Abstract
Influenza virus enters host cells by endocytosis. The low pH of endosomes triggers conformational changes in hemagglutinin (HA) that mediate fusion of the viral and endosomal membranes. We have used cryo-electron tomography to visualize influenza A virus at pH 4.9, a condition known to induce fusogenicity. After 30 min, when all virions are in the postfusion state, dramatic changes in morphology are apparent: elongated particles are no longer observed, larger particles representing fused virions appear, the HA spikes become conspicuously disorganized, a layer of M1 matrix protein is no longer resolved on most virions, and the ribonucleoprotein complexes (RNPs) coagulate on the interior surface of the virion. To probe for intermediate states, preparations were imaged after 5 min at pH 4.9. These virions could be classified according to their glycoprotein arrays (organized or disorganized) and whether or not they have a resolved M1 layer. Employing subtomogram averaging, we found, in addition to the neutral-pH state of HA, two intermediate conformations that appear to reflect an outwards movement of the fusion peptide and rearrangement of the HA1 subunits, respectively. These changes are reversible. The tomograms also document pH-induced changes affecting the M1 layer that appear to render the envelope more pliable and hence conducive to fusion. However, it appears desirable for productive infection that fusion should proceed before the RNPs become coagulated with matrix protein, as eventually happens at low pH.
INTRODUCTION
Influenza virus is an enveloped, single-stranded negative-sense RNA virus belonging to the Orthomyxoviridae family. Influenza A virions have three integral membrane proteins: an ion channel protein (M2) and two glycoproteins, hemagglutinin (HA), required for entry into host cells, and neuraminidase (NA), involved in the release of progeny virions from the host cell. Underneath the membrane, most virions have a layer of M1 matrix protein enclosing the viral ribonucleoprotein complexes (RNPs).
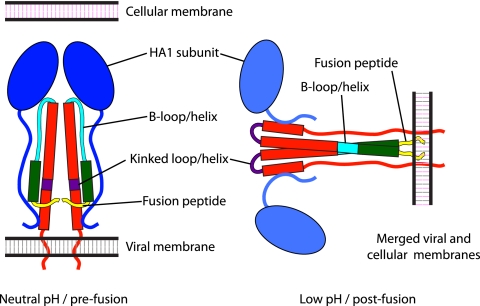
HA is a trimeric class I fusion protein synthesized as a precursor (HA0) that, to become fusion active, is cleaved by a cellular protease, yielding HA1 and HA2 subunits which remain disulfide linked (39). After the virus enters a host cell, HA is transformed by the low pH of the endosomes (4). The generally accepted model for these changes is based on X-ray crystallographic studies of the HA ectodomain at neutral (42) and low (2, 6, 10) pH. This model explains the low-pH-induced transition as an irreversible process (Fig. 1) in which (i) the HA1 membrane-distal domains dissociate (1, 15, 22), retaining their neutral pH structure; (ii) the fusion peptides are transferred to the membrane-distal region after a loop-to-helix transition in amino acids 55 to 76 of HA2 (known as the B loop); and (iii) there is a helix-to-loop transition in segment 106 to 112 of HA2 (the “kinked loop”) that positions the transmembrane domains and fusion peptides on the same side of the glycoprotein (19). The order in which these rearrangements take place, the existence of possible intermediate states, and the reversibility or otherwise of successive steps all remain to be established (11). However, mounting evidence points to the existence of reversible intermediate states, which would include an initial exposure of the fusion peptide and the kinked loop domain (25, 40) and a transition of the B loop to a loose conformation accompanied by a deformation of the HA1 subunit (43).
Fig 1.
Schematic representation of the conformations adopted by HA. Alpha-helices are represented by rectangles, loops by curved lines, and HA1 subunits by ovals. Color coding is as follows: HA1, blue; HA2, red, with amino acids 1 to 23 (fusion peptide) in yellow; amino acids 24 to 54, green; amino acids 55 to 76 (B loop/helix), cyan; and amino acids 106 to 112 (kinked loop/helix), purple.
Previously, cryo-electron microscopy (cryo-EM) has been used to image influenza virions at neutral pH (3, 14, 44) and low pH (23, 32, 36, 37). However, the coprojection of structures from all levels in these ∼100-nm-thick particles limited the interpretability of the resulting images. More recently, cryo-electron tomography (cryo-ET), which is capable of generating three-dimensional images of individual virions, has been used to compile a systematic account of the pleiomorphy exhibited in preparations of mostly spherical virions at neutral pH (17), to characterize filamentous viruses (7), and to investigate the initial steps of membrane fusion as mediated by influenza virus (24). In the present study, we used cryo-ET to characterize the structural changes undergone by virions of the X-31 strain of influenza A virus (serotype H3N2) when transferred to pH 4.9. Particular attention was paid to HA, whereby subtomogram averaging (12, 26, 41, 45, 47) was used to distinguish and characterize discrete conformational states of this fusogenic glycoprotein. We also observed major changes affecting the layer of matrix protein.
MATERIALS AND METHODS
Virus propagation and purification.
X-31 [A/Aichi/68 (H3N2)] influenza virus, grown in embryonated chicken eggs, purified according to standard procedures, and diluted to ∼2 mg/ml protein in 140 mM NaCl, 10 mM HEPES, pH 7.4, was purchased from Charles River Laboratories (North Franklin, CT).
Cryo-electron tomography.
