Abstract
The product of the human cytomegalovirus (HCMV) UL71 gene is conserved throughout the herpesvirus family. During HCMV infection, protein pUL71 is required for efficient virion egress and is involved in the final steps of secondary envelopment leading to infectious viral particles. We found strong indications for oligomerization of pUL71 under native conditions when recombinant pUL71 was negatively stained and analyzed by electron microscopy. Oligomerization of pUL71 during infection was further verified by native and reducing polyacrylamide gel electrophoresis (PAGE). By in silico analyses of the pUL71 sequence, we noticed a basic leucine zipper (bZIP)-like domain, which might serve as an oligomerization domain. We demonstrated the requirement of the bZIP-like domain for pUL71 oligomerization by coimmunoprecipitation and bimolecular fluorescence complementation using a panel of pUL71 mutants. These studies revealed that the mutation of two leucine residues is sufficient to abrogate oligomerization but that intracellular localization of pUL71 was unaffected. To investigate the relevance of the bZIP domain in the viral context, recombinant viruses carrying mutations identical to those in the panel of pUL71 mutants were generated. bZIP-defective viral mutants showed impaired viral growth, a small-plaque phenotype, and an ultrastructural phenotype similar to that of the previously described UL71 stop mutant virus. The majority of virus particles within the viral assembly compartment exhibited various stages of incomplete envelopment, which is consistent with the growth defect for the bZIP mutants. From these data we conclude that the bZIP-like domain is required for oligomerization of pUL71, which seems to be essential for correct envelopment of HCMV.
INTRODUCTION
Human cytomegalovirus (HCMV) is a member of the herpesvirus family and is a major human pathogen, causing serious illness in neonates as well as in immunocompromised adults (28). Although the family is diverse, all the herpesviruses share certain features. Each virion consists of an inner core containing the double-stranded DNA, an icosahedral nucleocapsid, an amorphous tegument, and an envelope with inserted viral glycoproteins.
HCMV has sequential regulation of viral gene expression, leading to induction and repression cycles occurring in the immediate-early (IE), early (E), and late (L) phases of viral replication (26). Immediate-early proteins induce expression of early proteins and mediate G1/S cell cycle arrest and host replication shutoff (4, 11). Early genes encode predominantly viral DNA replication factors, repair enzymes, or immune evasion proteins and induce the expression of late genes (27). Late proteins downregulate expression of early genes and are involved mainly in virion assembly.
DNA replication leads to the formation of concatemeric DNA, which must be resolved into unit-length genomes during packaging. Once the head-to-tail-linked concatemers are cleaved by the terminase (2), the unit-length genomes are encapsidated into preformed procapsids and organized into a highly condensed, ordered structure (38). After DNA packaging, the capsids undergo two sequential budding events, one during nuclear egress and the other during final envelopment at early endosomal cisternae (41). Since these two envelopment processes occur in different cellular compartments, different sets of viral proteins are involved (25).
Tegument proteins play important roles throughout the viral life cycle, in (i) translocation of incoming capsid to the nucleus, (ii) viral gene expression, (iii) nuclear egress of capsids, and (iv) final envelopment in the cytoplasm. By using mass spectrometry, Varnum et al. (43) demonstrated that tegument proteins represent 50% of all viral components.
The HCMV UL71 gene product is a tegument protein conserved throughout the herpesviruses. Two independent reports (34, 46) have shown previously that protein pUL71 is required for final envelopment of viral particles. Despite the important function of the protein, little is known so far about structure-function relationships. In this study we investigated the oligomerization of pUL71 and identified a basic leucine zipper (bZIP)-like domain that is required for the formation of higher-ordered forms of pUL71. Mutational analysis was applied to demonstrate that the bZIP-like domain is essential for the function of pUL71 during viral maturation.
MATERIALS AND METHODS
Cells and virus.
Human foreskin fibroblasts (HFFs), human embryonic kidney 293T (293T) cells (European Collection of Cell Cultures [ECCC], Salisbury, United Kingdom), HeLA cells (ECCC), and MRC-5 cells (ECCC) were grown in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, penicillin (5 U/ml), and streptomycin (50 μg/ml). HFF cells were used before passage 23 for infection experiments which were carried out with confluent cell monolayers. Infection of HFFs with HCMV AD169 was carried out as described before (37).
Insect cells (High Five 5B1-4) were grown in TC-100 medium (PAN Biotech) supplemented with glutamine (60 μg/ml) and 10% FCS. Recombinant baculovirus UL71 was generated by transposition into bacmid bMON14272 as described by Luckow et al. (22).
Plasmid construction and oligonucleotides.
Restriction enzymes were purchased from Fermentas (St. Leon-Rot, Germany) and used according to the instructions of the manufacturer. The pUL71 protein variants were generated by PCR amplification using the oligonucleotides and constructs listed in Table 1.
Table 1.
Oligonucleotide primers used in PCR
| Construct(s) | Sense primer (5′→3′) | Antisense primer (5′→3′) |
|---|---|---|
| Protein constructs | ||
| pUL71 | CGGGATCCATGCAGCTGGCCCAGC | CATCTAGATCACGCGGAGGACAGC |
| 71_wt-myc | TAGAATTCATGCAGCTGGCCCAGC | CAGGATCCTCACGCGGAGGACAGC |
| 71_L34A | GAGGACGTGGAGCTGCGCGAGCTGCAGGCGTTTGCTGACGAGAAC | AGTTCTCGTCCAGAAACGCCTGCAGCTCGCGAGCCTCCTCGTCC |
| 71_L41A | GAGGACGTGGAGGCTCGCGAGCTGCAGGCGTTTCTGGACGAGAAC | AGTTCTCGTCAGCAAACGCCTGCAGCTCGCGCAGCTCCTCGTCC |
| 71_L1-myc | GAGGACGTGGAGGCTCGCGAGCTGCAGGCGTTTGCTGACGAGAAC | AGTTCTCGTCAGCAAACGCCTGCAGCTCGCGAGCCTCCTCGTC |
| 71_L2-myc/71_L3-myc | CTTTAAGCAGGCTGAGATCACCCCGGCCGACGCTCGAACCTTTTCTC | CGAGAAAAGGTTCGAGCGTCGGCCGGGGTGATCTCAGCCTGCTTAAAG |
| 71_ΔLZ-myca | (1) CGGGATCCATGCAGCTGGCCCAGC | (1) ACGGATCCCTCCACGTCCTCGCTAG |
| (2) ACAGATCTACCTTTTCTCGCGACACG | (2) CATCTAGATCACGCGGAGGACAGC | |
| (3) CGGGATCCATGCAGCTGGCCCAGC | (3) CATCTAGATCACGCGGAGGACAGC | |
| Dimerization constructs | ||
| 71_wt-YC, 71_L1-YC, and 71_ΔLZ-YCa | (1) CACCAGATCTGATATCGGTGGCGGTGGCTCTGGAGGTGGTGGGTCCTACCCATACGATGTTCCAGATTACG | (1) AGTGCGGCCGCGTTAACTTACTTGTACAGCTCGTCCATGCC |
| (2) TACGGGATCCATGCAGCTGGCCCAGCGC | (2) TCAGGATCCCGCGGAGGACAGCAAGGCC | |
| (3) CACCAGATCTGATATCGGTGGCGGTGGCTCTGGAGGTGGTGGGTCCTACCCATACGATGTTCCAGATTACG | (3) TCAGGATCCCGCGGAGGACAGCAAGGCC | |
| 71_wt-YN | ||
| Baculovirus construct | ||
| pFast71 | TAGAATTCATGCAGCTGGCCCAGC | CAGGATCCTCACGCGGAGGACAGC |
A 3-step PCR procedure was used for construct 71_ΔLZ-myc and for the YC constructs. The primers used in each step are indicated.
Transfection of 293T and HeLa cells.
For transient expression, 293T cells or HeLa cells (5 × 106) were seeded onto 6-well plates containing coverslips. Cells with a confluence of 60% were transfected with plasmid constructs by using TurboFect according to the protocol of the manufacturer (Fermentas). At the appropriate time point, cells were fixed with 3% paraformaldehyde as described previously (37).
BAC mutagenesis.
