Abstract
Influenza A virus infection is a persistent threat to public health worldwide due to its ability to evade immune surveillance through rapid genetic drift and shift. Current vaccines against influenza A virus provide immunity to viral isolates that are similar to vaccine strains. High-affinity neutralizing antibodies against conserved epitopes could provide immunity to diverse influenza virus strains and protection against future pandemic viruses. In this study, by using a highly sensitive H5N1 pseudotype-based neutralization assay to screen human monoclonal antibodies produced by memory B cells from an H5N1-infected individual and molecular cloning techniques, we developed three fully human monoclonal antibodies. Among them, antibody 65C6 exhibited potent neutralization activity against all H5 clades and subclades except for subclade 7.2 and prophylactic and therapeutic efficacy against highly pathogenic avian influenza H5N1 viruses in mice. Studies on hemagglutinin (HA)-antibody complexes by electron microscopy and epitope mapping indicate that antibody 65C6 binds to a conformational epitope comprising amino acid residues at positions 118, 121, 161, 164, and 167 (according to mature H5 numbering) on the tip of the membrane-distal globular domain of HA. Thus, we conclude that antibody 65C6 recognizes a neutralization epitope in the globular head of HA that is conserved among almost all divergent H5N1 influenza stains.
INTRODUCTION
Since 1997, highly pathogenic avian influenza (HPAI) H5N1 virus has infected over 500 million poultry and an increasing number of humans in Asia, Europe, and Africa. As of 10 October 2011, 566 human H5N1 infections have been confirmed, resulting in 332 deaths (http://www.who.int/csr/disease/avian_influenza/country/en/). Although so far all human cases have most likely been transmitted from avian species, continuous reassortment and adaptation may evolve new H5N1 strains capable of human-to-human transmission. A wide spread of such new viruses could cause significant morbidity and mortality, since humans are immunologically naïve to H5N1 viruses.
In humans, HPAI H5N1 virus infection was characterized by severe pneumonia, lymphopenia, hypercytokinemia, and high viral loads in the respiratory tract (1, 6, 10, 26, 42). Viruses were often cultured from cerebrospinal fluid, fecal, throat, and serum specimens (6). Besides supportive care, current treatment relies mainly on antiviral drugs. However, some H5N1 isolates are resistant to the ion channel blockers amantadine and rimantadine (19). Although neuraminidase inhibitors such as oseltamavir and zanamavir are effective for treatment of seasonal influenza, it is less clear whether they are effective for treatment of H5N1 viruses. In animal studies, efficacy of neuraminidase inhibitors is demonstrated only when the drugs are given before or soon after the infection (26). In addition, oseltamavir-resistant H5N1 viruses have also been reported (7). Thus, alternative treatments that can rapidly control H5N1 virus dissemination in humans after symptoms emerge are urgently needed.
Antibody-based treatments using polyclonal antibodies and monoclonal antibodies (MAbs) have been effectively used prophylactically and therapeutically against many viral diseases, such as those caused by hepatitis A virus, hepatitis B virus, cytomegalovirus, rabies virus, varicella virus, and respiratory syncytial virus infection (31). In influenza, passive immunization by vertical acquisition of specific antibodies is also associated with influenza virus immunity in early infancy in humans (27, 30, 37, 38). Transfusion of human blood products from patients who recovered from the 1918 “Spanish flu” resulted in a 50% reduction in influenza mortality during the pandemic (21). Transfusion of convalescent-phase plasma from a patient recovered from H5N1 infection resulted in a dramatic reduction of viral loads and complete recovery (50). In addition, passive immunization using mouse, ferret, equine, and human antibodies effectively prevents and treats influenza infection in mice (3–5, 8, 9, 12, 18, 20, 23–25, 29, 32, 33, 35, 36, 39, 44, 48). More recently, Koudstaal et al. reported that a single injection with 15 mg/kg of a human monoclonal antibody resulted in much better prophylactic and therapeutic efficacy against lethal H5N1 and H1N1 challenge in mice than a 5-day treatment with oseltamivir at 10 mg/kg/day (18). Thus, collectively, these observations suggest that passive antibody therapy against influenza viruses is a viable option for the treatment of human cases of influenza infection.
On the basis of hemagglutinin (HA) sequences, 10 clades of H5N1 viruses have emerged in various species since 2000 (34). Among them, clade 2 is divided into 5 subclades, clade 7 is divided into 2 subclades, and subclade 2.3 is further divided into subclades 2.3.1, 2.3.2, 2.3.3, and 2.3.4 (34). So far the HPAI H5N1 viruses isolated from humans fall into clades 0, 1, 2, and 7, and most recent human isolates from China belong to subclade 2.3.4 (11, 34). Increasingly, subclade 2.3.4 is becoming one of the dominant strains in poultry and birds in Southeast and East Asia (16). One study has shown that at least four major antigenic groups exist among circulating human H5N1 isolates (16).
