Abstract
The establishment of HIV-1 latency can result from limiting levels of transcription initiation or elongation factors, restrictive chromatin modifications, transcriptional interference, and insufficient Tat activity. Since the viral protein Tat can counteract many of these factors, we hypothesized that the presence of exogenous Tat during infection might inhibit the establishment of latency. This was explored using a Jurkat model of latency establishment and reactivation. PCR and reverse transcriptase PCR (RT-PCR) confirmed the latent state in this model and showed evidence of transcriptional interference. To address our hypothesis, cells undergoing infection were first exposed to either purified recombinant Tat or a transactivation-negative mutant. Only the former resulted in a modest inhibition of the establishment of latency. Next, Jurkat cells stably expressing intracellular Tat were used in our latency model to avoid limitations of Tat delivery. Experiments confirmed that intracellular Tat expression did not affect the susceptibility of these cells to viral infection. Eight weeks after infection, Jurkat cells expressing Tat harbored up to 1,700-fold fewer (P < 0.01) latent viruses than Jurkat cells that did not express Tat. Additionally, Tat delivered by a second virus was sufficient to reactivate most of the latent population. Our results suggest that inhibition of the establishment of latent infection is theoretically possible. In a hypothetical scenario of therapy that induces viral gene expression during acute infection, activation of viruses which would otherwise have entered latency could occur while concurrent highly active antiretroviral therapy (HAART) would prevent further viral spread, potentially decreasing the size of the established latent reservoir.
INTRODUCTION
Despite the success of highly active antiretroviral therapy (HAART) for the treatment of HIV/AIDS, the presence of latently infected resting memory CD4+ T cells represents a major barrier to eradication of HIV-1 from an infected individual (45, 55). The latent reservoir is established during acute infection (10), and represents an archive of both wild-type (wt) and drug-resistant viruses (26). Since latent proviruses do not express viral gene products, they are shielded from both antiretroviral drugs and the host immune response, and the long-lived, latently infected host cell is not exposed to viral cytopathic effect. Reactivation of latently infected cells likely serves as the major source of viral rebound upon treatment interruption or failure (31). Thus, HIV-1 latency provides for lifelong persistence of infection and is the target of intense research effort. Several approaches attempting to reactivate the latent reservoir have been used in clinical trials, with the goal of rendering latently infected cells susceptible to immune clearance and/or viral cytopathicity. No clear, long-term benefits have yet been observed from this approach (reviewed in references 6, 22, and 59), although further trials are under way (12). Additional strategies concerning the latent reservoir are clearly needed.
HIV-1 gene expression is dependent upon the viral protein Tat. In the absence of Tat, transcription initiates normally at the 5′ long terminal repeat (LTR) but results in short, abortive viral transcripts due to RNA polymerase II (RNAPII) pausing shortly after promoter clearance. Tat is initially expressed from rare full-length transcripts that are multiply spliced, and it functions as a powerful activator of viral transcription. In contrast to classic DNA-binding transcription factors that control the initiation of transcription, Tat controls transcription at the level of RNAPII elongation through interaction with the TAR RNA (the first 59 nucleotides of each viral transcript) and the positive transcription elongation factor b (P-TEFb, composed of Cyclin T1 [CycT1] and cyclin-dependent kinase [CDK] 9). The recruitment of P-TEFb by Tat leads to several phosphorylation events carried out by CDK9 that convert the paused elongation complex to a highly processive form. CDK9 phosphorylates the negative elongation factors DSIF and NELF, converting DSIF to a positive elongation factor and causing NELF to dissociate from the complex. CDK9 also phosphorylates serine 2 of the RNAPII C-terminal domain heptapeptide repeat, allowing interaction with additional factors involved in productive transcription (reviewed in reference 44). The net result of these posttranslational modifications is synthesis of high levels of full-length viral transcripts.
The establishment of HIV-1 latency primarily results from one or more blocks at the transcriptional level, usually upon infection of an activated CD4+ T cell that transitions to a resting state (66). NF-κB and/or NFAT (depending on the cell type) is required for initiation of viral transcription through binding to κB sites on the 5′ LTR. These transcription factors can have their target DNA sequences occupied by transcriptional repressors or can be sequestered in the cytoplasm (especially in resting CD4+ T cells), limiting transcription initiation such that the virus enters latency (17). Mutations at κB or Sp1 sites on the 5′ LTR can also promote entry into latency (7). Additional blocks at the level of transcription initiation are imposed by specific epigenetic chromatin modifications at nucleosomes on the 5′ LTR, notably, deacetylation and methylation of histone N-terminal tails. These modifications both alter the physical conformation of chromatin and dictate which proteins can interact with chromatin (30), and they are believed to be a driving force in the establishment of HIV-1 latency (43, 60). DNA methylation has been associated with HIV-1 latency but likely enhances silencing of already-latent viruses rather than contributing to entry into latency (4, 33). P-TEFb is required for efficient elongation of viral transcripts. It is regulated by sequestration into the 7SK ribonucleoprotein (RNP) complex that includes 7SK small nuclear RNA (snRNA) and Hexim1; Hexim1 obstructs the ATP pocket of CDK9 (reviewed in reference 71). Additionally, limited levels of active P-TEFb have been associated with the establishment of latency in primary CD4+ T cells (60). Since most HIV-1 integration occurs within introns of actively expressed genes (24, 51, 53, 62), transcriptional interference can result. This phenomenon occurs when transcription from a cellular promoter antagonizes transcription initiation or elongation from the integrated viral promoter, and it is an important mechanism contributing to the establishment and maintenance of latency (17, 20, 25, 37, 53). Additional blocks to elongation can occur if overall Tat activity is insufficient, which can result from subthreshold Tat levels (for example, due to random fluctuations [63–65]) or mutations that attenuate Tat activity (69). Finally, posttranscriptional blocks to HIV-1 gene expression have been associated with latency, including insufficient nuclear export of unspliced viral mRNA (36) and silencing by cellular microRNAs (29), although whether these contribute to the establishment of latency is unclear.
There are several mechanisms by which Tat, if present in sufficient quantities, might counteract the establishment of HIV-1 latency by promoting transcriptional initiation or elongation. The transcriptionally active form of NF-κB, p50/p65, can be sequestered in the cytoplasm by IκBα. Tat itself can induce nuclear translocation of NF-κB p50/p65 (13), probably via direct interaction with PKR (double-stranded RNA-dependent protein kinase) which can result in the degradation of IκBα (14). In addition to promoting transcriptional initiation, NF-κB p50/p65 can displace HDAC1 (histone deacetylase 1)-bound p50/p50 homodimers from κB sites on the viral promoter. Restrictive chromatin modifications are also subject to regulation by Tat. Histone acetyltransferases (HATs), including p300, CBP, and PCAF, are recruited to the 5′ LTR by Tat (3, 28, 39), where they can reverse the effects of histone deacetylation. Nucleosome remodeling is induced by Tat via recruitment of the Ini1, BRG-1, and Brm components of the SWI/SNF chromatin remodeling complex (1, 38, 46, 58) and via recruitment of the histone chaperone hNAP-1 (61), relaxing chromatin structure and thereby permitting transcription. Tat can also overcome blocks to elongation by disruption of the 7SK RNP complex through direct displacement of Hexim1, resulting in increased nuclear levels of enzymatically active P-TEFb (2, 35, 41, 52). Recent findings show that Tat also stimulates elongation through recruitment of the elongation factor ELL2 (which aids elongation by appropriately aligning nascent mRNA in the RNAPII active site) via interaction with AFF4, resulting in cooperative stimulation of elongation between ELL2 and P-TEFb (27). Further, there is evidence that Tat can partially overcome transcriptional interference from some cellular genes, possibly by tipping the “balance of power” in favor of the 5′LTR rather than the cellular promoter (15, 20, 25). Finally, it was reported that CD4+ T cells from patients on suppressive HAART were enriched for viruses with attenuated transactivation activity resulting from mutations in Tat. This implies that decreased levels of Tat activity can contribute to the establishment of latency in vivo (69).