For the generation of bacterial artificial chromosome (BAC) mutant TBmut71-ΔLZ-BAC, the sequence coding for amino acids (aa) 34 to 41 of pUL71 was deleted, and for mutant TBmut71-L1-BAC, the codons for leucine residues at aa 34 and aa 41 of pUL71 were mutated to code for alanine. These mutations were introduced into the parental BAC TB40-BAC4 (34, 44) by Red-mediated mutagenesis as described previously by Tischer et al. and Schauflinger et al. (34, 40). The following primers were used for the mutagenesis: 71-ΔLZ_for (5′-GGCCGATTACGTGCTGCTGCAGCCTAGCGAGGACGTGGAGGACGAGAACTTTAAGCAGCTAGGATGACGACGATAAGTAGGG-3′) and 71-ΔLZ_rev (5′-GATCGGCCGGGGTGATCTCCAGCTGCTTAAAGTTCTCGTCCTCCACGTCCTCGCTAGGCTCAACCAATTAACCAATTCTGATTAG-3′) and 71-L1_for (5′-GGCCGATTACGTGCTGCTGCAGCCTAGCGAGGACGTGGAGGCCCGCGAGCTGCAGGCGTTTGCCGACGAGAAAGGATGACGACGATAAGTAGGG-3′) and 71-L1_rev (5′-GATCGGCCGGGGTGATCTCCAGCTGCTTAAAGTTCTCGTCGGCAAACGCCTGCAGCTCGCGGGCCTCCACGTCCAACCAATTAACCAATTCTGATTAG-3′). The revertant TBresc71-LZ-BAC was generated on the basis of TBmut71-L1-BAC using primers resc71-LZ-for (5′-GGCCGATTACGTGCTGCTGCAGCCTAGCGAGACGTGGAGCTCCGCGAGCTGCAGGCGTTTCTCGACGAGAAAGGATGACGACGATAAGTAGGG-3′) and resc71-LZ-rev (5′-GATCGGCCGGGGTGATCTCCAGCTGCTTAAAGTTCTCGTCGAGAAACGCCTGCAGCTCGCGGAGCTCCACGTCCAACCAATTAACCAATTCTGATTAG-3′).
All mutants were checked for correct mutagenesis by sequencing of the mutagenized region. TBmut71-L1-BAC contained an additional mutation of the codon for the aspartate residue at aa 42 to code for a glycine residue. Furthermore, all BAC DNAs were verified by restriction fragment length polymorphism analysis and agarose gel electrophoresis.
Virus reconstitution.
HCMV BAC DNA was isolated from bacteria by using a NucleoBond AX midi kit according to the instructions of the manufacturer (Macherey-Nagel, Düren, Germany) and utilized to reconstitute recombinant viruses. For reconstitution, MRC-5 cells were subjected to electroporation as described previously (6). HFFs were used for further virus propagation and preparation of virus stocks. The recombinant viruses recovered from TB40-BAC4, TBmut71-ΔLZ-BAC, TBmut71-L1-BAC, and TBresc71-LZ-BAC were designated the wild type, TBmut71-ΔLZ, TBmut71-L1, and TBresc71-LZ, respectively.
Human polyclonal antibody against pUL71.
HCMV pUL71-specific human polyclonal antibody pAbUL71 was purified from high-titer human serum (IgG-positive patient serum selected by a CMV diagnostic) by column affinity chromatography (with Affi-Gel 10 medium–Affi-Gel 15 medium–pUL71). Affinity purification was performed as described previously (10). The specificity of the purified pAbUL71 was determined using immunoblots with recombinant pUL71 (rpUL71). Immunoblot strips were reacted with Cytotect (Biotest, Dreieich, Germany), a hyperimmune serum against HCMV (HCMV positive; diluted 1:500); CMV pp52 (CH16; Santa Cruz Biotechnology, Heidelberg, Germany) monoclonal antibody against pUL44 (MAbUL44); and pAbUL71 (data not shown). Only Cytotect and the purified antibody pAbUL71 reacted on strips with rpUL71. As an additional control for the specificity of pAbUL71, immunoblot analyses with different viral proteins were performed. Immunoblot strips (kindly provided by Mikrogen Diagnostik, Neuried, Germany) containing recombinant polypeptides producing immediate-early protein 1 (IE1; 53 kDa), tegument protein pp150 (50 kDa), processivity factor pUL44 and single-stranded binding protein pUL57 (45 kDa), tegument protein pp65 (31 kDa), and glycoprotein B (gB1, 25 kDa; gB2, 18 kDa) reacted only with convalescence serum and specific antibody against pUL44 (data not shown). These observations demonstrate that pAbUL71 is a monospecific antibody against pUL71.
Purification of rpUL71.
High Five cells (3.6 × 108) were infected with recombinant baculovirus UL71 (multiplicity of infection [MOI], 2). The cells expressing rpUL71 were harvested 48 h postinfection (h p.i.). Sedimented cells were lysed in 50 ml anion exchange buffer (20 mM Bis-Tris, 200 mM NaCl, pH 6.5) containing 250 μM MgCl2, 200 μg DNase I, 200 μg RNase A, and 2.5 mM sucrose by sonification on ice. Undissolved material was removed by centrifugation (for 30 min at 10,000 × g and 4°C).
Further, as a first purification step, supernatant was precipitated with 45% (NH4)2SO4 and sediment was collected by centrifugation (for 30 min at 10,000 × g and 4°C) and dialyzed against anion exchange buffer. The sediment was passed through a 0.2-μm filter prior to being loaded onto an equilibrated anion exchange column (Resource Q) with a 1-ml bed volume (GE Healthcare, Munich, Germany). The purification was performed at 4°C using an ÄKTAFPLC fast protein liquid chromatography system (GE Healthcare, Munich, Germany). Elution was achieved using a linear NaCl gradient (from 0.2 to 1 M NaCl). Again, salt was removed by dialysis against anion exchange buffer. Next, fractions containing the protein were subjected to gel filtration carried out with a HiPrep 16/60 Sephacryl HR column (GE Healthcare, Munich, Germany) using the ÄKTAFPLC system. A total of 40 fractions were collected. All fractions were analyzed by SDS-PAGE followed by Coomassie blue staining or immunostaining throughout the whole purification procedure.
BiFC assay.
To generate the expression vectors for bimolecular fluorescence complementation (BiFC), fragments of the enhanced yellow fluorescent protein (EYFP) gene coding for the N-terminal 173 aa (YN) and the C-terminal 84 aa (YC), including sequences for c-myc and hemagglutinin (HA) epitope tags, respectively, were amplified from vectors pUC-SPYNE and pUC-SPYCE (45). The PCR products were cloned into eukaryotic expression vector pcDNA3.1 (Invitrogen). Sequences encoding pUL71 and pUL71 bZIP mutants were amplified and fused to the sequences encoding the N- and C-terminal EYFP fragments. The pUL71 sequences and EYFP fragments were connected by a linker peptide (GGGGSGGGS).
For BiFC, HeLa cells were grown on coverslips to 70% confluence. Prior to transfection, cells were starved for 12 h in medium containing 0.1% FCS. Cells were cotransfected with the expression vectors indicated for each experiment (125 ng for each pUL71 expression) using TurboFect. The fluorescence emissions were observed in paraformaldehyde (4%)-fixed cells 10 to 20 h after transfection by using a Zeiss Observer.Z1 fluorescence microscope. To compare the efficiencies of fluorescence complementation between the wild type and bZIP mutants, the cells were cotransfected with a pcDNA3.1-mCherry expression vector (250 ng). Cells were grown for 14 h at 37°C and 30 min at 32°C prior to fixation with 4% paraformaldehyde. The excitation of YFP and the mCherry emission were measured for each condition using fixed settings for the Zeiss Observer.Z1 fluorescence microscope and AxioVision software 4.8 (Carl Zeiss MicroImaging, Jena, Germany). The ratio between YFP and mCherry emissions was determined for randomly chosen single cells expressing mCherry on micrographs by using the colocalization module of the AxioVision software.
Western blotting.
Lysates of mock-infected, infected, or transfected cells were separated on polyacrylamide gel, transferred onto nitrocellulose sheets, and subjected to Western blot analysis as described previously (37). The anti-Xpress (Asp-Leu-Tyr-Asp4-Lys) antibody (Invitrogen, Karlsruhe, Germany), anti-myc tag mouse monoclonal antibody (MAb) against c-myc (EQKLISEEDL; Cell Signaling, Beverly, MA), and anti-HA purified rabbit sera against a synthetic peptide (YPYDVPDYA; ABR, Golden, CO), which were specific for the respective epitopes, were used as primary antibodies prior to incubation with horseradish peroxidase-conjugated anti-mouse F(ab′)2 fragments (diluted 1:5,000 in 3% bovine serum albumin [BSA]; Biozol, Eching, Germany). The membranes were reprobed with an antibody against β-actin (diluted 1:5,000; Biozol, Eching, Germany) and visualized as described above with peroxidase-conjugated secondary MAb to verify loading of equal amounts. Detection of protein bands was performed using enhanced chemiluminescence (ECL) reagent as recommended by the supplier (Thermo Fisher Scientific, Bonn, Germany).
Coimmunoprecipitation.
For coimmunoprecipitation (co-IP), mock-infected or AD169-infected cells (2 × 106) were harvested 72 h p.i. For co-IP with transfected cells, 293T cells (2 × 106) were transfected with two putative binding partners cloned into pcDNA3.1/HisB or pcDNA3.1/HisC (for the Xpress tag) and pHM1580 (for the myc tag) and collected 48 h after transfection. In detail, cells were transfected with pcDNA71 and with pcDNA71-myc, pcDNA71_ΔLZ-myc, pcDNA71_L1-myc, pcDNA71_L34A-myc, or pcDNA71_L41A-myc. Total cell extracts were harvested and solubilized in immunoprecipitation buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Tween 20, 5 mM EDTA) containing cOmplete protease inhibitor mix minitablets (Roche Diagnostics, Mannheim, Germany) prior to ultrasonic treatment. Insoluble material was sedimented for 30 min at 100,000 × g and 4°C. Comparable amounts of extracts and specific antibodies were used for precipitation as described previously (15). Immunoprecipitates were analyzed by SDS-PAGE prior to immunostaining.
