Abstract
A potential drawback of recent antiviral therapies based on the transgenic expression of artificial microRNAs is the ease with which viruses may generate escape mutations. Using a variation of the classic Luria-Delbrück fluctuation assay, we estimated that the spontaneous mutation rate in the artificial microRNA (amiR) target of a plant virus was ca. 6 × 10−5 per replication event.
TEXT
The rate of spontaneous mutation is a key parameter to understand the genetic structure of populations over time. Mutation represents the primary source of genetic variation on which natural selection and genetic drift operate. Although the exact value of mutation rate is important for several evolutionary theories, accurate estimates are available only for a reduced number of organisms (15). In the case of RNA viruses, mutation rates are orders of magnitude higher than those of their DNA-based hosts (7). These high mutation rates have important practical implications; for instance, for the long-term durability of vaccination strategies (6) and antiviral drugs (2), for the stability of live attenuated vaccines (26), for the eventual success of antiviral therapies based on the concept of lethal mutagenesis (1), or to determine the risk of new emerging viruses (14). The spontaneous mutation rate of a virus can be evaluated in vivo using a variety of experimental approaches. Among the most commonly used approaches are (i) estimating the frequency of mutants contained in a population generated from a single clone (7, 23), (ii) counting the number of mutant alleles accumulated in a locus which was protected against the action of purifying selection (20, 25), (iii) counting the number of lethal alleles present in a population (10), (iv) estimating the mean and variance in fitness declines among independent lineages during a mutation accumulation experiment and then applying the Mukai-Bateman method (8), or (v) using a fluctuation assay (19). Among all of these approaches, the last is considered the most flexible, robust, and reliable method (9). The fluctuation test, originally developed by Luria and Delbrück (19), allows estimation of the rate at which mutations arise in a genetic locus associated with an easy-to-score phenotype. The estimates obtained are independent of generation time and replication mode, factors that are not available for most RNA viruses. Advanced mathematical tools for the analysis of the distribution of the number of mutants across replicated cultures (the so-called Luria-Delbrück distribution) are readily available and easy to adapt to each experimental design (9).
The transgenic expression of 21-nucleotide (nt)-long artificial microRNAs (amiR) complementary to viral genomes has been proposed as a new antiviral strategy. Niu et al. (22) used the pre-miRNA159a precursor to engineer an amiR containing a sequence complementary to the RNA genome of Turnip mosaic potyvirus (TuMV). Transgenic expression of this amiR in Arabidopsis thaliana conferred high levels of specific resistance. Similarly, a gene-silencing mechanism (RNA interference [RNAi]) has been used in in vitro assays as antiviral therapeutics to inhibit the replication of several human viruses (5, 11, 16). However, a major issue of these amiR-based antiviral therapies has been the emergence of escape mutant viruses (3, 13, 17). These escape variants differ from the wild-type virus by at least one point mutation in the 21-nt target, leading to imperfect matching with the amiR. To evaluate the durability of amiR-mediated resistance in plants, Lafforgue et al. (17) performed an evolution experiment in which multiple independent lineages of TuMV were founded with an ancestral virus clone and allowed to evolve and diversify by serial passages in two different hosts. The first host was a wild-type A. thaliana and the second one was the partially resistant 10-4 transgenic A. thaliana line that expressed amiR at subinhibitory concentrations. Periodically, the evolving populations were used to challenge the resistance of the 12-4 transgenic A. thaliana line, which was fully resistant to the ancestral virus. It was found that all lineages that evolved in wild-type plants accumulated mutations in the amiR target and acquired the capacity to successfully infect 12-4 plants (17). The median time for lineages evolved in wild-type plants to break resistance was 14 passages, while lineages evolved in partially resistant plants only took 2 passages. The ease to break this resistance correlated with the existence of natural variation for the 21-nt target sequence (A. Lafforgue, S. F. Elena, unpublished results), suggesting that this genomic region shall not be under strong purifying selection.
The frequency at which mutations may be produced in the amiR target locus in evolving TuMV populations is fundamental to understanding the observed dynamics of resistance breaking. Here, we report the results of a fluctuation assay experiment designed to evaluate the spontaneous mutation rate at the amiR target locus of TuMV. In this case, the phenotype associated with the mutants was the ability to replicate in the 12-4 transgenic plants expressing the antiviral amiR. We used a modification of the analytical method proposed in reference 12 that provides improved accuracy and is especially well suited to large populations and/or high mutation rates. This method is a generalization of the statistical modeling developed by Lea and Coulson (18).
