Abstract
The replication of integrated human immunodeficiency virus type 1 (HIV-1) is dependent on the cellular cofactor cyclin T1, which binds the viral Tat protein and activates the RNA polymerase II transcription of the integrated provirus. The activation of resting CD4+ T cells upregulates cyclin T1 protein levels independently of an increase in cyclin T1 mRNA levels, suggesting a translational repression of cyclin T1 in resting CD4+ T cells. Hypothesizing that microRNAs (miRNAs) repress cyclin T1 translation in resting CD4+ T cells and that this inhibition is lifted upon cell activation, we used microarray expression analysis to identify miRNAs miR-27b, miR-29b, miR-150, and miR-223 as being significantly downregulated upon CD4+ T cell activation. The overexpression of these miRNAs decreased endogenous cyclin T1 protein levels, while treatment with the corresponding antagomiRs increased cyclin T1 protein levels. An miR-27b binding site within the cyclin T1 3′ untranslated region (3′UTR) was identified and confirmed to be functional after the mutation of key resides abrogated the ability of miR-27b to decrease the expression of a luciferase reporter upstream of the cyclin T1 3′UTR. Ago2 immunoprecipitation revealed an association with cyclin T1 mRNA that was decreased following treatment with miR-27b and miR-29b antagomiRs. Cells overexpressing miR-27b showed decreased viral gene expression levels of the HIV-1 reporter virus and a decreased replication of strain NL4.3; a partial rescue of viral transcription could be seen following the transfection of cyclin T1. These results implicate miR-27b as a novel regulator of cyclin T1 protein levels and HIV-1 replication, while miR-29b, miR-223, and miR-150 may regulate cyclin T1 indirectly.
INTRODUCTION
The replication of human immunodeficiency virus type 1 (HIV-1) is dependent on the expression of multiple cellular cofactors that, when present at limiting levels, can partly determine cellular permissivity to infection. For instance, resting CD4+ T cells contain low levels of several essential cofactors, including positive transcription elongation factor b (P-TEFb) (18, 19, 23). The transcription of integrated HIV-1 from the host genome is dependent on this complex, which is also essential for mediating the elongation of cellular RNA polymerase II (RNAP II) transcripts (37). P-TEFb is composed of cyclin-dependent kinase 9 (CDK9) as the catalytic subunit and one of three regulatory subunits: cyclin T1, T2A, or T2B (36). Cyclin T1-containing P-TEFb is the only form that supports HIV-1 transcription, as P-TEFb is recruited to nascent viral RNA by the direct binding of the viral transactivator protein Tat to the cyclin T1 subunit (4, 41, 50). P-TEFb hyperphosphorylates the C-terminal domain of RNA P II in addition to several negative elongation factors, thereby catalyzing a switch from abortive to fully processive transcriptional elongation (56). Cyclin T1 is therefore essential for the efficient transcription of the provirus, and HIV-1 replication is severely impaired in its absence (10, 11, 29, 54).
Upon CD4+ T cell activation or the differentiation of monocytes into macrophages, cyclin T1 protein levels dramatically increase, independently of changes in cyclin T1 mRNA levels (31, 32, 43), suggesting that cyclin T1 is posttranscriptionally repressed in resting CD4+ T cells and monocytes. We hypothesized that this repression might be mediated by microRNAs (miRNAs), as their function in posttranscriptional gene silencing has been well established, and it has been estimated that more than 50% of genes are subject to miRNA regulation (17). Furthermore, over 800 human miRNAs have been identified, and the functional validation of miRNA targets has indicated their involvement in a wide range of biological processes (13, 35, 48).
Following gene transcription by RNAP II, human primary miRNA transcripts are processed in the nucleus by the enzyme Drosha (6). The resulting pre-miRNAs are exported into the cytoplasm and cleaved by Dicer into the mature form, which is incorporated into the RNA-induced silencing complex (RISC). The miRNA-RISC then typically binds to the 3′ untranslated region (3′UTR) of a target mRNA, leading to translational repression by mechanisms still being elucidated (16). In the majority of cases, this is also accompanied by some level of miRNA-mediated mRNA degradation (21, 22, 30). While the entire length of an miRNA is usually not perfectly homologous to the target sequence, the so-called seed sequence of the miRNA, defined as nucleotides (nt) 2 to 8, almost always exhibits a high degree of base pair complementarity to the target and can be highly conserved across species. This observation forms the basis of in silico miRNA target prediction algorithms, which can be used to generate putative miRNA targets, albeit with a high false-positive rate, which makes experimental confirmation a necessity (2, 3).
Recent evidence has shown that the miRNA pathway has significant effects on HIV-1 replication (9). The small interfering RNA (siRNA) knockdown of the miRNA-processing enzyme Dicer considerably increases HIV-1 replication, indicating that miRNAs generally act to inhibit viral replication (46). Those same authors also found that HIV-1 infection of peripheral blood mononuclear cells (PBMCs) downregulates the miR-17-92 family, which was found to target PCAF, a cellular cofactor of Tat. The overexpression of miR-17-92 family members decreased both PCAF levels and HIV-1 replication. We previously identified an miRNA, miR-198, that has similar effects on decreasing HIV-1 replication but via the targeting of cyclin T1 (44). miR-198 is downregulated upon monocyte-to-macrophage differentiation, when cyclin T1 protein levels increase, and also binds to the cyclin T1 3′UTR. The overexpression of miR-198 in monocytes decreased endogenous cyclin T1 protein levels and viral replication in a monocytic cell line. However, we observed that miR-198 is present at only low levels in resting CD4+ T cells and is not downregulated following T cell activation, suggesting that this miRNA may play a limited role in the posttranscriptional repression of cyclin T1 in resting CD4+ T cells.