Growth analysis and plaque assay.
Analysis of viral growth kinetics and the plaque assay were performed as described previously (34). For the analysis of growth kinetics, HFFs were infected with wild-type, TBmut71-ΔLZ, TBmut71-L1, and TBresc71-LZ viruses at an MOI of 3. Supernatants were collected every 24 h and cleared of cell debris before the determination of virus yields on HFFs as described elsewhere (34).
For the plaque assay, confluent HFF cells were infected with the respective viruses. After 24 h of incubation at 37°C, the inoculum was replaced by a 0.6 to 0.7% methylcellulose overlay and incubation was continued at 37°C for 9 days. Infected cells were detected by staining for HCMV proteins IE1/IE2 and UL44. Images of randomly chosen plaques were taken for each virus by using a ×10 lens objective and the Axio Observer.Z1 fluorescence microscope (Zeiss). Plaque areas were determined by using the software ImageJ (http://rsbweb.nih.gov/ij/index.html).
Thin sectioning.
HFF cells were prepared for thin sectioning by two different methods 5 days p.i. For the high-pressure freezing method, cells were cultivated on carbon-coated sapphire disks (with a diameter of 3 mm; Engineering Office M. Wohlwend GmbH, Sennwald, Switzerland), infected at an MOI of 0.5, and prepared for electron microscopy exactly as described previously (34).
For the chemical fixation method, HFFs (106) were infected at an MOI of 3. At day 5 p.i., cells were fixed using 2.5% glutaraldehyde in HEPES buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) and consecutively with 1% osmium tetroxide, 0.1% tannin, and 1% Na2SO4, all diluted in HEPES buffer. After dehydration with ethanol (EtOH; sequential steps with 50%/70%/90%/100%) and a second staining with 0.2% uranyl acetate, cells were transferred into Beem capsules and embedded in 1,2-epoxypropane in glycid ether 100 after the addition of hardeners dodecenylsuccinic anhydride (DBA) and methylnadic anhydride (MNA). Polymerization was induced by heating for 3 days at 65°C. Thin sections were cut with an ultramicrotome (Leica, Wetzlar, Germany), and sections were transferred onto carbon-coated copper grids (Plano, Wetzlar, Germany) prior to final staining with 1% (wt/vol) uranyl acetate in 40% EtOH and lead citrate. Thin sections were analyzed using a TEM Tecnai G2 electron microscope (FEI Company, Eindhoven, Netherlands) operated at 120 kV. Micrographs were recorded with an Olympus Soft Imaging Solution 2,000-pixel (SIS 2K) digital camera at calibrated magnifications. The chemical fixation method was used for quantification of dense bodies.
Indirect carbon support films.
Carbon was evaporated from 1.0-mm carbon thread at a height of approximately 17 cm onto a glass slide. The carbon is reflected from the glass slide at an angle of 45° onto a freshly cleaved mica sheet in a coating unit with an operating vacuum of <10 Pa.
Electron microscopy for single particles.
Purified rpUL71 expressed by baculovirus (rpUL71 [fraction peak 3; maximum elution volume, 47.8 ml]) was incubated in phosphate buffer, pH 7.4, for different times, applied to the carbon on the mica, and negatively stained with 4% aqueous uranyl acetate (wt/vol) for 20 s according to the method of Valentine et al. (42). Analysis was performed using a TEM Tecnai G2 electron microscope (FEI Company, Eindhoven, Netherlands) operated at 120 kV. Micrographs were recorded with an SIS 2K digital camera at calibrated magnifications.
Biostatistical analysis.
In order to determine the efficiency of the bimolecular fluorescence emissions upon decrease in interaction, statistical analyses were performed. The results obtained from paired Student t tests were used to calculate significance. A P value of ≤0.05 was considered significant.
Bioinformatic tools.
For determination of putative oligomerization domains and other specific secondary structures, the following prediction programs were used: Motif Runner version 1.00 (http://www.generunner.net/) and Motif Scan (http://myhits.isb-sib.ch/cgi-bin/motif_scan) with HAMAP profiles, PROSITE patterns, the SMART tool, and the PFAM and FT-lines virtual motifs databases. For identification of coiled-coil motifs (CCM), the prediction program PCOILS (http://toolkit.tuebingen.mpg.de/pcoils) was used.
RESULTS
pUL71 forms oligomeric structures.
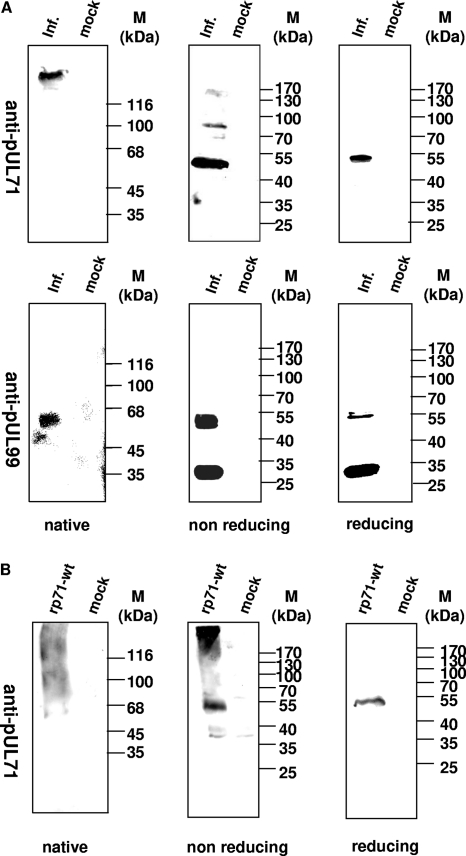
In order to analyze the oligomerization ability of pUL71, mock-infected and AD169-infected HFFs were harvested 72 h p.i. and extracts were separated under native conditions (harvesting with 0.1% sodium dodecyl sulfate [SDS] at room temperature and separation by native polyacrylamide gel electrophoresis [PAGE]), nonreducing conditions (harvesting with 0.1% SDS at room temperature and separation by SDS-PAGE), or reducing conditions (harvesting with 2% SDS and 5% β-mercaptoethanol (ME) for 5 min at 95°C and separation by SDS-PAGE) (Fig. 1). The immunostaining that followed was performed with antibodies against pUL71 or pp28, a prominent tegument protein.
Fig 1.
Analysis of pUL71 in infected cells. Extracts from AD169-infected (inf.) and mock-infected (mock) cells were harvested in 0.1% SDS and subjected to native 8% PAGE followed by immunostaining (native). Equivalent extracts were harvested in 0.1% SDS (nonreducing) or in 2% SDS–5% ME (reducing) prior to separation by SDS–8% PAGE. (A) Immunostaining was performed with pAbUL71 against pUL71. In addition, pp28 was detected using MAb5C3. (B) Extracts from 293T cells transfected with a construct expressing pUL71 (rpUL71) and mock-transfected cells (mock) were harvested in 0.1% SDS and subjected to native 8% PAGE followed by immunostaining (native). Equivalent extracts were harvested in 0.1% SDS (nonreducing) or in 2% SDS–5% ME (reducing) prior to separation by SDS–8% PAGE. Immunostaining was performed with pAbUL71 against pUL71. Molecular mass markers (M) are indicated on the right. Markers for native PAGE: β-galactosidase, 116 kDa; phosphorylase, 100 kDa; bovine serum albumin, 68 kDa; ovalbumin, 45 kDa; and carbonic anhydrase, 35 kDa.
No specific bands were found for mock-infected cells using the anti-UL71 antibody (Fig. 1A). In contrast, diverse bands were detected for AD169-infected cells. Under reducing conditions, we detected a single band at approximately 50 kDa, as described previously (34), which most likely represents the monomer of pUL71 (Fig. 1A, upper right panel). Under native conditions, however, a higher-molecular-mass form was detected, representing either an oligomeric form of pUL71 or a protein complex with other viral or cellular proteins (Fig. 1A, upper left panel). Identical extracts were treated under nonreducing conditions, where we detected a band at 50 kDa, as well as a second band approximately double that size and a faint band at 130 kDa, each representing a higher-ordered oligomeric form of pUL71 (Fig. 1A, upper middle panel).
As a control for our experiments, we analyzed the oligomerization of pp28 using identical conditions. As described previously by Seo and Britt (35), we detected several bands reflecting the monomeric form of pp28 at 28 kDa as well as multimeric forms at 55 kDa for AD169-infected cells under nonreducing and reducing conditions (Fig. 1A, lower middle and right panels). Under native conditions, we could detect only a single band, reflecting the higher-molecular-mass form of pp28 in AD169-infected cells (Fig. 1A, lower left panel). In addition, we performed identical analysis with expressed rpUL71 in order to address the question of whether solitary expressed protein has the same ability to form oligomers. Interestingly, besides the monomer, a high-molecular-mass form was detected in extracts treated under nonreducing conditions (Fig. 1B, middle panel). These results demonstrate the ability of pUL71 to form higher-ordered structures after solitary expression as well as during HCMV infection.
