Abstract
Three molecules have been identified as the main cellular factors required for binding and entry of human T-cell leukemia virus type 1 (HTLV-1): glucose transporter 1 (GLUT1), heparan sulfate (HS), and neuropilin 1 (NRP-1). However, the precise mechanism of HTLV-1 cell tropism has yet to be elucidated. Here, we examined the susceptibilities of various human cell lines to HTLV-1 by using vesicular stomatitis virus pseudotypes bearing HTLV-1 envelope proteins. We found that the cellular susceptibility to HTLV-1 infection did not correlate with the expression of GLUT1, HS, or NRP-1 alone. To investigate whether other cellular factors were responsible for HTLV-1 susceptibility, we conducted expression cloning. We identified two HS proteoglycan core proteins, syndecan 1 and syndecan 2, as molecules responsible for susceptibility to HTLV-1. We found that treatment of syndecan 1-transduced cells (expressing increased HS) with heparinase, a heparin-degradative enzyme, reduced HTLV-1 susceptibility without affecting the expression levels of HS chains. To further elucidate these results, we characterized the expression of HS chains in terms of the mass, number, and length of HS in several syndecan 1-transduced cell clones as well as human cell lines. We found a significant correlation between HTLV-1 susceptibility and the number of HS chains with short chain lengths. Our findings suggest that a combination of the number and the length of HS chains containing heparin-like regions is a critical factor which affects the cell tropism of HTLV-1.
INTRODUCTION
Human T-cell leukemia virus type 1 (HTLV-1) is the causative agent of adult T-cell leukemia (16, 49) and HTLV-1-associated myelopathy, also known as tropical spastic paraparesis (10, 24, 45). Previous investigations revealed that HTLV-1 infects not only human T lymphocytes and central nervous system cells but also cells of other tissues (6, 17, 21, 34, 51, 69). To date, glucose transporter 1 (GLUT1), neuropilin-1 (NRP-1), and heparan sulfate proteoglycans (HSPGs) have been implicated as being involved in HTLV-1 infection (reviewed in reference 12). The expression of GLUT1, which is responsible for viral binding and fusion mediated by the gp46 surface envelope (Env) protein (3, 26, 39), is ubiquitous, and it is also known that various cells express HSPGs. Although these findings might be able to explain the broad host cell range of HTLV-1, they are not sufficient to explain the variance of HTLV-1 cell tropism.
It was previously reported that HTLV-1 spreads from cell to cell via virological synapses (22, 38); however, recent studies by Pais-Correia and colleagues showed that the HTLV-1 virions retaining extracellular structures are important for HTLV-1 cell transmission (46). That study implied that the viral particle mediates HTLV-1 transmission and suggested the importance of the interaction between viral particles and the target cell surface. In this study, we investigated the susceptibilities of various human cell lines to cell-free HTLV-1 infection using highly infectious vesicular stomatitis virus (VSV) pseudotypes harboring the Env protein of HTLV-1. These pseudotype viruses express green fluorescent protein (GFP) in infected cells. We observed a >1,000-fold difference in the susceptibilities of these pseudotypes among the human cell lines. Interestingly, the levels of GLUT1 expression on the cell surface and the amount of cell surface heparan sulfate (HS) chains did not reflect susceptibility to HTLV-1 in human cell lines. In addition, the expression of NRP-1 mRNA also did not reflect susceptibility to HTLV-1. From these data, we suspected the existence of an unknown factor that affects the interaction between these three molecules and the viral particles and causes the differences in cell tropism of the HTLV-1 particle. Therefore, we used an expression cloning method to investigate potential candidates and found that two HS proteoglycan (HSPG) core proteins, syndecan 1 and 2 (SDC1 and SDC2), play a significant role in cellular susceptibility to HTLV-1.
MATERIALS AND METHODS
Cell lines.
The human T-cell line Molt-4 clone 8 (29), the HTLV-1-positive but HTLV-1 Env-negative T-cell line C8166 (53), the HTLV-1-producing T-cell lines C91/PL (50) and MT-2 (41), and K562 cells transduced with the CD4 gene and ecotropic murine leukemia virus receptor gene (K562/CD4/ecoR; referred to here as K4R cells) (58) were all maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). The human astrocytoma cell line U251MG (2), the human glioma cell line NP-2 (58), the feline kidney fibroblast cell line S+L-CCC clone 8C (8), 8C cell lines persistently infected with the strains 2M (cosmopolitan strain) (18) and MEL5 (Melanesian strain) (11) of HTLV-I (8C/HTLV-12M and 8C/HTLV-1MEL5) (19, 20), and a fetal lamb kidney cell line that was persistently infected with bovine leukemia virus (BLV), FLK (67), were all maintained in Eagle's minimum essential medium (MEM) supplemented with 10% FBS. The human osteosarcoma-derived cell line HOS (40), the human colon cancer cell line HT-29 (55), the human renal epithelial cell line expressing adenovirus E1a and SV40 large T antigen, 293T (14, 47), and the retrovirus-packaging cell lines Plat-E (42) and Phoenix-A (31) (kindly provided by T. Kitamura [Institute of Medical Science, The University of Tokyo, Tokyo, Japan] and G. Nolan [Stanford University, Stanford, CA], respectively) were maintained in Dulbecco's modified MEM supplemented with 10% FBS.
Preparation of VSVΔG*-G and VSVΔG* pseudotypes.
In this study, we used VSVΔG* pseudotypes bearing the VSV-G protein and BLV Env [VSVΔG*(HTLV-1MEL5), VSVΔG*(HTLV-12M), and VSVΔG*(BLV)] as the controls for VSVΔG* pseudotypes bearing HTLV-1 Env. Both of these viruses infect various mammalian cell lines similarly to HTLV-1, although their cellular receptors have yet to be identified. BLV and HTLV are ∼50% similar at the nucleotide level and share many common features; however, BLV is known to use a different cellular receptor (59).
Recombinant VSV, VSVΔG*-G, was kindly provided by M. A. Whitt (University of Tennessee, Memphis, TN) (62). VSVΔG*-G was inoculated into 293T cells that had been transfected with an expression plasmid of VSV G protein, pCAGGS/VSVG. Culture supernatants containing newly propagated VSVΔG*-G were harvested after incubation for 24 to 30 h at 37°C. 8C/HTLV-1MEL5, 8C/HTLV-12M, or FLK cells were plated on 100-mm plates and inoculated 1 day later with VSVΔG*-G at a multiplicity of infection of 5 to 10 (when titrated on 8C cells) for 1 to 2 h at 37°C. The cells were washed with medium, and then medium supplemented with 2% FBS and 50 mM HEPES (pH 7.1) was added. Culture supernatants containing propagated VSVΔG* pseudotypes bearing HTLV-1 Env [VSVΔG*(HTLV-1)] or BLV Env [VSVΔG*(BLV)] were harvested after 18 to 24 h of incubation at 33°C in a CO2 incubator. Culture supernatant containing the VSVΔG*-G or VSVΔG* pseudotype was clarified by low-speed centrifugation, and aliquots of the supernatant were stored at −80°C until use.
Prior to inoculation, VSVΔG*(HTLV-1) and VSVΔG*(BLV) samples were incubated with ∼2 to 10% anti-VSV serum, which was obtained from a goat that had been repeatedly immunized with VSV, for 20 min at 30°C to completely neutralize the remaining VSVΔG*-G. After 20 to 30 h of inoculation, the number of GFP-expressing cells was counted under a fluorescence microscope. Because of the absence of secondary infection by descendant VSVΔG* pseudotypes, the number of GFP-positive cells was the same as the number of infectious units (IUs) of VSVΔG* pseudotypes. The viral susceptibility of each cell was evaluated by the IUs obtained by inoculation with 1 ml VSVΔG* pseudotype. Serum (0.2%) from an HTLV-1-infected healthy carrier or a rat anti-gp46 monoclonal antibody (MAb), LAT-27 (a generous gift from Y. Tanaka, Ryukyu University, Nichihara, Okinawa, Japan) (15, 63), was used to neutralize VSVΔG*(HTLV-1). Anti-BLV serum (∼0.5 to 1%) from leukemic cattle was used to neutralize VSVΔG*(BLV). Neutralizations of VSVΔG*(HTLV-1) and VSVΔG*(BLV) by antisera and MAbs were carried out simultaneously with an anti-VSV serum treatment step; i.e., pseudotype viruses were treated with anti-VSV serum in the presence or absence of anti-HTLV-I (or BLV) serum for 20 min at 30°C. Formation of each pseudotype was confirmed by an almost complete neutralization using specific antisera or MAbs against each virus.
Molecular cloning and sequencing of cDNA responsible for increased HTLV-1 susceptibility.
Poly(A)+ RNA was extracted from the highly HTLV-1-susceptible U251MG cells using a FastTrack kit (Invitrogen, Carlsbad, CA). The RNA was then used to synthesize a cDNA library using the SuperScript Choice system for cDNA synthesis (Invitrogen). This cDNA library was cloned into the retroviral vector pMXpuro (44) according to the protocol of Kitamura and colleagues (32). The resultant expression cDNA library, pMXpuro/U251MG-cDNA, was then transfected into Plat-E cells (2 × 106 cells in a 60-mm culture plate) using the Fugene6 transfection reagent (Roche, Indianapolis, IN). Thirty-four hours after transfection, the culture supernatant containing the retroviral library was harvested and then inoculated into 1 × 106 K4R cells in the presence of Polybrene (10 μg/ml).
