligase, and the single-stranded binding (SSB) protein (lef-3) were absent from the M. demolitor transcriptome. Second, several nudivirus/baculovirus-like homologs (vlf-1, integrase, odv-e56, ac92, odv-e66, and HzNVorf9) were present in more than one contig and derive from duplicated genes (Table 1; also see Table S4 in the supplemental material). In most instances, we identified 2 or 3 distinct gene duplicates, but odv-e66 was represented in many different contigs (see Table S4). Although they were fragmented during assembly, we estimated that 8 or 9 paralogs of odv-e66 exist in the M. demolitor genome by counting the number of unique amino (9 peptide sequences) and carboxy (8 peptides) terminal ends present in our data set. We also were able to assemble 5 full-length transcripts (Table 1; also see Table S4). During our BLAST analyses, we noticed homology between vlf-1 and integrase (also known as vlf-1a and vlf-1b, respectively) and HzNVorf140 (16). To examine the evolution of BV integrase and vlf-1 duplicates in relation to nudivirus/baculovirus homologs, we conducted a phylogenetic analysis of these genes to assess whether the homologs identified in M. demolitor more likely arose prior to or after acquisition by the wasp. Our results strongly suggested that vlf-1 and integrase arose from an older duplication of vlf-1 in the nudivirus ancestor of BVs and that they have duplicated several times subsequently in M. demolitor (see Fig. S2).
Most nudivirus/baculovirus-like genes are strongly upregulated during MdBV replication.
Besides the breadth of gene sampling, a second strength of our sequencing approach was that it allowed us to quantify transcript abundance, because transcripts were sequenced in proportion to their representation in the original pool of mRNAs (32). By generating multiple independent samples and uniquely tagging each, we also could statistically compare the abundance of each transcript between our early pupal (prereplication) and adult (replication) treatments. Read mapping for each replicate was successful for 90.2% of reads and 15,135 of 32,817 loci were sufficiently abundant (RPKM ≥ 10) for further analysis. First, we compared the RPKM values in pupal and adult stages for the nudivirus/baculovirus-like transcripts we identified. Our results showed that most viral loci were expressed at significantly higher levels in adult ovaries (Table 1). Many of these loci, including the viral integrase and helicase, all of the pif genes, and all of the other predicted structural genes were expressed at near-zero levels in early pupae, which was fully consistent with their predicted functions in DNA replication, segment packaging, and virion formation. In contrast, lef-8, lef-9, lef-5, and vlf-1 were abundantly expressed in both stage 1 pupae and adults. Although values tended to be higher in adults, among sample variation for transcripts like lef-8 and lef-9 resulted in no statistical difference in expression between early pupae and adults.
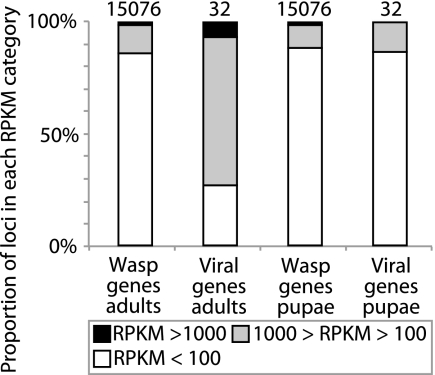
We then compared the RPKM values for the nudivirus/baculovirus genes to the RPKM values for all other loci, which we designated as wasp genes (Fig. 2). This analysis showed that a much larger proportion of the viral loci were either more highly (100 < RPKM < 1,000) or much more highly (RPKM > 1,000) expressed in adult than in stage 1 pupal ovaries (X2 = 54.3; P > 0.0001). In contrast, only 15% of wasp loci were highly expressed in adults, which was no different than the proportion of wasp loci that were highly expressed in stage 1 pupae (X2 = 0.9; P > 0.64). This result showed a strong bias in the proportion of nudivirus-like genes that are abundantly expressed during MdBV replication relative to wasp genes.
Fig 2.
Transcript abundance of all M. demolitor loci in adults and pupae, categorized into nudivirus-like genes and wasp genes. The total number of wasp or virus loci detected in the M. demolitor transcripts is shown above each bar. The viral gene category excludes the fragmented odv-e66 transcripts.
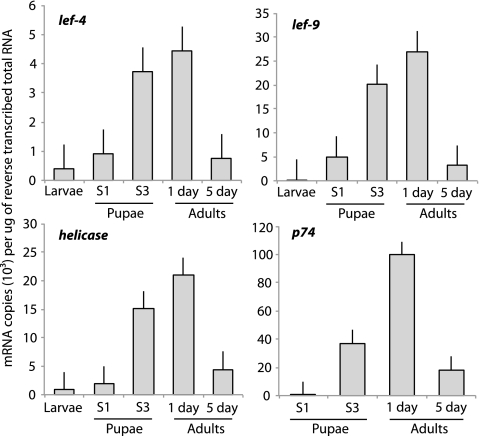
We selected two viral RNA polymerase subunits (lef-4, lef-9) plus two other nudivirus-like genes (helicase, p74) for analysis by qPCR. This allowed us to broaden the temporal scale of our expression data, while also providing results that could be compared to the RPKM data generated by sequencing. Overall, these results corroborated our sequencing data by showing that transcript abundance for each gene was highest in day 1 adults (Fig. 3). They also showed that while expression of lef-4, lef-9, and the helicase gene began in last-day larvae or stage 1 pupae, expression of p74 did not begin until pupae reached stage 3 (Fig. 3), when virions were first observed (Fig. 1). We noted that transcript abundance for each gene also declined between day 1 and day 5 adults (Fig. 3). Collectively, these data suggested lef-4, lef-9, and the helicase behave as early genes whose products are required for viral DNA replication or transcription of other viral genes, while p74 behaved as an MdBV “late” gene whose expression coincides with particle formation. Reduced transcript abundance of these genes between day 1 and day 5 adults also suggested that MdBV replication declines with wasp age.
Fig 3.
Mean transcript abundance ± SE of selected nudivirus-like genes from M. demolitor as determined by qPCR. The y axes of the four graphs show the copy number of the lef-4, lef-9, helicase, and p74 homologs per microgram of total RNA isolated from ovaries. The stages (S) of wasp development are presented along the x axis of each graph. The data for each sample time derive from three independently collected ovary samples.