Grids of influenza virus at pH 7.4 were prepared essentially as described previously (17). In brief, virus was mixed 1:1 with a suspension of 10-nm bovine serum albumin (BSA)-coated colloidal gold particles (Aurion, Wageningen, The Netherlands) to serve as fiducial markers. Four-microliter drops were then applied to R2/2 holey carbon grids (Quantifoil; SPI, West Chester, PA), thinned by blotting, and vitrified by plunge-freezing in liquid ethane with a Vitrobot (FEI, Hillsboro, OR). To prepare the low-pH samples, Quantifoil grids were placed for 5 min on 10-μl drops of the virus suspension mixed 1:1 with gold particles. The pH was then lowered to 4.9 by adding 5 μl of 130 mM NaCl, 50 mM sodium acetate, pH 4.6. In experiments addressing the reversibility of the low-pH-induced changes, 1.25 μl of 0.5 M HEPES at pH 8.1 was added after 5 min of incubation at pH 4.9 in order to bring the final pH to 7.4. All incubations were done at room temperature (22°C) and in a humid chamber to avoid possible effects from buffer evaporation. After incubation, viruses were vitrified as described above. The resulting grids were then transferred to a cryogenic specimen holder (type 626; Gatan, Warrendale, PA) for data acquisition. Single-axis tilt series were recorded on a Tecnai-12 transmission electron microscope (TEM) (FEI) operating at 120 keV. The microscope was equipped with an energy filter (GIF 2002; Gatan) operating in zero-loss mode with an energy slit width of 20 eV. Images were acquired on a 2,048- by 2,048-pixel charge-coupled device (CCD) camera (Gatan), using SerialEM (29) for automated tilting, tracking, focusing, and image recording. In each tilt series, images were recorded in 2° increments over an angular range of approximately −66° to +66°, at an effective magnification of 38,500× (0.78-nm pixel size). Electron dose per image was ∼1 e−/Å2, for a cumulative dose of ∼70 e−/Å2 per tilt series. Image defocus was set at −4 μm, corresponding to a first contrast transfer function zero at (3.7 nm)−1.
Tomogram reconstruction and image processing.
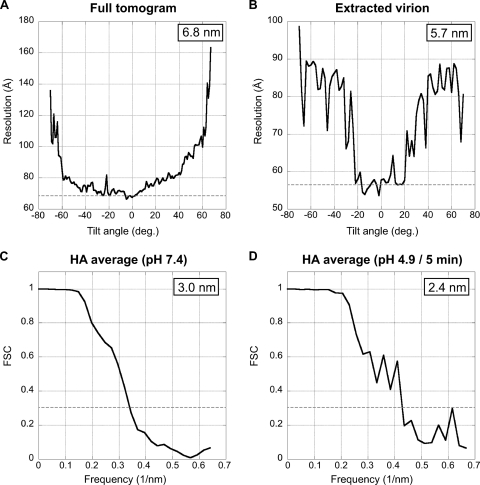
Data were processed using the Bsoft package (21). Projections were aligned using 10-nm gold particles as fiducial markers, and tomograms were then reconstructed. The in-plane resolution of the tomograms was estimated as ∼6.5 nm for full tomograms and 5.5 to 6.5 nm for individual virions as calculated by the NLOO-2D (noise-compensated leave-one-out in two dimensions) method (8) (Fig. 2A and B). Subtomograms containing individual virus particles were extracted and denoised by 20 iterations of anisotropic nonlinear diffusion (13) for further analysis.
Fig 2.
Resolution curves. (A and B) Variation of resolution with tilt angle, as measured by NLOO-2D for a full tomogram (A) and for an extracted virion (B). A cutoff threshold of 0.3 was used. (C and D) Fourier shell correlation (FSC) curves for the HA average from virions at neutral pH (A) and of the HA average (neutral pH-like) from pH 4.9/5-min virions with organized glycoprotein array and resolved M1 layer. Resolution (0.3 threshold) for each average is indicated.
HA averaging.
Individual HA molecules were located on the denoised virion-containing subtomograms and extracted from the corresponding raw subtomograms in volumes of 50 by 50 by 50 voxels. The center of each virion was used as a reference point to determine an initial z-axis orientation for each glycoprotein. HAs from neutral-pH virions and from each low-pH group were aligned and averaged independently, using appropriate missing-wedge masks. The same synthetic model, consisting of a cylinder of approximately the same size as the neutral-pH HA spike and two sheets of density representing the viral membrane and matrix layers, was used as an initial reference in each case. Alignment procedures were performed with routines from Bsoft, modified as needed and wrapped into Python scripts. To improve the resulting density maps, original data were masked after a stable solution was obtained (usually, after 3 or 4 rounds of iteration). After seven rounds of refinement with the masked data, glycoproteins whose direction differed by more than 45° from the initial one (16 to 36% of the HAs, depending on the group) were discarded. Classification and averaging were then performed by maximum likelihood as implemented in the Xmipp package (35). The resolutions of the averages were 2.4 to 3.2 nm for different data sets, as calculated in terms of Fourier shell correlation coefficients (0.3 threshold) (Fig. 2C and D). All averages were low pass-filtered, and 3-fold symmetry was applied. The top 75% of subvolumes from each class, as ranked by correlation coefficients, were used to calculate the final density maps.
RESULTS
Structural changes in influenza virus after 30 min at pH 4.9.
X-31 virions at neutral pH have been assigned to five morphological classes (17). The primary criteria were the presence or absence of a layer of the M1 matrix protein, underlying and resolved from the lipid bilayer of the viral envelope, and their internal contents, which consisted either of RNPs ∼14 nm in diameter and of variable length or a single solenoid with a diameter of ∼80 nm and a pitch of ∼10 nm. Solenoid-containing virions are quite common in certain strains of influenza virus, such as A/USSR/50/79 or A/Texas/1/77 (30), but they are rare with X-31, accounting for <2% of the virions observed in the earlier study (17) and a negligibly small fraction of the current batch. Accordingly, as starting material for acidification experiments, we could focus on the three RNP-containing classes of virions: class I (spherical, with visible M1 matrix layer), class II (elongated, with visible M1 layer), and class III (spherical, lacking a visible M1 layer).