The direct targeting of genomic HIV-1 RNA has also been suggested as a mechanism for miRNA-mediated effects on replication; miR-29a has been shown to directly target the 3′UTR of HIV-1 RNA and to enhance the interaction of viral RNA with epitope-tagged Ago2, a component of the RISC (34). Regardless of the mechanisms whereby miRNAs affect HIV-1 replication, PBMC profiling has defined groups of miRNAs consistently downregulated more than 2-fold in a majority of HIV-infected patients compared to uninfected donors (24), highlighting the potential importance of miRNAs in the context of HIV-1 infection. A role for miRNAs in the maintenance of HIV-1 latency has also been proposed. The transfection of antagomiRs against miR-28, miR-125b, miR-150, miR-223, and miR-382, which were all predicted to target viral RNA, reactivated virus from latently infected CD4+ T cells isolated from patients on suppressive highly active antiretroviral therapy (HAART) regimens (25).
In this study, we sought to identify miRNAs which inhibit cyclin T1 protein expression in resting CD4+ T cells, thereby contributing to the inhibition of HIV-1 replication in this nonpermissive milieu. We found that miR-27b, -29b, -150, and -223 are present at higher levels in resting than in activated CD4+ T cells and that these miRNAs all function to repress cyclin T1 protein levels. While it was previously reported that miR-29b, -150, and -223 inhibit HIV-1 replication (25, 34), to our knowledge, this is the first report detailing the ability of miR-27b to do so. Furthermore, we show that miR-27b inhibits viral replication via the direct targeting of cyclin T1 mRNA and, along with miR-29b, mediates the targeting of cyclin T1 mRNA to the RISC.
MATERIALS AND METHODS
CD4+ T cell isolation.
Resting CD4+ T cells were isolated from blood of healthy donors (Gulf Coast Regional Blood Center, Houston, TX) by performing Isolymph density centrifugation (Gallard-Schlesinger Industries, Inc.) to isolate PBMCs, followed by CD4+ T cell Isolation Kit II (Miltenyi Biotech), or by using the RosetteSep human CD4+ T cell enrichment cocktail (StemCell) on whole blood. Activated cells were then removed by using CD30 Microbeads (Miltenyi Biotech). Resting CD4+ T cell preparations were >90% pure (data not shown), as assessed by flow cytometry analysis of staining for the CD4 and CD3 T cell markers along with CD25 and CD69 for activation status (BD Biosciences). For cell activation, a portion of the isolated cells was activated with phytohemagglutinin (PHA) treatment (10 ng/ml) and cultured in RPMI supplemented with 10% fetal bovine serum (FBS) for 2 days.
miRNA expression profiling.
Total RNA was isolated from donor resting and activated CD4+ T cells by using the miRVana miRNA isolation kit (Applied Biosystems), and miRNA microarray analysis was performed by LC Sciences (Houston, TX). For donors 1 and 2 the version 7.1 platform was used to detect 321 unique miRNAs; for donors 3 and 4, version 12.0 was used to detect 695 miRNAs. Cy3 and Cy5 dye switching between donors was performed to eliminate biases.
miRNA transfections.
For protein and mRNA expression analyses, the indicated pre-miR miRNA precursor molecule or pre-miR negative control 1 (Applied Biosystems) was transfected into HeLa cells by using the siPORT NeoFX transfection agent (Applied Biosystems). Seventy-two hours later, lysates were harvested for Western blotting (see below). Alternatively, resting CD4+ T cells were isolated as described above and then transfected with the indicated anti-miR miRNA inhibitor or anti-miR negative control 1 (Applied Biosystems) using the human T cell Nucleofector kit (Lonza) and the program U-14. Lysates were harvested 48 h later.
Protein and RNA analyses.
Protein was analyzed by Western blotting using antibodies against cyclin T1 (catalog number SC-8127; Santa Cruz Biotechnology), α-tubulin (catalog number SC-5286; Santa Cruz Biotechnology), and β-actin (catalog number A2066; Sigma). ImageJ (39) was used to quantify bands; statistical analysis was performed by regression analysis on the geometric means of the cyclin T1/β-actin ratio. Both the confidence interval and the P value were used for determining significance. cDNA amplification of mRNA was performed by using the iScript cDNA synthesis kit (Bio-Rad), and quantitative real-time PCR was done by using iQ SYBR green Supermix (Bio-Rad) and the following primers: cyclin T1 forward primer 5′-GGCGTGGACCCAGATAAAG-3′, cyclin T1 reverse primer 5′-CTGTGTGAAGGACTGAATCAT-3′, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward primer 5′-CGCCAGCCGAGCCACATC-3′, and GAPDH reverse primer 5′-AATCCGTTGACTCCGACCTTCAC-3′. A cyclin T1 standard curve was generated by using a dilution series of a cyclin T1-containing plasmid.
Luciferase assays.
The cyclin T1 3′UTR-luciferase (luc) construct was described previously (44). The mutant cyclin T1 3′UTR-luc construct was generated by using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). Pre-miR-27b was transfected into HeLa cells along with the 3′UTR-luc constructs and pRL-TK expressing Renilla luciferase, as an internal transfection control (Promega), using Lipofectamine 2000 (Invitrogen). The Dual-Luciferase reporter assay system (Promega) was used to quantify both firefly and Renilla luciferase expression levels at 24 h posttransfection.
Ago2 immunoprecipitation.