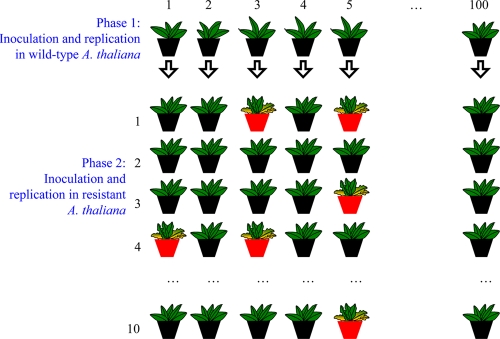
Figure 1 shows a scheme of the experimental design for this fluctuation assay. A large stock of infectious sap was obtained from Nicotiana benthamiana plants inoculated with a plasmid containing TuMV cDNA (4, 17). This amplification step was necessary to overcome the low-efficiency-infecting A. thaliana plants with the TuMV cDNA. Sap was obtained by grinding infected tissues in a mortar with liquid N2 and 20 volumes of extraction buffer (50 mM potassium phosphate, pH 7.0, 3% polyethylene glycol 6000). One hundred wild-type A. thaliana plants were inoculated with 5 μl sap containing 10% carborundum applied on three different leaves and gentle rubbing with a cotton swab (Fig. 1). After inoculation, plants were maintained in a growth chamber (16 h of light at 25°C and 8 h of dark at 24°C). TuMV replicated and systemically colonized the plants until reaching a population size of Ni, where the subscript denotes the ith plant. From each of these plants, virus was extracted from symptomatic tissue 14 days postinoculation (dpi), as described above. Heterogeneity in virus accumulation on leafs of different ages was minimized by pooling them into a single extraction. However, as only a fraction of the virus-infected host tissues was extracted, only a fraction (d) of total virus produced was obtained and successfully transmitted. This extract was divided into 10 parts, each of which was used to inoculate a 12-4-resistant plant as described above; i.e., there were 10 resistant plants per each susceptible one (Fig. 1). Fourteen dpi, the number of resistant plants on which infection was successfully established was recorded. From this vector of counts, R, a mutation rate was estimated using the following procedure. The number of mutants after growth and extraction from wild-type plants has a distribution whose probability generating function is (12):
where p = d/10 is the total dilution factor (dilution due to extraction, d, and dilution due to partitioning the extract into 10 parts), μ is mutation rate per amiR target locus, and z denotes the argument of the generating function h(·).
Fig 1.
Schematic representation of the fluctuation assay. A TuMV stock was produced from infected N. benthamiana plants that were previously inoculated with a TuMV clone. This stock was used to mechanically inoculate 100 wild-type A. thaliana plants (phase 1). During this phase, erroneous viral replication produces spontaneous mutants in the amiR 21-nt target that accumulate in the population. Fourteen dpi, virus was purified from each of these plants and used to inoculate batches of 10 A. thaliana 12-4 plants expressing the antiviral amiR (phase 2). During this phase, only those genomes carrying a mutation in the 21-nt target would eventually escape from the RNA silencing. Plants inoculated with these mutants will develop symptoms, whereas plants inoculated with the wild-type TuMV will not. The number of infected 12-4 plants showing symptoms of infection was recorded 14 dpi (red pots).
Infection is not a deterministic process, and a single virion has probability q of infecting a plant and 1 − q of not doing so. If the diluted inoculum contains m mutants, then the probability of not establishing an infection on the 12-4 resistant plants is (1 − q)m. The number of mutants is unknown and so is treated as a random variable, and the total probability of not establishing an infection is therefore (1 − q)m ϕ(m), where ϕ(m) is the mth coefficient in the expansion of h(z). The total probability of not establishing infection is therefore h(1 − q). Since the probability of establishing an infection is 50% when 1 − (1 − q)λ0.5 = ½, where λ0.5 is the median infectious dose, the parameter q may thus be calculated as q = 1 − 2−1/λ0.5.
The log-likelihood function for μ is thus l(μ|R, Ni, d, λ0.5) = logξ(i), where 100 × 10 = 1,000 is the total number of resistant plants used in the fluctuation assay and
This function is maximized at μ = μ̂, the maximum likelihood (ML) estimate of the mutation rate. Therefore, in addition to the vector R with the counts of infected 12-4 plants for each wild-type plant, the other relevant parameters to be experimentally determined are Ni (i = 1, …, 100), d, and λ0.5.