In these experiments, we distinguish particles according to two features: whether or not there is a resolved M1 layer (with this simplification, virions of classes I and II are grouped together [Table 1 and Fig. 3A to C]) and whether the array of glycoprotein spikes is “organized” or “disorganized.” In neutral-pH virions, the array is always organized in the sense that although the spikes do not form a crystalline lattice, they are close packed with an average center-to-center spacing of ∼11 nm, and the distinctive “peanut” morphology of HA spikes and “mushroom” morphology of NA spikes are clearly defined. In disorganized arrays (of virions exposed to low pH), HA spikes, which form ∼85% of the total, present mostly as blurred densities in which it is difficult to distinguish individual molecules.
Table 1.
Incidence of a visible M1 matrix layer and an organized glycoprotein array of influenza virions under three conditionsa
| Condition | % of virions (n) |
|||
|---|---|---|---|---|
| Resolved M1 layer |
No M1 layer seen |
|||
| Organized glycoprotein array | Disorganized glycoproteins | Organized glycoprotein array | Disorganized glycoproteins | |
| pH 7.4b | 89 (54) | 11 (7) | ||
| pH 4.9/5 min | 34 (61) | 21 (38) | 14 (25) | 31 (55) |
| pH 4.9/30 min | 20 (4) | 80 (16) | ||
When there is a partial M1 layer in pH 4.9/30-min virions, it is seen only in small patches; therefore, these virions were classified as M1 layer negative. Four tomograms were employed for the analysis of neutral-pH virions, 31 were used for the pH 4.9/5-min virions, and 3 were used for the pH 4.9/30-min virions. All pH 7.4 virions had dispersed RNPs. For pH 4.9/5-min virions, RNP organization was as follows: all virions with an M1 layer and an organized array of spikes had dispersed RNPs; ∼50% (n = 18) of virions with an M1 layer and disorganized spikes had dispersed RNPs, while the rest had coagulated RNPs; ∼30% (n = 8) of virions without a visible M1 layer and with an organized spike array had dispersed RNPs, while ∼70% had coagulated RNPs; and all virions without a visible M1 layer and with disorganized spikes had coagulated RNPs.
This distribution is largely consistent with the work of Harris et al. (17), who studied similarly prepared X-31 virions at neutral pH. If we ignore the sparsely populated classes IV and V, 95% of the remainder were reported to have a resolved M1 layer (81% of class I and 14% of class II) and 5% to lack this feature (class III).
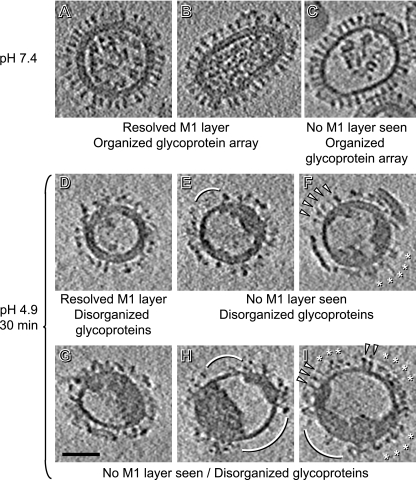
Fig 3.
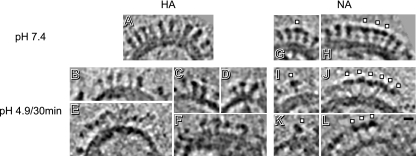
Central slices of tomograms of influenza virions. Data are shown for pH 7.4 (A to C) and after 30 min of incubation at pH 4.9 (D to I). Note the differences in glycoproteins, RNPs, and matrix. Arcs mark regions cleared of glycoprotein spikes. In panels F and I, arrowheads mark neutral-pH-like HAs and asterisks mark disorganized patches of glycoproteins. Bar, 50 nm.
To explore the effects of acid pH, we incubated the virus for 30 min at pH 4.9. Under these conditions, the virus is no longer fusogenic (23, 32) and HA should be in its postfusion state. In preparing these samples for microscopy, we encountered some aggregation, but many virions remained dispersed. Major changes were indeed observed in several respects (Fig. 3D to I and Table 1). (i) No elongated particles were seen, consistent with the work of Calder et al. (7). (ii) Approximately 20% of the particles were much larger, with diameters ranging from 145 to 200 nm (Fig. 4A to D), i.e., 1.5 to 2.0 times the average diameter for pH 7.4 virions (95 ± 14 nm) and corresponding to 2.2 to 4.0 times their average surface area. We conclude that they were produced by fusion of two or more virions (see also reference 7). (iii) All glycoprotein arrays were disorganized (Fig. 3D to I and 4A to D), leaving—in some virions—tracts of envelope seemingly devoid of spikes (arcs, Fig. 3E, H, and I and Fig. 4B to D). These bare patches were usually found at sites where there was no discernible M1 layer nor internal densities underlying the viral membrane (Fig. 3E and H and Fig. 4B and C; exceptions are shown in Fig. 3I and 4D). These changes affect mainly the HA spikes, as normal NA spikes with their long-stemmed “mushroom” shape are readily apparent (Fig. 5). As at neutral pH (17), most NAs are in local clusters (Fig. 5H, J, and L), although single NAs can also be found (Fig. 5G, I, and K). As for HAs, their characteristic “peanut” shape assumed at pH 7.4 is seen only in a few patches (Fig. 5B to D). Not all HA spikes on a given virion have the same morphology (Fig. 3F and I), and many HAs are difficult to identify as individual molecules (Fig. 3D to I, 4A to D, and 5E and F). (iv) An M1 layer is no longer seen in the majority (∼60%) of virions and is complete in only 20% (e.g., Fig. 3D and 4A) and partial in another 20% (e.g., Fig. 3E and F). (v) Most virions (∼90%) contained, instead of dispersed RNPs, a single coagulate at the periphery of the internal cavity, leaving large voids that account for most of its volume (Fig. 3E to I and 4B to D). In many cases, this coagulate also contains M1 protein from the envelope-lining layer (see below). Here, we use the term “coagulate” instead of “aggregate,” because the observed densities were homogeneously dense, lacking the local gaps that we would expect to see if RNPs simply aggregated but retained their structures (see Fig. 7). Of note, dispersed RNPs, when observed, were found only in virions with a complete M1 layer (Fig. 3D and 4A).