Genomic DNA extracted from isolated cell clones that showed an increased susceptibility to VSVΔG*(HTLV-1MEL5) compared with parental K4R cells was subjected to PCR to recover the introduced cDNAs. The following PCR primers were used, which were derived from the pMXpuro vector sequences flanking the inserted cDNAs: 5′-GGACCATCCTCTAGACTGCCGGATCCCAGTGTG-3′ and 5′-ATACTTCTGCCTGCTGGGGAGCCTGGGGAC-3′. PCR was performed using LA Taq polymerase (Takara Shuzo Co., Ltd., Ohtsu, Shiga, Japan) under the following conditions: 1 cycle at 94°C for 2 min, then 35 cycles at 98°C for 20 s and 68°C for 5 min, and one final cycle at 68°C for 5 min. PCR-amplified fragments were purified using a Geneclean II kit (Bio 101, Inc., La Jolla, CA) and then cloned into the pCR2.1 TA vector (Invitrogen). The resultant plasmids were sequenced using a dye primer and a DNA sequencer SQ5050 (Hitachi, Tokyo, Japan). The cloned sequences were analyzed for identity and similarity to other known sequences using the on-line BLAST program (28).
Reverse transcriptase PCR and real-time PCR.
SDC1, SDC2, SDC3, SDC4, glypican 1 (GPC1), and NRP-1 mRNAs were detected by reverse transcription-PCR (RT-PCR). Total RNA was extracted from 293T, C8166, HOS, K4R, Molt-4, and U251MG cells using an RNA extraction kit (Isogen; Nippongene Co., Ltd., Tokyo, Japan) according to the manufacturer's protocol. For each sample, 4 μg of total RNA was subjected to an RT reaction using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) according to the manufacturer's protocol. First-strand cDNA was then diluted to 50 μl using Tris-EDTA buffer (pH 8.0), and 1 μl was used as the template for PCR.
The relative NRP-1 mRNA levels were quantified in duplicate by quantitative PCR (qPCR) in the Mx3000P real-time PCR system (Agilent Technologies, CA) with brilliant SYBR green QPCR Master Mix (Agilent Technologies-Stratagene, CA), according to the manufacturer's instructions. qPCR mixtures were preincubated at 95°C for 10 min, followed by 40 cycles of PCR at 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s, using the specific primer pair NRP-1/sense (5′-CCCGCACCTCATTCCTACATC-3′; positions 1946 to 1966) and NRP-1/antisense (5′-CATTCATCCACCAAGTTCCCG-3′; positions 2311 to 2331) (GenBank accession no. NM_003873.5). The NRP-1 mRNA level in each cell line was normalized to human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA using a predesigned Perfect real-time primer set for GAPDH (primer set identification HA031578; Takara Shuzo) and reported as the change (fold) compared with that from K4R cell data using the comparative quantitation analysis MxPro QPCR software available with Mx3000P.
PCR was performed with each specific primer pair to amplify the corresponding sequence. The codes and sequences of the PCR primers for the SDC1, SDC2, SDC3, SDC4, GPC1, and GLUT1 genes areas follows: SDC1/sense, 5′-GGTCCGGGCAGCATGAGGCGCGCGGCGCTC-3′ (positions 288 to 317), and SDC1/antisense, 5′-CGCATGGCTCCCGCGTCAGGCATAGAATTC-3′ (positions 1218 to 1247) (GenBank accession no. NM_002997); SDC2/sense, 5′-ATGCGGCGCGCGTGGATCCTGCTCACCTT-3′ (positions 619 to 647), and SDC2/antisense, 5′-TTACGCATAAAACTCCTTAGTAGGTGCCT-T-3′ (positions 1195 to 1224) (GenBank accession no. NM_002998); SDC3/sense, 5′-CCGCCATGAAGCCGGGGCCGCCGCACCGTG-3′ (positions 50 to 79), and SDC3/antisense, 5′-CACTAGGCATAGAACTCCTCCTGCTTGTCA-3′ (positions 1356 to 1385) (GenBank accession no. AF248634); SDC4/sense, 5′-ATGGCCCCCGCCCGTCTGTTCGCGCTGCT-3′ (positions 27 to 55), and SDC4/antisense, 5′-TCACGCGTAGAACTCATTGGTGGGGGCTTT-3′ (positions 594 to 623) (GenBank accession no. XM_009530); GPC1/sense, 5′-ATGGAGCTCCGGGCCCGAGGCTGGTGGCTG-3′ (positions 222 to 251), and GPC1/antisense, 5′-TTACCGCCACCGGGGCCTGGCTACTGTAAG-3′ (positions 1869 to 1898) (GenBank accession no. NM_002081). The initiation codons (ATG) and termination codons (TCA, TTA, and CTA, corresponding to TGA, TAA, and TAG, respectively, in the sense frame) are underlined. PCR was performed using LA Taq polymerase (Takara Shuzo) as follows: 1 cycle at 94°C for 1.5 min; 35 cycles of 98°C for 20 s, 50°C for 20 s, and 68°C for 3 to 5 min; and a final extension at 68°C for 3 to 5 min. As a control for confirming the integrity and amount of mRNA in each sample, a GAPDH mRNA was amplified with a primer set (GAPDH-F, 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′, and GAPDH-R, 5′-CATGTGGGCCATGAGGTCCACCAC-3′) (GenBank accession no. M17851).
Establishment of cells expressing SDC1, SDC2, SDC3, SDC4, or GPC1.
The complete open reading frame (ORF) sequences of the human SDC1, SDC2, SDC3, SDC4, and GPC1 genes were obtained by PCR amplification of DNA samples from the pMXpuro/U251MG cDNA library or U251MG cDNA as a template. PCR products were cloned into the retroviral expression vector plasmid pMXpuro or pCXbsr (1). K4R cells were inoculated with retrovirus vector produced by Plat-E or by Phoenix-A cells transfected with the pMXpuro or pCXbsr plasmid containing the ORF sequence for the SDC1, SDC2, SDC3, SDC4, or GPC1 gene. The cells transduced with these vectors were selected by culture in medium containing puromycin at 1 μg/ml or blasticidin S at 10 μg/ml for 1 to 2 weeks. The cell lines thus established were named K4R/SDC1, K4R/SDC2, K4R/SDC3, K4R/SDC4, and K4R/GPC1, respectively. Cell lines transduced with pMXpuro or pCXbsr vector alone, namely, K4R/pMX and K4R/pCX, were established as controls. K4R/SDC1 cell clones were obtained by limiting dilution, and clonal cells were assessed for their susceptibilities to VSVΔG*(HTLV-1).
Detection of cell-free HTLV-1 infection.
Cell-free HTLV-1 produced by 8C/HTLV-1cells was prepared as described previously (15, 63). Target cells (105 cells/300 μl) were inoculated with 100 μl of cell-free HTLV-1, which had been treated for 10 min with or without anti-HTLV-I serum (1%) on ice, and then incubated for 24 h at 37°C in a CO2 incubator. DNAs of target cells were isolated using a Wizard DNA purification kit (Promega, Madison, WI) and dissolved in 50 μl of Tris-EDTA buffer, and 2 μl was used as the template for qPCR as described above. qPCR was carried out to quantify the HTLV-1 proviral DNA synthesis level using the HTLV-1 pX-specific primers designated pX-F (5′-CCCACTTCCCAGGGTTTGGACAGAG-3′; positions 7325 to 7349) and pX-R (5′-CTGTAGAGCTGAGCCGATAACGCG-3′; positions 7504 to 7577) (GenBank accession no. J02029), giving an amplified fragment which was a 203-bp sequence of HTLV-1 pX. qPCR mixtures were preincubated at 95°C for 10 min, followed by 40 to 50 cycles of 95°C for 30 s, 65°C for 30 s, and 72°C for 30 s. As a control for confirming the integrity and amount of DNA in each sample, a human β-globin gene was amplified with a primer set (PCO3, 5′-ACACAACTGTGTTCACTAGC-3′, and PCO4, 5′-CAACTTCATCCACGTTCACC-3′) (52). β-Globin gene amplifications were performed at an annealing temperature of 55°C, and all other conditions were the same as described above.
Syncytium assay.
To investigate the effect of HSPG for syncytium formation, HTLV-1 producing MT-2 cells were cocultured with target cells. Molt-4 cells were used as a positive control T-cell line. Two days after, cells were fixed with 2.5% paraformaldehyde, and the number of syncytia (more than two cell diameters) was counted.
Treatment of cells with heparitinase I and heparinase.
Heparitinase I and heparinase (Seikagaku Kogyo, Tokyo, Japan) were serially diluted with phosphate-buffered saline (PBS). Serially diluted heparitinase I and heparinase were diluted 10-fold with Opti-MEM I reduced-serum medium (Gibco BRL). Cells were incubated with diluted heparitinase I- or PBS-containing Opti-MEM at 37°C for 1 h.