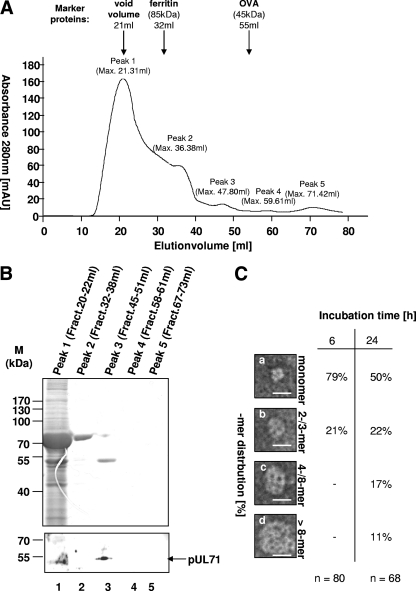
The detection of the higher-molecular-mass structures indicated possible oligomerization of pUL71. To verify this observation, baculovirus-expressed recombinant protein rpUL71 was purified by a combination of anion exchange and gel permeation chromatography. In the gel permeation analysis, five different fractions termed peak 1, peak 2, peak 3, peak 4, and peak 5 were collected and concentrated before being taken for further analysis (Fig. 2A). Recombinant pUL71 was detected only in fraction peak 3 (45 ml) but not in the other peak fractions after SDS-PAGE and immunoblotting using pAbUL71 (Fig. 2B). In contrast to molecular mass markers ovalbumin (Stokes radius [rs]= 3.05 nm), eluting at 55 ml, and ferritin (rs = 6.10 nm), eluting at 32 ml, purified rpUL71 had a molecular mass of between 45 and 85 kDa and most likely represents a monomeric form of rpUL71. The purified protein of fraction peak 3 was dialyzed in phosphate buffer, pH 7.4, incubated for 6 and 24 h, and negatively stained prior to being subjected to electron microscopy analysis. The masses of the species found by electron microscopy were estimated using the volume-mass relationship described by Zipper et al. (49), relating 1.37 nm3 to 1 kDa. Species of different sizes that were calculated to be monomeric (Fig. 2C, panel a), as well as higher-molecular-mass forms of the protein (Fig. 2C, panels b to d), were detected. During 6 h of incubation, only monomers and di-/trimers of rpUL71 accumulated; a longer incubation time of 24 h led to the formation of higher-molecular-mass forms (Fig. 2C). These results show the ability of rpUL71-Bac to form higher-ordered structures on its own.
Fig 2.
Oligomerization potential of rpUL71. (A) Gel permeation chromatography analysis of rpUL71. rpUL71 purified in a single step was subjected to chromatography with a HiLoad 16/60 Superdex prep column using an ÄKTAFPLC system, and data were recorded at an absorbance of 280 nm. The column was calibrated with the following molecular mass markers: void volume (21 ml), ferritin (rs = 6.10 nm; elution volume, 32 ml), and ovalbumin (OVA; rs = 3.05 nm; elution volume, 55 ml). Five peaks were distinguished and termed peak 1 (maximum elution volume, 21.31 ml), peak 2 (maximum elution volume, 36.38 ml), peak 3 (maximum elution volume, 47.80 ml), peak 4 (maximum elution volume, 59.61 ml), and peak 5 (maximum elution volume, 71.42 ml). mAU, milli-absorbance units. (B) Analysis of the peak fractions by SDS-PAGE. The fractions peak 1 to peak 5 were separated by SDS-PAGE prior to Coomassie blue staining and immunoblot analysis with pAbUL71. Molecular mass markers are indicated on the left, and the position of rpUL71 is indicated on the right. (C) Electron microscopy analysis of purified rpUL71 from peak 3 negatively stained with 4% uranyl acetate. Molecular mass (in kilodaltons) can be calculated from particle size on the basis of the findings of Zipper et al. (49). Different high-molecular-mass forms of rpUL71, monomeric forms (a), dimers or trimers (b), and higher oligomeric forms (c and d), were obtained from the sample. The scale bars correspond to 20 nm. Incubation in phosphate buffer for 6 h led to predominantly monomeric forms (a) and dimers or trimers (b), but no higher-ordered structures (c and d) could be found. Incubation for 24 h led to predominantly monomeric forms (a), but dimers or trimers (b) and higher-ordered structures (c and d) could also be found.
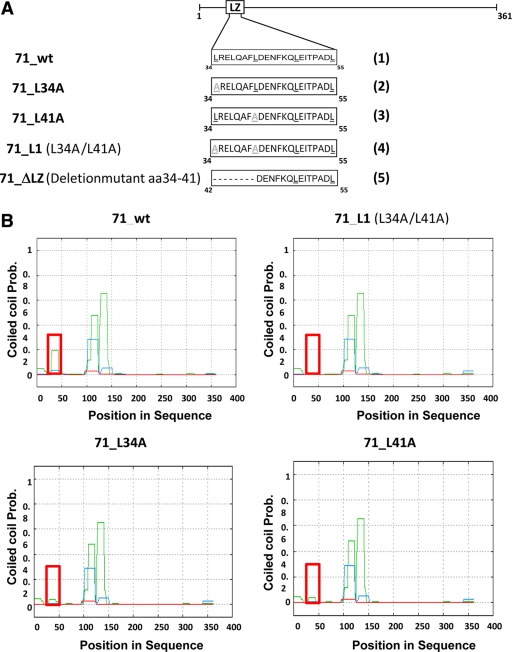
To further characterize oligomerization of pUL71, we used different programs for identification of oligomerization domains. The only motif found with potential to influence oligomerization is the leucine zipper motif (aa 34 to 55), also called the bZip domain, which was first described by Landschulz et al. (20) as an interaction domain in DNA binding protein GCN4 in yeast. O'Shea et al. (30, 31) described the motif to be composed of at least four heptads with hydrophobic amino acids at positions 1 and 4 forming a hydrophobic core. In other words, the leucine zipper motif facilitates dimerization through parallel coiled-coil motifs and binds DNA through its hydrophobic domain. In more recent publications, this motif is no longer considered to be restricted to DNA binding but is generally accepted as a protein-protein interaction domain (24, 32). To illustrate the domain in pUL71, we presented aa 34 to 55 in an alpha-helical wheel (data not shown). The classical bZIP motif is disturbed by asparagine (N44) on position 4 (3). However, we find a rather large hydrophobic face (L55, L37, L48, L41, L34, P52, and F45) consisting of the leucines on position 1 and some other hydrophobic amino acids on positions 4 and 5 of the heptads (data not shown). Therefore, we conclude that this motif, although not recognized as a classical bZIP motif, still has potential to mediate hydrophobic interactions that could be responsible for oligomerization of pUL71. Therefore, we will hereafter refer to the motif as a bZIP-like motif.
Determination of the oligomerization domain of pUL71.
To identify whether the bZIP-like motif functions as an oligomerization domain of pUL71 in vitro, different mutants as well as the full-length protein were analyzed in silico prior to protein-protein interaction studies.
First, a set of alterations affecting either individual amino acids or the whole motif was introduced into UL71. Five different constructs were analyzed: wild-type UL71, designated 71_wt (construct 1 in Fig. 3A); three point mutation constructs, 71_L34A (construct 2), 71_L41A (construct 3), and 71_L1 (carrying L34A and L41A mutations; construct 4); and deletion mutant 71_ΔLZ (with the first half of the bZIP deleted; construct 5). The constructs were first analyzed using secondary structure prediction software PCOILS, coils, Psipred, and Quick2D. As shown in Fig. 3B, the exchange of two leucines with alanines at amino acids 34 and 41 resulted in the loss of the predicted coiled-coil domain in the bZIP-like motif. While we see a peak of 0.2 on the probability scale (using PCOILS sliding window 14) for 71_wt (Fig. 3B), none is detected in prediction for 71_L1 (Fig. 3B) and a reduced peak is seen in prediction for 71_L34A and 71_L41A (Fig. 3B).
Fig 3.
Mutational analysis of the bZIP-like domain in pUL71. (A) Schematic representation of the leucine zipper-like motif (white bars). Exchanged amino acids are indicated in gray. Constructs 71_L34A, 71_L41A, and 71_L1 carry one or two single-nucleotide exchanges starting on heptad position 1 of the bZIP motif. Mutant 71_ΔLZ harbors a deletion of amino acids 34 to 41 of pUL71, representing the N-terminal part of the bZIP-like motif. Underlining indicates positions of the leucines (black) or the mutated leucines (grey). (B) Analysis of coiled-coil structures using PCOILS software. The position of the leucine zipper-like motif within the amino acid sequence is indicated by the red square. PCOILS uses sliding windows of 14 (green), 21 (blue), and 28 (red).
The loss of the predicted coiled-coil structure through the amino acid exchange hints at a corresponding loss in the interacting abilities of those mutants. To test this hypothesis, the constructs were applied to in vitro assays.