Given the above, we hypothesized that activation of viral gene expression during infection, by exogenous Tat, would inhibit the establishment of latent infection. We found that purified recombinant Tat had only a limited impact on the number of latently infected cells, while Tat that was provided intracellularly strongly inhibited the entry of HIV-1 into latency.
(Work by D.A.D. was performed in partial fulfillment of the requirements for the Ph.D. degree from the Faculty of Graduate Studies and Research, McGill University, Montréal, Québec, Canada.)
MATERIALS AND METHODS
Cells and viruses.
Jurkat (clone E6-1), Jurkat-tat (8), JLTRG-R5 (42), and Tzm-bl cells were obtained through the NIH AIDS Research and Reference Reagent Program. Jurkat and JLTRG-R5 cells were maintained in RPMI 1640 medium (Invitrogen), Jurkat-tat cells were maintained in RPMI 1640 with 800 μg/ml G-418, and Tzm-bl and 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) (Invitrogen). All cells were supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin/streptomycin. The HIV-1 molecular clones pNL4-3 and pNL4-3-ΔE-EGFP (70) were obtained through the NIH AIDS Research and Reference Reagent Program. The following mutations were individually introduced into pNL4-3-ΔE-EGFP using the Stratagene QuikChange II XL site-directed mutagenesis kit: H13L (CAT to TTA), C22G (TGT to GGA) and T23N (ACC to AAC). Pseudovirus was produced by cotransfection of 9 × 106 293T cells with 6.25 μg pVPack-VSV-G (Stratagene)—a vesicular stomatitis virus G protein (VSV-G) envelope-encoding construct—in combination with 18.75 μg of pNL4-3-ΔEnv-EGFP (or tat mutant derivatives) using Lipofectamine 2000 (Invitrogen). Replication-competent virus was similarly produced, using 25 μg of pNL4-3 or pBR-NL4-3-IRES-dsRed. Supernatants were harvested at 48 h posttransfection, clarified by centrifugation for 5 min at 470 × g, and passed through a 0.45-μm-pore-size filter. Virus was treated with 50 U/ml benzonase (Sigma) in the presence of added 10× benzonase buffer (10× buffer contains 500 mM Tris-HCl, pH 8.0, 10 mM MgCl2, and 1 mg/ml bovine serum albumin [BSA]) at 37°C for 20 min to digest contaminating plasmid DNA (49).
Cell culture model of latency establishment and reactivation.
Jurkat or Jurkat-tat cells (3.75 × 105) were infected with 150 ng p24 of VSV-G-pseudotyped NL4-3-ΔE-EGFP (or the tat H13L derivative) in 24-well plates. At 16 h postinfection (p.i.), cells were centrifuged at 470 × g for 5 min, virus-containing medium was removed, and cells were resuspended in 1 ml fresh medium. Cells were cultured for up to 56 days p.i. and split with fresh medium as needed. Beginning 2 days p.i. and on subsequent days, the percentages of active, total, and silent infection was determined as follows. One-third of each well was treated for 24 h with tumor necrosis factor alpha (TNF-α) (20 ng/ml) to reactivate silent/latent virus, one-third of each well was subject to control treatment (RPMI only, since TNF-α stocks were made in RPMI), and medium was added to replenish the remaining one-third of each well. At 24 h after TNF-α or control treatment, cells were fixed in 1% paraformaldehyde (PFA) for 20 min. Flow cytometry was performed using a BD FACSCalibur instrument (Becton Dickinson), and data were analyzed with FCS Express software, by gating for live cells by forward and side scatter and then quantifying the number of cells positive for expression of the virally encoded enhanced green fluorescent protein (EGFP). The percentage of EGFP-positive cells after control treatment represents active infection, the percentage after TNF-α treatment represents total infection, and the value obtained by subtracting the value for active infection from that for total infection represents latent infection.
For reactivation of latent virus (see Fig. 6), archived populations of latent cells, generated as described above, were infected 62 days after the original infection that established the latent population. Latently infected cells (or uninfected Jurkat cells) (2 × 105) were infected with pBR-NL4-3-IRES-dsRed (600 ng p24 per million cells) by spinoculation at 1,500 × g for 2 h at 37°C. Following spinoculation, cells were allowed to rest for 1 h; virus was then removed, and fresh medium was added. At 72 h p.i., cells were fixed and analyzed by flow cytometry as described above.
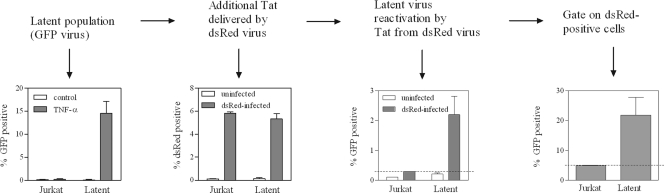
Fig 6.
Delivery of Tat by a second virus reactivates latent virus. (First panel) Populations of latently infected Jurkat cells at 62 days following infection with NL4-3-ΔEnv-EGFP (tat H13L). (Second panel) Latently infected populations of Jurkat cells were infected with replication-competent pBR-NL4-3-IRES-dsRed, to deliver additional Tat. (Third panel) Reactivation of latent virus was determined by measuring levels of EGFP-positive cells following infection with the second virus. (Fourth panel) dsRed-positive cells were gated, and levels of latent virus reactivation in this subpopulation were measured.
PCR, reverse transcriptase PCR (RT-PCR), and real-time PCR. (i) PCR for integrated viral DNA.
Cellular DNA was extracted with a DNeasy blood and tissue kit (Qiagen). A previously described nested Alu-gag real-time PCR (68) was modified as described below for use in endpoint PCR with Platinum Taq (Invitrogen). The first-round reaction (performed in both the presence and absence of an Alu-specific primer) was performed using 26 ng DNA, 2 mM MgCl2, and 200 μM deoxynucleoside triphosphates (dNTPs) in a total volume of 20 μl, using the primers Alu-F (5′-GCCTCCCAAAGTGCTGGGATTACAG-3′) and gag-R (5′-GTTCCTGCTATGTCACTTCC-3′). Cycling conditions were 95°C for 2 min and 15 cycles of 95°C for 15 s, 50°C for 15 s, and 72°C for 3.5 min. A 4-μl sample of the resulting first-round product was used as a template for the second-round nested reaction in the presence of 5 mM MgCl2 (final concentration, including carryover from first round) and 200 μM added dNTPs. Cycling conditions were 95°C for 2 min and 25 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. Second-round primers were LTR-F (5′-TTAAGCCTCAATAAAGCTTGCC-3′) and LTR-R (5′-GTTCGGGCGCCACTGCTAGA-3′). β-Globin was amplified as an internal control using primers BetaGlo-F (5′-GGTACGGCTGTCATCACTTAGAC-3′) and BetaGlo-R (5′-AACGGCAGACTTCTCCTCAG-3′).