Several BV-associated genes of unknown origin are present among M. demolitor transcripts.
Previous studies of C. congregata and C. inanitus BVs provided a list of genes that are of uncertain origin (virus or wasp), but most likely have a virus-related function. The first category of genes in this regard consisted of orthologs for any of the 28 non-nudivirus genes recently identified by proteomic analysis of CiBV virions (54). We identified 7 orthologs in total with transcript abundance for 6 being more abundant in adult ovaries, as expected for predicted structural genes (Table 2). The 58b ortholog we identified did not differ in abundance between pupae and adults, and transcript abundance of 58b also did not differ between prereplication- and active-replication-stage ovaries in C. inanitus (54), suggesting that it is not a virion component. Unlike CiBV, the 35a locus we identified consisted of 3 transcripts, suggesting the duplication of this gene in M. demolitor (Table 2). The second category of genes consists of those located in C. congregata in a region of a wasp chromosome that contains many nudivirus-like genes: a nudivirus cluster (9). We identified 3 orthologs similar to genes located in this cluster in C. congregata, and all are highly upregulated in adult ovaries (Table 2).
Table 2.
M. demolitor transcripts from ovaries of unknown origin with virus-related genomic positions or functionsa
| Locus | Ortholog name | Accession no. | nr BLAST hits | Adult mean RPKM (n = 6) | Pupae mean RPKM (n = 6) | P value |
|---|---|---|---|---|---|---|
| CiBV particle proteins | ||||||
| 17a | 17a | CBA62651.1 | Hypothetical protein (C. congregata) | 2,309 | 2 | 2.2e−05 |
| 2287 | 27b | CBH32500.1 | Hypothetical protein Cc50C22.7 (C. congregata) | 1,823 | 1 | 2.2e−05 |
| 1266 | 30b | CBA62633.1 | Hypothetical protein (C. congregata) | 210 | 0 | 8.5E−09 |
| 417 | 35a | CBA62630.1 | None | 129 | 0 | 5.8E−09 |
| 420 | 35a | CBA62630.1 | None | 249 | 0 | 9.2E−08 |
| 615 (transcript 4, 9) | 35a | CBA62630.1 | None | 277 | 0 | 2.7E−05 |
| 576 | 97a | CBA62572.1 | None | 334 | 0 | 7.5E−08 |
| 937 | 97b | CBA62575.1 | Disintegrin and metalloproteinase (Acromyrmex echinator) | 309 | 0 | 0.002 |
| Nudivirus cluster genes from CcBVb | ||||||
| 370 | Cc50C22.3 | CAR82241.1 | Hypothetical protein Cc50C22.3 (C. congregata) | 362 | 0 | 5.1e−08 |
| 3938 | Cc50C22.5 | CAR82243.1 | Hypothetical protein Cc50C22.5 (C. congregata) | 300 | 0 | 2.0e−05 |
| 1624 | 431 | 0 | 6.5e−09 | |||
| 4428 | 601 | 0 | 3.1e−08 | |||
| 2935 | 908 | 0 | 4.4e−08 | |||
| 5909 | Cc50C22.6 | CAR82244 | Hypothetical protein Cc50C22.6 (C. congregata) | 242 | 0 | 1.3e−05 |
| 2485 | 1,230 | 1 | 6.1e−06 |
Expression levels are indicated as in Table 1.
These genes were fragmented in the M. demolitor assembly.
Multiple wasp genes are strongly upregulated during MdBV replication.
The second priority in mining our data set was to identify wasp genes that were differentially expressed in our pupal and adult samples. We assigned all differentially expressed wasp loci to a category of genes with potential roles in BV replication. Excluding all of the nudivirus/baculovirus-like or BV-associated genes discussed above, 960 loci in our data set had mean RPKM values that differed 2-fold or more between our pupal and adult samples (two-sided t test, degrees of freedom [df] = 10, using a Bonferroni-corrected P value of <3.3 × 10−6). A total of 920 were upregulated in adults while 40 were downregulated. We further narrowed this list by focusing on transcripts with mean RPKM values that were ≥50 in adult calyces and that were >50-fold more abundant in adults than in pupae. This yielded 165 loci that we classified as strong candidates for having direct or indirect roles in MdBV replication. As shown in Table 3, 41 of these loci shared significant identity with known genes, 30 shared identity with other hypothetical genes in databases, and 94 were unique hypothetical genes. Among the known loci, 3 genes strongly upregulated in adult ovaries contained BTB/POZ domains characteristic of some zinc finger proteins, which suggested that they are unique transcription factors (61). Four other genes also had domains typical of transcription factors (zinc finger and leucine zipper domains), including locus 2319, which was highly similar to a nuclear receptor transcription factor in ants and other insects, and locus 3605, which contained a nuclear hormone receptor domain similar to tailless from Apis mellifera.
Table 3.