Ago2 and control IgG immunoprecipitations were performed as described previously (45), with the following modifications. Five micrograms of EIF2C2 (Ago2) monoclonal antibody (catalog number H00027161-M01; Abnova) or isotype control mouse IgG1κ monoclonal antibody (catalog number ab18447; Abcam) was bound to 300 μl of protein A Dynabeads (Invitrogen) for 8 h with rotation at 4°C. Dynabeads were then washed twice with 0.02% Tween 20 in phosphate-buffered saline (PBS) and once with NT2 buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40). Jurkat cells were lysed in freshly prepared polysome lysis buffer (5 mM MgCl2, 100 mM KCl, 10 mM HEPES [pH 7], 0.5% Nonidet P-40, 1 mM dithiothreitol [DTT], 100 U/ml RNasin Plus RNase inhibitor [Promega], 1× protease inhibitor cocktail [Sigma], 400 μM vanadyl ribonucleoside complexes [NEB]). Jurkat cells were also transfected with anti-miR miRNA inhibitors or anti-miR negative control 1 (Applied Biosystems) by using Oligofectamine (Invitrogen) and were lysed 72 h later. All lysates were frozen at −80°C and then thawed and cleared by centrifugation at 10,000 × g for 10 min. A total of 2.5 mg of the protein lysate was mixed with supplemented NT2 buffer (NT2 buffer plus 200 U/ml RNasin Plus, 1 mM DTT, and 15 mM EDTA) to a total volume of 1 ml, loaded onto the antibody-bound Dynabeads, and rotated for 4 h at 4°C. During the first magnetic separation of the beads, the supernatant was taken as the flowthrough fraction. Dynabeads were then washed four times with 0.02% Tween 20 in PBS. Beads were then split into fractions for protein and RNA analyses. SDS loading buffer was used to elute proteins from the beads, and the lysis buffer included with the miRVana miRNA isolation kit (Applied Biosystems) was used to extract RNA directly from the bead-bound material. Western blotting was performed by using the same Ago2 antibody, and quantitative reverse transcription-PCR (qRT-PCR) to measure mRNA levels was performed as described above, along with the following primers: PCAF forward primer 5′-TGCTGTCAGTATTTTAACACCC-3′ and PCAF reverse primer 5′-GCACTAAACTGGAATCCCAAG-3′ (46). qRT-PCR to measure miRNA levels was performed by using the TaqMan microRNA reverse transcription kit, followed by TaqMan microRNA assays with Universal PCR Master Mix (Applied Biosystems). Equal volumes of prepared RNA were used for the reverse transcription reaction, except for the flowthrough samples, where equal concentrations of total RNA were used, as RNA levels were much higher and could be reliably measured by using spectrometry.
HIV-1 assays.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4.3-Luc (R− E−), from Nathaniel Landau, and pNL4.3, from Malcolm Martin. For viral transcription and replication assays with HeLa and TZM-bl cells, cells were first transfected with the indicated miRNA, followed by infection with vesicular stomatitis virus G protein (VSV-G)-pseudotyped NL4.3-Luc or NL4.3 virus 2 days later. For some assays, cells were also transfected with a cyclin T1 siRNA (5′-GCAGCGTCTTAACGTCTCA-3′) from Dharmacon or AllStars negative-control siRNA (Qiagen) and then infected with the strains indicated and washed with PBS 6 h later to remove virus. Cells were harvested 72 h later, and luciferase expression was measured by using the Single Luciferase system (Promega). The viral supernatant was spun down at 1,000 × g, and p24 levels were quantified by an enzyme-linked immunosorbent assay (ELISA) (Zeptometrix). For viral transcription assays with CD4+ T cells, primary resting cells were isolated, as described above, from four healthy blood donors. miRNAs, pNL4.3-Luc, and pCMV-RL (Renilla luciferase) were transfected by using nucleofection, and luciferase expression was measured 2 days later by using the Dual Luciferase system (Promega). Statistical analysis was performed by using a repeated-measures analysis of variance (ANOVA) (no between-subject/experiment factors and two repeated variables).
RESULTS
miR-27b, -29b, -150, and -223 are downregulated in activated CD4+ T cells and decrease cyclin T1 protein expression levels.
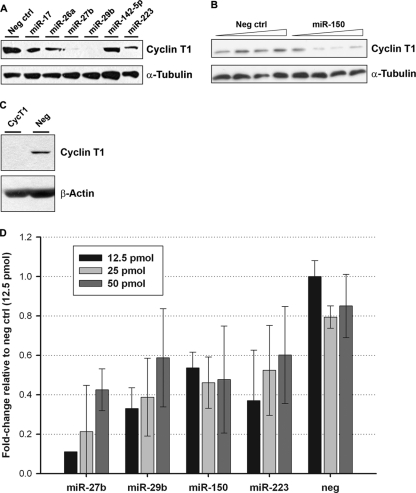
The expression levels of miRNAs that potentially repress cyclin T1 translation in resting CD4+ T cells should be inversely correlated with the expression level of the protein, which is low in resting cells and high in activated cells. In order to identify cyclin T1-repressing candidate miRNAs, we performed miRNA expression profiling of resting and activated CD4+ T cells isolated from four healthy blood donors (see Tables S1 and S2 in the supplemental material). For miRNAs downregulated in the majority of donors, we assessed their ability to bind to the cyclin T1 3′UTR using the target prediction algorithms RNA22 (33) and RNAhybrid (40). Synthetic precursors corresponding to candidate miRNAs were then transfected into HeLa cells to screen for their ability to repress endogenous cyclin T1 protein expression, along with miR-142-5p, which is not predicted to target the cyclin T1 3′UTR or coding sequence, and a nontargeting negative-control miRNA. Data from a representative experiment are shown in Fig. 1A; compared to the transfection of the negative control or miR-142-5p, the overexpressions of miR-27b, -29b, and -223 were consistently able to downregulate endogenous cyclin T1 protein levels in HeLa cells. The overexpression of miR-150 also decreased cyclin T1 protein levels (Fig. 1B). As expected, the decrease in cyclin T1 protein levels following miRNA treatment was generally less than that induced by cyclin T1 siRNA (Fig. 1C). These four miRNAs also decreased levels of cyclin T1 mRNA when overexpressed in HeLa cells (Fig. 1D); this pattern is consistent with the mRNA destabilization observed for most miRNA-mediated silencing (21).