(ii) RT-PCR for transcriptional interference.
Using a strategy similar to one previously reported (17), RT-PCR was performed to detect transcriptional interference arising from latent viruses integrated into actively transcribing host genes. The forward primer anneals to U3 sequence, i.e., upstream of the HIV-1 transcription start site, whereas the reverse primer anneals downstream of U5 near the viral primer binding site (PBS), implying that mRNA containing U3-PBS sequence originated from cellular promoters. Cellular RNA was extracted with an RNeasy minikit (Qiagen) and treated with Turbo DNase (Ambion) to remove contaminating genomic DNA. RT-PCR was performed using a SuperScript III one-step RT-PCR kit (Invitrogen) and 50 ng RNA, with primer TI-F1 (5′-CACACACAAGGCTACTTCCCT-3′) or TI-F2 (5′-GCGAGCCCTCAGATGCTAC-3′) and primer TI-R (5′-CTTTCGCTTTCAAGTCCCTGTTC-3′). Cycling conditions were 55°C for 15 min, 94°C for 2 min, and 32 cycles of 94°C for 20 s, 60°C for 20 s, and 68°C for 20 s. Reactions were also performed with Platinum Taq only without reverse transcriptase (“no RT” reactions), to ensure that amplified products were derived exclusively from mRNA. GAPDH mRNA was amplified as an internal control with primers GAPDH-F (5′-AGGTCGGAGTCAACGGATTTGG-3′) and GAPDH-R (5′-GATGGCAACAATATCCACTTTACCA-3′).
(iii) RT-PCR for viral genomic RNA.
Viral RNA was extracted from supernatants of infected cells using a QIAamp viral RNA minikit (Qiagen). Cycling conditions were the same as for the transcriptional interference RT-PCR, above, but for 30 cycles, using primers total-F (5′-CCGTCTGTTGTGTGACTCTGG-3′) and total-R (5′-GAGTCCTGCGTCGAGAGATCT-3′) (67). Products of all PCR and RT-PCRs were visualized on 1% agarose TAE (Tris-acetate-EDTA) gels.
(iv) Real-time PCR for total viral DNA.
Cellular DNA was extracted from infected Jurkat or Jurkat-tat cells with a DNeasy blood and tissue kit (Qiagen). PCR was performed with Platinum qPCR supermix-UDG (Invitrogen) on a Corbett Rotor-Gene 6000 thermocycler. Cycling conditions were 50°C for 2 min, 95°C for 1 min, and 45 cycles of 95°C for 3 s and 60°C for 30 s, using 65 ng DNA per 20-μl reaction. Primers and probes were total-F, total-R, and total-probe (5′-FAM-TCTAGCAGTGGCGCCCGAACAGG-BHQ1-3′) (67).
Cloning, expression, and purification of recombinant Tat.
Tat86 derived from HIV-1IIIB was amplified by RT-PCR from mRNA of Jurkat-tat cells using primers that introduced a 5′ NcoI restriction site (with internal ATG start codon), as well as a 3′ (C-terminal) 6×His tag followed by two stop codons and an AscI restriction site. This PCR product was cloned into the NcoI and AscI sites of a previously used pDEST14 (Invitrogen) expression vector (i.e., the sequence between the vector's attR recombination sites had previously been removed). The resulting construct was confirmed by sequencing and by detection of expressed Tat86-6×His by Western blotting with an anti-Tat monoclonal antibody (NIH AIDS Research and Reference Reagent Program catalog number 1974). C22G and T23N Tat mutations were introduced separately, using a QuikChange II site-directed mutagenesis kit (Stratagene). Expression of recombinant Tat was induced with 1 mM IPTG (for 3 h beginning at an optical density at 600 nm [OD600] of 0.4), and pellets were frozen until further use. Purification was performed as previously reported (56), with several modifications as described below. All solutions were buffered with HEPES at pH 8.0, and siliconized materials were not used until collection of eluted proteins. For each 0.5-liter starting culture, 15 ml lysis buffer (with 20 mM imidazole) was added and sonication was performed on two 7.5-ml samples. Sonication conditions proved to be critical to successful protein purification and were optimized as suggested previously (19); each 7.5 ml of lysate was subject to 10 rounds of sonication for 20 s at 20% amplitude, with a 40-s rest between rounds. Ni-NTA agarose beads (Qiagen) were added to clarified lysates (2 ml of 50% slurry for each 0.5-liter original culture) in the batch method, for 1 h at 4°C. Using 5-ml columns, beads were washed with 40 ml of wash buffer containing Triton X-100, followed by 20 ml of wash buffer without Triton X-100, 5 ml of wash buffer containing 0.5 M NaCl and 100 mM imidazole, and 5 ml of wash buffer containing 0.15 M NaCl and 200 mM imidazole. Recombinant Tat was eluted with 5 ml of elution buffer containing 0.15 M NaCl and 500 mM imidazole. Eluted proteins were dialyzed against 2.5 liters of dialysis buffer (20 mM HEPES, pH 8.0, 0.15 M NaCl, 1 mM dithiothreitol [DTT]) in 3.5K MWCO slide-a-lyzer dialysis cassettes (Thermo Scientific) for 18 h at 4°C. Protein concentration was calculated from absorbance at 280 nm using appropriate extinction coefficients (0.798 and 0.803 for the T23N and C22G variants, respectively, of Tat86-6×His in reducing conditions). 6×His tags were not removed since their presence does not affect Tat's biological activities (56).
Use of recombinant Tat in cell culture.
The HIV-1 LTR/Tat-dependent reporter cell lines Tzm-bl and JLTRG-R5 were treated with recombinant Tat proteins to determine biological activity. Tzm-bl cells were treated in 96-well plates with 2.5 μg of Tat (C22G or T23N) per well for 4 h in serum-free Opti-MEM reduced serum medium (Invitrogen), in the presence or absence of a protein transfection reagent (Bioporter Quickease protein delivery kit; Genlantis), and then a 1× volume of DMEM containing 20% fetal bovine serum (FBS) was added. At 24 h after treatment, luciferase activity was determined using a Bright-Glo luciferase assay system (Promega). JLTRG-R5 cells were treated in 24-well plates with 12.5 μg Tat (C22G or T23N) as described above, in the presence or absence of a protein transfection reagent, and then 1× volume RPMI containing 20% FBS was added. JLTRG-R5 cells were additionally treated with 125 μg Tat (T23N only, due to insufficient yield of C22G) in the absence of protein transfection reagent. At 24 h after treatment, JLTRG-R5 cells were fixed in 1% PFA, and the percentage of GFP-positive cells was determined by flow cytometry as described above. To determine the effects of purified Tat on the establishment of latency, Jurkat cells were treated during infection as follows. Beginning 16 h p.i. (as described above for the Jurkat latency model) cells were spun at 470 × g for 5 min and resuspended in 0.5 ml serum-free media containing 12.5 μg Tat (C22G or T23N) in the presence of a protein transfection reagent. At 4 h later, 0.5 ml RPMI containing 20% serum was added. This treatment was repeated after 24 and 48 h (i.e., on days 2 and 3 p.i.), for a total of three treatments. Cultures were maintained until 23 days p.i. Similar experiments were performed with a single, 24-h treatment of 125 μg Tat (T23N only) beginning at 16 h p.i., in the absence of protein transfection reagent. All experiments with recombinant Tat were performed at least three times, using at least three different purifications of each protein.