M. demolitor transcripts identified as highly or very highly expressed in day 1 adult ovariesa
| Locusa | Category | PFAM domains | Best nr hit | Identity | Number of loci |
|---|---|---|---|---|---|
| 1068 | Variable membrane protein | Variable membrane protein precursor, Trichomonas vaginalis G3 | 19% | 1 | |
| 5485 | ELO domain-containing protein (long chain fatty acid elongation) | ELO: GNS1/SUR4 family, 2.3e−52 | Similar to CG6921-PA, Tribolium castaneum | 63% | 1 |
| 1253 | Aggregation factor | Agrin-like isoform 1, Apis mellifera | 65% | 1 | |
| 4897 | DNA Pol B2 domain-containing protein | DNA_pol_B_2: DNA polymerase type B, organellar and viral, 3.9e−07 | DNA Pol B2 domain-containing protein, Glyptapanteles flavicoxis | 73% | 1 |
| 2979 | BTB/POZ domain-containing protein | BTB: BTB/POZ domain, 2.4e−21 | Hypothetical protein, Trichoplax adhaerens | 30% | 3 |
| 4014 | BTB: BTB/POZ domain, 2.6e−18 | Hypothetical protein, Tribolium castaneum | 29% | ||
| 7763 | BTB: BTB/POZ domain, 4e−23 | Hypothetical protein, Tribolium castaneum | 35% | ||
| 3605 | Nuclear hormone receptor ligand-binding domain-containing protein | Hormone_recep: Ligand-binding domain of nuclear hormone receptor, 4e−10 | Tailless, Apis mellifera | 53% | 1 |
| 2319 | Nuclear receptor transcription factor | zf-C4, Zinc finger, C4 type (two domains), 1.5e−14 | Nuclear receptor subfamily 2 group E member 1, Acromyrmex echinatior | 91% | 1 |
| 1232 | Transcription factor | bZIP_2: Basic region leucine zipper, 3.7e−16; bZIP_1: bZIP transcription factor, 2.6e−06 | Hypothetical protein, Apis mellifera | 33% | 1 |
| 4701 | Zinc finger domain-containing protein | zf-C2H2: Zinc finger, C2H2 type, 3.8e−15, 4 copies; zf-met: Zinc finger of C2H2 type, 2e−08 | Similar to PR-domain containing protein 13, Apis mellifera | 44% | 1 |
| 38 | Ankyrin repeat protein | Ank: Ankyrin repeat, 5.7e−48, 4 copies | Hypothetical protein, Cotesia congregata | 34% | |
| 212 | Ank_2: Ankyrin repeats (3 copies), 3.2e−17, 2 copies | Hypothetical protein, “Candidatus Amoebophilus asiaticus” 5a2 | 24% | ||
| 3477 | Ank_2: Ankyrin repeats (3 copies), 2.3e−23, 2 copies | Hypothetical protein, “Candidatus Amoebophilus asiaticus” 5a2 | 34% | ||
| 4062 | Ank: Ankyrin repeat, 1.5e−14 | Ankyrin repeat protein, Trichomonas vaginalis | |||
| 1302 | Ankyrin repeat protein, Wolbachia-like | Ank_2: Ankyrin repeats (3 copies), 7.2e−33, 3 copies | Ankyrin repeat protein, Wolbachia endosymbiont wVitA of Nasonia vitripennis phage WOVitA1 | 26% | |
| 785 | Retroviral aspartyl protease domain-containing protein | RVP: Retroviral aspartyl protease, 3.3e−08 | Hypothetical protein, Tribolium castaneum | 29% | 1 |
| 1134 | Putative metalloprotease | Peptidase_M13: Peptidase family M13, 2.4e−58 | Neprilysin 2, Apis mellifera | 71% | 4 |
| 1422 | Peptidase_M13_N: Peptidase family M13, 2.8e−09 | Similar to zinc metalloprotease, Nasonia vitripennis | 68% | ||
| 1485 | Peptidase_M13_N: Peptidase family M13, 3.9e−17 | Similar to zinc metalloprotease, Nasonia vitripennis | 62% | ||
| 2862 | PFAM Peptidase_M13_N, Peptidase family M13, 1.1e−07 | Membrane metalloendopeptidase-like 1, Harpegnathos saltator | 57% | ||
| 4662 | Putative uridine phosphorylase | Uridine phosphorylase 1-like, Apis mellifera | 52% | 1 | |
| 493 | Sodium/calcium exchanger | Sodium/calcium exchanger 3, Camponotus floridanus | 86% | 2 | |
| 682 | Sodium/calcium exchanger 3, Camponotus floridanus | 88% | |||
| 4183 | Sorbitol dehydrogenase | ADH_zinc_N: Zinc-binding dehydrogenase, 1.7e−21; ADH_N: Alcohol dehydrogenase GroES-like domain, 5e−20 | Sorbitol dehydrogenase, Harpegnathos saltator | 74% | 1 |
| 1017 | Transposase domain-containing protein | Transp_Tc5_C: Tc5 transposase C-terminal domain, 4.9e−07; DDE_1: DDE superfamily endonuclease, 2.3e−06; HTH_Tnp_Tc5: Tc5 transposase DNA-binding domain, 4.2e−06 | Hypothetical protein, Solenopsis invicta | 27% | 1 |
| 1337 | Transposase domain-containing protein | DDE_1: DDE superfamily endonuclease, 1.8e−14; HTH_Tnp_Tc5: Tc5 transposase DNA-binding domain, 1.1e−05; Transp_Tc5 _C: Tc5 transposase C-terminal domain, 5.6e−05 | Hypothetical protein, Solenopsis invicta | 28% | 1 |
| Not listed | Hypothetical protein | 94 | |||
| Not listed | Conserved hypothetical protein | 30 |
All loci listed were differentially expressed in pupae and adult ovaries (P < 3.3 × 10−6). All loci also met the criteria of having (i) RPKM values that were >50-fold higher in adults than pupae and (ii) RPKM values that were >50 in adults (see Results).
Another set of transcripts that were highly abundant in adult calyces encoded predicted ankyrin repeat proteins. All but one of these were most similar to ankyrin repeat genes from microorganisms, which suggested the possibility of horizontal acquisition by the wasp. Each was also distinct from known IκB-like (i.e., vankyrin) genes in the encapsidated genome of MdBV and other BVs whose functions include the inhibition of host NF-κB transcription factors (49). Ankyrin repeat domains are well known to mediate protein-protein interactions, but the function of the ankyrin repeat genes that these transcripts are most similar to is unknown. Finally, 5 upregulated transcripts encoded predicted proteases. Locus 1134 had high similarity to neprilysin, a metalloprotease found in the virus-like particles of the ichneumonid wasp Venturia canescens (5). The other four predicted proteases were also metalloprotease family members, which together with the ortholog for CiBV 97b (shown in Table 2) suggested a potentially critical role for metalloproteases in the formation of BV virions or virion function in parasitized host insects.
The M. demolitor transcriptome contains several DNA polymerase genes.