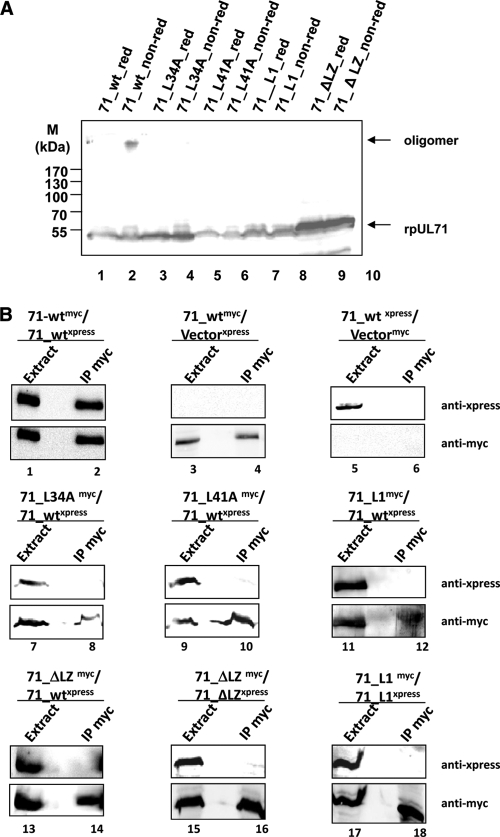
Cell extracts expressing wild-type pUL71, 71_L34A, 71_L41A, 71_L1, or 71_ΔLZ were treated under reducing (red) or nonreducing (non-red) conditions prior to immunostaining with pAbUL71 (Fig. 4A). We detected oligomers only in cells expressing the wild-type protein (Fig. 4A, lanes 1 and 2).
Fig 4.
Analyzing the oligomerization potential of pUL71. (A) 293T cells were transfected with pcDNA71 (encoding 71_wt), pcDNA71_L34A (encoding 71_L34A), pcDNA71_L41A (encoding 71_L41A), pcDNA71_L1 (encoding 71_L1), or pcDNA71_ΔLZ (encoding 71_ΔLZ). Cell extracts were harvested 48 h posttranfection and subjected to immunostaining with the Xpress antibody. Molecular mass markers (M) are indicated on the left. (B) 293T cells were transfected with a combination of pcDNA71-xpress (encoding 71-wt-Xpress) and pcDNA71-myc (encoding 71_wt-myc), pcDNA71_L34A (encoding 71_L34A), pcDNA71_L41A (encoding 71_L41A), pcDNA71_L1-myc (encoding 71_L1-myc), or pcDNA71_ΔLZ-myc (encoding 71_ΔLZ-myc). As a control for self-interaction, cells were transfected with a combination of pcDNA71_L1-xpress (encoding 71_L1-Xpress) and pcDNA71_L1-myc (encoding 71_L1-myc) or pcDNA71_ΔLZ-xpress (encoding 71_ΔLZ-Xpress) and pcDNA71_ΔLZ-myc (encoding 71_ΔLZ-myc). Mock-infected cells or cells transfected with pcDNA71 or pcDNA71-myc together with the empty vector pHM1580 (Vectormyc) or pcDNA3.1+ (Vectorxpress) also served as controls. Cells were harvested 48 h after transfection and subjected to precipitation against MAbMyc_r. Extract and precipitate (IP myc) fractions were analyzed using the Xpress antibody (upper panels) and MAbMyc_r (lower panels). Between the left and right lanes in each panel, an additional lane was left empty to prevent contamination upon loading.
For coimmunoprecipitation, a specific antibody against the myc tag (MAbMyc_r) of 71_wt-myc, 71_L34A-myc, 71_L41A-myc, 71_L1-myc, and 71_ΔLZ-myc was used prior to immunostaining against the Xpress tag of 71_wt-Xpress. Cells were additionally transfected with the empty vector pHM1580 (Vectormyc) or pcDNA3.1+ (Vectorxpress) (Fig. 4B, lanes 3 to 6). The lysate fractions were stained with both antibodies to test for the expression of myc-tagged and Xpress-tagged proteins. We detected interaction only between wild-type proteins (Fig. 4B, lanes 1 and 2). No specific proteins were precipitated from cell extracts transfected with empty vectors. These results demonstrated that the oligomerization domain of pUL71 is required for its homodimerization.
In order to test whether the bZIP-mediated effect is due to self-interaction, coimmunoprecipitation experiments were also performed with constructs 71_L1-myc and 71_ΔLZ-myc prior to staining with antibody against the Xpress tag of 71_L1-Xpress and 71_ΔLZ-Xpress, leading to comparable results (Fig. 4B, lanes 15 to 18).
To verify our results from the coimmunoprecipitation studies, a BiFC technique was used. This technique allows the visualization of protein complex formation in living cells and has already been used to characterize several protein interactions, including those between bZIP motifs (18). In our assay, protein interactions were detected by the formation of a fluorescent complex by two nonfluorescent fragments of YFP, which were fused to pUL71. We used two overlapping fragments of YFP, one containing the N-terminal 173 amino acids together with a myc epitope (YN) and the other containing the C-terminal 84 amino acids fused to the HA epitope (YC) (44). The YN and the YC sequences were fused to wild-type UL71 and to bZIP-like motif mutant UL71 sequences, separated by a linker sequence.
We first examined the suitability for BiFC by determining the levels of transient expression of our constructs to exclude the possibility that the fusion of the YN or YC fragment or the mutation of the bZIP-like motif affected protein stability. We cotransfected HeLa cells with expression vectors encoding pUL71 BiFC fusion constructs and green fluorescent protein (GFP) and determined protein levels 16 h posttransfection by Western blot analysis. Fusion of either the YN fragment or the YC fragment and mutation of the leucine zipper-like motif did not affect the level of protein expression (data not shown). Therefore, we excluded that differences in protein expression impact BiFC efficiency. Second, we examined intracellular localization of our BiFC constructs in comparison to untagged pUL71 by using immunofluorescence. Intracellular localization of pUL71 is mainly undisturbed by addition of the YFP fragments or mutations (data not shown). However, we noticed that fusion of the YFP fragments to pUL71 can cause atypical cytoplasmic accumulation in some cases when the protein is highly overexpressed. Such accumulations could virtually be eliminated by shortening the time of expression and reducing the amount of DNA with which cells were transfected. From these experiments, we concluded that our constructs and conditions are suitable for BiFC analysis.
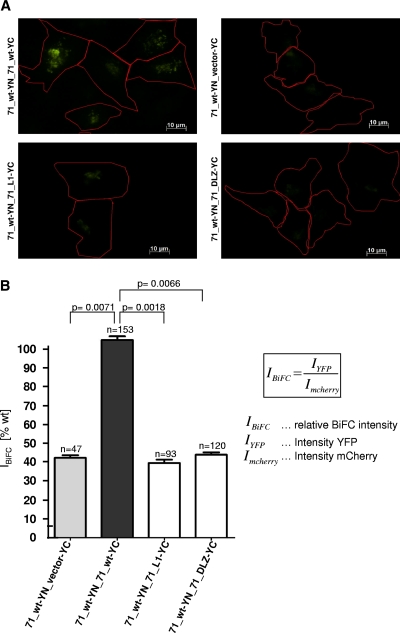
To investigate whether pUL71 interacts with itself to form oligomers, we cotransfected HeLa cells with expression vectors encoding 71_wt-YN and 71_wt-YC and examined the cells by fluorescence microscopy within 12 h of transfection. YFP fluorescence in cells coexpressing 71_wt-YN and 71_wt-YC constructs was found to be perinuclear in the cytoplasm (Fig. 5A). Expression of either 71_wt-YN or 71_wt-YC alone did not produce detectable fluorescence (data not shown). Mutation or deletion of the bZIP-like motif visibly impaired YFP fluorescence (Fig. 5A).
Fig 5.
Analyzing oligomerization potential of pUL71 using BiFC. Cells were grown on coverslips prior to transfection with a combination of pcDNA71_YN (encoding 71_wt-YN) and construct pcDNA-YC (Vector-YC), pcDNA71_YC (encoding 71_wt-YC), pcDNA71_L1-YC (encoding 71_L1-YC), or pcDNA71_ΔLZ-YC (encoding 71_ΔLZ-YC). Additionally, cells were transfected with a construct expressing mCherry. Cells were fixed 14 h after transfection and analyzed using immunofluorescence. (A) Immunofluorescence data. YFP fluorescence is shown in green, and cell outlines are indicated in red using AxioVision software. (B) Quantification of BiFC results. IBiFC is set to 100% for the 71_wt-YN/71_wt-YC interaction. Other interactions are set in ratio. Error bars on the histogram represent standard deviations of results from three independent experiments. The significance of results was determined using the paired Student t test. n, number of analyzed cells.
To quantify the interaction efficiency of pUL71 in cells, we cotransfected cells with another plasmid expressing mCherry, allowing the normalization of YFP emission by using the emission of mCherry. In order to perform the efficiency analysis, the BiFC cells were grown on coverslips prior to transfection with the respective combinations of transient BiFC expression constructs together with the construct expressing mCherry. Cells were grown under conditions described above prior to fixation for immunofluorescence analysis.