In this study, using a highly sensitive HA and neuraminidase (NA) pseudotype-based neutralization (PN) assay and molecular cloning techniques, we have developed stable Drosophila S2 cell transfectants secreting three fully human monoclonal antibodies from the memory B cells of a convalescent individual who had been infected with an H5N1 virus from subclade 2.3.4 (50). One such antibody, 65C6, exhibited potent neutralization activity against all clades and subclades of H5N1 strains except subclade 7.2 as well as prophylactic and therapeutic efficacy against highly pathogenic avian influenza H5N1 viruses in mice. Studies on HA-antibody complexes by electron microscopy and epitope mapping indicate that antibody 65C6 binds to a conformational epitope comprising amino acid residues at positions 118, 121, 161, 164, and 167 on the tip of the membrane-distal globular domain of HA.
MATERIALS AND METHODS
Ethics statement.
The study protocol for humans was approved by the Ethical Committee of the Shenzhen Donghu Hospital. The patients provided written informed consent for research use of blood samples. All animal experiments were carried out at biosafety level 3 (BSL3) containment facilities complying with the Ethics Committee regulations of the Institut Pasteur, Paris, France, in accordance with EC directive 86/609/CEE and were approved by the Animal Ethics Committee of the Institut Pasteur in Cambodia (permit number VD100820). Before each inoculation or euthanasia procedure, the mice were anesthetized by intraperitoneal (i.p.) injection of pentobarbital sodium, and all efforts were made to minimize suffering.
Human H5N1 influenza case.
The adult blood donor in this study was diagnosed with HPAI H5N1 virus infection and treated with convalescent-phase plasma donated by a previously H5N1-infected and recovered individual in June 2006 at the Shenzhen Donghu Hospital, Shenzhen, China (50). The blood samples were drawn during early convalescence (6 months after illness onset). The plasma was collected, and the peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation. Both plasma and cell samples were stored at −80°C until use (see below).
Animals.
All animal protocols were approved by the Institutional Animal Care and Use Committee at the Pasteur Institute of Cambodia. Female BALB/c mice (Mus musculus) at the age of 6 to 8 weeks were purchased from Charles River Laboratories (L'Arbresle, France) and housed in microisolator cages ventilated under negative pressure with HEPA-filtered air and a 12/12-hour light/dark cycle. Virus challenge studies were conducted in BSL3 facilities at the Pasteur Institute of Cambodia. Before each inoculation or euthanasia procedure, the mice were anesthetized by i.p. injection of pentobarbital sodium (75 mg/kg; Sigma), and all efforts were made to minimize suffering.
Cell lines.
The packaging cell line 293T was maintained in complete Dulbecco modified Eagle medium (DMEM) medium [i.e., high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml),) and streptomycin (100 μg/ml); Invitrogen Life Technologies] containing 0.5 mg/ml of G418. Madin-Darby canine kidney (MDCK) cell lines were maintained in complete DMEM. Drosophila S2 cells were maintained in complete medium [i.e., Express Five serum-free medium (SFM) with 10% FBS, 50 U/ml penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine] at 28°C.
Viruses.
HPAI H5N1 viruses A/Shenzhen/406H/06 and A/Cambodia/P0322095/05 were originally isolated from human patients at the Donghu Hospital in Shenzhen, China, and at the Pasteur Institute of Cambodia, respectively (2, 50). Viruses were propagated in MDCK cells, and virus-containing supernatants were pooled, clarified by centrifugation, and stored in aliquots at −80°C. The 50% tissue culture infective dose (TCID50) was determined by serial titration of viruses in MDCK cells and was calculated by the method of Reed and Muench (28). To determine the 50% mouse lethal doses (MLD50) of the viruses, groups of 5 mice were inoculated intranasally (i.n.) with serial 10-fold dilution of virus. After the inoculation, mice were monitored daily for clinical signs for 14 days. Mice that lost more than 35% of their original body weight were euthanized and counted as dead. The MLD50 was calculated by the method of Reed and Muench (28). All research with HPAI H5N1 viruses was conducted under BSL3 laboratory containment.
Generation of an HA/NA pseudotype panel.