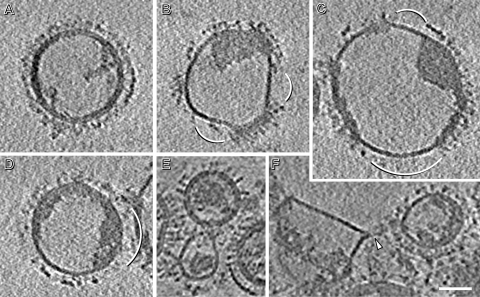
Fig 4.
(A to D) Central slices of tomograms showing large particles representing fused virions. They were observed after incubating influenza virions for 30 min at pH 4.9. Diameters of these particles range from 145 to 200 nm. (E and F) Central slices of tomograms of influenza virions after 5 min of incubation showing two possible examples of prefusion events taking place between them. In panel F, an arrowhead marks a funnel-shaped deformation of one of the virions. Bar, 50 nm.
Fig 5.
Details from tomographic slices of influenza virus glycoproteins. Data from pH 7.4 (A, G, and H) and after 30 min of incubation at pH 4.9 (B to F and I to L) are compared. (A to F) HA clusters. While at pH 7.4 HAs have a characteristic “peanut” shape, after 30 min at pH 4.9 only some maintain a neutral-pH-like shape (B to D), but most of the glycoprotein array is disorganized (E and F). (G, I, and K) Single NAs. (H, J, and L) NA clusters. White squares mark individual NAs. Bar, 10 nm.
Fig 7.
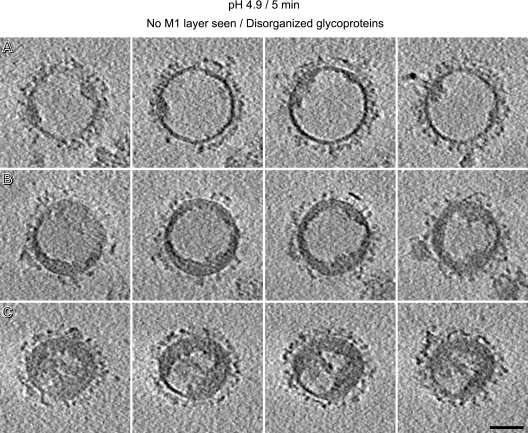
Tomographic slices of the pH 4.9/5-min virions with nonresolved M1 layer and disorganized glycoproteins. Virions correspond to the ones shown in Fig. 6J to L (A to C, respectively). Bar, 50 nm.
Structural changes in influenza virus after 5 min at pH 4.9.
Next, we sought to capture fusion intermediates. In order to identify conditions under which a substantial fraction of virions and HA molecules would have embarked on fusion but not yet reached the postfusion state, we referred to studies in which fluorescence dequenching measurements were used to show that the fusion activity of X-31 virions was partly but not largely reduced by preincubation for 5 min at pH 5.0 (23). Accordingly, we shortened the incubation time at pH 4.9 to 5 min. We also tried 15 min of incubation, but as the 5-min incubation appeared more promising, we focused our efforts on it. The outcome (see below) appears to justify this choice of experimental conditions.
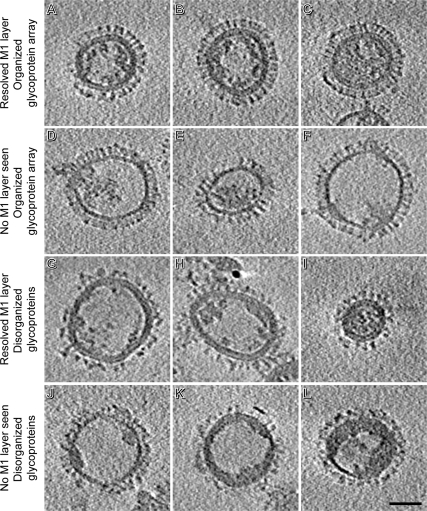
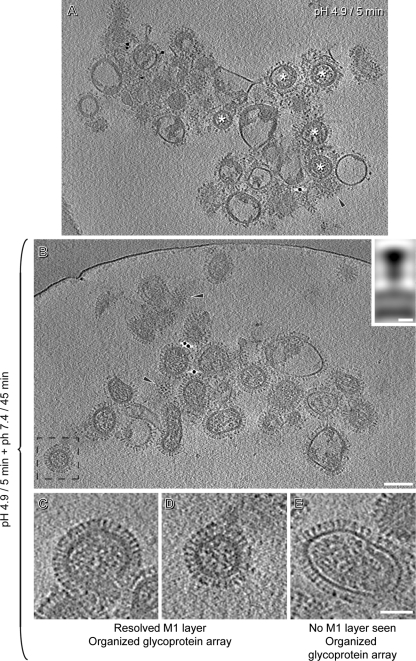
Some of these virions retained neutral-pH-like morphologies (Fig. 6A to C), others resembled pH 4.9/30-min virions (Fig. 6J to L), and still others had differently altered morphologies (Fig. 6D to I). Whereas at neutral pH all virions had organized glycoprotein arrays and all pH 4.9/30-min virions had disorganized arrays, pH 4.9/5-min virion preparations had roughly equal numbers of virions of the two kinds (Table 1). Moreover, whereas a large majority (80 to 90%) of neutral-pH virions have visible M1 layers and many fewer (∼20%) pH 4.9/30-min virions have this feature, pH 4.9/5-min virions are again intermediate in this regard (∼55%). Finally, RNP morphologies in pH 4.9/5-min virions were also intermediate with respect to those from neutral-pH virions (which all had dispersed RNPs) and pH 4.9/30-min virions (∼90% of which had coagulates), with about equal numbers of particles exhibiting dispersed RNPs (∼47%) and coagulated RNPs (∼53%).