Flow-cytometric analysis.
To examine the cell surface expression of GLUT1 and HSPG, 6 × 105 cells per sample were prepared for flow-cytometric analysis. Adherent cells were detached from tissue culture dishes using Versene (0.2% EDTA in PBS). The cells were then suspended in 30 μl of PBS containing 3% bovine serum albumin (BSA) and 0.1% sodium azide (PBS-BSA-NaN3) and incubated on ice for 30 to 40 min with 1 μg of anti-GLUT1 mouse MAb, clone 202915 (R&D Systems), 2 μg of anti-GLUT1 (N-20) goat polyclonal antibody (PAb) (Santa Cruz), 1.2 μg of anti-SDC1 core protein mouse MAb, clone B-B4 (Immunotech, Marseille, France), or 0.3 μg of anti-HS mouse MAb clone F58-10E4 (Seikagaku Kogyo Co. Ltd., Tokyo, Japan). The cells were washed with PBS containing 1% FBS and 0.1% sodium azide (PBS-FBS-NaN3), suspended in 30 to 40 μl of PBS-BSA-NaN3 containing fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse immunoglobulin (IgG) (1:30; MBL, Nagoya, Japan) or FITC-conjugated rabbit anti-goat IgG (1:30; ICN/Cappel, Costa Mesa, CA) secondary Ab, and incubated on ice for 30 to 40 min. The cells were washed with PBS-FBS-NaN3 and suspended in 300 to 600 μl of PBS containing 1% paraformaldehyde, and then flow-cytometric analysis (FCM) was performed using a flow cytometer, model Cyto ACE-100 (Jasco, Tokyo, Japan). As a control, the cells were treated with the secondary Ab alone and were subjected to FCM. To detect the HS neo-epitopes (ΔHS) generated at the terminal stubs of the HS by digestion with heparitinase I, 1 × 106 to 2 × 106 cells were resuspended in 100 to 200 μl of Opti-MEM reduced-serum medium (Gibco BRL) and incubated with heparitinase I (10 mU/ml) (Seikagaku Kogyo Co. Ltd.) for 1 h at 37°C. Six hundred thousand cells were then washed with PBS, suspended in 30 μl of PBS-BSA-NaN3, and stained with 0.3 μg of anti-Δ-HS mouse MAb, clone F69-3G10 (10 μg/ml) (Seikagaku Kogyo Co. Ltd.), as described above.
RESULTS
Preparation of VSV pseudotypes bearing HTLV-1 Env proteins.
VSVΔG*-G, a recombinant VSV carrying a GFP reporter gene, efficiently infected C8166 human T cells, as indicated by the GFP-positive cells detected by fluorescence microscopy (Fig. 1A). This infectivity was almost completely inhibited by the pretreatment of VSVΔG*-G with anti-VSV serum. VSVΔG*-G was then inoculated into 8C/HTLV-12M or 8C/HTLV-1MEL5 cells to produce the VSVΔG* pseudotypes, VSVΔG*(HTLV-12M), or VSVΔG*(HTLV-1MEL5), respectively. VSVΔG*(HTLV-12M) and VSVΔG*(HTLV-1MEL5) were capable of infecting approximately 20 to 30% of C8166 human cells even after pretreatment with anti-VSV serum (Fig. 1A), and the infectivity was drastically reduced by pretreatment of the pseudotypes with a MAb specific for the HTLV-1 Env gp46 (data not shown) or with 0.2% anti-HTLV-1 human serum (Fig. 1A) but not with anti-BLV antisera (data not shown). We also confirmed that VSVΔG*-G infectivity was not significantly affected by treatment with anti-HTLV-1 antisera or anti-BLV antisera (data not shown). As expected from viral interference, VSVΔG*(HTLV-1) was nearly undetectable in cell lines expressing HTLV-1 Env (MT-2 and C91/PL) (Fig. 1B). These data indicated that infectious pseudotypes bearing HTLV-1 Env could be produced.
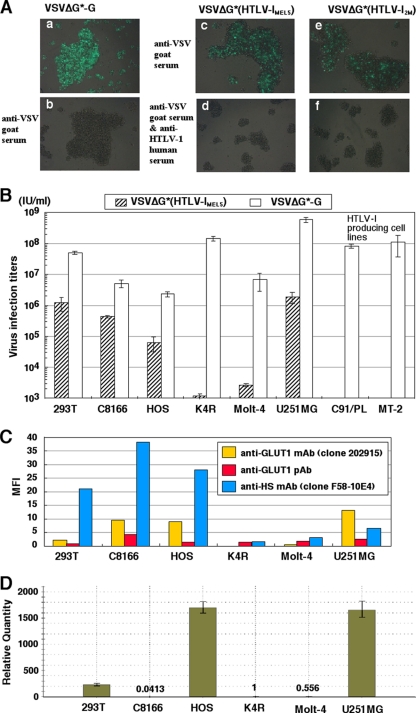
Fig 1.
Susceptibility of cell lines to VSVΔG* pseudotypes bearing the HTLV-1 Env protein. (A) C8166 cells were inoculated with VSVΔG*-G in the absence (a) or presence (b) of anti-VSV goat serum (1%). C8166 cells were also inoculated with VSVΔG*(HTLV-1MEL5) and VSVΔG*(HTLV-12M) pseudotypes treated with anti-VSV goat serum (5%), in the absence (c and e) or presence (d and f) of anti-HTLV-1 human serum (0.2%). One day after inoculation, the cells were observed by fluorescence microscopy. (B) Titration of the VSVΔG*(HTLV-1MEL5) and VSVΔG*-G using various human cell lines. VSVΔG*(HTLV-1MEL5) treated with anti-VSV goat serum (see Materials and Methods) and VSVΔG*-G were serially diluted 10-fold, and cells were inoculated to determine the respective titers. Each titer was calculated from the number of GFP-positive cells in endpoint dilutions. (C) Expression of GLUT1 and HS chains on the surface of various cell lines. Expression of GLUT1 and HS chains in human cell lines was examined by FCM. Cells were stained with anti-GLUT1 mouse MAb 202915, anti-HS mouse MAb F58-10E4, or anti-GLUT1 goat PAb and then stained with an FITC-conjugated rabbit anti-mouse IgG or an FITC-conjugated rabbit anti-goat IgG secondary Ab. Mean fluorescent intensities detected by the anti-GLUT1 MAb, the anti-GLUT1 PAb, and the anti-HS mouse MAb were determined by subtracting the MFI of the negative controls (secondary Ab alone). (D) Relative mRNA expression levels of NRP-1 in various cell lines were measured by real-time RT-PCR and the values were normalized to expression levels of GAPDH mRNA.
Susceptibilities of different human cell lines to HTLV-1 pseudotypes and their expression levels of GLUT1, NRP-1, and cell surface HS chains.
We investigated the susceptibility of several human cell lines to the HTLV-1 pseudotype and VSVΔG*-G, including 293T, C8166, HOS, Molt-4, U251MG, and the CD4-transduced K562 cell clone K4R. These cells were inoculated with VSVΔG*(HTLV-1MEL5) or VSVΔG*G, and GFP expression was monitored the next day (Fig. 1B). These cell lines varied in permissiveness to the HTLV-1 pseudotype. In particular, U251MG cells were approximately 1,600-fold more susceptible than K4R cells (Fig. 1B). This U251MG cell line has been shown to be highly susceptible to HTLV-1 virions in a cell-free infection system (15). In contrast, the susceptibility of U251MG cells to VSVΔG*-G was only 4-fold higher than that of K4R cells (Fig. 1B).
FCM revealed that the levels of cell surface GLUT1 expression detected with anti-GLUT1 mouse MAb (r = 0.52, P = 0.29) and anti-GLUT1 goat PAb (r = 0.01, P = 0.34) were not significantly correlated with susceptibility to the HTLV-1 pseudotype among these cell lines (Fig. 1C). Although we observed the highest reactivity of anti-GLUT1 mouse MAb to the U251MG cells and the lowest reactivity to the K4R cells, Kinet et al. (30a) reported that this MAb does not detect endogenous GLUT1 but rather interacts with a cell surface protein which is associated with GLUT1 overexpression in transformed cell lines. In addition, the cell surface HS expression level, determined by FCM using the MAb F58-10E4, which reacts with epitopes containing N-sulfated glucosamine residues present in many types of HS (4, 65), did not significantly correlate with susceptibility to the HTLV-1 pseudotype (Fig. 1C). Moreover, the mRNA expression levels of NRP-1 did not correlate with the susceptibilities to the HTLV-1 pseudotype in six different cell lines (r = 0.41, P = 0.42) (Fig. 1D). Thus, the range in susceptibility to HTLV-1 among these cell lines could not be explained by the respective expression levels of GLUT1, cell surface HS, or NRP-1 independently. Although another explanation in which the expression balance of GLUT-1, HS, and NRP-1 affects the HTLV-1 susceptibilities would be possible, we adopted a working hypothesis that an unknown cellular factor may also be involved.
Identification of molecules responsible for the increased susceptibility to HTLV-1 pseudotype.