Table S1 in the supplemental material lists a panel of 20 H5 and 1 H1 HAs constructed and used in this study. The H5 panel covers all 10 clades and all 5 subclades of clade 2 of H5 HA. Among H5 HAs, (sub)clades 0, 1, 2.1, 2.2, 2.3, and 7 were derived from human isolates and the rest were derived from avian species. The methods used to generate the codon optimized-H5 and H1 HAs and Flag epitope-tagged N1 NA [A/Thailand/1(KAN)-1/04] (22) and the methods used to produce influenza virus HA and NA pseudotypes and the pseudotype-expressing vesicular stomatitis virus G protein (VSV-G) control were as described before (43).
PN assay.
The pseudotype-based neutralization (PN) assay used to screen neutralizing-antibody activity in convalescent-phase plasma was carried out as described previously (43). Neutralizing-antibody activity in culture supernatants from immortalized B cells and from Drosophila S2 transfectants (see below) was screened by PN assay essentially as described before (43). Briefly, supernatants were incubated with the HA and NA pseudotypes (A/Shenzhen/406H/06) for 1 h at 37°C prior to addition to MDCK cells. After overnight incubation, cells were washed with phosphate-buffered saline (PBS) and cultured in complete DMEM. Relative luciferase activity (RLA) was measured at 48 h by a BrightGlo luciferase assay according to the manufacturer's instruction (Promega). The percentage of inhibition was calculated as (RLA in pseudotypes and medium control − RLA in pseudotypes and culture supernatants)/RLA in pseudotypes and medium control.
To test the neutralization activities of purified human monoclonal antibodies, serially 3-fold-diluted antibodies 65C6, 100F4, and 3C11were incubated with the pseudotypes for 1 h at 37°C prior to addition to MDCK cells. After overnight incubation, cells were washed and cultured in complete DMEM. RLA was measured as described above. The 95% inhibitory concentration (IC95) was calculated as the amount of a given antibody that resulted in 95% reduction of luciferase activity.
HI assays.
Viruses were diluted to 8 HA units and incubated with an equal volume of serially diluted antibody 65C6 at room temperature. An equal volume of 0.5% horse red blood cells was added to the wells, and incubation was continued on a gently rocking plate for 30 min at room temperature. Cell button formation was scored as evidence of hemagglutination inhibition (HI).
Construction of Drosophila S2 cell expression cassettes containing the constant regions of the heavy and light chains of human immunoglobulin.
To facilitate molecular cloning of human monoclonal antibodies, total RNA was isolated from Epstein-Barr virus (EBV)-transformed human B cells and cDNA was generated by reverse transcription. Gene segments encoding the constant regions of the human κ1, λ1, and γ1 chains were PCR amplified using pairs of primers listed in Table S2 in the supplemental material and inserted into the TA vector for sequencing. The correct sequences of the human κ1 and γ1 chains were inserted into the BglII and PmeI sites of the pMT/Bip vector (Invitrogen), and the correct sequence of the humanλ1 chain was inserted into the XhoI and PmeI sites of the pMT/Bip vector. The resulting vector cassettes were designated pMT/Bip/κ1 constant, pMT/Bip/λ1 constant, and pMT/Bip/γ1 constant, respectively.
Generation of stable Drosophila S2 cell transfectants that produce human monoclonal antibodies.
In this study, we did two independent EBV transformation experiments. In the first experiment we screened 8,000 wells in 96-well plates, and in the second experiment we screened an additional 8,000 wells in 96-well plates. In both experiments CD22+ cells were isolated from the aforementioned patient PBMCs with bead-conjugated anti-human CD22 antibody (Miltenyi) as described in the manufacturer's instructions. Isolated CD22+ cells (30 cells per well) were cultured in complete RPMI 1640 supplemented with 10% FBS and immortalized using EBV in the presence of CpG oligonucleotide 2006 and irradiated allogeneic PBMCs as described by Traggiai et al. (41). Culture supernatants were harvested on day 14 and screened for their ability to neutralize H5N1 pseudotype A/Shenzhen/406H/06 by PN assay (see above). In the first experiment, cells in the positive wells were harvested and limiting-dilution assay was performed for 5 cells per well (the first round of subcloning). Cells in the positive wells after the first-round subcloning were harvested, and again limiting dilution assay was performed for 1 cell per well (the second round of subcloning). RNA samples from cells in the positive wells after the second-round subcloning were isolated using the RNeasy Plus minikit (Qiagen Corporation). In the second experiment, cells in the positive wells were harvested and limiting-dilution assay was performed for 5 cells per well (the first round of subcloning). RNA samples from cells in the positive wells after the first round of subcloning were isolated. cDNA was generated by reverse transcription. Variable heavy (VH) and Vκ and Vλ gene segments of human immunoglobulin were then PCR amplified using a set of degenerate primer pairs as described by Tiller et al. (40) (see Table S2 in the supplemental material). The amplified VH, Vκ, and Vλ gene segments were inserted into vector cassettes pMT/Bip/γ1 constant, pMT/Bip/κ1 constant, and pMT/Bip/λ1 constant, respectively, to generate heavy and κ and λ light chain gene libraries (Fig. 1A) (13).