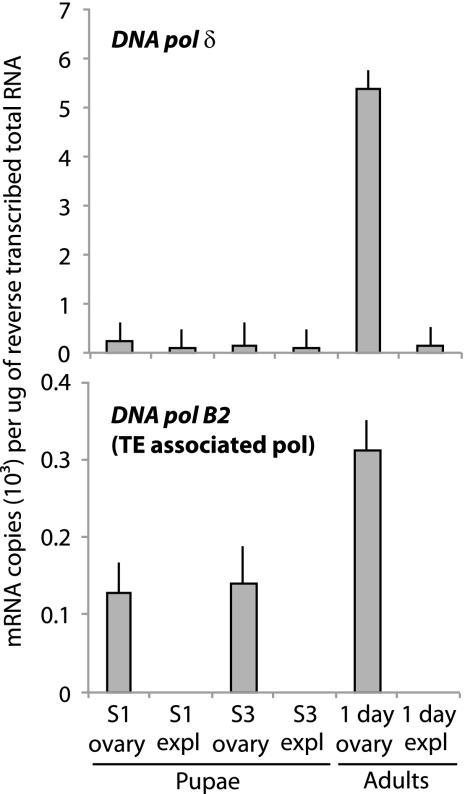
The absence of nearly all nudivirus/baculovirus genes considered essential for replication from our data set clearly suggested that replication depends on the DNA synthesis machinery of the wasp. No genes with clear roles in DNA synthesis met our criteria for inclusion in Table 3. Nonetheless, our data set contained all expected insect DNA polymerases (25), including the family B (replication) DNA polymerases α, δ, and ε and the family A mitochondrial DNA polymerase γ (Table 4). The mean RPKM value for dnapol γ was higher in our adult than in our pupal samples. In contrast, RPKM values for dnapol α were higher in pupae, while no significant differences were detected between treatments for dnapol δ and ε (Table 4). We also identified transcripts containing a DNA polymerase B2 domain, including several whose best BLAST hit was to a DNA polymerase B2-containing ORF from another BV-carrying wasp (Glyptapanteles flavicoxis) (15). Interestingly, the G. flavicoxis ORF is located in a Maverick transposable element-like sequence and also flanks a GiBV proviral segment (15). A tblastn search of the G. flavicoxis homolog against all transcripts identified 52 hits in the M. demolitor data set (see Table S5 in the supplemental material). Most of these were less than full length, suggesting fragmentation during assembly and that fewer copies of this gene actually exist in the M. demolitor genome. The 5 transcripts listed in Table 4 were the most intact because their DNA pol B2 domains were still recognizable by HMM searches. Locus 4897, hereafter named DNA pol B2, was also the only transcript with an intact DNA pol B2 domain whose mean RPKM value was higher in adults than in pupae (Table 4). Because of their higher RPKM values in adults, we selected dnapol δ and DNA pol B2 for further study by comparing their abundance in ovaries versus the wasp body without ovaries. Our results showed that the copy number of dnapol δ was very low in the ovaries and bodies of stage 1 and 3 pupae but that the copy number was significantly higher in ovaries than in the bodies of day 1 adults (Fig. 4). For DNA pol B2, we detected no expression in pupal- or adult-stage bodies, but in ovaries expression was also distinctly higher in day 1 adults than in stage 1 and 3 pupae (Fig. 4).
Table 4.
DNA polymerase domain-containing transcripts from M. demolitor ovariesa
| Locus | PFAM DNA polymerase domain | Best nr hit | Accession no. | Best nr hit species | Identity | Adult mean RPKM (n = 6) | Pupal mean RPKM (n = 6) | P value |
|---|---|---|---|---|---|---|---|---|
| 4754 | DNA_pol_B, DNA_pol_B_exo1 | DNA polymerase delta subunit | BAJ78765.1 | Vespa analis insularis | 77% | 58 | 46 | NS |
| 1052 | DNA_pol_B, DNA_pol_B_exo1 | DNA polymerase alpha subunit | XP_001121438.2 | Apis mellifera | 50% | 70 | 51 | 0.03 |
| 2371 | DNA_pol_B, DNA_pol_B_exo1 | DNA polymerase zeta subunit | XP_001607949.1 | Nasonia vitripennis | 50% | 45 | 35 | NS |
| 1137 | DNA_pol_B, DNA_pol_B_exo1, DNA_pol_B_exo2 | DNA polymerase epsilon subunit | XP_393171.4 | Apis mellifera | 68% | 23 | 21 | NS |
| 921 | DNA_pol_A | DNA polymerase subunit gamma-1, mitochondrial | EFN71060.1 | Camponotus floridanus | 62% | 15 | 38 | 0.001 |
| 1212 | DNA_pol_A | DNA polymerase theta subunit | EGI59836.1 | Acromyrmex echinator | 48% | 12 | 4 | 9.7e−06 |
| 15870 | DNA_pol_B2 | Hypothetical protein | XP_001599083.1 | Nasonia vitripennis | 45% | very low | very low | |
| 24190 | DNA_pol_B2 | Hypothetical protein | XP_001600116.1 | Nasonia vitripennis | 54% | very low | very low | |
| 4358 | DNA_pol_B2 | DNA pol B2 domain-containing protein | ACE75264.1 | Glyptapanteles flavicoxis | 70% | 16 | 5 | 2.0e−08 |
| 4897 | DNA_pol_B2 | DNA pol B2 domain-containing protein | ACE75264.1 | Glyptapanteles flavicoxis | 76% | 94 | 0 | 1.5e−09 |
| 23368 | DNA_pol_B2 | DNA pol B2 domain-containing protein | ACE75264.1 | Glyptapanteles flavicoxis | 65% | very low | very low | |
| 13723 | DNA_pol_B2 | DNA pol B2 domain-containing protein | ACE75264.1 | Glyptapanteles flavicoxis | 68% | 5 | 0 | 2.3e−06 |
| 7467 | DNA_pol_B2 | DNA pol B2 domain-containing protein | ACE75264.1 | Glyptapanteles flavicoxis | 59% | 14 | 3 | 3.3e−06 |
Expression levels are indicated as in Table 1.
Fig 4.
Mean transcript abundance ± SE of a predicted M. demolitor dna pol δ and DNA pol B2 as determined by qPCR. The x and y axes of each graph are labeled as described in Fig. 3; expl, whole animals with ovaries explanted. The data for each sample time derives from three independently collected ovary samples.
Genes with roles in locus-specific DNA amplification are upregulated in adult wasp ovaries.