Statistical analyses.
Unpaired two-tailed t tests or a one-way analysis of variance (ANOVA) were used to test for statistically significant differences as indicated in the figure legends. When significant differences were found by ANOVA, Bonferroni's multiple comparison posttest was used to determine where such differences exist. All statistical analyses were performed with GraphPad Prism 5.0 software.
RESULTS
Characterization of a Jurkat-based model of HIV-1 latency establishment and reactivation.
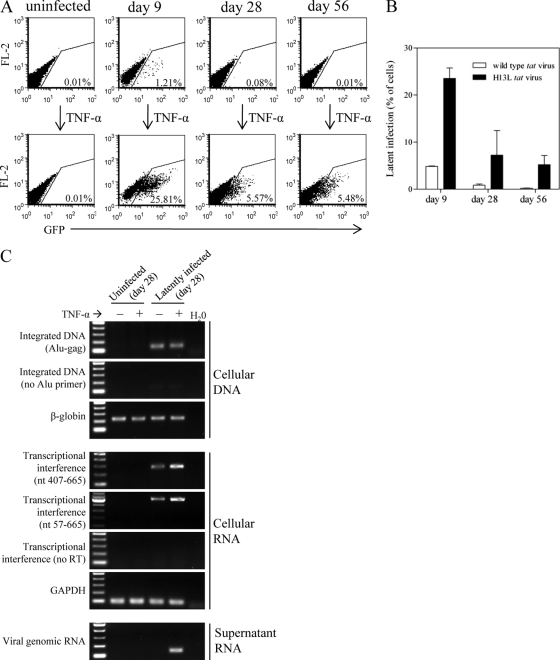
We describe a model of latency establishment and reactivation using CD4+ Jurkat T cells, which generates heterogeneous populations of latently infected cells carrying full-length viral genomes and which does not select for or against initial patterns of viral gene expression (for example, by restricting the study to only GFP+ or GFP− populations via cell sorting). Jurkat cells were infected with NL4-3-Δenv-EGFP and cultured in the absence of selection so as to include all integration events and initial viral gene expression profiles. Following an extended culture period, productively infected cells die by cytopathic effect, leaving only uninfected and latently infected cells. At various times postinfection (p.i.), silent/latent viruses were reactivated with TNF-α and identified by flow cytometry for virus-derived EGFP expression, and they could be detected at frequencies as low as 0.002%. Levels of latent infection are calculated by subtracting levels of active infection (% EGFP+ cells following control treatment) from levels of total infection (% EGFP+ cells following TNF-α treatment). Since relatively low levels of latency were obtained with virus expressing wt tat (Fig. 1B), experiments were also conducted with attenuated tat viruses carrying the H13L mutation (18), which decreases the affinity of Tat for CDK9 (47). This mutation decreases transactivation activity by approximately 40% (data not shown) and has been previously used to generate higher levels of latency (34, 43, 60). Latent infections were 10- to 20-fold more abundant when the attenuated tat virus was used (Fig. 1A and B).
Fig 1.
Model of HIV-1 latency establishment and reactivation. Jurkat cells were infected with NL4-3-ΔE-EGFP or the attenuated tat (H13L) derivative and cultured for up to 8 weeks. Productively infected cells die by cytopathic effect, leaving only uninfected and latently infected cells, which can be reactivated with TNF-α and quantified by flow cytometry for viral EGFP. (A) Representative flow cytometry results for one of three independent experiments with attenuated tat virus. (B) Results of three independent experiments with attenuated or wild-type tat viruses; results represent mean ± standard deviation (SD). (C) PCR and RT-PCR characterization of uninfected or latently infected cells 28 days p.i. Integrated DNA was detected with Alu-gag primers; reactions with no Alu primer serve as a control to confirm that the Alu-gag band is derived from integrated DNA. Two different products of transcriptional interference were detected by RT-PCR, containing viral U3-PBS sequence that is derived from host cell promoters; “no RT” reactions confirm that U3-PBS products are derived from mRNA. Viral genomic RNA was absent in supernatants of control-treated latently infected cells but was detected following reactivation of latent virus by treatment with TNF-α.
The latent state was further characterized at 28 days p.i. by PCR and RT-PCR (Fig. 1C). First, equal levels of integrated viral DNA were present in latently infected cells lacking viral gene expression and TNF-α-treated cells showing EGFP expression. Second, mRNA species containing viral sequence but derived from cellular promoters were present in latently infected (and TNF-α-treated) cells. These transcripts contained U3-PBS sequence spanning nucleotides 407 to 665 and 57 to 665. Viral mRNA does not contain the U3 sequence since the transcription start site is located immediately downstream of U3 at nucleotide 454, implying that these U3-PBS mRNAs originated from host promoters. Such transcriptional interference has previously been reported to be an important mechanism involved in the establishment and maintenance of HIV-1 latency (17, 20, 25, 37, 53). Finally, viral genomic RNA was absent from the supernatants of latently infected cells but was detected in supernatants of these cells following treatment with TNF-α.
Characterization of recombinant Tat proteins in cell culture.
We modified a previously reported protocol to produce Tat that retains a wide array of biological activities and contains low levels of endotoxin (56). We took advantage of the T23N Tat variant, which exhibits greater binding to CDK9 and therefore higher transactivation activity than the wild type (48), since this should yield the greatest effect in terms of LTR transactivation. Additionally, we used the C22G Tat mutant, which cannot interact with CDK9 (21, 72) and is therefore transactivation negative. Two Tat/LTR-dependent reporter cell lines—the HeLa-based Tzm-bl and the Jurkat-based JLTRG-R5—were used to test the transactivation activities of purified T23N or C22G Tat. Direct addition of Tat to JLTRG-R5 cells resulted in detectable activity only at high Tat concentration. However, T23N but not C22G Tat was active in these cells at moderate concentrations when a protein transfection reagent was used, with a maximum of <10% of cells showing detectable Tat activity (Fig. 2A and B). T23N but not C22G Tat had detectable but low activity when added directly to Tzm-bl cells at moderate concentrations but exhibited high levels of activity when a protein transfection reagent was used (Fig. 2C).
Fig 2.
Biological activity of recombinant Tat proteins. Purified T23N (increased transactivation variant) and C22G (transactivation-negative) Tat proteins were added to the LTR/Tat-dependent reporter cell lines JLTRG-R5 (A and B) or Tzm-bl (C), in the presence or absence of a protein transfection reagent. Reporter activity (GFP or luciferase) was determined 24 h after Tat addition. (A) Representative flow cytometry results from one of three independent experiments with JLTRG-R5 cells. Cells were treated with 12.5 μg/ml recombinant Tat using a protein transfection reagent. (B) Results of three independent experiments in JLTRG-R5 cells treated with moderate (12.5 μg/ml) or high (125 μg/ml) concentrations of Tat. C22G Tat was not used for 125-μg/ml Tat treatments. (C) Results of three independent experiments in Tzm-bl cells treated with 12.5 μg/ml Tat. Infection with NL4-3 (50 ng p24 per well; 24 h infection) serves as a reference for levels of luciferase activity. Results in panels B and C represent means ± standard deviations (SD). A protein transfection reagent was used as indicated.