Fig 6.
Central slices of tomograms of influenza virions after 5 min of incubation at pH 4.9. Virions are classified according to whether or not there is a resolved M1 layer and whether the array of glycoprotein spikes is organized or disorganized. Bar, 50 nm.
Precursor-product relationships.
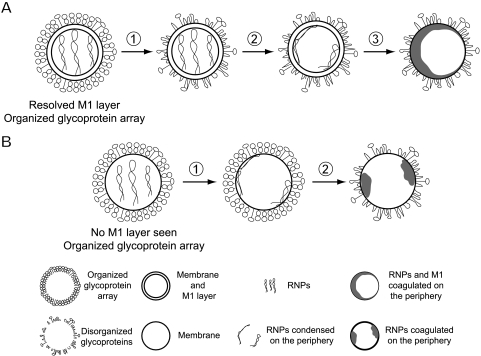
Like neutral-pH virions, the populations observed after 5 min or 30 min of acidification are heterogeneous. Nevertheless, the data allowed us to draw the following conclusions as to the course of events. We assume that pH 4.9/5-min virions with a resolved M1 layer came from neutral-pH virions that also had this feature. Similarly, pH 4.9/5-min virions lacking an M1 layer but with organized spikes should come from their neutral-pH counterparts. On the other hand, the preponderance of matrix layer-negative virions after 5 min at pH 4.9, compared with those at neutral pH (45% versus 11%), argues that some of them originally had an M1 layer. This trend—a stripping or at least a major redistribution of the M1 layer—carries further in pH 4.9/30-min preparations (∼80% versus 11%). As for pH 4.9/5-min virions lacking the M1 layer and with disorganized spikes, we propose that those with relatively little internal density (27% of these virions, e.g., in Fig. 6J) derive from virions that lacked an M1 layer, while the rest (73%, e.g., in Fig. 6K and L) derive from virions that formerly had an M1 protein layer and in which the RNPs and the M1 protein have coagulated together. According to these assignments, 78% of pH 4.9/5-min virions came from virions containing M1 layers, matching reasonably well the estimated ∼90% in the initial population. The inferred courses of events for different kinds of virions, when transferred to pH 4.9, are sketched out in Fig. 10 and discussed further below.
Fig 10.
Schematic of sequential changes in influenza virions induced by low pH. (A) After incubation at pH 4.9, neutral-pH virions with a resolved M1 layer first lose the organization of their glycoprotein array (step 1), followed by adherence of RNPs to the periphery (step 2) and, finally, the condensation of RNPs and M1 into a single coagulate (step 3). (B) In neutral-pH virions lacking an M1 layer and with organized glycoproteins, RNPs move to the periphery (step 1), followed by a disordering of the glycoprotein array and condensation of the RNPs at the periphery (step 2).
As M1 is proposed, during an infection, to dissociate from the RNPs soon after they are released into the cytoplasm (20, 27, 28), we considered the possibility that M1 and RNPs might coagulate separately but rejected this idea because we did not observe virions to have two separate internal “lumps” (Fig. 7).
Detection of intermediate HA conformations.
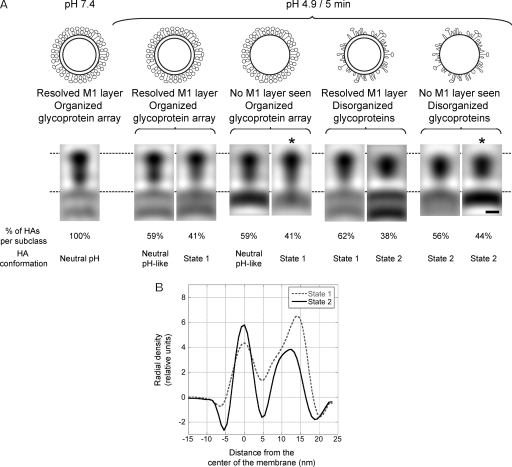
In order to probe for possible conformational changes in the HA spikes, we extracted sets of HA-containing subtomograms from the pH 4.9/5-min virions. These data were aligned, classified, and averaged independently within each class (Table 2). In Fig. 8A, sagittal sections through these density maps are compared with each other and with the average HA structure obtained from virions at pH 7.4 (38). All HAs from neutral-pH virions had the same conformation, i.e., they formed a single class. This structure shows distinguishable membrane-proximal (formed by HA2 and the N- and C-terminal segments of HA1) and membrane-distal (comprising residues 50 to 261 of HA1) domains, and it is ∼15 nm long from the membrane surface to the outer tip and ∼6 nm wide at the level of the membrane-distal domains, as measured from the averages. Some HAs (35%) on pH 4.9/5-min virions retained this conformation, but others exhibited two other structures that we call “state 1” (37%) and “state 2” (28%). In state 1, the membrane-proximal density of the peanut narrows and the “waist” between its two lobes is less evident, but the overall length of HA and the maximum width of the membrane-distal domain are very similar to those of the neutral-pH conformation. In state 2, however, the molecule is shorter (∼13.5 nm) and wider (∼7.5 nm) than in the neutral-pH structure and it is no longer bilobed. Density profiles along the axis of a state 1 HA and a state 2 HA (Fig. 8B) convey the difference in length between them. The percentage of HAs assigned to each subclass correlated with visual estimates of how each pH 4.9/5-min group was affected by the pH (Fig. 8).