Prior studies suggest two models for BV replication. The first is that replication occurs by locus-specific amplification of the integrated proviral genome (30) in a manner akin to selective amplification of genes by diverse eukaryotes, including insects (50). The second is that a larger progenitor molecule(s) first excises from the wasp genome and is then amplified by a recombination-dependent or rolling circle mechanism (35, 40, 41, 50). In both scenarios, subsequent excision/circularization of the genomic DNAs that are packaged into virions is thought to occur by site-specific recombination across direct repeat junctions flanking individual segments (6, 8, 40).
Results presented in Fig. 1 clearly showed that amplification of MdBV proviral segment B precedes the formation of episomal B, but these assays do not distinguish between locus-specific amplification of the proviral genome versus excision and amplification of a larger precursor molecule. However, if locus-specific amplification is important, we would expect that M. demolitor contains the corresponding essential genes, because of their conserved roles in organisms that range from Drosophila melanogaster to Tetrahymena (50). Moreover, we would expect these transcripts to be more abundant in our adult than in our pupal samples. Of 16 essential genes in D. melanogaster, we identified 14 in M. demolitor of which 9 were more strongly expressed in adult ovaries (Table 5). In Drosophila, these genes are involved in chorion gene amplification, which is also a possibility in M. demolitor, and further functional experiments are needed to determine the role of locus-specific amplification genes in BV DNA replication. Discerning candidate genes with essential roles in rolling circle or recombination-dependent replication is more difficult. Recombination-dependent replication of lambda phage depends upon several essential genes that include the red α exonuclease and single-strand binding (SSB) protein. In turn, all baculoviruses encode homologs of red α exonuclease called alkaline nuclease (alk-exo), while both baculoviruses and at least some nudiviruses contain the SSB protein homolog lef-3 (34, 39; J. Burand, personal communication). However, we failed to identify homologs of either factor in the M. demolitor ovary transcriptome.
Table 5.
M. demolitor transcripts from ovaries that are homologous to genes from D. melanogaster with essential roles in locus-specific amplificationa
| Drosophila amplification gene | M. demolitor ortholog | Adult mean RPKM (n = 6) | Pupal mean RPKM (n = 6) | P value | Function in gene amplification |
|---|---|---|---|---|---|
| ORC1 | 1371 | 24 | 11 | 9.1e−06 | Binds origin DNA |
| ORC2 | 5864 | 24 | 9 | 2.5e−05 | Binds origin DNA |
| Cyclin E | 2123 | 72 | 37 | 0.0002 | Required for S phase of endocycle |
| Chiffon (Dbf4) | 1535 | 153 | 86 | 0.004 | ORCb binding |
| Mus101 | 6591 | 32 | 16 | 0.0003 | Replication, repair |
| E2F1 | 1016 | 89 | 66 | NS | S-phase transcription factor, ORC binding |
| E2F2 | S-phase transcription factor | ||||
| DP | 4839 | 82 | 76 | NS | Replication complex assembly |
| 3522 | 152 | 157 | NS | ||
| Humpty dumpty | 4200 | 14 | 6 | 5.4e−06 | Unknown essential function, repair |
| Mcm6 | 1678 | 81 | 72 | NS | Replication complex |
| Geminin | Negative regulator of gene amplification | ||||
| Myb | 6479 | 9 | 44 | 0.0002 | Myb complex, origin binding |
| Mib130 | 2019 | 93 | 61 | 0.006 | Myb complex, origin binding |
| 2603 | 120 | 72 | 0.006 | ||
| Mib120 | 1644 | 31 | 29 | NS | Myb complex, origin binding |
| Mib40 | 3127 | 248 | 153 | 0.005 | Myb complex, origin binding |
| Caf1 | 1532 | 134 | 154 | NS | Myb complex |
Expression levels are indicated as in Table 1.
ORC, origin recognition complex.
DISCUSSION
EST sequencing has long been the core method of approach for novel transcript discovery, but sequencing capacity limits the number of genes and gene variants this approach can identify and provides no information about transcript abundance (32). Ultra-high-throughput sequencing of cDNA is a far more comprehensive way to measure transcriptome composition, but its application to the study of viruses has primarily focused on characterizing within host population diversity (12, 17, 48, 56). However, this approach can also be extremely valuable for understanding viral-host transcriptome interactions (36, 59), especially in situations like the study of BVs where high throughput sequencing is likely the only way to determine how much of an ancestral nudivirus genome remains transcriptionally functional or to comprehensively identify wasp genes with potential roles in replication.
While all BVs initiate replication in the wasp pupal stage, the exact timing of key events during this life stage varies among species (2, 43, 57, 58). We therefore had to first establish the timing of major replication events in M. demolitor before initiating transcriptome studies. Our results show that amplification of genomic DNA containing proviral segment B begins in stage 2 pupae while virion formation begins in stage 3. MdBV replication also continues in newly emerged adults although declines in segment B abundance suggests that replication may slow or cease with age. These data justified our selection of day 1 pupae and day 1 adults as sources of ovary RNA before and during MdBV replication. A second consideration in regard to our results is the phylogenetic placement of M. demolitor relative to the two other species of wasps (C. congregata and C. inanitus) for which detailed data on BV replication are available (2, 10, 30, 35, 40, 41). All BV-carrying wasps reside within the microgastroid complex which is divided into six subfamilies (55). Both C. congregata and M. demolitor reside in the largest of these subfamilies, the Microgastrinae, but belong to the two genera that are the most distantly related to one another in this group (33). C. inanitus by contrast resides in the Cheloninae, which is the subfamily that is most distantly related to the Microgastrinae (33). Thus, comparisons between M. demolitor and C. congregata provide insights into BV replication that spans the breadth of the largest subfamily of BV-carrying braconids (Microgastrinae). Comparison of results generated in C. congregata and M. demolitor to C. inanitus in turn provides data that spans the breadth of the microgastroid complex.