Purified Tat protein modestly inhibits the establishment of HIV-1 latency.
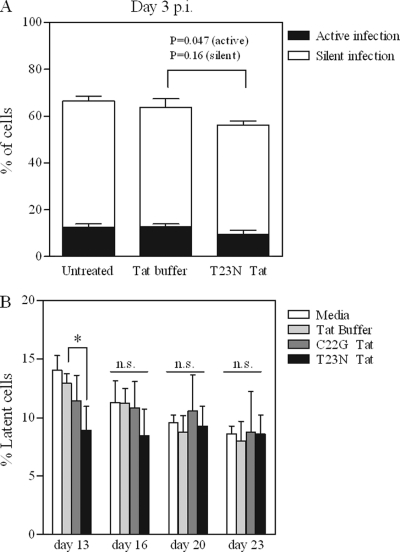
We wished to determine whether purified Tat protein could inhibit the establishment of HIV-1 latency, when added during infection of CD4+ T cells. Initial experiments were carried out by treating Jurkat cells directly (no protein transfection reagent) with a high concentration of T23N Tat for 24 h, beginning 16 h p.i. (since before this time most integration has not occurred) (16). Following Tat treatment, levels of active and silent infection were determined by control or TNF-α treatment followed by flow cytometry for virus-derived EGFP. As shown in Fig. 3A, for cells treated with T23N Tat, a modest but significant reduction in active infection (26% decrease; P = 0.047) was observed compared to control Tat buffer-treated cells. An 8% decrease in silent infection levels was observed for these treatments, although this was not statistically significant (P = 0.16). Analysis at later times or with longer treatment durations was not possible, since this high-concentration Tat treatment resulted in cell death after several days.
Fig 3.
Modest inhibition of the establishment of latency by purified Tat. (A) Jurkat cells were infected with attenuated tat virus as described for Fig. 1. A single 24-h treatment with high concentration (125 μg/ml) T23N Tat, in the absence of a protein transfection reagent, was begun starting at 16 h p.i. At 48 h p.i., cells were treated with TNF-α (or control) to reactivate silently integrated virus. At 72 h p.i., levels of total, active, and silent virus were determined by flow cytometry for viral EGFP as follows: total infection = % GFP+ cells after TNF-α treatment; active infection = % GFP+ cells after control treatment; silent/latent infection = active infection subtracted from total infection. Unpaired two-tailed t tests were performed on T23N-treated versus control Tat buffer-treated cells, for both active infection and silent infection. (B) Jurkat cells were infected with attenuated tat virus as described for Fig. 1. Beginning at 16 h p.i., cells were treated with 12.5 μg/ml C22G or T23N Tat in the presence of a protein transfection reagent for 24 h. This treatment was repeated twice (i.e., starting 24 and 48 h after the first treatment began) for a total of three 24-h treatments. Latent virus was reactivated with TNF-α and quantified by flow cytometry for viral EGFP on days 13, 16, 20, and 23 p.i. One-way ANOVA was performed on the results for each treatment day; Dunnett's multiple comparison post test was used to compare Tat-treated versus buffer-treated cells when significant differences were found by one-way ANOVA. *, P < 0.05; n.s., not significant. All results represent mean ± standard deviation (SD) of results of three independent experiments.
Therefore, similar experiments were carried out with moderate Tat concentrations. A protein transfection reagent was used to introduce T23N or C22G Tat into Jurkat cells daily for each of 3 days p.i., and cultures were continued for up to 23 days to allow for death of productively infected cells and quantification of latent infections. At 13 days p.i., T23N-treated cells harbored 31% fewer latent infections than cells treated with control Tat buffer (P < 0.05); however, this trend was not statistically significant at later times (Fig. 3B).
Intracellular Tat expression does not alter the susceptibility of Jurkat cells to infection.
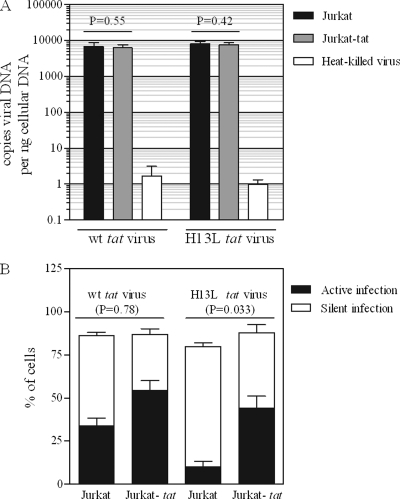
Since our results indicated that addition of purified Tat might inhibit the establishment of latency (Fig. 3) but fewer than 10% of CD4+ Jurkat-based reporter cells exhibited detectable Tat activity (Fig. 2A and B), we hypothesized that a more efficient delivery of Tat might circumvent this issue. Therefore, we took advantage of Jurkat cells that stably express Tat86 (8). To rule out any differences in susceptibility to infection between Jurkat and Jurkat-tat cells, we infected cells under identical conditions with viruses carrying wt or attenuated (H13L) tat genes and confirmed by real-time PCR that viral DNA levels at 18 h p.i. in both cell lines were equivalent (P = 0.55 and P = 0.42 for wt and H13L tat viruses, respectively) (Fig. 4A). To confirm that there are no cell line-specific differences in the capacity for integration and subsequent viral gene expression between Jurkat and Jurkat-tat cells (with the obvious exception of intracellular Tat expression in the latter), we treated cells with TNF-α for 24 h, beginning at 18 h p.i., to induce expression of any silently integrated viral genomes. As expected, total (TNF-α treated) viral gene expression was slightly lower in Jurkat cells infected with attenuated tat virus (8% less than infection of Jurkat-tat cells; P = 0.033) due to the absence of wt Tat (Fig. 4B). However, our results show that the capacity for total viral gene expression in Jurkat and Jurkat-tat cells is equivalent when wt Tat derived from either the cell or the virus is present (P = 0.78). Collectively, these results imply that no Tat-independent differences exist between these two cell lines in terms of their abilities to support viral infection. Therefore, these cells were used to determine the effects of intracellularly expressed Tat on the establishment of HIV-1 latency.
Fig 4.
Jurkat and Jurkat-tat cells are equally susceptible to viral infection. (A) Jurkat or Jurkat-tat cells were infected with VSV-G-pseudotyped NL4-3-ΔE-EGFP (or the H13L tat derivative; 400 ng p24 per 106 cells) for 18 h. Real-time PCR was performed on cells collected 18 h p.i. to determine levels of viral DNA. Heat-killed virus serves as a control for residual plasmid contamination from transfection; mock infections were carried out with heat-killed virus under identical conditions. t tests were used to compare levels of viral DNA in Jurkat versus Jurkat-tat cells. Results represent mean ± standard deviation (SD) of results of two independent experiments. (B) Infections were carried out as for panel A. At 18 h p.i., cells were treated with TNF-α or control, and viral EGFP was measured by flow cytometry 24 h later. t tests were used to compare total (active + silent) infection levels for Jurkat versus Jurkat-tat cells (total infection = % GFP+ cells after TNF-α treatment; active infection = % GFP+ cells after control treatment; silent infection = active infection subtracted from total infection). Results represent mean ± SD of results of four independent experiments.