The YFP intensity (IYFP) in individual cells from acquired micrographs was measured relatively to mCherry expression (ImCherry) according to the formula in Fig. 5B by using Zeiss AxioVision software 4.8 to allow quantitative comparison of BiFC intensity (IBiFC) levels between different cells. Results for mutants 71_L1 and 71_ΔLZ are represented in a histogram (Fig. 5B). The IBiFC for the interaction between 71_wt-YN and 71_wt-YC was set to 100%. Relative to the interaction between the wild-type fusion proteins, BiFC efficiency for two bZIP-like motif mutants, 71_L1 and 71_ΔLZ, was determined to be approximately 40%. This reduced IBiFC was comparable to the BiFC efficiency for the vector expressing the YC fragment only. The significance of the results was determined using the paired Student t test (Fig. 5B). The findings indicate that an alteration in the bZIP-like motif results in a loss of interaction between the two proteins and therefore in a decrease in YFP emission.
Mutation of the bZIP-like domain results in incomplete viral envelopment.
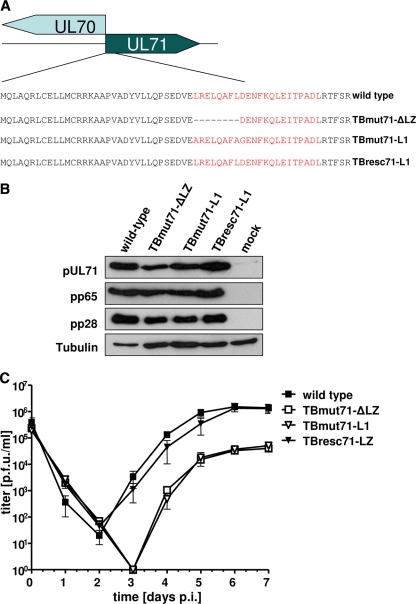
In order to investigate the importance of pUL71 oligomerization for viral replication, two mutant viruses harboring mutations of the bZIP-like motif were generated. Constructs pcDNA71_ΔLZ or pcDNA71_ΔLZ-YC and pcDNA71_L1 or pcDNA71_L1-YC, each tested in vitro, were used to introduce the leucine zipper deletion mutation (ΔLZ) and the leucine-to-alanine changes at aa 34 and 41 into BAC TB40-BAC4 (36). Markerless BAC mutagenesis resulted in mutant TBmut71-ΔLZ-BAC and in mutant TBmut71-L1-BAC (Fig. 6A). By repairing the mutations of TBmut71-L1-BAC, a revertant BAC mutant, TBresc71-LZ-BAC, was generated which is distinguishable from BAC TB40-BAC4 in harboring silent mutations. Wild-type virus (TB40-BAC4), mutant viruses TBmut71-ΔLZ and TBmut71-L1, and the revertant virus TBresc71-LZ were reconstituted from the respective BAC DNAs in fibroblasts. We first investigated the expression of pUL71 in our mutant viruses to exclude that the mutation of the bZIP-like motif results in altered expression levels. Lysates of HFFs infected with wild-type, TBmut71-ΔLZ, TBmut71-L1, and TBresc71-LZ viruses were subjected to Western blot analysis (Fig. 6B). pUL71 protein levels similar to those for wild-type and TBresc71-LZ viruses were observed for both TBmut71-ΔLZ and TBmut71-L1. The lysates were additionally probed for viral proteins pp65 and pp28, whose levels were equal in all lysates. These data suggested that pUL71 is normally expressed by the bZIP-like motif mutant viruses.
Fig 6.
Construction of recombinant HCMV UL71 mutant genomes and representation of virus growth kinetics. (A) To investigate the function of the bZIP domain of pUL71 in the viral context, two different HCMV pUL71 mutant viruses as well as a revertant virus were constructed. The bZIP-like motif and the mutations are shown in red. (B) Protein expression of the different viral mutants 5 days postinfection. Immunostaining was performed using a polyclonal anti-pUL71 antibody (12), MAb65-33 (kindly provided by W. Britt, University of Alabama) against pp65, and MAb5C3 against pp28. (C) Growth kinetics of wild-type and mutant viruses. HFFs were infected using an MOI of 3 with the wild type, TBmut71-ΔLZ, TBmut71-L1, and TBresc71-LZ. Supernatants were harvested at the indicated time points, and progeny virus yields in the supernatants were determined by titration. Day 0 values represent the inoculum. Error bars in the histogram indicate the standard deviations of results from three independent experiments.
To determine the role of the bZIP-like motif in viral replication, growth properties of TBmut71-ΔLZ and TBmut71-L1 viruses in HFFs were evaluated and compared to those of wild-type and TBresc71-LZ viruses (Fig. 6C). In replication kinetics determined using an MOI of 3, virus shedding into the supernatant started at 3 days p.i. for wild-type and TBresc71-LZ viruses and was delayed for TBmut71-ΔLZ and TBmut71-L1 viruses to 4 days p.i. At all time points, virus yields in the supernatants of TBmut71-ΔLZ- and TBmut71-L1-infected cells were reduced by up to 2 logs compared to wild-type or revertant virus yields. The revertant virus TBresc71-LZ exhibited the same growth characteristics as wild-type virus (Fig. 6C).
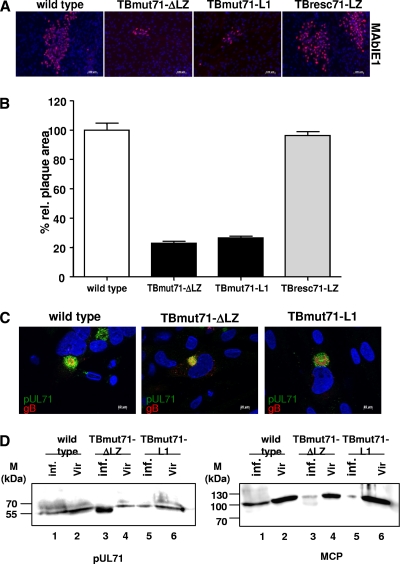
In order to analyze the cell-to-cell spread abilities of our mutant viruses compared to those of the wild type and the revertant virus, plaque sizes after incubation for 9 days p.i. were measured. Plaque sizes of TBmut71-ΔLZ and TBmut71-L1 were significantly smaller than those of wild-type and revertant viruses (Fig. 7A). Infection with bZIP mutant viruses produced very small plaques, with an area ca. 25% of that of plaques produced by wild-type and revertant virus infection (Fig. 7B). These results showed that a functional bZIP-like motif is required for efficient viral replication and cell-to-cell spread.
Fig 7.
Analysis of cell-to-cell spread and intracellular localization of pUL71. (A) HFFs were infected with 100 PFU of the wild type, TBmut71-ΔLZ, TBmut71-L1, and TBresc71-LZ grown in overlay medium containing methyl cellulose (Methocel). The cells were fixed and subjected to immunofluorescence at day 9 p.i. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole), and infected cells were stained with MAbIE1 (red) against IE1. Representative micrographs of plaques produced by the wild type, TBmut71-ΔLZ, TBmut71-L1, and TBresc71-LZ are shown. (B) The plaque areas of individual plaques for each indicated virus were analyzed using a ×10 objective lens and the Axio Observer.Z1 fluorescence microscope. Plaque areas of at least 50 plaques of each virus were measured by the program ImageJ. The mean percentages of the areas and standard errors relative to the mean area of the wild-type virus plaques (set at 100%) are given. The experiment was repeated at least two times. The significance of the results compared to data for the wild type was determined by using an unpaired Student t test (TBmut71-ΔL1, P < 0.0001; TBmut71-L1, P < 0.0001; and TBresc71-LZ, P < 0.476). (C) HFFs were infected at an MOI of 1 with the wild type, TBmut71-ΔLZ, or TBmut71-L1. The spatial distribution of viral proteins was analyzed 120 h p.i. by confocal microscopy using a polyclonal anti-pUL71 antibody (12) (green) and MAb 3F12 (Virusys Corporation) directed against gB (red). (D) Association of bZIP mutants with purified extracellular virions. Infected cell extracts (lanes 1, 3, and 5) and extracellular virions (lanes 2, 4, and 6) were analyzed by immunoblotting with pAbUL71 and antibody against MCP. The molecular mass standards (M) are indicated on the left.
To analyze the intracellular distribution of pUL71 in relationship to the localization of other viral proteins, confocal microscopy was performed. HFFs were infected with wild-type, TBmut71-ΔLZ, and TBmut71-L1 viruses at an MOI of 1 until being fixed for immunofluorescence at 120 h p.i. In cells infected with wild-type virus, both pUL71 and glycoprotein B (gB) are localized to the assembly compartment (AC) in the typical juxtanuclear pattern (Fig. 7C, left). gB localized in a broad circle (donut shape) within the spherical pUL71 pattern. In both mutant TBmut71-ΔLZ-infected cells (Fig. 7C, middle) and mutant TBmut71-L1-infected cells (Fig. 7A, right), an alteration of the AC was observed. Whereas pUL71 localization was not significantly affected in cells infected with the bZIP mutants compared to those infected with wild-type virus, gB was more dispersed in mutant-infected cells and accumulated in many cells in vesicular structures at the periphery of the AC, demonstrating a bZIP mutant phenotype comparable to the phenotype of a UL71 stop mutant (34). Large vesicular structures were observed in 25% of mutant-virus-infected cells, where virus particles appeared to accumulate as indicated by pp150 signals. These data show that the AC structure is altered when the bZIP-like motif of pUL71 is destroyed.