Fig 1.
miRNAs downregulated after CD4+ T cell activation repress cyclin T1. (A) HeLa cells were transfected with the indicated miRNAs for 72 h. Cyclin T1 protein expression was then examined by immunoblotting. (B) HeLa cells were transfected with increasing doses of miR-150 or the negative control as described above. (C) HeLa cells were transfected with cyclin T1 siRNA for 48 h. Cyclin T1 protein knockdown was then visualized by immunoblotting. (D) Cyclin T1 mRNA levels were measured by qPCR 72 h after miRNA transfection in HeLa cells. Threshold cycle (CT) values were normalized to GAPDH levels in the same samples; data for all samples are presented relative to the cyclin T1-to-GAPDH CT ratio for the negative-control miRNA.
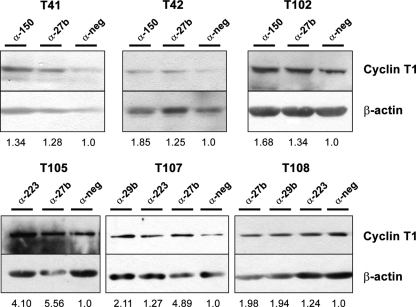
We next analyzed the effect of the inhibition of the expression of these miRNAs on primary resting CD4+ T cells. After immunoblotting to visualize cyclin T1 protein levels, we used densitometry analysis to quantify cyclin T1 bands relative to β-actin loading controls and found that antagomiR treatment increased cyclin T1 levels by approximately 1.25- to 4-fold in resting cells (Fig. 2); miR-27b was particularly effective at increasing cyclin T1 protein levels in multiple donors and was statistically significant over the negative control, while the other miRNAs displayed a similar trend but did not quite reach statistical significance. Given that the average miRNA is estimated to mediate a relatively modest decrease in the level of its target protein (3, 42) and that cyclin T1 protein levels are typically very low in resting cells, these results suggested that miR-27b, -29b, -150, and -223 are indeed acting in vivo to suppress cyclin T1 expression in resting CD4+ T cells.
Fig 2.
Inhibition of miRNAs increases cyclin T1 levels in resting CD4+ T cells. Resting CD4+ T cells were isolated from additional healthy blood donors and transfected with antagomiRs. Two days later, cyclin T1 protein levels were analyzed by immunoblotting. Numbers represent intensities of cyclin T1 bands, analyzed by using ImageJ software, normalized to those of the β-actin loading control. Anti-miR-27b treatment induced a statistically significant increase in the cyclin T1/β-actin ratio (P = 0.006), while the other antagomiRs showed similar trends but were just shy of statistical significance, likely due to the smaller number of donors (n = 2, 3, and 3 for miR-29b, -150, and -223, respectively).
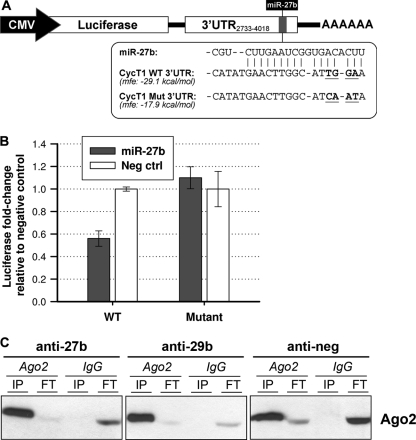
miR-27b directly binds the cyclin T1 3′UTR.
We identified a putative binding site within the cyclin T1 3′UTR for miR-27b and sought to validate it via mutational analysis. We generated a reporter construct containing the firefly luciferase gene upstream of a ∼1,300-bp portion of the cyclin T1 3′UTR encompassing the putative miR-27b binding site (Fig. 3A). Four consecutive mutations were introduced into the 3′UTR predicted to be bound by the seed sequence of miR-27b, thereby increasing the estimated minimum free energy of binding. Constructs containing the wild-type or mutant cyclin T1 3′UTR portions were then cotransfected with synthetic miR-27b into HeLa cells, and luciferase expression was measured. While the wild-type plasmid expressed ∼40% less luciferase upon cotransfection with miR-27b, the mutant plasmid was not responsive to miR-27b (Fig. 3B). This finding indicated that the four residues mutated within the cyclin T1 3′UTR are necessary for miR-27b binding, as the disruption of this site abrogates the miR-27b-mediated inhibition of luciferase expression. Using luciferase reporter plasmids, we were unable to verify cyclin T1 3′UTR or coding region binding sites for miR-29b, -150, and -223, suggesting that these three miRNAs may regulate cyclin T1 indirectly.
Fig 3.
Association of miRNAs with the cyclin T1 3′UTR and RISC. (A) Schematic of the luciferase-cyclin T1 3′UTR construct. The plasmid contains CMV-driven firefly luciferase followed by nucleotides 2733 to 4018 of the 4,580-nt cyclin T1 3′UTR and a poly(A) tail. The putative miR-27b binding site falls at residues 3783 to 3803. The mutated residues are highlighted in boldface type and lie within the nucleotides bound by the seed sequence of miR-27b; the minimum free energy (mfe) of binding, as predicted by RNA22, was increased for the mutant. (B) Wild-type (WT) and mutant luciferase-cyclin T1 3′UTR plasmids were cotransfected into HeLa cells along with miR-27b or a negative-control miRNA and a Renilla luciferase plasmid to assess transfection efficiency. Luciferase expression was measured 24 h later, and firefly luciferase expression levels were normalized to Renilla luciferase expression levels. The relative luciferase values for treatment with the negative-control miRNA were then set at 1.0 for cotransfection with both the wild-type and mutant plasmids. (C) Western blotting of Ago2 and IgG immunoprecipitations performed by using Jurkat cell lysates harvested 72 h post-antagomiR transfection. FT, flow-through.