Table 2.
Summary of the number of virions and HAs selected for the averaging of HAs of influenza virions under different pH conditionsa
| Condition | M1 layer | Organized glycoprotein array | No. of virions (%) | No. of selected HAs | No. of selected HAs/virion |
|---|---|---|---|---|---|
| pH 7.4 | + | + | 20 | 800 | ∼40 |
| pH 4.9/5 min | + | + | 61 (34) | 3,035 | ∼50 |
| − | + | 25 (14) | 1,086 | ∼43 | |
| + | − | 38 (21) | 1,385 | ∼36 | |
| − | − | 55 (31) | 1,777 | ∼32 | |
| Total | 179 (100) | 7,283 | ∼41 | ||
| pH 4.9/5 min + pH 7.4/45 min | + | + | 13 | 292 | ∼22 |
Two different tomograms were employed for the subtomogram averaging of HA from neutral-pH virions (∼10 virions/tomogram), 31 were used for the pH 4.9/5-min virions (∼6 virions/tomogram), and 5 were used for the pH 4.9/5-min plus pH 7.4/45-min virions (∼3 virions/tomogram).
Fig 8.
Subtomogram averaging showing altered conformations of HA ectodomains. (A) (Top) Schematic diagram of virion types analyzed. (Bottom) Sagittal sections through averaged density maps of the HA glycoprotein of influenza virions at pH 7.4 and from the different pH 4.9/5-min groups of virions. They exhibit three conformations: neutral-pH conformation, state 1, and state 2. (B) Axial density profiles of the HAs marked with asterisks in panel A. Note the difference in length between state 1 and state 2. Bar, 5 nm.
Reversibility of the changes in HA after 5 min at pH 4.9.
Reversibility of the initial changes in HA at low pH has been shown by biochemical methods for HA expressed at the surface of eukaryotic cells (25) and by X-ray crystallographic analysis of purified HA (43). In order to test for reversibility in intact spikes on virions, we used cryo-ET to compare pH 4.9/5-min viruses with the same sample after a subsequent incubation for 45 min at pH 7.4. As illustrated in Fig. 9A, ∼50% of the pH 4.9/5-min virions (from a total of 115) showed disorganized glycoproteins: this was as expected (see above). However, after the postincubation at pH 7.4, all virions (n = 150) exhibited an organized glycoprotein array (Fig. 9B), with most or all of the HAs reverting to their neutral-pH conformation (Fig. 9C to E), as confirmed by subtomogram averaging (inset in Fig. 9B).
Fig 9.
Reversibility of the low-pH-induced changes in HA. (A) Tomographic slice of virions after 5 min at pH 4.9. White asterisks mark some of the virions that are less affected by the low pH. (B) Tomographic slice of virions after 5 min at pH 4.9 and 45 min at pH 7.4. The inset shows a sagittal section through the averaged density map of HAs from these virions. Black arrowheads mark tangential sections through viruses in panels A and B in order to compare the organization of their glycoproteins. (C to E) Central slices of tomograms of influenza virions after 5 min at pH 4.9 and 45 min at pH 7.4. The glycoprotein arrays of these virions are as organized as they are at neutral pH. Panel D is an enlargement of the boxed virion in panel B. Bars, 100 nm in panels A and B, 5 nm in inset of panel B, and 50 nm in panels C to E.
DISCUSSION
The pleiomorphism of influenza virus long hampered detailed study by cryo-EM. Cryo-ET, however, has made possible three-dimensional (3D) structural analyses of individual virions (16). Here we employed this approach to investigate the structural changes undergone by influenza virions when switched to low pH. Although our initial focus was on the HA ectodomains, the observations also revealed major changes in virion size (from virion fusion) and shape (disappearance of elongated virions) and in the organization of internal material, i.e., coagulation of M1 protein and RNPs. It is noteworthy that—even within the same morphological class—all virions did not respond in the same way to the acidification treatments. The reason for this is unclear, although it could be due to differing amounts of the M2 ion channel protein in the viral membrane, which could lead to slower or faster decrease of the pH inside a virion. (Measurements of the average content of M2 per virion by two independent biochemical methods yielded values ranging from 14 to 68 molecules [46] but no information as to whether the M2 complement is fixed or variable on a given virion.)
Transition pathways between the neutral-pH virions and the pH 4.9/5-min virions.