In order to analyze whether pUL71 was incorporated into virions in the bZIP-defective mutants, extracts and purified extracellular virions from wild-type, TBmut71-ΔLZ, and TBmut71-L1 virus-infected cells were subjected to immunoblotting with pAbUL71 and, as a control, with MAb28-4 against monocyte chemoattractant protein (MCP) (Fig. 7D). These analyses demonstrated that the alteration of the bZIP-like motif has no effect on virion incorporation.
To investigate whether the observed growth impairment of the bZIP-defective mutant viruses is based on a defect in morphogenesis, electron microscopy was performed. It has been shown previously that secondary envelopment is severely impaired in the absence of pUL71 (34). Thus, we focused mainly on the cytoplasmic stages of virus morphogenesis. Two different methods to prepare the cells for electron microscopy were used, chemical fixation (16) and high-pressure freezing. For the latter, HFFs were grown on sapphire disks, infected with the wild type, TBmut71-ΔLZ, or TBmut71-L1 at an MOI of 0.5, cryofixed 5 days postinfection by high-pressure freezing, and further processed for transmission electron microscopy (TEM) analysis.
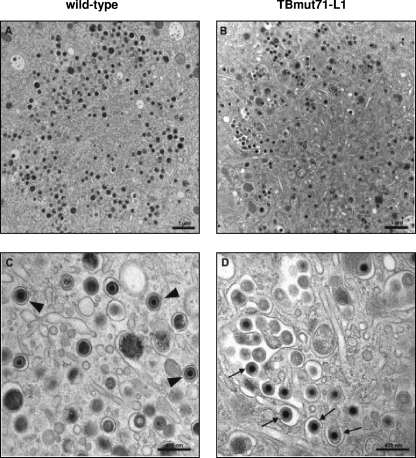
In contrast to nuclear capsid maturation profiles, which were indistinguishable among wild-type, TBmut71-ΔLZ, and TBmut71-L1 viruses (data not shown), secondary envelopment patterns differed (Fig. 8). A large number of cytoplasmic nucleocapsids in the assembly compartments of cells infected with TBmut71-L1 (Fig. 8B and D), as well as those of cells infected with TBmut71-ΔLZ (data not shown), were not yet fully enveloped and underwent a process of envelopment very often at membranes of enlarged vesicles or tubules. Notably, multiple budding events were observed at those enlarged vesicles (Fig. 8D). In cells infected with the wild-type virus, however, an enlargement of vesicles utilized for budding of nucleocapsids was rarely observed and a large number of completely enveloped virus particles was found (Fig. 8A and C).
Fig 8.
Electron microcopy analysis of ultrathin sections of HFFs infected with the wild type (A and C) or with TBmut71-L1 (B and D) at 5 days postinfection. Cells were prepared for electron microscopy by high-pressure freezing and freeze substitution. (A) Overview of a typical cytoplasmic assembly compartment of a wild-type HCMV-infected cell. (C) Higher magnification of a section of the assembly compartment of a wild-type virus-infected cell. Many wild-type virus particles are fully enveloped within single vesicles (arrowheads). (B) In TBmut71-L1 virus-infected cells, enlarged vesicles and accumulated virus particles can be observed in the assembly compartment. (D) These larger vesicles are used as multiple budding sites by TBmut71-L1 virus particles. Many virus particles have not completed their envelopment (arrows). Scale bars: 1 μm (A and B) and 400 nm (C and D).
To quantify the differences in secondary envelopment, completely enveloped and budding virus particles were counted in micrographs taken of randomly selected regions of the assembly compartments of wild-type, TBmut71-ΔLZ, and TBmut71-L1 virus-infected cells. As summarized in Table 2, both bZIP-defective viruses exhibited a clear reduction in the number of enveloped particles compared to wild-type virus. Incomplete envelopment was observed approximately four times more frequently for nucleocapsids of the bZIP-defective viruses than for those of the wild-type virus. In addition, the number of dense bodies from cells infected with either mutant virus was dramatically reduced to approximately 70 to 90% (Table 3). Taken together, our results show that alterations in the bZIP-like motif lead to viruses with incomplete secondary envelopment of HCMV particles.
Table 2.
Quantification of HCMV envelopment in the assembly compartment 5 days p.i.
| Virus | No. of cells analyzed | % Enveloped particlesa | % Budding particlesb | % Naked particlesc |
|---|---|---|---|---|
| Wild type | 27 | 86.64 ± 14.82 | 12.95 ± 14.68 | 0.41 ± 1.66 |
| TBmut71-ΔLZ | 30 | 20.81 ± 8.57 | 75.74 ± 8.39 | 3.45 ± 5.22 |
| TBmut71-L1 | 17 | 18.89 ± 7.51 | 79.71 ± 7.30 | 1.40 ± 3.02 |
Percentage (mean ± SD) of enveloped virus particles among total cytoplasmic particles.
Percentage (mean ± SD) of particles with virus capsids attached to membranes among total cytoplasmic particles.
Percentage (mean ± SD) of particles with virus capsids not connected to membranes (naked capsids) among total cytoplasmic particles.
Table 3.
Quantificationa of dense bodies
| Virus | Total no. of viral particles | No. (%) of dense bodies |
|---|---|---|
| Wild type | 629 ± 80.4 | 286 ± 17.3 (45.7 ± 2.7) |
| TBmut71-ΔLZ | 285 ± 26.9 | 41.8 ± 3.3 (14.7 ± 1.2) |
| TBmut71-L1 | 253 ± 44.0 | 4.6 ± 4.2 (1.8 ± 1.7) |
Values are means ± SD.
DISCUSSION
Human cytomegalovirus pUL71 is a tegument protein and is highly conserved throughout all herpesviruses analyzed up to now. However, the homology at the amino acid level is not significant. Interestingly, alphaherpesvirus homologs, e.g., pseudorabies virus (PrV) UL51 (21), herpes simplex virus 1 (HSV-1) UL51 (7), bovine herpesvirus (BHV) UL51 (14), and Marek's disease virus 2 UL51 (17), have similar sizes of approximately 28 to 30 kDa, whereas HCMV pUL71 has a molecular mass of approximately 50 kDa (34).
The present study demonstrated by using the affinity-purified monospecific antibody pAbUL71 that pUL71 has the ability to form oligomers. These results were confirmed by gel filtration followed by single-particle analysis of baculovirus-expressed rpUL71. In silico analyses revealed a leucine zipper (bZIP)-like motif at the N terminus of pUL71. For oligomerization, it has been shown previously that bZIP domains consist of alpha-helices that are bound together via a coiled-coil structure (5). In addition, analysis using the program PCOILS (13) predicted for the bZIP region of pUL71 a high probability of folding into coiled-coil structures. Interestingly, this structural feature is no longer predicted after exchange of the leucine residues at amino acid positions 34 and 41 for alanine. In addition, coimmunoprecipitation revealed that this exchange is sufficient to prevent self-interaction of pUL71. Self-interaction of pUL71 and the involvement of the bZIP region were confirmed by a quantitative BiFC assay. bZIP mutants exhibited a reduction in BiFC efficiency of 60% compared to wild-type pUL71. To further investigate the function of the bZIP region in the viral context, two viral mutants containing either a deletion of the first half of the bZIP or the mutation of the two leucines at positions 34 and 41 were constructed. Both mutants showed a small-plaque phenotype, with ca. 80% reduced plaque sizes compared to wild-type and revertant virus plaques and a reduction of viral yields in the supernatant of approximately 30-fold. A very similar phenotype has been described for a UL71 stop mutant in the same viral background (34). A small-plaque phenotype and impaired virus release have also been reported previously for HSV-1 and PrV mutants lacking the homolog of pUL51 (1, 19, 29). In addition, analysis by confocal microscopy demonstrated identical localization patterns for bZIP-defective pUL71 mutants and the wild type, while the structure of the AC itself was changed. Similarly altered morphology was reported for the UL71 stop mutants (34, 46). The AC is thought to mediate tegumentation and final envelopment, because many structural proteins (such as tegument and envelope proteins) have been found in ACs (8, 23, 33, 39). Tooze et al. (41) showed by using the endocytic tracer horseradish peroxidase that the envelope of mature virions is derived from tubular endosomes. Recently, Das and Pellet (9) presented data indicating that HCMV-induced cytoplasmic remodeling is required not only for formation of the AC but in addition for new functions of this compartment. The underlying mechanism is not entirely clear; however, the recycling of endosomes plays an important role in HCMV maturation. The phenotype found for our mutants of UL71 could be due to a loss of protein function. This protein may be important for mediating envelopment by bridging viral glycoproteins and capsid proteins. The loss of this function may result in a less condensed structure of vesicles in the AC.