Association of cyclin T1 mRNA with Ago2 is dependent on miR-27b and -29b.
To biochemically confirm that cyclin T1 mRNA is targeted by the RISC, we performed Ago2 immunoprecipitations in Jurkat cells, using nonimmune mouse IgG antibody as an isotype control. After verifying that Ago2 complexes were specifically pulled down by the Ago2 antibody and not isotype control IgG (Fig. 3C), cyclin T1 mRNA was measured by quantitative PCR (qPCR) and found to be associated with Ago2 (data not shown). The presence of coimmunoprecipitating miR-27b and PCAF mRNA, a positive control previously shown to be targeted by the miR-17-92 family (46), was also verified by qPCR (data not shown). Ago2 immunoprecipitation was then performed by using lysates from Jurkat cells transfected with antagomiRs against the cyclin T1-inhibitory miRNAs (Fig. 3C). The inhibition of miR-27b and miR-29b decreased the association of cyclin T1 mRNA with Ago2 (Table 1), while miR-150 and miR-223 had no effect in repeated experiments, further suggesting that miR-150 and miR-223 may be acting indirectly on the cyclin T1 protein.
Table 1.
Copies of immunoprecipitated cyclin T1 mRNA following antagomiR treatmenta
| Anti-miR | No. of copies of cyclin T1 normalized to no. of copies in starting lysate |
|
|---|---|---|
| Ago2 IP | IgG IP | |
| Anti-miR-27b | 1,420 | 250 |
| Anti-miR-29b | 135 | 959 |
| Negative control | 3,175 | 846 |
Following Ago2 or IgG immunoprecipitation (IP), cyclin T1 mRNA was quantified by qPCR, and numbers of copies were calculated against a cyclin T1 standard curve and then normalized to the number of cyclin T1 copies in the starting lysate (data from a representative experiment are shown).
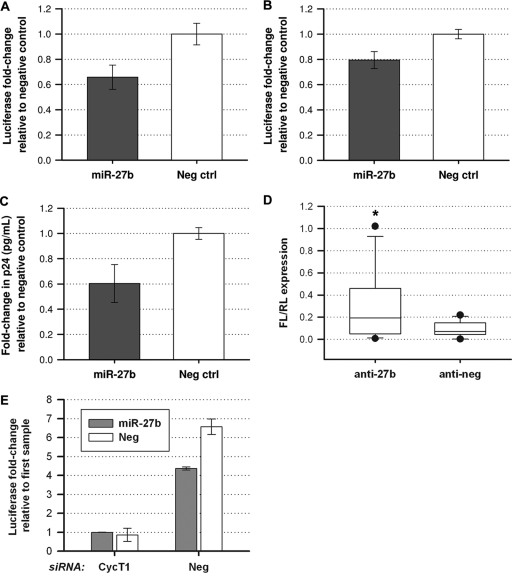
miR-27b inhibits HIV-1 replication.
As miR-29b, -150, and -223 were previously reported to inhibit HIV-1 replication (25, 34), we examined the ability of miR-27b to inhibit HIV-1 long terminal repeat (LTR)-driven gene expression using a luciferase reporter virus, NL4.3-Luc. Synthetic miR-27b was transfected into HeLa cells, which were then infected with a VSV-G-pseudotyped NL4.3-Luc virus. The overexpression of miR-27b decreased the gene expression of NL4.3-Luc by ∼35% in HeLa cells (Fig. 4A). We also used TZM-bl cells to evaluate whether miR-27b can inhibit replication-competent NL4.3 (Fig. 4B). TZM-bl cells are modified HeLa cells which express CD4, CXCR4, and CCR5 for viral entry and also contain an integrated luciferase reporter under the control of the viral LTR. After transfecting miR-27b into TZM-bl cells, infection with NL4.3 was performed. The viral gene expression level, as measured by the LTR-activated expression of luciferase, was decreased ∼20% in TZM-bl cells, while the viral production level was decreased by ∼40%, as measured by a p24 ELISA of the culture supernatant (Fig. 4C).
Fig 4.
miR-27b inhibits NL4.3 replication. (A) HeLa cells were transfected with miR-27b 48 h prior to infection with NL4.3-Luc pseudotyped with VSV-G. At 72 h postinfection, the luciferase activity of the cell lysates was measured. The values presented were normalized to those for the negative-control miRNA treatment. (B) TZM-bl cells were transfected with miR-27b 2 days prior to infection with full-length laboratory-adapted HIV-1 strain NL4.3. Luciferase activity was measured at 72 h postinfection. (C) Virus production in infected TZM-bl cells was measured by a p24 ELISA of the supernatant and was shown to be reduced by miR-27b overexpression. (D) Primary resting CD4+ T cells were transfected with antimiR-27b or a negative-control antagomiR, the NL4.3-Luc proviral plasmid, and a transfection control plasmid expressing Renilla luciferase (RL). Luciferase expression was measured 2 days later, and the firefly luciferase (FL) expression level was normalized to the Renilla luciferase expression level. Data shown represent data from 4 independent blood donors, transfected in triplicate. Error bars represent the 10th and 90th percentiles, and the dots represent the minimum and maximum of each data set (*, P < 0.05; the interaction of the repeated variables, treatment and repeat, was shown to be not significant [P = 0.0787], under Box's conservative epsilon). (E) HeLa cells were cotransfected with siRNA against cyclin T1 or a negative-control siRNA in addition to miR-27b or a negative-control miRNA on day 0 (for immunoblotting demonstrating the efficacy of cyclin T1 siRNA [Fig. 1C]). On day 2, cells were transfected with the NL4.3-Luc proviral plasmid, and luciferase expression was measured 72 h later.