First, the concentration of the TuMV genomic [TuMV(+)] RNA strand in the original stock as well as resulting from each of the 100 wild-type A. thaliana plants (e.g., Ni) was measured by absolute reverse transcription (RT)-quantitative PCR (qPCR) using an external standard as described in reference 21. In short, the standard curve was constructed using 1/5-fold dilution intervals of TuMV(+) RNA in the range of 1.28 × 108 to 4 × 104 molecules. Aliquots of 100 ng total RNA were reverse transcribed in triplicate in the presence of 250 nM primer PI (5′-TAACCCCTTAACGCCAAGTAAG-3′; sequence complementary to TuMV GenBank accession no. AF530055.2, positions 9599 to 9620) with Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Fermentas) in 20-μl reaction mixtures for 10 min at 25°C, 45 min at 42°C, and 5 min at 50°C. Reactions were stopped by heating the mixtures at 72°C for 15 min. Sequence-specific qPCR analyses were performed with 2 μl reverse transcription products in 20 μl final volume using the Maxima SYBR green master mix reagent (Fermentas) and primers PI and PII (5′-CAATACGTGCGAGAGAAGCACAC-3′; sequence homologous to TuMV positions 9448 to 9470) at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Considering the total aerial plant masses (see below), measured Ni values ranged from 3.845 × 1010 to 3.429 × 1011, with an average value of 1.226 × 1011 TuMV(+) RNA molecules per plant (95% confidence interval [95% CI] around the mean, 1.110 × 1011 to 1.341 × 1011).
Second, the dilution factor d was approximated as the fraction of wild-type plant tissue used to generate the sap that was later used to inoculate the corresponding set of 12-4 resistant plants. On average, the aerial part of the infected wild-type plants weighted 0.922 ± 0.090 g (±1 the standard error of the mean [SEM]) and the average weight of the tissue ground to produce the 100 inocula was 0.122 ± 0.007 g, which corresponds to a dilution factor of d = 0.132 ± 0.021.
Third, a dose infectivity assay was used to evaluate λ0.5. To do so, the original TuMV stock was serially diluted with 1/5-fold intervals in the range of 1/1 to 1/500, and each dilution was used to inoculate sets of 10 plants. Twenty dpi, the number of symptomatic plants was recorded. Infectivity data were subjected to a probit analysis that rendered an estimate of the median infectious dose of λ0.5 = 8.826 × 106 TuMV genomes per 12-4 plant (95% CI, 4.543 × 106 to 1.779 × 107; goodness of fit test, χ2 = 2.694; 5 degrees of freedom [d.f.]; P = 0.747).
Finally, the fluctuation assay rendered the following results. From a total of 100 wild-type A. thaliana plants used as a source of TuMV inocula, only 11 contained escape mutants that produced at least one 12-4 plant infected (five cases of 1/10 and two cases each of 2/10, 3/10, and 5/10). Feeding all data into the ML algorithm, the estimate of the mutation rate for the amiR target locus was μ̂ = 5.545 × 10−5 mutations per replication event (95% CI, 2.886 × 10−5 to 9.507 × 10−5). Since the amiR target is 21-nt long, this estimate can be expressed in a more common per-nucleotide scale as 2.640 × 10−6 substitutions per site and per replication event (s/n/r) (95% CI, 1.374 × 10−6 to 4.527 × 10−6). This empirical estimate is between 17 to 30 times lower than the value suggested by the simulations performed by Lafforgue et al. (17) which, given the many assumptions behind the simulations, can be considered a reasonable discrepancy.
Direct estimates of mutation rates for plant RNA viruses are scarcer than for their animal and bacterial counterparts. The first estimate ever reported for a plant virus was for Tobacco mosaic virus and it was ca. 1.8 × 10−5 s/n/r (20). Later on, the mutation rate for Tobacco etch virus (TEV) was estimated to range between 2.960 × 10−5 (23) and 4.754 × 10−6 s/n/r (25). Our data for TuMV are in good agreement with those reported for TEV, another potyvirus. Furthermore, all these estimates are well within the range of 10−6 to 10−4 that was recently reported for several animal RNA viruses and bacteriophages (24). Altogether, the recent estimates obtained for plant RNA viruses and the reanalysis made of previous data (24) suggest that the mutation rate of RNA viruses may be lower than previously proposed (7).