Our observations that the exchange of two amino acids of the bZIP motif lead to growth impairment suggest that this is caused by a defect of viral assembly. HCMV assembly was analyzed by electron microscopy of ultrathin sections of cells infected with either wild-type or bZIP-defective viruses. Although capsid formation was not affected by the defects, the number of incompletely enveloped particles in the cytoplasm increased, thus leading to a decrease of mature virions. These results are in line with the recently described phenotypes of the pUL71 stop mutants and a mutant generated by transposon insertion (34, 46, 48). Interestingly, the number of dense bodies was dramatically reduced for the bZIP deletion or leucine exchange mutant. This outcome was unexpected, because the most abundant tegument protein in dense bodies, pp65 (25), is not affected by the introduced mutations. In addition to a reduction of viral titers, an accumulation of capsid in the perinuclear space was observed in cells infected with an HSV-1 UL51 deletion mutant (19). Since pUL71 and its homologs are tegument proteins, the latter observation may be due to a secondary effect of impaired viral maturation.
Our results presented here are remarkable, because the bZIP motif of HCMV pUL71 itself is sufficient to prevent correct final envelopment of viral particles while it does not affect expression levels and localization of pUL71. For HSV-1, it has been reported previously that a bZIP motif mediates docking of the terminase to the portal protein during viral DNA packaging (47). Many reports on the requirement of the bZIP motif for multimerization of transcription factors are available. Our data are, to our knowledge, the first report of an important function of a leucine zipper in triggering correct envelopment of human cytomegalovirus.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) BO 1214/15-1 and Graduiertenkolleg (GRK1121, project A9). C.S.M. is a member of the ZIBI graduate school.
We thank the imaging unit (ZBS4) of the Robert Koch Institute for technical support, i.e., ultrathin sectioning and providing of the electron microscope. We gratefully acknowledge R. Schilf and I. Woskobjnik for technical assistance and thank C. Priemer for assistance concerning cell culture. We thank W. Britt (University of Alabama) for providing the monoclonal antibodies MAb28-4 against MCP and MAb65-33 against pp65, and R. Waadt for providing the BiFC plasmids and for his technical support. E.B. thanks D. Krüger for continuing support.
Footnotes
Published ahead of print 28 December 2011
REFERENCES
- 1. Barker DE, Roizman B. 1990. Identification of three genes nonessential for growth in cell culture near the right terminus of the unique sequences of long component of herpes simplex virus 1. Virology 177:684–691 [DOI] [PubMed] [Google Scholar]
- 2. Bogner E. 2002. Human cytomegalovirus terminase as a target for antiviral chemotherapy. Rev. Med. Virol. 12:115–127 [DOI] [PubMed] [Google Scholar]
- 3. Bornberg-Bauer E, Rivals E, Vingron M. 1998. Computational approaches to identify leucine zippers. Nucleic Acids Res. 26:2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bresnahan WA, Boldogh I, Thompson AE, Albrecht T. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150–160 [DOI] [PubMed] [Google Scholar]
- 5. Busch DJ, Sassone-Corsi P. 1990. Dimers, leucine zippers and DNA-binding domains. Trends Genet. 6:36–40 [DOI] [PubMed] [Google Scholar]
- 6. Chevillotte M, et al. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 83:2480–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daikoku T, Ikenoya K, Yamada H, Goshima F, Nishiyama Y. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J. Gen. Virol. 79:3027–3031 [DOI] [PubMed] [Google Scholar]
- 8. Das S, Pellet PE. 2007. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology 361:263–273 [DOI] [PubMed] [Google Scholar]
- 9. Das S, Pellet PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J. Virol. 85:5864–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dittmer A, Bogner E. 2005. Analysis of the quaternary structure of the putative HCMV portal protein pUL104. Biochemistry 44:759–765 [DOI] [PubMed] [Google Scholar]
- 11. Dittmer D, Mocarski ES. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giesen K, Radsak K, Bogner E. 2000. The potential terminase subunit pUL56 of HCMV is translocated into the nucleus by its own NLS and interacts with importin α. J. Gen. Virol. 81:2231–2244 [DOI] [PubMed] [Google Scholar]
- 13. Gruber M, Söding J, Lupas AN. 2006. Comparative analysis of coiled-coil prediction methods. J. Struct. Biol. 155:140–145 [DOI] [PubMed] [Google Scholar]
- 14. Hamel F, Boucher H, Simard C. 2002. Transcriptional and translational expression kinetics of the bovine herpesvirus 1 UL51 homologue gene. Virus Res. 84:125–134 [DOI] [PubMed] [Google Scholar]
- 15. Hwang J-S, Bogner E. 2002. ATPase activity of the terminase subunit pUl56 of human cytomegalovirus. J. Biol. Chem. 277:6943–6948 [DOI] [PubMed] [Google Scholar]
- 16. Hwang J-S, et al. 2007. Identification of acetylated, tetrahalogenated benzimidazole D-ribonucleotides with enhanced activity against human cytomegalovirus. J. Virol. 81:11604–11611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Izumiya Y, et al. 1998. Identification and transcriptional analysis of the homologues of the herpes simplex virus type 1 UL41 to UL51 genes in the genome of nononcogenic Marek's disease virus serotype 2. J. Gen. Virol. 79:1997–2001 [DOI] [PubMed] [Google Scholar]
- 18. Kerppola TM. 2009. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem. Soc. Rev. 38:2876–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klupp BG, et al. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Landschulz WH, Johnson PF, McKnigh SL. 1988. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science 240:1759–1764 [DOI] [PubMed] [Google Scholar]
- 21. Lenk M, Visser N, Mettenleiter TC. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J. Virol. 71:5635–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luckow VA, Lee SC, Barry GF, Olins PO. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mach M, Kropff B, Kryzaniak M, Britt W. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martínez-Turiño S, Hernández C. 2011. A membrane-associated movement protein of Pelargonium flower break virus shows RNA-binding activity and contains a biologically relevant leucine zipper-like motif. Virology 413:310–319 [DOI] [PubMed] [Google Scholar]
- 25. Mettenleiter TC. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167–180 [DOI] [PubMed] [Google Scholar]
- 26. Mocarski ES. 1993. Cytomegalovirus biology and replication, p 173–226 In Roizman B, Whitley R, Lopez C. (ed), The human herpesviruses. Raven Press, New York, NY [Google Scholar]
- 27. Mocarski ES, Courcelle CT. 2001. Cytomegaloviruses and their replication, p 2629–2673 In Knipe DM, et al. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 28. Mocarski ES, Jr, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2771 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 29. Nozawa N, et al. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of viral particles. J. Virol. 79:6947–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Shea EK, Klemm JD, Kim PS, Alber T. 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254:539–544 [DOI] [PubMed] [Google Scholar]
- 31. O'Shea EK, Rutkowski R, Kim PS. 1989. Evidence that the leucine zipper is a coiled coil. Science 243:538–542 [DOI] [PubMed] [Google Scholar]
- 32. Reinke AW, Grigoryan G, Keating A. 2011. Identification of bZIP interaction partners of viral proteins HBZ, MEQ, BZLF1, and K-bZIP using coiled-coil arrays. Biochemistry 49:1985–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schauflinger M, et al. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J. Virol. 85:3821–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seo JY, Britt WJ. 2008. Multimerization of tegument protein pp28 within the assembly compartment is required for cytoplasmic envelopment of human cytomegalovirus. J. Virol. 82:6272–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sinzger C, et al. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359–368 [DOI] [PubMed] [Google Scholar]
- 37. Smuda C, Bogner E, Radsak K. 1997. The human cytomegalovirus glycoprotein B gene (ORF UL55) is expressed early in the infectious cycle. J. Gen. Virol. 78:1981–1992 [DOI] [PubMed] [Google Scholar]
- 38. Spaete RR, Mocarski ES. 1985. The alpha sequence of the cytomegalovirus genome functions as a cleavage/packaging signal for herpes simplex virus defective genomes. J. Virol. 54:817–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Theiler RN, Compton T. 2002. Distinct glycoprotein O complexes arise in a post-Golgi compartment of cytomegalovirus infected cells. J. Virol. 76:2890–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tischer BC, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 42:191–197 [DOI] [PubMed] [Google Scholar]
- 41. Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163–178 [PubMed] [Google Scholar]
- 42. Valentine R, Shapiro C, Stadtman ER. 1968. Regulation of glutamine synthetase: XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7:2143–2152 [DOI] [PubMed] [Google Scholar]
- 43. Varnum SM, et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Waadt R, et al. 2008. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56:505–516 [DOI] [PubMed] [Google Scholar]
- 45. Walter M, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40:428–438 [DOI] [PubMed] [Google Scholar]
- 46. Womack A, Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio 1:e00282–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang K, Will E, Baines JD. 2009. The putative leucine zipper of the UL6-encoded portal protein of herpes simplex virus 1 is necessary for interaction with pUL15 and pUL28 and their association with capsids. J. Virol. 83:4557–4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zipper P, Kratky O, Herrmann R, Hohn T. 1971. An X-ray small angle study of the bacteriophage fr and R17. Eur. J. Biochem. 18:1–9 [DOI] [PubMed] [Google Scholar]