To investigate cellular factors other than GLUT-1, HS, and NRP-1 affecting the HTLV-1 susceptibility, we attempted to identify the responsible factor from U251MG cells, which were permissive to the HTLV-1 pseudotype. We generated a pMX vector-based retroviral expression cDNA library from U251MG cells and transduced the library into K4R cells, which were least HTLV-1 susceptible. cDNA library-expressing K4R cells were seeded into 96-well plates (20 to 40 cells/well), and cultured for 2 weeks. Cells were then screened for their susceptibilities to VSVΔG*(HTLV-1MEL5). Eighty-three of 5,088 wells showed an increased frequency of GFP-positive cells in the first round of screening and were then subjected to a second round of screening. In that screening, two wells, designated w16 and w40, showed a >3-fold increase in GFP-positive cells compared to other wells, and those cells were subsequently cloned. Cell clones derived from each well, K4R/cDNA-w16-c and K4R/cDNA-w40-c, demonstrated 40-fold-higher susceptibilities to the HTLV-1 pseudotype than the parental K4R cells (Fig. 2A and B).
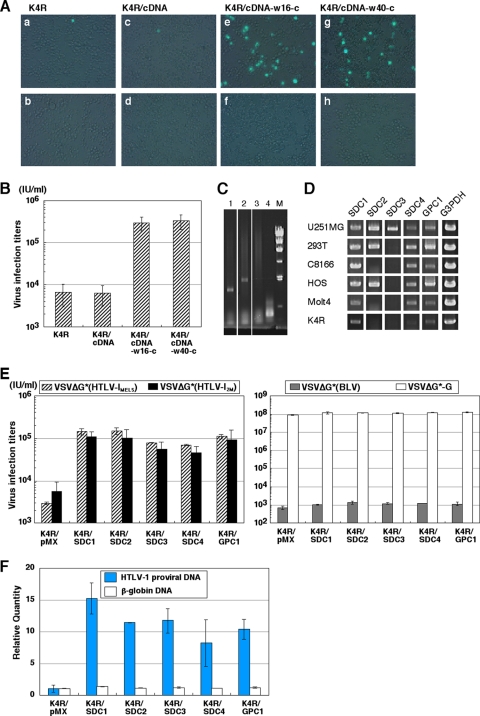
Fig 2.
K4R cell clones transduced with the U251MG cDNA library showed an enhanced susceptibility to the HTLV-1 pseudotypes. (A) Detection of GFP-positive cells inoculated with the VSVΔG*(HTLV-1MEL5). Parental K4R cells (a and b), K4R cells transduced with the U251MG cDNA library (K4R/cDNA cells) (c and d), and two K4R/cDNA cell clones showing increased HTLV-1-susceptibility (K4R/cDNA-w16-c [e and f] and K4R/cDNA-w40-c cells [g and h]) were inoculated with VSVΔG*(HTLV-1MEL5) treated with anti-VSV goat serum (final concentration of 10%) in the absence (a, c, e, and g) or presence (b, d, f, and h) of anti-HTLV-I serum (final concentration of 0.2%). (B) Enhanced susceptibilities of two K4R/cDNA cell clones to VSVΔG*(HTLV-1MEL5) infection. VSVΔG*(HTLV-1MEL5) infectious titers of each cells calculated from the number of GFP-positive cells in endpoint dilutions are shown. (C) Detection of introduced cDNAs harbored in the K4R/cDNA-w16-c and K4R/cDNA-w40-c cells. Genomic DNA from K4R/cDNA-w16-c, K4R/cDNA-w40-c, and parental K4R cells and pMXpuro plasmid DNA were subjected to PCR using specific primers for the pMXpuro vector to detect transduced cDNA. Lanes: 1, K4R/cDNA-w16-c; 2, K4R/cDNA-w40-c; 3, parental K4R; 4, pMXpuro plasmid DNA; M, molecular size marker (λ DNA Hind III digest). (D) Expressions of SDCs and GPC1 in various human cell lines. Cellular RNA extracted from human cell lines for the expression of SDC1, SDC2, SDC3, SDC4, and GPC1 genes was amplified by RT-PCR. GAPDH mRNA was amplified as a control. (E) Enhanced susceptibilities of K4R cells transduced with HSPG core protein genes to the HTLV-1 pseudotypes. Cells were inoculated with serially diluted VSVΔG*(HTLV-1MEL5), VSVΔG*(HTLV-12M), and VSVΔG*(BLV) pseudotypes treated with anti-VSV goat serum (see Materials and Methods) or VSVΔG*-G. Three independent experiments were done, and the data are from one representative experiment of the three. (F) Enhanced susceptibilities of K4R cells transduced with HSPG core protein genes to the cell-free HTLV-1. Cells were inoculated with cell-free wild-type HTLV-12M, and the formation of HTLV-1 DNA was monitored by real-time PCR using the pX region primer 24 h postinfection. β-Globin DNA in each sample was amplified as a control. Three independent experiments were done, and the data are from one representative experiment of the three.
To identify the molecule(s) responsible for the increased susceptibility to the HTLV-1 pseudotype, genomic DNA was extracted from K4R/cDNA-w16-c and K4R/cDNA-w40-c and subjected to PCR amplification using pMX vector-specific primers. Two fragments of 1.3 and 2.0 kb were amplified from w16-c and w40-c clones, respectively (Fig. 2C), and subsequently cloned into a pCR2.1 vector to determine their nucleotide sequences. Contrary to our expectations, K4R/cDNA-w16-c contained a cDNA corresponding to nt 49 to 1331 of the human SDC1 gene transcript variant 2 (GenBank accession no. NM_002997), and K4R/cDNA-w40-c contained cDNA corresponding to nt 297 to 2312 of the human SDC2 gene (GenBank accession no. NM_002998).
The SDC family core proteins have previously been reported to exhibit cell type-specific distributions (5, 9, 30). In addition to SDCs, cell surface HSPGs include GPC (5, 9, 30). We therefore examined the expression levels of several cell surface HSPG core protein mRNAs by RT-PCR and found that each cell line expressed different combinations of HSPG core protein mRNAs (Fig. 2D). The U251MG cell line, which was most susceptible to HTLV-1 pseudotype infection, expressed all four SDC core proteins and GPC-1 mRNAs, whereas the K4R cells, which were least susceptible, expressed only SDC1, SDC4, and GPC1 mRNAs.
To confirm whether the transduction of these HSPG core protein genes was responsible for the increased susceptibility to the HTLV-1 pseudotype, we established K4R cells transduced with SDC1, SDC2, SDC3, SDC4, and GPC1 cDNA, referred to as K4R/SDC1, K4R/SDC2, K4R/SDC3, K4R/SDC4, and K4R/GPC1 cells, respectively. We then examined their susceptibilities to HTLV-1 pseudotypes. All of these transduced cells exhibited a >20-fold increase in susceptibility to both VSVΔG*(HTLV-1MEL5) and VSVΔG*(HTLV-12M) pseudotypes compared to the transduced control cells (K4R/pMX) (Fig. 2E). In contrast, susceptibilities of these transduced cells to both VSVΔG*-G and VSVΔG*(BLV) pseudotypes were within 2-fold of the transduction control cells (K4R/pMX) (Fig. 2E).
Next we examined the susceptibilities of K4R/SDC1, K4R/SDC2, K4R/SDC3, K4R/SDC4, and K4R/GPC1 cells to cell-free wild-type HTLV-1. These cells were inoculated with HTLV-12M, and the proviral DNA was detected 24 h postinfection by qPCR. Consistent with the results obtained with HTLV-1 pseudotypes (Fig. 2E), the susceptibility of K4R cells to cell-free HTLV-1 was increased markedly by the transduction of HSPG core protein genes (Fig. 2F).
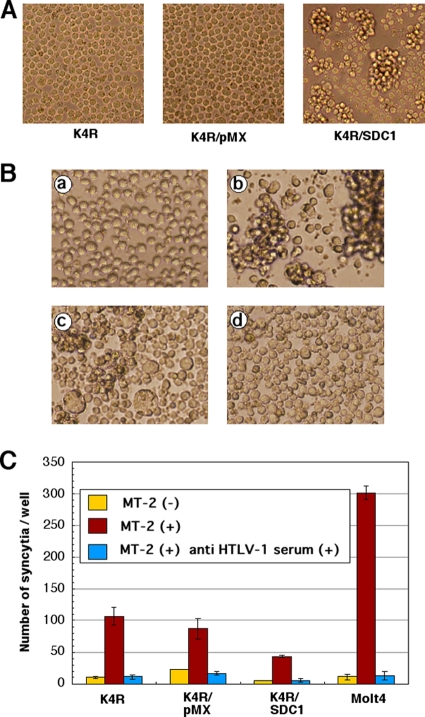
Expression of HSPG core proteins in K4R cells induces cell aggregation but does not increase cell fusion activity to HTLV-1-producing MT-2 cells.