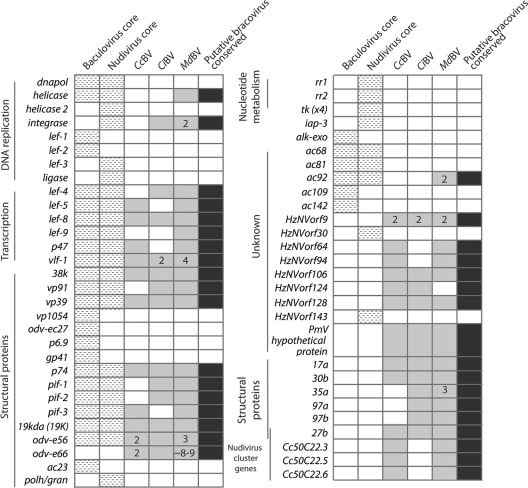
With this background in mind, our data together with results from C. congregata and C. inanitus provide sufficient information to propose a conserved gene set that we hypothesize is present in all BV-carrying wasps. We present this gene set in Fig. 5 together with the 30 core genes present in all known baculoviruses and the 20 members of this baculovirus core set that are present in nudiviruses (23, 39, 51). Our list indicates that all of the nudivirus-like transcripts identified from C. congregata and/or C. inanitus (9) are expressed in M. demolitor ovaries with one exception (HzNVorf124). While homologs of vp91, vlf-1, lef-4, pif-1, and pif-2 were identified in C. inanitus but not C. congregata, their presence in M. demolitor suggests these genes are conserved among all BV-carrying braconids. Likewise, while one or more subunits of the nudivirus/baculovirus RNA polymerase complex (lef-4, lef-9 and p47), late transcription factor (vlf-1) and capping enzyme (lef-5) are missing from the C. congregata or C. inanitus data sets, the presence of each subunit in M. demolitor strongly suggests that all BV-carrying wasps likely have the genes necessary to form a complete RNA polymerase complex. Wetterwald et al. (54) identified 28 non-nudivirus-like proteins from CiBV virions, but the presence of orthologs for only 6 of these factors in M. demolitor suggests most of these proteins are not conserved among BVs generally (Fig. 5). We identified only two other nudivirus or baculovirus-like genes in the M. demolitor transcriptome (helicase and ac92) unknown from C. congregata or C. inanitus. The most notable absences from our proposed BV gene set are a baculovirus/nudivirus-like dnapol, primase (lef-1), primase-associated factor (lef-2), or single-stranded binding protein (lef-3), which together with helicase are potentially important in regulating viral DNA replication (39).
Fig 5.
Predicted BV conserved gene set in relation to the core genes for all known baculoviruses and nudiviruses. Hatched boxes represent core constituents of either baculovirus or nudivirus genomes. Light gray boxes show homologs identified by transcript sequencing of ovaries from Cotesia congregata, which carries CcBV; Chelonus inanitus, which carries CiBV; and M. demolitor, which carries MdBV. Dark gray boxes show the predicted conserved genes for BVs. Numbers within boxes indicate transcripts for which more than 1 locus was identified (duplicated genes).
In undertaking our study, we did not expect to identify so few additional nudivirus- or baculovirus-like genes relative to previously generated EST data (9). The depth of our data set overall suggests that we identified most nudivirus-like genes transcribed in M. demolitor ovaries. However, our approaches also have some weaknesses that could result in some nudivirus-like genes not being identified. First, our data as well as prior EST studies rely on homology for the identification of nudivirus-like genes, but only four nudivirus genomes are available in the literature (51). Given the low similarity of the nudivirus-like genes identified from M. demolitor, C. congregata, and C. inanitus relative to the corresponding homologs in pathogenic nudiviruses, some genes of nudivirus origin could be overlooked because they are too poorly conserved to recognize. Second, it is possible that BVs contain genes of nudivirus origin that are absent in the genomes of the nudiviruses that have been sequenced to date. For example, the predicted 27b gene likely derives from a nudivirus, because it is located in a nudivirus gene cluster in C. congregata and is found in C. inanitus virions, yet there is no 27b homolog among known nudivirus genomes (9, 15).
Third, while our sequencing approach generated much greater coverage than previous EST studies, misassembly issues that result in failure to identify particular transcripts can occur. Ironically, using our method of assembly, this problem most often arises in the case of very-high-coverage transcripts, which can result in fragmentation during assembly. A case in point was our initial failure to identify a transcript in M. demolitor for vp39, which is the most abundantly expressed baculovirus-like gene in M. demolitor ovaries during MdBV replication. However, this initial failure also led us to carefully reanalyze our entire data set using computational approaches that correct for misassembly of high-coverage transcripts. This resulted in the identification of vp39, but we also identified only one other gene (17a) of unknown origin but potential importance in BV replication. Thus, we conclude that misassembly of high-coverage transcripts was not a pervasive problem in our data set. In contrast, our deep sequencing approach is very sensitive to the detection of transcripts of low and intermediate abundance. For example, eukaryotic DNA polymerases are well known to be transcribed at low levels, but we identified homologs for all of the expected eukaryotic dnapol genes in M. demolitor but no nudivirus-like dnapol. We also manually searched our data set using motifs that are highly conserved in all dnapol genes, but this approach did not identify any nudivirus-like dnapol. While we cannot categorically exclude the possibility that M. demolitor contains a nudivirus-like dnapol, our results overall indicate such a gene is not transcribed.
With these caveats in mind, we conclude the genes presented in Fig. 5 represent most of the nudivirus/baculovirus-related genes that are transcriptionally functional in M. demolitor during BV replication. Wasp genome sequence and data on the position of nudivirus-like genes in clusters will be one way that other nudivirus-like genes can be identified (15). Another could be through additional transcriptome data from wasps in subfamilies of the micrograstroid complex outside of the Microgastrinae and Cheloninae.
The presence of a nudivirus/baculovirus DNA helicase in BV-carrying wasps suggests that a small portion of the ancestral viral DNA replication machinery may remain functional during BV DNA replication, but the absence of a viral dnapol and primase suggests that the amplification of proviral loci depends upon wasp gene products. As previously noted, our data set identifies several wasp dnapol genes, but their expression profiles do not correlate well with our observations of the timing of MdBV DNA replication. This is not unexpected for the polymerase B2 domain-containing transcript we named DNA pol B2, which may not be a functional polymerase. Its location in a TE also suggests that it may not have a stable presence in the M. demolitor genome, as would be expected for a gene that is essential to the persistence of a symbiosis that has existed for 100 million years. In contrast, the differential expression of several genes with known functions in locus-specific amplification suggest that these factors could potentially play roles in regulating BV replication. These locus-specific amplification genes could also be important in regulating the amplification of chorion or other wasp genes that are similarly expressed in ovaries during MdBV replication.