We performed a morphological classification of pH 4.9/5-min virions and correlated these classes with those seen in the starting material at neutral pH and with pH 4.9/30-min virions. A summary of inferred transition pathways is given in Fig. 10. In virions that initially have a resolved M1 layer, the low pH first affects the HA ectodomains, which become disorganized (Fig. 10A, step 1); then the RNPs condense onto the M1 layer (step 2); and finally an M1 layer is no longer seen (step 3). The internal material finishes up as a dense coagulate, with most of the space inside the viral envelope void of protein density. We infer that the coagulate usually consists of RNPs plus M1 protein. This inference is consistent with reports that RNPs are still associated with some M1 protein just after being released into the cytoplasm (5, 28). It is not clear whether the RNPs break down or retain their structures, but the continuous nature of the coagulated density suggests the former, i.e., we would expect to see gaps between RNPs that were simply aggregated. In the minority of virions that initially lack a matrix layer, the RNPs lose their neutral pH organization faster, as they condense onto the inner surface of the envelope (Fig. 10B, step 1) before finally coagulating (step 2).
Influence of the M1 layer on form determination and consequences of its relaxation.
M1 protein has been proposed to be a determinant in the formation of filamentous virions (33, 34). The shape of elongated or filamentous virions is presumably imposed by the membrane-lining lattice of M1 protein. On the other hand, the low-energy state of membrane vesicles corresponds to spherical morphology. It appears likely that the disappearance of elongated virions at low pH corresponds to their converting to spherical shape. In the same vein, the sphericity of the giant particles produced by virion-virion fusion (Fig. 4) presumably results from remodeling from the hourglass morphology that would initially be realized. In both instances, spherical morphology is likely to result from relaxation of M1-envelope interactions as well as M1-M1 interactions (18) in response to low pH.
Initial changes in HA upon exposure to low pH.
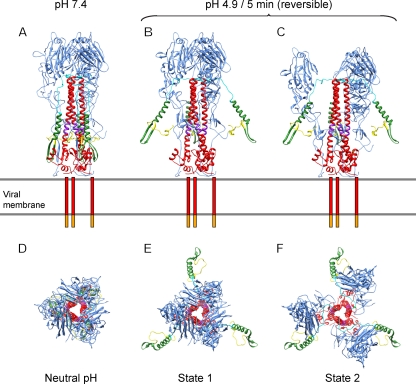
Since the characterization by X-ray crystallography of the neutral- and low-pH forms of HA (2, 6, 42), there has been progress—albeit still far from complete—toward understanding the complex process of virally induced membrane fusion. Here, subtomogram averaging has allowed us to address the effects of low pH on HA and thus to detect two intermediate conformations on the fusion pathway (Fig. 11). Both changes are reversible. Although the resolution is limited to 2.5 to 3.0 nm, the changes are large enough to be clearly demonstrated. The limited resolution also constrains the detail in which they may be interpreted. Thus, the reversible movement of the B loop to a relaxed form and the deformation of the HA1 subunit described by Xu and Wilson (43) that probably precede the state 1 and state 2 conformations are too subtle to be detected. With that caveat, we suggest the following: upon exposure to low pH, the fusion peptide and part of HA2 corresponding to approximately residues 24 to 54 (shown in yellow and green, respectively) move outwards to reach the state 1 conformation (Fig. 11B and E). This would expose both the fusion peptide and the kinked loop (residues 106 to 112 of HA2, in purple) (25). Next, the HA1 domains (blue) move downwards and outwards to reach the state 2 conformation (Fig. 11C and F) without “unclamping” the B loop. The existence of reversible intermediate conformations of HA could increase the success rate of the fusion process by allowing HA to have multiple attempts to reach the cellular membrane.
Fig 11.
Model of the possible intermediate states of HA prior to fusion. Lateral (A to C) and top (D to F) views of the models. The viral membrane is shown as two gray lines, the transmembrane domains are represented as red bars, and the cytoplasmic tails are represented as orange bars. Color coding is as follows: HA1 domain, blue; HA2 domain, red, with amino acids 1 to 23 (fusion peptide) in yellow; amino acids 24 to 54, green; amino acids 55 to 76 (B loop), cyan; and amino acids 106 to 112 (kinked loop), purple.
The rearrangement of HA1 proposed here differs from that which takes place as the molecule transitions, irreversibly, into the postfusion conformation (9, 15). After state 2, although HA is presumably still present in the viral envelope, it is hard to make out individual molecules in the tomograms: this appears to reflect the dissociation of HA1 membrane-distal domains and their dispersal, albeit still connected by flexible linkers to the core of the trimeric molecule.
Implications for the infection process.
These observations bear on the events that take place after a virion has entered an endosome, leading to release of viral RNPs into the cytoplasm. We envisage that, upon exposure to low pH, HA undergoes the reversible conformation changes described above. As the pH in the endosome drops, it is accompanied by an M2-mediated pumping of protons into the virion (31), eliciting a change in intermolecular interactions in the M1 layer and weakening its binding to the viral membrane. These changes render the envelope more pliable and hence more fusion-compatible (as hypothesized by Lee [24]). At the same time, disruption of M1-RNP interactions would free the RNPs to be discharged into the host cell cytoplasm after membrane fusion (20). The dense nature of coagulates seen after 30 min at pH 4.9 suggests that the RNPs trapped in them may no longer be infectious. Accordingly, we propose that soon after the M1 layer has been disrupted, HAs should complete their conformational changes and induce fusion in order to avoid coagulation of the RNPs.
ACKNOWLEDGMENT
This research was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health.