We observed that K4R cells transduced with HSPG core protein genes formed cell clumps, as shown in Fig. 3A. These cellular aggregations were observed in all species of SDCs and GPC1 (data not shown), indicating that the expression of HSPGs induced cell-cell adhesion. Similar to our observation, HS-dependent cell-cell adhesion has been reported in B lymphoid cell lines and cells transduced with SDC1 (36, 56, 60). To determine whether this increased cell-cell contact mediated by HSPG is related to the fusion activity with an HTLV-1-producing cell line, we conducted a syncytium assay as described in Materials and Methods. We observed syncytia in the coculture of HTLV-1-producing MT-2 cells with both K4R cells (Fig. 3B) and Molt-4 T cells (data not shown). The cell fusion was effectively inhibited by anti-HTLV-1 serum (Fig. 3C). The number of syncytia in the coculture of K4R/SDC1 with MT-2 cells was lower than that in the coculture of MT-2 cells with parental K4R cells or vector-transduced K4R cells (Fig. 3C). These findings indicate that the increased cell-cell adhesion mediated by cell surface expression of HSPGs does not contribute to membrane fusion between target cells and HTLV-1-producing cells.
Fig 3.
Expression of HSPG core proteins induces cell aggregation but does not increase cell fusion activity with HTLV-1-producing MT-2 cells. Cell surface expression of HSPG core protein does not increase cell fusion activity when the protein is incubated with HTLV-1-producing cells. The expression of HSPG core protein (A) K4R cells transduced with the HSPG core protein gene frequently showed cell clumps. Under normal culture conditions, K4R/SDC1 cells but not parental or vector-transduced K4R cells showed the formation of aggregates. (B) Syncytium formation in K4R cells cocultured with MT-2 cells. K4R cells (a), MT-2 cells (b), and cocultured K4R and MT-2 cells in the absence (c) or presence (d) of anti-HTLV-I serum (final concentration at 1%) were fixed 4 days after coculture. (C) No augmentation of syncytium formation in K4R cells by overexpression of SDC1 was observed. The syncytium assay was conducted by coculturing 2 × 104 target cells (K4R, K4R/pMX, K4R/SDC1, and Molt-4 cells with MT-2 cells [5 × 103] in a 96-well plate at 37°C for 2 days in the absence or presence of anti-HTLV-1 serum [final concentration, 0.5%]). The number of syncytia was counted as the number of large cells whose diameter was greater than twice that of normal cells (1.5 ± 0.5 μm). Two independent experiments were done, and the data are from one representative experiment of the two.
Reduced susceptibility to VSVΔG*(HTLV-1) following heparitinase I or heparinase treatment.
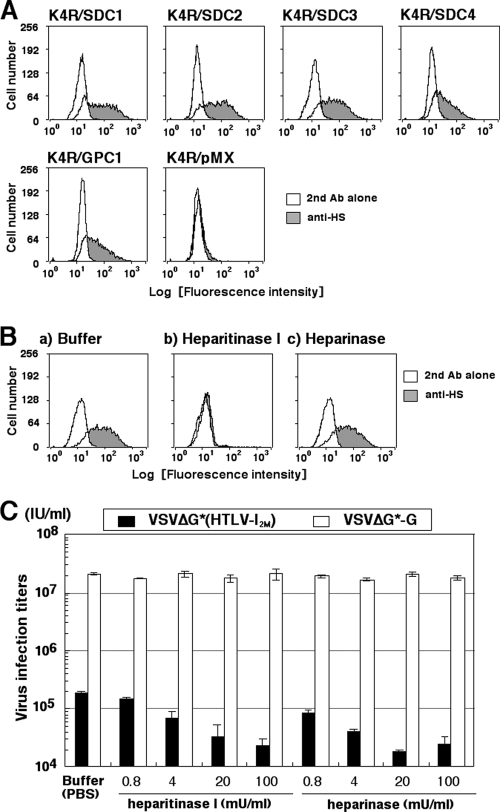
K4R cells transduced with HSPG core proteins (SDC1, SDC2, SDC3, SDC4, and GPC1) showed increased cell surface expression of HS, as detected by an anti-HS MAb, F58-10E4 (Fig. 4A). To evaluate the role of HS in the increased susceptibility to the HTLV-1 pseudotype, the transduced cells were treated with the HS-degradative enzyme heparitinase I and the heparin-degradative enzyme heparinase. Almost all of the HS chains detected by the MAb F58-10E4 on K4R/SDC1 cells were trimmed by heparitinase I treatment (Fig. 4B). The susceptibility of the heparitinase I-treated K4R/SDC1 cells was reduced to about one-tenth of that of the untreated cells (Fig. 4C). Although the susceptibilities to the pseudotyped virus were similarly reduced by heparinase treatment (Fig. 4C), most of the HS chains detected by the MAb F58-10E4 remained on the K4R/SDC1 cells (Fig. 4B). These results indicate that HS chains, which remained on the cell surface after heparinase treatment, did not support HTLV-1 infection of K4R/SDC1 cells. In other words, the reduced region of HS chains, which could not be detected by the MAb F58-10E4, played a critical role in increasing the susceptibility to HTLV-1. Heparinase cleaves heparin or highly sulfated heparin-like structural regions of HS chains at the alpha-glucosaminide linkage to 2-O-sulfo-l-iduronic acid, in which 2-deoxy-2-sulfoamino-(6-O-sulfo)-d-glucose participates (37). Therefore, there are two possible explanations for this phenomenon: (i) highly sulfated heparin-like structure regions of HS chains digested by heparinase exist on only a few HS chains, or (ii) such regions exist on only the terminal region of HS chains, which is distant from the core protein. In support of the latter explanation, it was recently reported that there are highly sulfated structures in the distal ends of HS chains (61, 68). Thus, we posited that HTLV-1 interacted with these heparinase-sensitive heparin-like regions, which are located at the distal, nonreducing end of HS in K4R/SDC1 cells.
Fig 4.
Cell surface expression of HS detected by anti-HS MAb F58-10E4 did not affect HTLV-1 pseudotype infection. (A) Cell surface HS chains in K4R cells transduced with the SDC1, SDC2, SDC3, SDC4, or GPC1 core protein gene were stained with F58-10E4, treated with FITC-conjugated anti-mouse IgG, and analyzed by FCM. (B) Cell surface HS was examined by treating cells with buffer, heparitinase I (10 mU/ml), or heparinase (20 mU/ml) were stained with F58-10E4 and FITC-conjugated anti-mouse IgG and then analyzed by FCM. (C) K4R/SDC1 cells treated with heparitinase I or heparinase at the indicated concentrations were seeded into 96-well plates (4 × 104 cells/well) before inoculation with VSVΔG*(HTLV-12M) or VSVΔG*-G. One day after inoculation, the GFP-positive cells were counted under a fluorescence microscope. Three independent experiments were done, and the data are from one representative experiment of the three.
Formula for expressing the relationship between cell surface expression of HS and HTLV-1 susceptibility.
To examine how the expressed form of the HS chains influences the susceptibilities of the cells to HTLV-1 infection, we established several K4R/SDC1 cell clones and examined their susceptibilities to HTLV-1 pseudotypes in relation to HS chain contents. To define the contents of the HS chains on the cells, the total amount and the number of HS chains were detected separately by FCM using two different MAbs, F58-10E4 and F69-3G10, respectively.
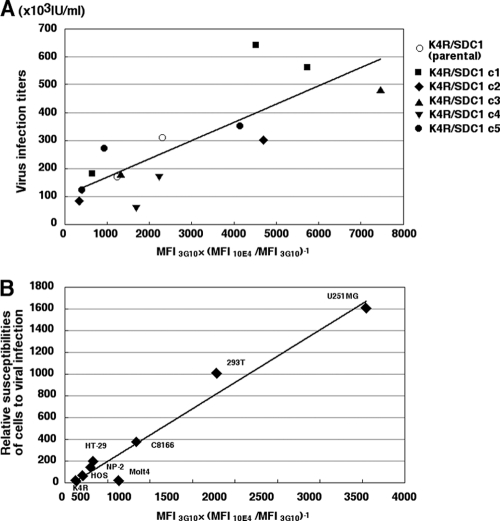
The total amount of expressed HS chains (the net level of HS per cell) was assessed by using the MAb F58-10E4, which is expected to trace the mass of HS chains (4), and the results were expressed as the mean fluorescence intensity (MFI10E4). Initially, we noticed that the cell surface expression of HS in each clone and parental K4R/SDC1 cells fluctuated depending on the test occasion (Table 1). Given that HS expression has been reported to vary in Molt-4 cells in relation to the cell growth (27), the observed fluctuation of HS expression in each clone might reflect the subtle differences in culture conditions. We repeated the experiments at least twice for each clone and found that the MFI10E4 of the K4R/SDC1 clones moderately and negatively correlated with susceptibilities to VSVΔG*(HTLV-12M) (r = −0.562; P = 0.036) (Table 1).
Table 1.