A second important finding of our study is that several more nudivirus/baculovirus homologs are duplicated in the M. demolitor genome than previously found for C. congregata and C. inanitus (Fig. 5). Whether these gene variants have different functions is unknown, but their presence is notable because many genes in the encapsidated genomes of BVs are also duplicated (13, 15, 19, 21). In the case of MdBV, virtually all duplicated genes in the encapsidated genome are expressed in parasitized hosts (11), while experimental studies indicate that some gene family members are functionally distinct (7, 29, 49). Sequence analysis of gene families in other BVs also provide evidence for functional specialization (21, 37). Genes belonging to the vlf-1 family of genes in BVs (vlf-1, integrase, and HzNVorf140) could be functionally specialized to partition the roles attributed to VLF-1 in baculoviruses: viral integrase, transcription factor, structural protein, and packaging of DNA into capsids (39).
Our understanding of how host and symbiont genes interact to maintain eukaryotic-microbial symbioses is in its infancy. While most studies of beneficial symbionts focus on bacteria (22, 31), the symbiotic relationship between BVs and parasitoid wasps provides a powerful alternative system for understanding how pathogenic viruses can evolve to form a beneficial association with a eukaryotic host and how virus and wasp genes interact to developmentally synchronize a function like replication that each entity depends upon for survival. The use of massively parallel sequencing is an essential first step to producing a data set that captures a large percentage of the expressed regions of the M. demolitor genome. Designing experiments that functionally test the role of specific viral and wasp genes is obviously a critical next step, as are experiments to distinguish between whether locus-specific amplification or an excision/amplification model underlies replication by BVs more generally.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by grants from the U.S. Department of Agriculture and National Science Foundation to M.R.S.
We also appreciate the feedback of two anonymous reviewers whose suggestions improved the manuscript.
Footnotes
Published ahead of print 11 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ahrens CH, Rohrmann GF. 1996. The DNA polymerase and helicase genes of a baculovirus of Orgyia pseudosugata. J. Gen. Virol. 77:825–837 [DOI] [PubMed] [Google Scholar]
- 2. Albrecht U, et al. 1994. Polydnavirus of the parasitic wasp Chelonus inanitus (Braconidae): characterization, genome organization and time point of replication. J. Gen. Virol. 75:3353–3363 [DOI] [PubMed] [Google Scholar]
- 3. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Annaheim M, Lanzrein B. 2007. Genome organization of the Chelonus inanitus polydnavirus: excision sites, spacers and abundance of proviral and excised segments. J. Gen. Virol. 88:450–457 [DOI] [PubMed] [Google Scholar]
- 5. Asgari S, Reineke A, Beck M, Schmidt O. 2002. Isolation and characterization of a neprilysin-like protein from Venturia canescens virus-like particles. Insect Mol. Biol. 11:477–485 [DOI] [PubMed] [Google Scholar]
- 6. Beck MH, Inman RB, Strand MR. 2007. Microplitis demolitor bracovirus genome segments vary in abundance and are individually packaged in virions. Virology 359:179–189 [DOI] [PubMed] [Google Scholar]
- 7. Beck MH, Strand MR. 2007. A novel polydnavirus protein inhibits the insect prophenoloxidase activation pathway. Proc. Natl. Acad. Sci. U. S. A. 104:19267–19272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beck MH, Zhang S, Bitra K, Burke GR, Strand MR. 2011. The encapsidated genome of Microplitis demolitor bracovirus integrates into the host Pseudoplusia includens. J. Virol. 85:11685–11696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bézier A, et al. 2009. Polydnaviruses of braconid wasps derive from an ancestral nudivirus. Science 323:926–930 [DOI] [PubMed] [Google Scholar]
- 10. Bézier A, Herbinière J, Lanzrein B, Drezen JM. 2009. Polydnavirus hidden face: the genes producing virus particles of parasitic wasps. J. Invertebr. Pathol. 101:194–203 [DOI] [PubMed] [Google Scholar]
- 11. Bitra K, Zhang S, Strand MR. 2011. Transcriptomic profiling of Microplitis demolitor bracovirus reveals host, tissue and stage-specific patterns of activity. J. Gen. Virol. 92:2060–2071 [DOI] [PubMed] [Google Scholar]
- 12. Brackney DE, Beane JE, Ebel GD. 2009. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 5:e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen YF, et al. 2011. Deep sequencing of Cotesia vestalis bracovirus reveals the complexity of a polydnavirus genome. Virology 414:42–50 [DOI] [PubMed] [Google Scholar]
- 14. Cheng CH, et al. 2002. Analysis of the complete genome sequence of the Hz-1 virus suggests that it is related to members of the Baculoviridae. J. Virol. 76:9024–9034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desjardins CA, et al. 2008. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 9:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drezen JM, Herniou EA, Bézier A. 2012. Evolutionary progenitors of bracoviruses, p 15–32 In Beckage NE, Drezen J-M. (ed), Parasitoid viruses: symbionts and pathogens. Academic Press, London, United Kingdom [Google Scholar]
- 17. Eckerle LD, et al. 2010. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6:e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Espagne E, et al. 2004. Genome sequence of a polydnavirus: insights into symbiotic virus evolution. Science 306:286–289 [DOI] [PubMed] [Google Scholar]
- 20. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedman R, Hughes AL. 2006. Pattern of gene duplication in the Cotesia congregata Bracovirus. Infect. Genet. Evol. 6:315–322 [DOI] [PubMed] [Google Scholar]
- 22. Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. U. S. A. 108:2849–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herniou EA, Olszewski JA, Cory JS, O'Reilly DR. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211–234 [DOI] [PubMed] [Google Scholar]
- 24. Honeybee Genome Sequencing Consortium 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443:931–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hubscher U, Maga G, Spadari S. 2002. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71:133–163 [DOI] [PubMed] [Google Scholar]