Intracellular expression of Tat during infection strongly inhibits the establishment of HIV-1 latency.
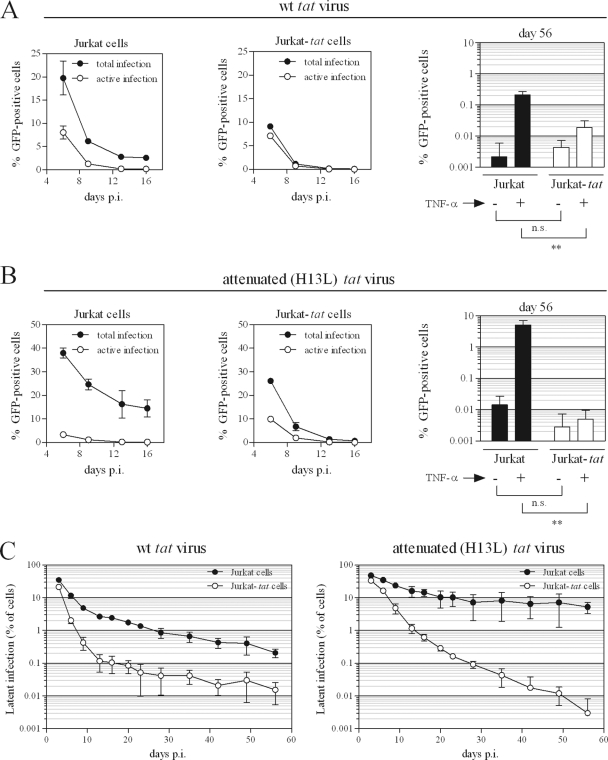
After infection in the presence or absence of intracellularly expressed Tat, levels of active, total, and silent viral gene expression were monitored over the course of 56 days (as in Fig. 1A and B). Actively expressing viral genomes led to the death of infected cells, so that over time, only uninfected and latently infected cells were present. As shown in Fig. 5A, the intracellular expression of Tat caused rapid declines in levels of active and total infection over the first 1 to 2 weeks of infection; this trend is especially pronounced in H13L tat virus-infected cells (Fig. 5B). At 56 days p.i., levels of active infection (control-treated cells) were extremely low in all cases and were not significantly different whether or not intracellular Tat was expressed (P = 0.47 and P = 0.22 for infection with wt or H13L tat virus infection, respectively). However, levels of total infection (TNF-α-treated cells) were significantly lower at 56 days p.i. when intracellular Tat was expressed (P = 0.0066 and P = 0.0098 for wt or H13L tat virus infection, respectively). Levels of latent infection throughout the duration of the experiment are shown in Fig. 5C. For infection with wt tat virus, Jurkat-tat cells harbored 13.5-fold fewer (P = 0.0060) latent infections by day 56 compared to Jurkat cells. When H13L tat virus was used, Jurkat-tat cells harbored >1,700-fold fewer (P = 0.0096) latent infections by day 56 compared to Jurkat cells.
Fig 5.
Potent inhibition of the establishment of latency by intracellularly expressed Tat. (A and B) Jurkat or Jurkat-tat cells were infected with wt (A) or attenuated (B) tat virus as described for Fig. 1. Levels of total and active infection over the first 16 days of infection (left and middle panels) and on day 56 (right panels) are shown. Total infection = % GFP+ cells after TNF-α treatment; active infection = % GFP+ cells after control treatment; silent infection = active infection subtracted from total infection. t tests were used to compare levels of GFP-positive cells at 56 days p.i. following TNF-α or control treatment, for Jurkat versus Jurkat-tat cells. **, P < 0.01; n.s., not significant. (C) Levels of latent infection throughout 56 days are shown for wt (left panel) or attenuated (right panel) tat virus infection of Jurkat and Jurkat-tat cells. All results represent mean ± standard deviation (SD) of results of three independent experiments. t tests were used to compare latent infection levels at 56 days p.i. for Jurkat versus Jurkat-tat cells; P = 0.0060 for wt tat virus; P = 0.0096 for attenuated tat virus.
Tat delivered by a second virus is sufficient to reactivate the majority of latently infected cells.
It is reasonable to speculate that counteracting the mechanisms responsible for maintenance of latency might be more difficult after latency has already been established, as opposed to counteracting these mechanisms while infection is taking place, and we wished to determine whether Tat added via superinfection might be able to reactivate latent virus. We therefore used the replication-competent reporter virus pBR-NL4-3-IRES-dsRed to deliver Tat into latently infected cells. Populations of latently infected cells generated as described in Fig. 1 were infected with this second virus at 62 days following the initial infection that established the latent population. Infection levels were monitored by flow cytometry for dsRed encoded by the second virus, and reactivation of latent virus was determined by measurement of viral EGFP levels. The results (Fig. 6) show that Tat delivered in this manner was able to reactivate the vast majority of the latent population for those cells to which Tat was delivered by the second virus.
DISCUSSION
Latently infected resting memory CD4+ T cells are of critical importance given that the latent reservoir is established early during acute infection (10), is not susceptible to antiretroviral therapy or host immune attack, and serves as the major source of viral rebound upon treatment interruption or failure (31). Although several approaches to reactivate latent reservoirs have been used in clinical trials and further trials are under way (6, 12, 22, 59), additional strategies to combat HIV-1 latency would be invaluable. The viral Tat protein might, when present, counteract many of the mechanisms involved in the establishment of HIV-1 latency. Tat might do so by (i) inducing nuclear translocation of active NF-κB (13, 14), (ii) recruiting HATs (3, 28, 39), members of the SWI/SNF chromatin remodeling complex (1, 38, 46, 58), and histone chaperones (61), (iii) directly displacing Hexim1 from the 7SK RNP complex, thereby increasing levels of active P-TEFb (2, 35, 41, 52), (iv) recruiting the elongation factor ELL2 (27), and (v) overcoming transcriptional interference from some cellular promoters (15, 20, 25). Therefore, we wished to determine whether exogenous Tat, if present during infection, could inhibit the establishment of HIV-1 latency.
We first characterized a model of latency establishment and reactivation that generates heterogeneous populations of latently infected cells (Fig. 1). In this model, productively infected cells die by cytopathic effect, similar to in vivo infection, while only uninfected and latently infected cells propagate. Potential integration site biases were excluded by the absence of any selection, such that all potential latent infection events can be represented in the population. Since TNF-α has been shown to reactivate latent viruses in all Jurkat models of HIV-1 latency (9), this was used in our model to quantify numbers of latent infections. Gating 50,000 live cells by flow cytometry permitted the detection of latent viruses to a frequency of 0.002%. The use of viruses with the attenuated H13L tat mutation increased the number of latent infections by 10- to 20-fold (Fig. 1B).