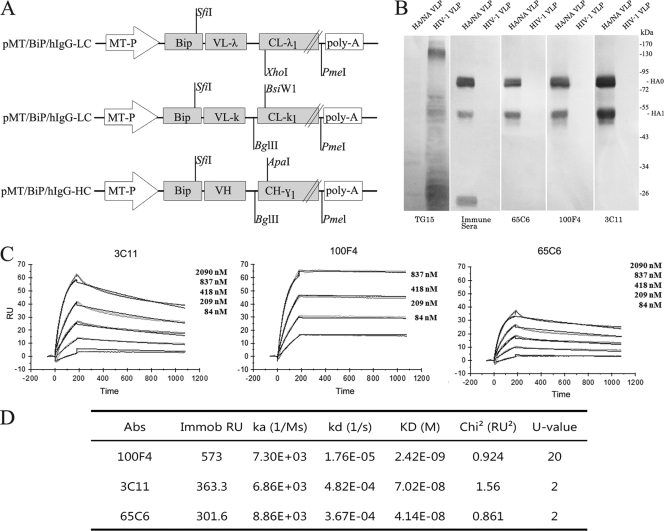
Fig 1.
Characterization of human monoclonal antibodies 65C6, 3C11, and 100F4. (A) Schematic diagram of Drosophila S2 cell expression vectors (pMT/Bip) for the heavy chain of IgG1 and the lambda and kappa light chains of antibody molecules. (B) Western blot analysis of binding specificities of antibodies 65C6, 3C11, and 100F4. TG15 is a control antibody that recognizes an epitope in the HP2 domain of HIV-1 gp41. (C) Antibodies 3C11, 65C6, and 100F4 and irrelevant IgG1 antibody were immobilized on a CM5 sensor chip for surface plasmon resonance (SPR) kinetic binding analysis with the H5 HA proteins. (D) Calculated values of the on-rate constant (ka), off-rate constant (kd), and KD of antibodies 3C11, 65C6, and 100F4.
To identify the correct heavy-chain genes, each heavy-chain gene from the heavy-chain library paired with the κ and λ chain library was cotransfected into Drosophila S2 cells using a calcium phosphate precipitation method. After cells were induced with 5 μM CdCl2 for 72 h, supernatants were harvested to screen neutralization activity. The heavy-chain genes from transfected cells whose supernatants showed the highest neutralization activity were sequenced and used for subsequent cotransfection (see below).
Conversely, to identify the correct light-chain genes for pairing, each light-chain gene from the κ or λ chain library paired with the heavy-chain library was cotransfected into Drosophila S2 cells using the same calcium phosphate precipitation method. After cells were induced with 5 μM CdCl2 for 72 h, supernatants were harvested to screen neutralization activity. The κ and λ chain genes from transfected cells whose supernatants showed the highest neutralization activity were then sequenced and used for subsequent cotransfection (see below).
To generate stable Drosophila S2 cell transfectants, the heavy- and light-chain gene constructs identified above along with selection vector pCoBlast (containing the blasticidin resistance gene) were cotransfected into Drosophila S2 cells to generate stable transfectant cell lines. As a control Drosophila S2 cells were also cotransfected with IgG1 heavy-chain and κ light-chain genes encoding a human monoclonal antibody (TG15) which recognizes the HR2 domain of HIV-1 gp41 (51). After overnight incubation, cells were washed once with PBS and cultured in complete High Five medium (Invitrogen). Seventy-two hours later, the selection marker blasticidin was added at 25 μg/ml, and stable S2 transfectant cell lines were generated in 2 weeks. To test production of human monoclonal antibodies, transfected cells were induced with 5 μM CdCl2 for 3 days and the supernatants were tested for neutralization activity. Stable transfectant clones were generated by limiting-dilution assay. After individual S2 clones were established, culture supernatants were tested for their ability to neutralize H5N1 pseudotypes. S2 clones designated 65C6, 3C11, and 100F4, which exhibited good neutralization activity, were chosen for human monoclonal antibody production (see below). The VH, Vκ, and Vλ gene segments of these S2 clones were analyzed for homology to known human V, D, and J genes using the database at http://www.ncbi.nlm.nih.gov/projects/igblast/.