Susceptibility of K4R/SDC1 cell clones to the HTLV-1 pseudotype and their HS chain content
| Expta | Clone | Infectious titer of pseudotype virus (IU/ml) | MFI determined by FCM |
MFI3G10 × (MFI10E4/MFI3G10)−1 | ||
|---|---|---|---|---|---|---|
| MFI10E4 | MFI3G10 | MFI10E4/MFI3G10 | ||||
| 1 | K4R/SDC1 (parental) | 310,000 | 18 | 204 | 0.088 | 2,312 |
| 3 | K4R/SDC1 (parental) | 170,000 | 53 | 256 | 0.207 | 1.237 |
| 1 | Clone 1 | 640,000 | 29 | 362 | 0.080 | 4,519 |
| 2 | Clone 1 | 560,000 | 28 | 401 | 0.070 | 5,743 |
| 4 | Clone 1 | 180,000 | 78 | 227 | 0.344 | 661 |
| 1 | Clone 2 | 300,000 | 39 | 429 | 0.091 | 4,719 |
| 3 | Clone 2 | 81,000 | 48 | 135 | 0.356 | 380 |
| 1 | Clone 3 | 480,000 | 21 | 396 | 0.053 | 7,467 |
| 3 | Clone 3 | 180,000 | 39 | 226 | 0.173 | 1,310 |
| 3 | Clone 4 | 62,000 | 37 | 251 | 0.147 | 1,703 |
| 4 | Clone 4 | 170,000 | 72 | 401 | 0.180 | 2,233 |
| 2 | Clone 5 | 350,000 | 25 | 322 | 0.078 | 4,147 |
| 3 | Clone 5 | 270,000 | 69 | 258 | 0.267 | 965 |
| 4 | Clone 5 | 120,000 | 59 | 159 | 0.371 | 428 |
| rb | −0.562 | 0.650 | −0.690 | 0.816 | ||
| P | 0.036 | 0.012 | 0.0062 | 0.00037 | ||
Experiments 1 to 4 were carried out with the same stock of VSVΔG*(HTLV-12 M).
Coefficient of correlation relative to the infectious titer of the HTLV-I pseudotype.
Next, the number of HS chains was assessed by using MAb F69-3G10, which specifically recognizes the HS neo-epitopes (ΔHS) generated at the terminal stubs of HS in heparitinase I-treated cells. Since HS chains are unbranched disaccharide polymers, and heparitinase I cuts the α-N-acetyl-d-glucosaminidic linkage in HS chains of HSPGs, one ΔHS epitope was generated at the stub of each HS chain (43). Thus, F69-3G10 staining was the representative of the number of HS chains expressed on the cell surface. We observed a moderate correlation between the number of HS chains deduced by MFI3G10 on the heparitinase I-treated K4R/SDC1 cell clones and the susceptibilities to VSVΔG*(HTLV-12M) (r = 0.650; P = 0.012) (Table 1). We assumed that the value we calculated by dividing the total mass of HS chains expressed (MFI10E4) by the total number of HS chains (MFI3G10) would reflect the mean length of the HS chains on the cells. The value of MFI10E4/MFI3G10 in K4R/SDC1 clones showed a significant negative correlation with the susceptibilities to VSVΔG*(HTLV-12M) (r = −0.690; P < 0.01) (Table 1), suggesting that short HS chains might contribute to the more efficient entry of the HTLV-1 pseudotype. Furthermore, an index for short HS chains could be derived by the following formula: number of HS chains (MFI3G10) × reciprocal length of HS chains (MFI10E4/MFI3G10). The value of MFI3G10 × (MFI10E4/MFI3G10)−1 in K4R/SDC1 clones significantly correlated with the susceptibility to the HTLV-1 pseudotype (r = 0.816; P < 0.001) (Table 1 and Fig. 5A). Next, we examined whether the susceptibility to the HTLV-1 pseudotype in 8 different human cell lines correlated with the following parameters: number of SDC1 molecules (MFIB4), amount of HS chains, number of HS chains, mean length of HS chains, and short-HS-chain index, deduced by the following formula: MFI3G10 × (MFI10E4/MFI3G10)−1. We found that the values of MFIB4, MFI10E4, MFI3G10 andMFI10E4/MFI3G10 for each cell line did not significantly correlate with susceptibility to the HTLV-1 pseudotype, whereas the value of MFI3G10 × (MFI10E4/MFI3G10)−1 did significantly correlate (r = 0.98, P < 0.0001) (Table 2 and Fig. 5B). These results suggest that the amount of HS chains or the number of HS chains alone did not determine the cellular susceptibility to the HTLV-1 pseudotype.
Fig 5.
The linear relationship between the susceptibilities to HTLV-1 pseudotype infection and short HS chain index [MFIΔHS × (HFIHS/MFIΔHS)−1]. (A) Susceptibilities of K4R/SDC1 parental cells and derived clones to the VSVΔG*(HTLV-12M) were plotted against short-HS-chain indexes (Table 1). (B) Relative susceptibilities of human cell lines to the VSVΔG*(HTLV-1MEL5) were plotted against short-HS-chain indexes (Table 2). The lines show correlation.
Table 2.
Relative susceptibility of human cell lines to the HTLV-1 pseudotype and their HS chain content
| Cell line | Relative susceptibility to HTLV-1 pseudotypea | MFI determined by FCM |
MFI10E4/MFI3G10 | MFI3G10 × (MFI10E4/MFI3G10)−1 | ||
|---|---|---|---|---|---|---|
| MFIB4 | MFI10E4 | MFI3G10 | ||||
| U251MG | 1,600 | 150 | 6.5 | 152 | 0.043 | 3,554 |
| 293T | 1,000 | 25 | 21 | 194 | 0.108 | 1792 |
| C8166 | 370 | 210 | 38 | 180 | 0.211 | 853 |
| HT-29 | 190 | 157 | 167 | 241 | 0.693 | 348 |
| NP-2 | 130 | 28 | 77 | 153 | 0.503 | 304 |
| HOS | 53 | 26 | 28 | 78 | 0.359 | 217 |
| Molt4 | 2.3 | 6 | 3 | 44 | 0.068 | 645 |
| K4R | 1.0 | 0.1 | 1.5 | 14 | 0.107 | 131 |
| rb | 0.37 | −0.24 | 0.41 | −0.42 | 0.98 | |
| P | 0.36 | 0.57 | 0.31 | 0.30 | <0.0001 | |
Relative susceptibilities of cells to VSVΔG*(HTLV-1MEL5).
Coefficient of correlation relative to the relative susceptibility of the HTLV-I pseudotype.
DISCUSSION
In the present study, we first examined the susceptibilities of different cell types by using highly infectious VSV pseudotypes harboring the Env protein of HTLV-1 and found that there was no significant correlation between the cellular susceptibility to HTLV-1 infection and the expression of GLUT1, HS, or NRP-1, which have been identified as the main cellular factors for binding and entry of HTLV-1 (13, 39, 48). To investigate whether other cellular factors were responsible for HTLV-1 susceptibility, we carried out expression cloning to identify cellular molecules that are associated with HTLV-1 susceptibility, other than GLUT1, HSPG, and NRP-1. Consequently, the cell surface HSPG core proteins SDC1 and SDC2 were identified as molecules that are responsible for HTLV-1 susceptibility.
Enhanced susceptibilities of K562 cells transduced with HSPG core genes were also previously observed in human immunodeficiency virus type 1 and human papillomavirus type 11 (54, 57). In these experiments, transduction of SDC1 is more effective in increasing the susceptibility of cells to viral infection than that of GPC1, and Shafti-Keramat and colleagues observed a correlation between HS expression and the relative expression level of HSPGs (54, 57). Although we observed an enhanced susceptibility of K4R cells transduced with HSPG core genes to cell-free HTLV-1 infection, the correlation between various expression levels of cell surface HS and susceptibilities to HTLV-1 among other cell lines (Fig. 1C) remained unclear. We believe that the discrepancy of the HTLV-1 cell tropism may be explained in the context of HS expression, since there is considerable variation in the structure of HS on the cell surface, including differences in sugar components, patterns of sulfation and HS chain length (7, 25, 33). As a clue to address this issue, we found that K4R/SDC1 cells treated with the heparin-degradative enzyme heparinase showed reduced HTLV-1 susceptibility without significantly alteration of the expression of HS chains. This finding indicated that most of the HS chains detected by MAb F58-10E4 were not responsible for HTLV-1 susceptibility.
To investigate in-depth how HS expression affects HTLV-1 susceptibility, we established the K4R/SDC1 clones expressing HS chains at various levels and examined their susceptibilities to the HTLV-1 pseudotype. Our findings suggest that the number of HS chains, but not their mass, is one of the factors affecting HTLV-1 susceptibility. To clarify why the mass of HS chains did not affect the susceptibility of HTLV-1 infection, we investigated the HS chain length as one of the parameters reflecting the amount of HS. However, it was difficult to accurately determine the HS chain length in various compositions of HS chains; therefore, we calculated the value of the mean length of HS chains by dividing the mass by the number of HS chains. This value, represented by MFI10E4/MFI3G10, was negatively correlated with the HTLV-1 susceptibility of each K4R/SDC1 cell clone. Next, we considered that shorter HS chains might contribute to the efficiency of HTLV-1 entry, and that the number of shorter HS chains may be important for HTLV-1 susceptibility. The value obtained by multiplying the number of HS chains (MFI3G10) by the inverse mean length of HS chains (MFI10E4/MFI3G10) significantly correlated with the HTLV-1 susceptibility in not only K4R/SDC1 clones but also in various human cell lines. This short-HS-chain index, given by MFI3G10 × (MFI10E4/MFI3G10)−1, will increase when the cells express larger numbers of shorter HS chains, whereas the value will decrease when the cells express fewer and longer HS chains. This observation indicates that human cell lines expressing not only larger numbers of HS chains but also shorter lengths of HS chains were more susceptible to HTLV-1 infection.