A more detailed characterization of our model by PCR and RT-PCR included detection of integrated DNA, products of transcriptional interference, and viral genomic RNA (Fig. 1C). Unexpectedly, transcriptional interference appeared to increase rather than decrease upon reactivation of latent viruses by TNF-α treatment. While the reasons for this observation are unknown, it is possible that TNF-α induction of a large number of cellular genes results in a global increase in transcriptional interference, despite viral reactivation. Likewise one might hypothesize that after induction by TNF-α treatment, viral transcription could initiate at additional, upstream transcription start sites prior to the classic “+1” site. This is reasonable to speculate, given that dispersed transcription initiation from multiple sites spanning up to 100 nucleotides occurs at many vertebrate promoters (reviewed in reference 32), although we are unaware of any evidence for this at the HIV-1 5′ LTR. Therefore, RT-PCR was repeated using a forward primer annealing ∼400, rather than ∼50, nucleotides upstream of the transcription start site, and again we observed an increase in transcriptional interference following TNF-α treatment. This likely rules out transcription initiation from dispersed upstream sites. These observations highlight the idea that transcriptional interference—a series of related mechanisms operating at the transcriptional level (reviewed in references 40 and 54)—plays a complex role in the establishment and maintenance of latency, as reported in more detailed studies of this phenomenon (20, 25, 37, 53).
To determine whether exogenous Tat might inhibit the establishment of HIV-1 latency, purified Tat proteins were first assayed for biological activity on two reporter cell lines. While a Tat variant with increased transactivation activity (T23N) resulted in high-level LTR-driven gene expression when introduced into Tzm-bl cells (Fig. 2C), treatment of more biologically relevant CD4+ JLTRG-R5 T cells indicated that a maximum of <10% of treated cells displayed Tat-mediated transactivation activity (Fig. 2A and B). This indicated that any impact purified Tat might have on the establishment of latent infections could be limited to a subpopulation of cells, i.e., those cells which contained biologically active exogenous Tat. This appears to have been the case, as the results of Fig. 3 suggest that treatment of Jurkat cells with purified Tat protein during infection had only moderate effects at the population level. Only T23N Tat-treated cells, and not C22G Tat-treated cells, exhibited a decrease in latent infection levels, a likely reflection of the affinity of each Tat variant studied for P-TEFb. Unfortunately it was not possible to determine whether this limited effect was due to insufficient overall Tat activity as opposed to the absence of Tat in ∼90% of cells undergoing viral infection. This is because when latent infection levels were measured at various days p.i., it was not possible to determine which individual cells might have contained exogenous Tat during the prior treatment period. It might be that the establishment of latency was inhibited to a greater extent in cells which contained exogenous Tat but that this effect was diluted by the majority of cells that were not affected.
To avoid the concerns associated with delivery of purified Tat, Jurkat-tat cells were used to provide Tat intracellularly during infection and throughout the subsequent period of culture. After confirming that the expression of intracellular Tat did not alter the susceptibility of Jurkat-tat cells to infection (Fig. 4), we looked for effects of intracellular Tat on the establishment of latency. During the first days following infection, intracellular Tat led to rapidly reduced levels of active and total viral infection (Fig. 5A and B, Jurkat-tat versus Jurkat), due to greater initial viral gene expression and the resulting deaths of productively infected cells (data not shown). The full impact of intracellular Tat expression on the establishment of HIV-1 latency can best be appreciated in the results of Fig. 5C. While a 13.5-fold reduction in latency levels was observed for wt tat virus infections by day 56 p.i., a >3-log decrease in the number of latent infections was observed for attenuated tat virus infections. The substantially greater effect observed for attenuated tat virus infection is likely due to the higher number of potential latent infections associated with this virus (Fig. 1B) that were able to be inhibited by the Tat provided intracellularly.
It is likely that any attempt to limit the establishment of the latent reservoir will need to focus not only on inhibition of future latent infection but also on the presence of cells that are already latently infected, regardless of how early such an attempt is made. Further, it is probable that reactivation of an already-silenced virus might be more difficult than inhibiting the establishment of latency, since in the latter case, silencing has not yet been “locked in.” Therefore, we delivered Tat to latently infected cells by infection with a second virus, which was distinguishable from the latent virus population by use of a fluorescence marker. Figure 6 shows that reactivation of latent virus was achieved as a result of the Tat that was delivered by the second virus (second and third panels). By comparing population-wide levels of latency (Fig. 6, first panel) to levels of latent virus reactivation in cells that contained the second virus (Fig. 6, last panel), it can be appreciated that the vast majority of latent viruses are potentially able to be reactivated when additional Tat is delivered.
A limitation of our work is that while our experiments used CD4+ Jurkat T cells, important differences might exist in primary cells. For example, induction of NF-κB is sufficient to reactivate virus in Jurkat models of latency, but evidence suggests that induction of NFAT and/or P-TEFb is required to reactivate virus in primary cell models of latency (5, 9, 60).
Current reactivation strategies focus on already-established latent reservoirs, typically in patients past the stage of acute infection. These existing approaches could be complemented by strategies which attempt to limit the initial size of the latent reservoir. This idea has been discussed in recent studies, which have suggested that early treatment initiation during acute infection might decrease the size of established latent reservoirs (11, 23, 50, 57). Resolving whether this actually does occur has been highlighted as a question of importance (59). Our results suggest that inhibition of the establishment of latent infection events is theoretically possible. A hypothetical therapy option during acute infection could include treatment with compounds that aim to counteract the factors involved in the establishment of latency. These compounds could induce PKC activity or increase the available P-TEFb pool, or they could inhibit restrictive chromatin modifications. In such a scenario, activation of viruses which would otherwise have entered latency could occur, while concurrent HAART would prevent further viral spread, potentially decreasing the size of the established latent reservoir.
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institutes of Health Research (CIHR). D.A.D. is the recipient of a predoctoral fellowship from CIHR.