Footnotes
Published ahead of print 18 January 2012
REFERENCES
- 1. Barbey-Martin C, et al. 2002. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology 294:70–74 [DOI] [PubMed] [Google Scholar]
- 2. Bizebard T, et al. 1995. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376:92–94 [DOI] [PubMed] [Google Scholar]
- 3. Booy FP, Ruigrok RW, van Bruggen EF. 1985. Electron microscopy of influenza virus. A comparison of negatively stained and ice-embedded particles. J. Mol. Biol. 184:667–676 [DOI] [PubMed] [Google Scholar]
- 4. Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26(Suppl. 4):D49–D53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bukrinskaya AG, Vorkunova NK, Kornilayeva GV, Narmanbetova RA, Vorkunova GK. 1982. Influenza virus uncoating in infected cells and effect of rimantadine. J. Gen. Virol. 60:49–59 [DOI] [PubMed] [Google Scholar]
- 6. Bullough PA, Hughson FM, Skehel JJ, Wiley DC. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37–43 [DOI] [PubMed] [Google Scholar]
- 7. Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 107:10685–10690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cardone G, Grünewald K, Steven AC. 2005. A resolution criterion for electron tomography based on cross-validation. J. Struct. Biol. 151:117–129 [DOI] [PubMed] [Google Scholar]
- 9. Carr CM, Chaudhry C, Kim PS. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. U. S. A. 94:14306–14313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen J, Skehel JJ, Wiley DC. 1999. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil. Proc. Natl. Acad. Sci. U. S. A. 96:8967–8972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cross KJ, Langley WA, Russell RJ, Skehel JJ, Steinhauer DA. 2009. Composition and functions of the influenza fusion peptide. Protein Pept. Lett. 16:766–778 [DOI] [PubMed] [Google Scholar]
- 12. Forster F, Medalia O, Zauberman N, Baumeister W, Fass D. 2005. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl. Acad. Sci. U. S. A. 102:4729–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135:239–250 [DOI] [PubMed] [Google Scholar]
- 14. Fujiyoshi Y, Kume NP, Sakata K, Sato SB. 1994. Fine structure of influenza A virus observed by electron cryo-microscopy. EMBO J. 13:318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Godley L, et al. 1992. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68:635–645 [DOI] [PubMed] [Google Scholar]
- 16. Grünewald K, et al. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396–1398 [DOI] [PubMed] [Google Scholar]
- 17. Harris A, et al. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl. Acad. Sci. U. S. A. 103:19123–19127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harris A, Forouhar F, Qiu S, Sha B, Luo M. 2001. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 289:34–44 [DOI] [PubMed] [Google Scholar]
- 19. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Helenius A. 1992. Unpacking the incoming influenza virus. Cell 69:577–578 [DOI] [PubMed] [Google Scholar]
- 21. Heymann JB, Cardone G, Winkler DC, Steven AC. 2008. Computational resources for cryo-electron tomography in Bsoft. J. Struct. Biol. 161:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kemble GW, Bodian DL, Rose J, Wilson IA, White JM. 1992. Intermonomer disulfide bonds impair the fusion activity of influenza virus hemagglutinin. J. Virol. 66:4940–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korte T, Ludwig K, Booy FP, Blumenthal R, Herrmann A. 1999. Conformational intermediates and fusion activity of influenza virus hemagglutinin. J. Virol. 73:4567–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee KK. 2010. Architecture of a nascent viral fusion pore. EMBO J. 29:1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leikina E, Ramos C, Markovic I, Zimmerberg J, Chernomordik LV. 2002. Reversible stages of the low-pH-triggered conformational change in influenza virus hemagglutinin. EMBO J. 21:5701–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117–130 [DOI] [PubMed] [Google Scholar]
- 28. Martin K, Helenius A. 1991. Transport of incoming influenza virus nucleocapsids into the nucleus. J. Virol. 65:232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 [DOI] [PubMed] [Google Scholar]
- 30. Murti KG, Brown PS, Bean WJ, Jr, Webster RG. 1992. Composition of the helical internal components of influenza virus as revealed by immunogold labeling/electron microscopy. Virology 186:294–299 [DOI] [PubMed] [Google Scholar]
- 31. Pinto LH, Holsinger LJ, Lamb RA. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517–528 [DOI] [PubMed] [Google Scholar]
- 32. Puri A, Booy FP, Doms RW, White JM, Blumenthal R. 1990. Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J. Virol. 64:3824–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roberts PC, Compans RW. 1998. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. U. S. A. 95:5746–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roberts PC, Lamb RA, Compans RW. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127–137 [DOI] [PubMed] [Google Scholar]
- 35. Scheres SH, Melero R, Valle M, Carazo JM. 2009. Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure 17:1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shangguan T, et al. 1998. Morphological changes and fusogenic activity of influenza virus hemagglutinin. Biophys. J. 74:54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stegmann T, Booy FP, Wilschut J. 1987. Effects of low pH on influenza virus. Activation and inactivation of the membrane fusion capacity of the hemagglutinin. J. Biol. Chem. 262:17744–17749 [PubMed] [Google Scholar]
- 38. Steven AC, Cardone G, Butan C, Winkler DC, Heymann JB. 2011. Three-dimensional structures of pleiomorphic viruses from cryo-electron tomography, p 62–80 In Agbandje-McKenna M, McKenna R. (ed), Structural virology. The Royal Society of Chemistry, Cambridge, United Kingdom [Google Scholar]
- 39. White JM, Delos SE, Brecher M, Schornberg K. 2008. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 43:189–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White JM, Wilson IA. 1987. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J. Cell Biol. 105:2887–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. White TA, et al. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6:e1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366–373 [DOI] [PubMed] [Google Scholar]
- 43. Xu R, Wilson IA. 2011. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 85:5172–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamaguchi M, Danev R, Nishiyama K, Sugawara K, Nagayama K. 2008. Zernike phase contrast electron microscopy of ice-embedded influenza A virus. J. Struct. Biol. 162:271–276 [DOI] [PubMed] [Google Scholar]
- 45. Zanetti G, Briggs JA, Grünewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zebedee SL, Lamb RA. 1988. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol. 62:2762–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu P, et al. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852 [DOI] [PubMed] [Google Scholar]