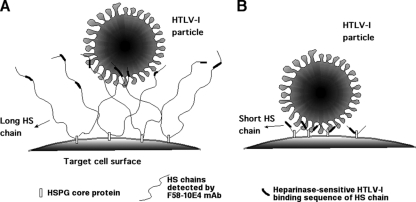
Although we determined only the relative length in this study, the average length of HS chains has been reported to be between 50 and 200 disaccharide repeats, corresponding to 40 to 160 nm in length (64). These HS chains are assumed to adopt extended structures when they are in aqueous solution because their extensive sulfation makes them hydrophilic (23). From this point of view, our findings provide a possible model to explain the difference in HTLV-1 susceptibility between cells expressing different-length HS chains. HTLV-1 particles may get closer to the target cell surface when they bind to heparin-like regions in short HS chains (Fig. 6). In other words, the high expression level of HS chains that are detected by the MAb F58-10E4 impedes the entry of HTLV-1. Recently, Jastrebova and colleagues reported that contiguous sulfated domains were the most abundant in the shortest HS chains and that short HS chains promoted fibroblast growth factor 2 (FGF2) cellular signaling more efficiently than longer HS chains by the formation of ternary complexes (FGF2-HS-FGF receptor) (25). It has been reported that HSPGs and NRP-1 function cooperatively during the initial binding of HTLV-1 to the cell surface (35) and that heparin induces the dimerization of NRP-1 (66), increasing ligand binding to NRP-1. These shorter HS chains might be able to produce receptor complexes (HTLV-1 Env-HS-NRP-1-GLUT1) more efficiently than that of longer HS chains by attracting the HTLV-1 particles to the target cell surface.
Fig 6.
Hypothetical model showing interactions of HTLV-1 particles and cell surface HSPGs bearing long- or short-chain HS. (A) Cell lines expressing longer HS chains, such as NP-2 cells; (B) cell lines expressing shorter HS chains, such as U251MG cells.
In this study, we observed that K4R cells expressing HSPGs aggregated in cultures, suggesting that the cell surface expression of HSPGs creates an environment to mediate the cell-to-cell transmission of HTLV-1 particles trapped on the surface of HTLV-1 producing cells. Intriguingly, the increased cell-cell adhesion of target cells induced by HSPGs does not augment membrane fusion with HTLV-1-producing cells. These results suggest that cell surface expression of HSPGs promotes the target cells to receive the HTLV-1 particles without cell fusion. In fact, it has been reported that HS chain expression decreases HTLV-1 Env-mediated syncytium formation (48). Because fused cells are likely to die in vitro, HSPGs may promote the survival of HTLV-1 infected cells.
In conclusion, our findings suggest that the interactions of HTLV-1 with SDCs bearing short HS chains containing heparin-like structures are important for efficient HTLV-1 infection. The combination of cell surface HS chain structure and the number of HS chains (the number of SDC core proteins) is one of the key factors affecting the cell tropism of HTLV-1. Future experiments are needed to correlate the differences in HS expression in target cells of HTLV-1, such as primary T cells and cells in other target tissues, with the cellular tropism of HTLV-1 in vivo. Nonetheless, the findings obtained in this study may provide the basis for developing effective therapies against HTLV-I infection and an index for assessing the prognosis of disease associated with HTLV-1 infections.
ACKNOWLEDGMENTS
We thank M. A. Whitt for kindly supplying the VSV G-expressing pCAGGS/VSV-G plasmid and VSVΔG*G; G. Nolan (Stanford University, Stanford, CA) and T. Kitamura (Institute of Medical Science, The University of Tokyo, Tokyo, Japan) for kindly providing the Phoenix-A and Plat-E retrovirus-packaging cell lines, respectively; Y. Tanaka (Division of Immunology, Faculty of Medicine, University of the Ryukyus, Okinawa, Japan) for kindly supplying the anti-HTLV-1 gp46 MAb, LAT-27; and S. Takase-Yoden (Faculty of Engineering and Graduate School of Engineering, Soka University, Tokyo, Japan), T. Miyazawa and P. Gee (Institute for Virus Research, Kyoto University, Kyoto, Japan), R. Watanabe (Department of Veterinary Medicine, Yamaguchi University, Yamaguchi, Japan), M. Saha (University Health Network, Toronto, Canada), and J. Komano (AIDS Research Center National Institute of Infectious Diseases, Tokyo, Japan) for their suggestions and critical reading of the manuscript. We also thank M. Nakazawa and A. Nakamura (Graduate School of Medicine, Gunma University, Gunma, Japan) for their assistance with the statistical analyses.
This work was supported in part by grants-in-aid from the Japanese Society for the Promotion of Science, the Japan Health Sciences Foundation, and CREST.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Akagi T, Shishido T, Murata K, Hanafusa H. 2000. v-Crk activates the phosphoinositide 3-kinase/AKT pathway in transformation. Proc. Natl. Acad. Sci. U. S. A. 97:7290–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bigner DD, et al. 1981. Heterogeneity of Genotypic and phenotypic characteristics of fifteen permanent cell lines derived from human gliomas. J. Neuropathol. Exp. Neurol. 40:201–229 [DOI] [PubMed] [Google Scholar]
- 3. Coskun AK, Sutton RE. 2005. Expression of glucose transporter 1 confers susceptibility to human T-cell leukemia virus envelope-mediated fusion. J. Virol. 79:4150–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. David G, Bai XM, Van der Schueren B, Cassiman JJ, Van den Berghe H. 1992. Developmental changes in heparan sulfate expression: in situ detection with mAbs. J. Cell Biol. 119:961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. David G, et al. 1990. Molecular cloning of a phosphatidylinositol-anchored membrane heparan sulfate proteoglycan from human lung fibroblasts. J. Cell Biol. 111:3165–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Revel T, et al. 1993. In vitro infection of human macrophages with human T-cell leukemia virus type 1. Blood 81:1598–1606 [PubMed] [Google Scholar]
- 7. Esko JD. 1991. Genetic analysis of proteoglycan structure, function and metabolism. Curr. Opin. Cell Biol. 3:805–816 [DOI] [PubMed] [Google Scholar]
- 8. Fischinger PJ, Peebles PT, Nomura S, Haapala DK. 1973. Isolation of RD-114-like oncornavirus from a cat cell line. J. Virol. 11:978–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gallagher JT, Turnbull JE, Lyon M. 1990. Heparan sulphate proteoglycans. Biochem. Soc. Trans. 18:207–209 [DOI] [PubMed] [Google Scholar]
- 10. Gessain A, et al. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407–410 [DOI] [PubMed] [Google Scholar]
- 11. Gessain A, Boeri E, Yanagihara R, Gallo RC, Franchini G. 1993. Complete nucleotide sequence of a highly divergent human T-cell leukemia (lymphotropic) virus type I (HTLV-I) variant from Melanesia: genetic and phylogenetic relationship to HTLV-I strains from other geographical regions. J. Virol. 67:1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ghez D, Lepelletier Y, Jones KS, Pique C, Hermine O. 2010. Current concepts regarding the HTLV-1 receptor complex. Retrovirology 7:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghez D, et al. 2006. Neuropilin-1 is involved in human T-cell lymphotropic virus type 1 entry. J. Virol. 80:6844–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graham FL, et al. 1975. Studies on in vitro transformation by DNA and DNA fragments of human adenoviruses and simian virus 40. Cold Spring Harbor Symp. Quant. Biol. 39:637–650 [DOI] [PubMed] [Google Scholar]
- 15. Haraguchi Y, et al. 1994. Detection of neutralizing antibodies against human T-cell leukemia virus type 1 using a cell-free infection system and polymerase chain reaction. Int. J. Cancer 59:416–421 [DOI] [PubMed] [Google Scholar]
- 16. Hinuma Y, et al. 1981. Adult T-cell leukemia: antigen in an ATL cell line and detection of antibodies to the antigen in human sera. Proc. Natl. Acad. Sci. U. S. A. 78:6476–6480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho DD, Rota TR, Hirsch MS. 1984. Infection of human endothelial cells by human T-lymphotropic virus type I. Proc. Natl. Acad. Sci. U. S. A. 81:7588–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoshino H, et al. 1983. Establishment and characterization of 10 cell lines derived from patients with adult T-cell leukemia. Proc. Natl. Acad. Sci. U. S. A. 80:6061–6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoshino H, Nakamura T, Tanaka Y, Miyoshi I, Yanagihara R. 1993. Functional conservation of the neutralizing domains on the external envelope glycoprotein of cosmopolitan and melanesian strains of human T cell leukemia/lymphoma virus type I. J. Infect. Dis. 168:1368–1373 [DOI] [PubMed] [Google Scholar]