While we had previously shown that the inhibition of miR-27b in primary resting CD4+ T cells increased cyclin T1 protein levels, we wanted to determine if anti-miR-27b treatment could also increase HIV gene expression levels in these nonpermissive cells. Therefore, we cotransfected resting CD4+ T cells with anti-miR-27b, pNL4.3-Luc, and a cytomegalovirus (CMV)-driven Renilla luciferase plasmid as a transfection control (Fig. 4D). Although primary resting CD4+ T cells are difficult to transfect, we did observe low levels of luciferase expression and found that they underwent a slight but statistically significant increase upon anti-miR-27b treatment, indicating that inhibiting miR-27b does increase HIV transcription.
In order to establish that the miR-27b-mediated repression of HIV-1 replication is due to its ability to repress cyclin T1 protein levels and not its effects on other mRNA targets, we cotransfected HeLa cells with cyclin T1 siRNA and miR-27b or the appropriate negative controls (Fig. 4E). Two days later, NL4.3-Luc proviral DNA was transfected into the cells. Decreased luciferase expression levels were observed for cells treated with cyclin T1 siRNA versus negative-control siRNA, as expected for the inhibition of this critical viral cofactor. When cells were treated with negative-control siRNA, decreased luciferase expression levels were also seen when cells were cotransfected with miR-27b versus negative-control miRNA. However, in cells treated with cyclin T1 siRNA, the cotransfection of miR-27b did not decrease luciferase expression more than did cotransfection with the negative-control miRNA, suggesting that miR-27b inhibits HIV-1 transcription in a cyclin T1-dependent manner.
DISCUSSION
It is well known that CD4+ T cells undergo dramatic changes at both the transcriptional and protein expression levels upon the activation of the T cell receptor (14, 20), driven by signaling cascades that turn on gene regulation programs that can accommodate the needs of a rapidly proliferating cell (26). HIV-1 appears to have evolved to take advantage of this generalized growth-conducive environment, in that the replication efficiency is much higher in activated CD4+ T cells as well as in activated macrophages. Cyclin T1 mRNA levels remain relatively constant during CD4+ T cell activation or monocyte-to-macrophage differentiation, whereas cyclin T1 protein levels undergo a large increase (31, 32, 43). Given that miRNAs are an elegant means of posttranscriptional control, we sought to identify miRNAs that target cyclin T1 mRNA. Previously, we identified miR-198 as a cyclin T1-targeting miRNA active in undifferentiated monocytes. However, the ability of miR-198 to target cyclin T1 appeared to be cell type specific, as levels were very low in resting CD4+ T cells and did not change in response to cell activation. Here we have identified miR-27b, -29b, -150, and -223 as contributors to the posttranscriptional gene repression of cyclin T1 in resting CD4+ T cells. We found that these miRNAs are downregulated upon cellular activation and that their overexpression decreased cyclin T1 protein levels in HeLa cells, while their inhibition increased cyclin T1 protein levels in resting CD4+ T cells. An miR-27b binding site within the cyclin T1 3′UTR was identified, and cyclin T1 mRNA immunoprecipitated with Ago2-containing complexes in a manner partially dependent on both miR-27b and miR-29b. The overexpression of miR-27b also negatively affected HIV-1 replication.
We previously showed that a large portion of cellular gene expression induced by T cell activation is in fact dependent upon cyclin T1 (53). We show here that the inhibition of individual miRNAs can increase cyclin T1 protein levels in resting CD4+ T cells, indicating that miR-27b, -29b, -150, and -223 significantly contribute to repression of cyclin T1 in vivo. After these cyclin T1-targeting miRNAs are downregulated in the activated T cell, presumably newly translated cyclin T1 protein supports both increased cellular and proviral transcription as one-half of the core P-TEFb complex. Given that a cyclin T1 knockdown is not cytotoxic in various cell lines (10, 29, 54), presumably because of a functional redundancy with cyclin T2 (38), therapeutic interventions using miR-27b, -29b, -150, or -223 may be possible in the future. However, it should be noted that the increase in cyclin T1 protein levels fell short of that seen upon cell activation, arguing that additional miRNAs or other mechanisms may be contributing to the inhibition of cyclin T1 translation in resting CD4+ T cells. Furthermore, while it is clear that these miRNAs inhibit cyclin T1 expression, it is likely that they also act on other cellular proteins that may also be cofactors of HIV-1. To examine the full contribution that miRNAs make to restricting HIV-1 replication in resting CD4+ T cells, Ago2 immunoprecipitation could be used in combination with deep sequencing to identify the complete set of mRNAs targeted by miRNAs (the so-called “targetome”), as has recently been done for other cell types (8, 15, 45).