ACKNOWLEDGMENTS
This work was supported by grants BFU2009-06993 from the Spanish Ministerio de Ciencia e Innovación, RGP12/2008 from the Human Frontier Science Program Organization, and PROMETEO2010/019 from Generalitat Valenciana to S.F.E.; by CSIC grant 2010TW0015 to J.-A.D.; and by U.S. National Institutes of Health grants R01GM079843-01 and ARRA PDS#35063 and EC grant FP7231807 to P.J.G. F.M. was supported by a fellowship from Universidad Politénica de Valencia, J.H. was supported by a fellowship from the Spanish Ministerio de Ciencia e Innovación, and J.M.C. was contracted under the CSIC JAE-Doc program.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Anderson JP, Daifuku R, Loeb LA. 2004. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 58:183–205 [DOI] [PubMed] [Google Scholar]
- 2. Arribas M, Cabanilles L, Lázaro E. 2011. Identification of mutations conferring 5-azacytidine resistance in bacteriophage Q.β. Virology 417:343–352 [DOI] [PubMed] [Google Scholar]
- 3. Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. 2003. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77:11531–11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen CC, et al. 2003. Identification of Turnip mosaic virus isolates causing yellow stripe and spot on calla lily. Plant Dis. 87:901–905 [DOI] [PubMed] [Google Scholar]
- 5. Coburn GA, Cullen BR. 2002. Potent and specific inhibition of Human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davenport MP, Loh L, Petravic J, Kent SJ. 2008. Rates of HIV immune escape and reversion: implications for vaccination. Trends Microbiol. 16:561–566 [DOI] [PubMed] [Google Scholar]
- 7. Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 96:13910–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elena SF, Moya A. 1999. Rate of deleterious mutation and the distribution of its effects on fitness in vesicular stomatitis virus. J. Evol. Biol. 12:1078–1088 [Google Scholar]
- 9. Foster PL. 2006. Methods for determining spontaneous mutation rates. Methods Enzymol. 409:195–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gago S, Elena SF, Flores R, Sanjuán R. 2009. Extremely high mutation rate of a hammerhead viroid. Science 323:1308. [DOI] [PubMed] [Google Scholar]
- 11. Ge Q, MT, et al. 2003. RNA interference of Influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc. Natl. Acad. Sci. U. S. A. 100:2718–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerrish PJ. 2008. A simple formula for obtaining markedly improved mutation rate estimates. Genetics 180:1773–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gitlin L, Andino R. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 79:1027–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holmes EC. 2009. The evolutionary genetics of emerging viruses. Annu. Rev. Ecol. Evol. Syst. 40:353–372 [Google Scholar]
- 15. Kondrashov FA, Kondrashov AS. 2010. Measurements of spontaneous rates of mutation in recent past and the near future. Philos. Trans. R. Soc. Lond. B 365:1169–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Krönke J, et al. 2004. Alternative approaches for efficient inhibition of Hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 78:3436–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lafforgue G, et al. 2011. Tempo and mode of plant RNA virus escape from RNA interference-mediated resistance. J. Virol. 85:9686–9695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lea DE, Coulson CA. 1949. The distribution of the numbers of mutants in bacterial populations. J. Genet. 49:264–285 [DOI] [PubMed] [Google Scholar]
- 19. Luria SE, Delbrück M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malpica JM, et al. 2002. The rate and character of spontaneous mutation in an RNA virus. Genetics 162:1505–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martínez F, Sardanyés J, Elena SF, Daròs JA. 2011. Dynamics of a plant RNA virus intracellular accumulation: stamping machine vs. geometric replication. Genetics 188:637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niu Q, et al. 2006. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24:1420–1428 [DOI] [PubMed] [Google Scholar]
- 23. Sanjuán R, Agudelo-Romero P, Elena SF. 2009. Upper limit mutation rate estimation for a plant RNA virus. Biol. Lett. 5:394–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J. Virol. 84:9733–9748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tromas N, Elena SF. 2010. The rate and spectrum of spontaneous mutations in a plant RNA virus. Genetics 185:983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vignuzzi M, Wendt E, Andino R. 2008. Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 14:154–161 [DOI] [PubMed] [Google Scholar]