- 20. Hoshino H, Shimoyama M, Miwa M, Sugimura T. 1983. Detection of lymphocytes producing a human retrovirus associated with adult T-cell leukemia by syncytia induction assay. Proc. Natl. Acad. Sci. U. S. A. 80:7337–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoxie JA, Matthews DM, Cines DB. 1984. Infection of human endothelial cells by human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. U. S. A. 81:7591–7595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Igakura T, et al. 2003. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science 299:1713–1716 [DOI] [PubMed] [Google Scholar]
- 23. Imberty A, Lortat-Jacob H, Perez S. 2007. Structural view of glycosaminoglycan-protein interactions. Carbohydr. Res. 342:430–439 [DOI] [PubMed] [Google Scholar]
- 24. Jacobson S, Raine CS, Mingioli ES, McFarlin DE. 1988. Isolation of an HTLV-1-like retrovirus from patients with tropical spastic paraparesis. Nature 331:540–543 [DOI] [PubMed] [Google Scholar]
- 25. Jastrebova N, Vanwildemeersch M, Lindahl U, Spillmann D. 2010. Heparan sulfate domain organization and sulfation modulate FGF-induced cell signaling. J. Biol. Chem. 285:26842–26851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin Q, Agrawal L, VanHorn-Ali Z, Alkhatib G. 2006. Infection of CD4+ T lymphocytes by the human T cell leukemia virus type 1 is mediated by the glucose transporter GLUT-1: evidence using antibodies specific to the receptor's large extracellular domain. Virology 349:184–196 [DOI] [PubMed] [Google Scholar]
- 27. Jones KS, Petrow-Sadowski C, Bertolette DC, Huang Y, Ruscetti FW. 2005. Heparan sulfate proteoglycans mediate attachment and entry of human T-cell leukemia virus type 1 virions into CD4+ T cells. J. Virol. 79:12692–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kikukawa R, et al. 1986. Differential susceptibility to the acquired immunodeficiency syndrome retrovirus in cloned cells of human leukemic T-cell line Molt-4. J. Virol. 57:1159–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim CW, Goldberger OA, Gallo RL, Bernfield M. 1994. Members of the syndecan family of heparan sulfate proteoglycans are expressed in distinct cell-, tissue-, and development-specific patterns. Mol. Biol. Cell 5:797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a. Kinet S, et al. 2007. Isolated receptor binding domains of HTLV-1 and HTLV-2 envelopes bind Glut-1 on activated CD4+ and CD8+ T cells. Retrovirology 4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kinsella TM, Nolan GP. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405–1413 [DOI] [PubMed] [Google Scholar]
- 32. Kitamura T, et al. 1995. Efficient screening of retroviral cDNA expression libraries. Proc. Natl. Acad. Sci. U. S. A. 92:9146–9150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kjellen L, Lindahl U. 1991. Proteoglycans: structures and interactions. Annu. Rev. Biochem. 60:443–475 [DOI] [PubMed] [Google Scholar]
- 34. Koyanagi Y, et al. 1993. In vivo infection of human T-cell leukemia virus type I in non-T cells. Virology 196:25–33 [DOI] [PubMed] [Google Scholar]
- 35. Lambert S, et al. 2009. HTLV-1 uses HSPG and neuropilin-1 for entry by molecular mimicry of VEGF165. Blood 113:5176–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Langford JK, Stanley MJ, Cao D, Sanderson RD. 1998. Multiple heparan sulfate chains are required for optimal syndecan-1 function. J. Biol. Chem. 273:29965–29971 [DOI] [PubMed] [Google Scholar]
- 37. Linker A, Hovingh P. 1984. Structural studies on heparin. Tetrasaccharides obtained by heparinase degradation. Carbohydr. Res. 127:75–94 [DOI] [PubMed] [Google Scholar]
- 38. Majorovits E, et al. 2008. Human T-lymphotropic virus-1 visualized at the virological synapse by electron tomography. PLoS One 3:e2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manel N, et al. 2003. The ubiquitous glucose transporter GLUT-1 is a receptor for HTLV. Cell 115:449–459 [DOI] [PubMed] [Google Scholar]
- 40. McAllister RM, et al. 1971. Cultivation in vitro of cells derived from a human osteosarcoma. Cancer 27:397–402 [DOI] [PubMed] [Google Scholar]
- 41. Miyoshi I, Kubonishi I, Yoshimoto S, Shiraishi Y. 1981. A T-cell line derived from normal human cord leukocytes by co-culturing with human leukemic T-cells. Gann 72:978–981 [PubMed] [Google Scholar]
- 42. Morita S, Kojima T, Kitamura T. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066 [DOI] [PubMed] [Google Scholar]
- 43. Nackaerts K, et al. 1997. Heparan sulfate proteoglycan expression in human lung-cancer cells. Int. J. Cancer 74:335–345 [DOI] [PubMed] [Google Scholar]
- 44. Onishi M, et al. 1996. Applications of retrovirus-mediated expression cloning. Exp. Hematol. 24:324–329 [PubMed] [Google Scholar]
- 45. Osame M, et al. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031–1032 [DOI] [PubMed] [Google Scholar]
- 46. Pais-Correia AM, et al. 2009. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat. Med. 16:83–89 [DOI] [PubMed] [Google Scholar]
- 47. Pear WS, Nolan GP, Scott ML, Baltimore D. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U. S. A. 90:8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pinon JD, et al. 2003. Human T-cell leukemia virus type 1 envelope glycoprotein gp46 interacts with cell surface heparan sulfate proteoglycans. J. Virol. 77:9922–9930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poiesz BJ, et al. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 77:7415–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Popovic M, et al. 1983. Isolation and transmission of human retrovirus (human T-cell leukemia virus). Science 219:856–859 [DOI] [PubMed] [Google Scholar]
- 51. Richardson JH, Edwards AJ, Cruickshank JK, Rudge P, Dalgleish AG. 1990. In vivo cellular tropism of human T-cell leukemia virus type 1. J. Virol. 64:5682–5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saiki RK, et al. 1985. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354 [DOI] [PubMed] [Google Scholar]
- 53. Salahuddin SZ, et al. 1983. Restricted expression of human T-cell leukemia–lymphoma virus (HTLV) in transformed human umbilical cord blood lymphocytes. Virology 129:51–64 [DOI] [PubMed] [Google Scholar]
- 54. Saphire AC, Bobardt MD, Zhang Z, David G, Gallay PA. 2001. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 75:9187–9200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schultz RM, Woods WA, Chirigos MA. 1975. Detection in colorectal carcinoma patients of antibody cytotoxic to established cell strains derived from carcinoma of the human colon and rectum. Int. J. Cancer 16:16–23 [DOI] [PubMed] [Google Scholar]
- 56. Sebestyen A, et al. 2000. Syndecan-1-dependent homotypic cell adhesion in HT58 lymphoma cells. Tumour Biol. 21:349–357 [DOI] [PubMed] [Google Scholar]
- 57. Shafti-Keramat S, et al. 2003. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 77:13125–13135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Soda Y, et al. 1999. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem. Biophys. Res. Commun. 258:313–321 [DOI] [PubMed] [Google Scholar]
- 59. Sommerfelt MA, Weiss RA. 1990. Receptor interference groups of 20 retroviruses plating on human cells. Virology 176:58–69 [DOI] [PubMed] [Google Scholar]
- 60. Stanley MJ, Liebersbach BF, Liu W, Anhalt DJ, Sanderson RD. 1995. Heparan sulfate-mediated cell aggregation. Syndecans-1 and -4 mediate intercellular adhesion following their transfection into human B lymphoid cells. J. Biol. Chem. 270:5077–5083 [DOI] [PubMed] [Google Scholar]
- 61. Staples GO, Shi X, Zaia J. 2010. Extended N-sulfated domains reside at the nonreducing end of heparan sulfate chains. J. Biol. Chem. 285:18336–18343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Takada A, et al. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 94:14764–14769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tanaka Y, Zeng L, Shiraki H, Shida H, Tozawa H. 1991. Identification of a neutralization epitope on the envelope gp46 antigen of human T cell leukemia virus type I and induction of neutralizing antibody by peptide immunization. J. Immunol. 147:354–360 [PubMed] [Google Scholar]
- 64. Turnbull J, Powell A, Guimond S. 2001. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 11:75–82 [DOI] [PubMed] [Google Scholar]
- 65. van den Born J, et al. 2005. Novel heparan sulfate structures revealed by monoclonal antibodies. J. Biol. Chem. 280:20516–20523 [DOI] [PubMed] [Google Scholar]
- 66. Vander Kooi CW, et al. 2007. Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U. S. A. 104:6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Van Der Maaten MJ, Miller JM. 1975. Replication of bovine leukemia virus in monolayer cell cultures. Bibl. Haematol. 1975:360–362 [DOI] [PubMed] [Google Scholar]
- 68. Wu ZL, Lech M. 2005. Characterizing the non-reducing end structure of heparan sulfate. J. Biol. Chem. 280:33749–33755 [DOI] [PubMed] [Google Scholar]
- 69. Yamada M, Watabe K, Saida T, Kim SU. 1991. Increased susceptibility of human fetal astrocytes to human T-lymphotropic virus type I in culture. J. Neuropathol. Exp. Neurol. 50:97–107 [DOI] [PubMed] [Google Scholar]