miR-29b is part of the miR-29 family, comprised of miR-29a, -29b, and -29c, which all share the same seed region. All miR-29 family members were proposed previously to target a well-conserved site in the HIV-1 3′UTR and were shown to inhibit VSV-G-pseudotyped NL4.3-Luc virus production (34). miR-29a was also shown to target HIV-1 mRNA to the RISC. In this study, we show that miR-29b does the same for cyclin T1 mRNA, although we were unable to verify a binding site within the cyclin T1 mRNA sequence. However, we believe that miR-29b may bind directly to the cyclin T1 mRNA, as sequence-specific targeting to the RISC is mediated solely by small noncoding RNAs, making an miRNA without transcript complementarity an unlikely recruitment partner. It is very possible that our methodology was too limited and thus failed to identify an existing miR-29b binding site. miR-150 and miR-223 have also been proposed to target the HIV-1 3′UTR and were shown to be downregulated following CD4+ T cell activation (25), which we confirmed in our miRNA microarray. Furthermore, these two miRNAs modestly increased viral production in primary resting CD4+ T cells transfected with pNL4.3 (25). Our results suggest that miR-150 and miR-223 also affect HIV-1 replication by targeting cyclin T1, likely in an indirect fashion. miR-150 is highly expressed in hematopoietic cells (55) and has been shown to target the transcription factor c-Myb (52). As we were unable to show that miR-150 targets cyclin T1 directly, either by the identification of a binding site or by alterations in the Ago2 immunoprecipitation of the cyclin T1 transcript, we hypothesized that miR-150 may be affecting cyclin T1 via c-Myb. However, a c-Myb siRNA knockdown did not decrease cyclin T1 protein levels (data not shown), indicating that another mechanism may be at work.
To our knowledge, this is the first report describing the ability of miR-27b to inhibit HIV-1 replication. The transfection of miR-27b was able to decrease gene expression from the NL4.3-Luc reporter virus and the replication of full-length NL4.3. Anti-miR-27b treatment of primary resting CD4+ T cells led to a modest but statistically significant increase in NL4.3-Luc gene expression levels, although we noticed a substantial amount of variation between replicates derived from the same donor but transfected individually with anti-miR-27b (but not the negative-control antagomiR). This may be attributable to the stochastic expression of Tat and its amplifying effects on the positive-feedback loop of HIV transcription. It has been shown that clonal populations of Jurkat cells containing an integrated Tat feedback loop (HIV LTR-green fluorescent protein [GFP]-internal ribosome entry site [IRES]-Tat) exhibit varied GFP expression levels that can be predicted by a stochastically fluctuating model of Tat expression, wherein random increases in Tat levels can lead to the prolonged transactivation of HIV gene expression (51). It is possible that anti-miR-27b treatment, in combination with a randomly higher Tat level in an individual sample, may serve to drive NL4.3-Luc expression to a much higher level than that of the biologically matched replicates, accounting for the wide range of luciferase values for the anti-miR-27b data set. While cyclin T1 is the first miR-27b target to be specifically shown to have functional significance in CD4+ T cells, other targets validated in different tissues have been reported (1, 12, 27, 28, 47, 49). However, there is no obvious connection between any of these identified targets and HIV-1 replication. Furthermore, the best matches from the miR-27b target prediction analysis do not have any obvious connections to known HIV-1 cofactors. As has been reported for the other three miRNAs examined here, it is also possible that miR-27b might bind directly to the 3′UTR of HIV-1 genomic RNA, thereby targeting it for RISC-mediated translational repression. We have not identified any miR-27b binding sites in the 3′UTR of the NL4.3-based strains used in our experiments, and we believe that the effect of miR-27b on viral replication occurs indirectly, via the inhibition of cyclin T1 and possibly other proteins.
The miR-27 family (miR-27a and miR-27b, differing by only one nucleotide at the 3′ end) was recently implicated in the replication processes of two viruses found in New World primates and mice. Herpesvirus saimiri expresses a noncoding RNA, HSUR1 (herpesvirus saimiri U-rich RNA 1), which downregulates miR-27 expression, resulting in low levels of miR-27 in herpesvirus-transformed marmoset T cells (7). The knockdown of HSUR1 leads to a corresponding increase in miR-27 levels, while the mutation of sites complementary to HSUR1 within miR-27 abrogates its immunoprecipitation with small nuclear ribonucleoproteins where HSUR1 localizes. miR-27 is also downregulated via an unknown mechanism following murine cytomegalovirus infection in a variety of cell lines and in primary murine macrophages (5); the overexpression of miR-27 leads to greatly decreased viral replication. It is currently unknown which targets of miR-27 drive these two viruses to decrease levels of miR-27 as a means of increasing protein expression levels of critical viral cofactors. While it is very likely that a variety of miR-27 targets contribute to viral replication, we suggest that increases in cyclin T1 levels, as a transcriptional elongation factor, would be beneficial for supporting general viral replication.
Supplementary Material
ACKNOWLEDGMENTS
2D10 cells were a gift from Jonathan Karn (Case Western University). We thank Sona Budhiraja for technical assistance and Claudia Kozinetz and Laura Ferlic-Stark for help with statistical analysis.
K.C. and T.-L.S. conceived, performed, and analyzed experiments, and A.P.R. conceived and analyzed experiments. K.C. and A.P.R. wrote the manuscript.
This study was supported by funding from the National Institutes of Health (grants AI007471-14 to K.C. and AI036211 and DAO30166-01 to A.P.R.). This project was also supported by the BCM Cytometry and Cell Sorting Core with funding from the NIH (NCRR grant S10RR024574, NIAID grant AI036211, and NCI grant P30CA125123).