- 26. Jehle JA. 2010. Nudiviruses, p 153–170 In Asgari S, Johnson KN. (ed), Insect virology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 27. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, et al. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu Z, Beck MH, Strand MR. 2010. Egf1.5 is a second phenoloxidase cascade inhibitor encoded by Microplitis demolitor bracovirus. Insect Biochem. Mol. Biol. 40:497–505 [DOI] [PubMed] [Google Scholar]
- 30. Marti D, Grossniklaus-Burgin C, Wyder S, Wyler T, Lanzrein B. 2003. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. I. Ovary morphogenesis, amplification of viral DNA and ecdysteroid titres. J. Gen. Virol. 84:1141–1150 [DOI] [PubMed] [Google Scholar]
- 31. McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21:1366–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5:621–628 [DOI] [PubMed] [Google Scholar]
- 33. Murphy N, Banks JC, Whitfield JB, Austin AD. 2008. Phylogeny of the parasitic microgastroid subfamilies (Hymenoptera: Braconidae) based on sequence data from seven genes, with an improved time estimate of the origin of the lineage. Mol. Phylogenet. Evol. 47:378–395 [DOI] [PubMed] [Google Scholar]
- 34. Okano K, Vanarsdall AL, Rohrmann GF. 2004. Characterization of a baculovirus lacking the alkaline nuclease gene. J. Virol. 78:10650–10656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pasquier-Barre F, et al. 2002. Polydnavirus replication: the EP1 segment of the parasitoid wasp Cotesia congregata is amplified within a larger precursor molecule. J. Gen. Virol. 83:2035–2045 [DOI] [PubMed] [Google Scholar]
- 36. Peng X, et al. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 1:e00206–e00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Provost B, et al. 2004. Bracoviruses contain a large multigene family coding for protein tyrosine phosphatases. J. Virol. 78:13090–13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rice P, Longden I, Bleasby A. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 39. Rohrmann GF. 2008. Baculovirus molecular biology. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD [Google Scholar]
- 40. Savary S, Beckage N, Tan F, Periquet G, Drezen JM. 1997. Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp Cotesia congregata (Braconidae, Microgastinae) at potential recombinase binding sites. J. Gen. Virol. 78:3125–3134 [DOI] [PubMed] [Google Scholar]
- 41. Savary S, Drezen JM, Tan F, Beckage NE, Periquet G. 1999. The excision of polydnavirus sequences from the genome of the wasp Cotesia congregata (Braconidae, microgastrinae) is developmentally regulated but not strictly restricted to the ovaries in the adult. Insect Mol. Biol. 8:319–327 [DOI] [PubMed] [Google Scholar]
- 42. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 43. Stoltz DB, Vinson SB. 1979. Viruses and parasitism in insects. Adv. Virus Res. 24:125–171 [DOI] [PubMed] [Google Scholar]
- 44. Strand MR. 2010. Polydnaviruses, p 171–197 In Asgari S, Johnson KN. (ed), Insect virology. Caister Academic Press, Norwich, United Kingdom [Google Scholar]
- 45. Strand MR, Johnson JA, Culin JD. 1988. Developmental interactions between the parasitoid Microplitis demolitor (Hymenoptera: Braconidae) and its host Heliothis virescens (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 81:822–830 [Google Scholar]
- 46. Strand MR, McKenzie DI, Grassl V, Dover BA, Aiken JM. 1992. Persistence and expression of Microplitis demolitor polydnavirus in Pseudoplusia includens. J. Gen. Virol. 73:1627–1635 [DOI] [PubMed] [Google Scholar]
- 47. Strand MR, Noda T. 1991. Alterations in the haemocytes of Pseudoplusia includens after parasitism by Microplitis demolitor. J. Insect Physiol. 37:839–850 [Google Scholar]
- 48. Szpara ML, Parsons L, Enquist LW. 2010. Sequence variability in clinical and laboratory isolates of herpes simplex virus 1 reveals new mutations. J. Virol. 84:5303–5313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thoetkiattikul H, Beck MH, Strand MR. 2005. Inhibitor kappaB-like proteins from a polydnavirus inhibit NF-kappaB activation and suppress the insect immune response. Proc. Natl. Acad. Sci. U. S. A. 102:11426–11431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tower J. 2004. Developmental gene amplification and origin regulation. Annu. Rev. Genet. 38:273–304 [DOI] [PubMed] [Google Scholar]
- 51. Wang Y, Jehle JA. 2009. Nudiviruses and other large, double-stranded circular DNA viruses of invertebrates: new insights on an old topic. J. Invertebr. Pathol. 101:187–193 [DOI] [PubMed] [Google Scholar]
- 52. Webb BA, et al. 2006. Polydnavirus genomes reflect their dual roles as mutualists and pathogens. Virology 347:160–174 [DOI] [PubMed] [Google Scholar]
- 53. Werren JH, et al. 2010. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327:343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wetterwald C, et al. 2010. Identification of bracovirus particle proteins and analysis of their transcript levels at the stage of virion formation. J. Gen. Virol. 91:2610–2619 [DOI] [PubMed] [Google Scholar]
- 55. Whitfield JB. 2002. Estimating the age of the polydnavirus/braconid wasp symbiosis. Proc. Natl. Acad. Sci. U. S. A. 99:7508–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wright CF, et al. 2011. Beyond the consensus: dissecting within-host viral population diversity of foot-and-mouth disease virus by using next-generation genome sequencing. J. Virol. 85:2266–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wyder S, et al. 2002. Characterization of Chelonus inanitus polydnavirus segments: sequences and analysis, excision site and demonstration of clustering. J. Gen. Virol. 83:247–256 [DOI] [PubMed] [Google Scholar]
- 58. Wyler T, Lanzrein B. 2003. Ovary development and polydnavirus morphogenesis in the parasitic wasp Chelonus inanitus. II. Ultrastructural analysis of calyx cell development, virion formation and release. J. Gen. Virol. 84:1151–1163 [DOI] [PubMed] [Google Scholar]
- 59. Yuan JF, et al. 2009. Global transcriptional response of pig brain and lung to natural infection by pseudorabies virus. BMC Microbiol. 9:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zollman S, Godt D, Prive GG, Couderc JL, Laski FA. 1994. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 91:10717–10721 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.