We thank Tamara Bar-Magen for providing assistance with protein purification and Maureen Oliveira, Susan Colby-Germinario, and Cesar Collazos for providing technical assistance. pBR-NL4-3-IRES-dsRed was kindly provided by J. Münch and F. Kirchhoff.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F. 2006. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barboric M, et al. 2007. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 35:2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benkirane M, et al. 1998. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J. Biol. Chem. 273:24898–24905 [DOI] [PubMed] [Google Scholar]
- 4. Blazkova J, et al. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bowman MC, Archin NM, Margolis DM. 2009. Pharmaceutical approaches to eradication of persistent HIV infection. Expert Rev. Mol. Med. 11:e6. [DOI] [PubMed] [Google Scholar]
- 7. Burnett JC, et al. 2010. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 84:5958–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caputo A, Sodroski JG, Haseltine WA. 1990. Constitutive expression of HIV-1 tat protein in human Jurkat T cells using a BK virus vector. J. Acquir. Immune Defic. Syndr. 3:372–379 [PubMed] [Google Scholar]
- 9. Chan JK, Greene WC. 2011. NF-κB/Rel: agonist and antagonist roles in HIV-1 latency. Curr. Opin. HIV AIDS 6:12–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chun TW, et al. 1998. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 95:8869–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chun TW, et al. 2007. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 195:1762–1764 [DOI] [PubMed] [Google Scholar]
- 12. Cohen J. 2011. The emerging race to cure HIV infections. Science 332:784–785, 787–789 [DOI] [PubMed] [Google Scholar]
- 13. Demarchi F, d'Adda di Fagagna F, Falaschi A, Giacca M. 1996. Activation of transcription factor NF-κB by the Tat protein of human immunodeficiency virus type 1. J. Virol. 70:4427–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demarchi F, Gutierrez MI, Giacca M. 1999. Human immunodeficiency virus type 1 tat protein activates transcription factor NF-κB through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR. J. Virol. 73:7080–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. De Marco A, et al. 2008. Intragenic transcriptional cis-activation of the human immunodeficiency virus 1 does not result in allele-specific inhibition of the endogenous gene. Retrovirology 5:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donahue DA, et al. 2010. Stage-dependent inhibition of HIV-1 replication by antiretroviral drugs in cell culture. Antimicrob. Agents Chemother. 54:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duverger A, et al. 2009. Determinants of the establishment of human immunodeficiency virus type 1 latency. J. Virol. 83:3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emiliani S, et al. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feliu JX, Cubarsi R, Villaverde A. 1998. Optimized release of recombinant proteins by ultrasonication of E. coli cells. Biotechnol. Bioeng. 58:536–540 [DOI] [PubMed] [Google Scholar]
- 20. Gallastegui E, Millan-Zambrano G, Terme JM, Chavez S, Jordan A. 2011. Chromatin reassembly factors are involved in transcriptional interference promoting HIV latency. J. Virol. 85:3187–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garber ME, et al. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Geeraert L, Kraus G, Pomerantz RJ. 2008. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu. Rev. Med. 59:487–501 [DOI] [PubMed] [Google Scholar]
- 23. Gianella S, et al. 2011. Effect of early antiretroviral therapy during primary HIV-1 infection on cell-associated HIV-1 DNA and plasma HIV-1 RNA. Antivir. Ther. 16:535–545 [DOI] [PubMed] [Google Scholar]
- 24. Han Y, et al. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han Y, et al. 2008. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4:134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han Y, Wind-Rotolo M, Yang H-C, Siliciano JD, Siliciano RF. 2007. Experimental approaches to the study of HIV-1 latency. Nat. Rev. Microbiol. 5:95–106 [DOI] [PubMed] [Google Scholar]
- 27. He N, et al. 2010. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol. Cell 38:428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hottiger MO, Nabel GJ. 1998. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J. Virol. 72:8252–8256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang J, et al. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 30. Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 31. Joos B, et al. 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. U. S. A. 105:16725–16730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Juven-Gershon T, Kadonaga JT. 2010. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339:225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim YK, Mbonye U, Hokello J, Karn J. 2011. T-cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 410:896–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krueger BJ, Varzavand K, Cooper JJ, Price DH. 2010. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One 5:e12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. 2006. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lenasi T, Contreras X, Peterlin B. 2008. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mahmoudi T, et al. 2006. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J. Biol. Chem. 281:19960–19968 [DOI] [PubMed] [Google Scholar]
- 39. Marzio G, Tyagi M, Gutierrez MI, Giacca M. 1998. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl. Acad. Sci. U. S. A. 95:13519–13524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. 2007. Transcriptional interference: an unexpected layer of complexity in gene regulation. J. Cell Sci. 120:2755–2761 [DOI] [PubMed] [Google Scholar]
- 41. Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. 2010. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 6:e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ochsenbauer-Jambor C, Jones J, Heil M, Zammit KP, Kutsch O. 2006. T-cell line for HIV drug screening using EGFP as a quantitative marker of HIV-1 replication. Biotechniques 40:91–100 [DOI] [PubMed] [Google Scholar]
- 43. Pearson R, et al. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82:12291–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peterlin BM, Brogie JE, Price DH. 2012. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 3:92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pomerantz RJ. 2002. Reservoirs of human immunodeficiency virus type 1: the main obstacles to viral eradication. Clin. Infect. Dis. 34:91–97 [DOI] [PubMed] [Google Scholar]
- 46. Pumfery A, et al. 2003. Chromatin remodeling and modification during HIV-1 Tat-activated transcription. Curr. HIV Res. 1:343–362 [DOI] [PubMed] [Google Scholar]
- 47. Reza SM, Rosetti M, Mathews MB, Pe'ery T. 2003. Differential activation of Tat variants in mitogen-stimulated cells: implications for HIV-1 postintegration latency. Virology 310:141–156 [DOI] [PubMed] [Google Scholar]
- 48. Reza SM, et al. 2003. A naturally occurring substitution in human immunodeficiency virus Tat increases expression of the viral genome. J. Virol. 77:8602–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sastry L, Xu Y, Cooper R, Pollok K, Cornetta K. 2004. Evaluation of plasmid DNA removal from lentiviral vectors by benzonase treatment. Hum. Gene Ther. 15:221–226 [DOI] [PubMed] [Google Scholar]
- 50. Schmid A, et al. 2010. Profound depletion of HIV-1 transcription in patients initiating antiretroviral therapy during acute infection. PLoS One 5:e13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schroder AR, et al. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521–529 [DOI] [PubMed] [Google Scholar]
- 52. Sedore SC, et al. 2007. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 35:4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shan L, et al. 2011. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J. Virol. 85:5384–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shearwin KE, Callen BP, Egan JB. 2005. Transcriptional interference—a crash course. Trends Genet. 21:339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen L, Siliciano RF. 2008. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection. J. Allergy Clin. Immunol. 122:22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Siddappa NB, et al. 2006. Transactivation and signaling functions of Tat are not correlated: biological and immunological characterization of HIV-1 subtype-C Tat protein. Retrovirology 3:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Strain MC, et al. 2005. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J. Infect. Dis. 191:1410–1418 [DOI] [PubMed] [Google Scholar]
- 58. Treand C, et al. 2006. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 25:1690–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trono D, et al. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329:174–180 [DOI] [PubMed] [Google Scholar]
- 60. Tyagi M, Pearson RJ, Karn J. 2010. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J. Virol. 84:6425–6437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vardabasso C, Manganaro L, Lusic M, Marcello A, Giacca M. 2008. The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology. 5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. 2007. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 17:1186–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182 [DOI] [PubMed] [Google Scholar]
- 64. Weinberger LS, Dar RD, Simpson ML. 2008. Transient-mediated fate determination in a transcriptional circuit of HIV. Nat. Genet. 40:466–470 [DOI] [PubMed] [Google Scholar]
- 65. Weinberger LS, Shenk T. 2007. An HIV feedback resistor: auto-regulatory circuit deactivator and noise buffer. PLoS Biol. 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Williams S, Greene W. 2007. Regulation of HIV-1 latency by T-cell activation. Cytokine 39:63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yamamoto N, et al. 2006. Analysis of human immunodeficiency virus type 1 integration by using a specific, sensitive and quantitative assay based on real-time polymerase chain reaction. Virus Genes 32:105–113 [DOI] [PubMed] [Google Scholar]
- 68. Yu JJ, et al. 2008. A more precise HIV integration assay designed to detect small differences finds lower levels of integrated DNA in HAART treated patients. Virology 379:78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yukl S, et al. 2009. Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 387:98–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang H, et al. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou Q, Yik JH. 2006. The yin and yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70:646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu Y, et al. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]