Footnotes
Published ahead of print 28 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Akhtar N, et al. 2010. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 62:1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. 2009. Lost in translation: an assessment and perspective for computational microRNA target identification. Bioinformatics 25:3049–3055 [DOI] [PubMed] [Google Scholar]
- 3. Baek D, et al. 2008. The impact of microRNAs on protein output. Nature 455:64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barboric M, Peterlin BM. 2005. A new paradigm in eukaryotic biology: HIV Tat and the control of transcriptional elongation. PLoS Biol. 3:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buck AH, et al. 2010. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA 16:307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cazalla D, Yario T, Steitz J. 2010. Down-regulation of a host microRNA by a herpesvirus saimiri noncoding RNA. Science 328:1563–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chi SW, Zang JB, Mele A, Darnell RB. 2009. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460:479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiang K, Rice AP. Mini ways to stop a virus: microRNAs and HIV-1 replication. Future Virol. 6:209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM. 2004. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/cyclinT1). J. Virol. 78:2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coburn GA, Cullen BR. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Crist CG, et al. 2009. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. U. S. A. 106:13383–13387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Croce CM. 2009. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10:704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diehn M, et al. 2002. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc. Natl. Acad. Sci. U. S. A. 99:11796–11801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dolken L, et al. 2010. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 7:324–334 [DOI] [PubMed] [Google Scholar]
- 16. Fabian MR, Sonenberg N, Filipowicz W. 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79:351–379 [DOI] [PubMed] [Google Scholar]
- 17. Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garriga J, et al. 1998. Upregulation of cyclin T1/CDK9 complexes during T cell activation. Oncogene 17:3093–3102 [DOI] [PubMed] [Google Scholar]
- 19. Ghose R, Liou LY, Herrmann CH, Rice AP. 2001. Induction of TAK (cyclin T1/P-TEFb) in purified resting CD4+ T lymphocytes by combination of cytokines. J. Virol. 75:11336–11343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grant MM, Scheel-Toellner D, Griffiths HR. 2007. Contributions to our understanding of T cell physiology through unveiling the T cell proteome. Clin. Exp. Immunol. 149:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo H, Ingolia NT, Weissman JS, Bartel DP. 2010. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hendrickson DG, et al. 2009. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 7:e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herrmann CH, Carroll RG, Wei P, Jones KA, Rice AP. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Houzet L, et al. 2008. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology 5:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang J, et al. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 26. Huang Y, Wange RL. 2004. T cell receptor signaling: beyond complex complexes. J. Biol. Chem. 279:28827–28830 [DOI] [PubMed] [Google Scholar]
- 27. Jennewein C, von Knethen A, Schmid T, Brune B. 2010. MicroRNA-27b contributes to lipopolysaccharide-mediated peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA destabilization. J. Biol. Chem. 285:11846–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karbiener M, et al. 2009. MicroRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 390:247–251 [DOI] [PubMed] [Google Scholar]
- 29. Li Z, et al. 2005. Specific inhibition of HIV-1 replication by short hairpin RNAs targeting human cyclin T1 without inducing apoptosis. FEBS Lett. 579:3100–3106 [DOI] [PubMed] [Google Scholar]
- 30. Lim LP, et al. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773 [DOI] [PubMed] [Google Scholar]
- 31. Liou LY, Haaland RE, Herrmann CH, Rice AP. 2006. Cyclin T1 but not cyclin T2a is induced by a post-transcriptional mechanism in PAMP-activated monocyte-derived macrophages. J. Leukoc. Biol. 79:388–396 [DOI] [PubMed] [Google Scholar]
- 32. Marshall RM, Salerno D, Garriga J, Grana X. 2005. Cyclin T1 expression is regulated by multiple signaling pathways and mechanisms during activation of human peripheral blood lymphocytes. J. Immunol. 175:6402–6411 [DOI] [PubMed] [Google Scholar]
- 33. Miranda KC, et al. 2006. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126:1203–1217 [DOI] [PubMed] [Google Scholar]
- 34. Nathans R, et al. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 34:696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. 2010. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10:111–122 [DOI] [PubMed] [Google Scholar]
- 36. Peng J, Zhu Y, Milton JT, Price DH. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297–305 [DOI] [PubMed] [Google Scholar]
- 38. Ramakrishnan R, Yu W, Rice AP. 2011. Limited redundancy in genes regulated by cyclin T2 and cyclin T1. BMC Res. Notes 4:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rasband WS. 1997–2009. ImageJ. US National Institutes of Health, Bethesda, MD: http://rsb.info.nih.gov/ij/ [Google Scholar]
- 40. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. 2004. Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rice AP, Herrmann CH. 2003. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr. HIV Res. 1:395–404 [DOI] [PubMed] [Google Scholar]
- 42. Selbach M, et al. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455:58–63 [DOI] [PubMed] [Google Scholar]
- 43. Sung TL, Rice AP. 2006. Effects of prostratin on cyclin T1/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology 3:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sung TL, Rice AP. 2009. miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1. PLoS Pathog. 5:e1000263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan LP, et al. 2009. A high throughput experimental approach to identify miRNA targets in human cells. Nucleic Acids Res. 37:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Triboulet R, et al. 2007. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 315:1579–1582 [DOI] [PubMed] [Google Scholar]
- 47. Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. 2006. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 66:9090–9098 [DOI] [PubMed] [Google Scholar]
- 48. Umbach JL, Cullen BR. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y, Rathinam R, Walch A, Alahari SK. 2009. ST14 (suppression of tumorigenicity 14) gene is a target for miR-27b, and the inhibitory effect of ST14 on cell growth is independent of miR-27b regulation. J. Biol. Chem. 284:23094–23106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462 [DOI] [PubMed] [Google Scholar]
- 51. Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182 [DOI] [PubMed] [Google Scholar]
- 52. Xiao C, et al. 2007. miR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131:146–159 [DOI] [PubMed] [Google Scholar]
- 53. Yu W, et al. 2008. Cyclin T1-dependent genes in activated CD4 T and macrophage cell lines appear enriched in HIV-1 co-factors. PLoS One 3:e3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu W, Wang Y, Shaw CA, Qin XF, Rice AP. 2006. Induction of the HIV-1 Tat co-factor cyclin T1 during monocyte differentiation is required for the regulated expression of a large portion of cellular mRNAs. Retrovirology 3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. 2007. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. U. S. A. 104:7080–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou Q, Yik JH. 2006. The yin and yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70:646–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.