Production and purification of human monoclonal antibodies by stable S2 clones.
WAVE bioreactor 20/50EHT systems with a WAVEPOD process control unit (GE Healthcare) were used for antibody production (L. Wang et al., unpublished data). Briefly, 150 ml (1 × 106 to 2 × 106 cells per ml) of stable S2 clones 65C6, 3C11, and 100F4 were inoculated into 2-liter cell bags along with 300 ml prewarmed fresh SFM. Two days later, 400 ml fresh SFM was added to the cell bags. The agitation settings initially were 22 rpm (rocking rate) and 8° (angle) and were increased to 26 rpm and 9° on day 3. Oxygen was provided via headspace aeration using room air at a flow rate of 0.15 liter/minute, and the pH was between 6.0 and 6.3 during the cultivation. Six days after the initial culture perfusion was started, the perfusion rate was increased from 0.3 to 1.5 culture volumes (CV)/day to keep the residual glucose concentration at 4 g/liter. Ten days after initial culture, 5 μM cadmium chloride was added to the culture medium to induce antibody production. Five days after the induction, culture supernatants were collected.
Collected supernatants were clarified by centrifugation at 12,000 × g for 10 min at 4°C and filtered through a 0.45-μm filter. All filtered supernatants were concentrated 5-fold using the QuixStand benchtop system with a 50,000 nominal molecular weight cutoff (NMWC) hollow-fiber cartridge (model UFP-50-C-4MA). Concentrated supernatants were once again centrifuged at 12,000 × g for 10 min at 4°C and filtered at 0.45 μm. Finally, samples mixed with 1 mM phenylmethylsulfonyl fluoride (PMSF) were loaded onto a HiTrap protein G HP 5-ml column. The eluted fractions were exchanged using a HiTrap desalting column. All equipment was purchased from GE Healthcare. Antibody concentrations were determined by enzyme-linked immunosorbent assay (ELISA) (see below).
ELISA.
An ELISA kit specifically measuring human IgG was purchased from Mabtech AB (Sweden). First, anti-human IgG antibody (1 μg/ml) in phosphate-buffered saline (PBS) at pH 7.4 was applied to microtiter wells overnight at 4°C. Unbound protein was discarded. The coated plate was washed with PBS and blocked with 0.1% bovine serum albumin (BSA), according to the manufacturer's recommendation. Serially 10-fold diluted culture supernatants collected from S2 transfectant clones or purified human monoclonal antibodies were added to wells along with human antibody standards at concentrations of from 0.1 to 500 ng/ml. After incubation at room temperature for 2 h, the wells were washed 5 times using PBS-Tween 20 (PBST), and alkaline phosphatase (ALP)-conjugated anti-human IgG antibody diluted to 1:1,000 in PBST plus 0.1% BSA was added and incubated for 1 h at room temperature. After this, the wells were washed 5 times with PBS-Tween 20, and the substrate p-nitrophenylphosphate was added for optical density measurement at 405 nm.
Western blotting.
To determine the binding specificities of human monoclonal antibodies, HIV-1 and H5N1 virus-like particle (VLP) samples were incubated for 15 min at 90°C in loading buffer containing SDS and 0.6 M dithiothreitol (DTT) and loaded for 12% SDS-PAGE. Gels were transferred onto polyvinylidene difluoride (PDVF) membranes, and the membranes were blocked with Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% nonfat dry milk for 1 h at room temperature. The membranes were subsequently incubated with 3 ml (0.5 μg/ml) of purified human monoclonal antibodies TG15, 3C11, 100F4, and 65C6 for 2 h at room temperature. After being washed twice with TBST, the membranes were probed with ALP-conjugated goat anti-human IgG antibody (Southern Biotech).
SPR analysis of monoclonal antibodies.
Surface plasmon resonance (SPR) analysis was performed on a Biacore T100 (Biacore AB, Sweden) according to the manufacturer's instructions. Antibodies 3C11, 65C6, and 100F4 and an irrelevant IgG1 antibody were immobilized on a CM5 sensor chip using an amine coupling kit. Serial dilutions (from 2090 nM to 84 nM) of soluble recombinant HA derived from the A/Anhui/05/01 strain were injected at a constant flow rate of 50 μl/min for 180 s at 25°C. Dissociation was facilitated with glycine HCl (pH 2.5) for 900 s, followed by regeneration using HPS-EP (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.005% [vol/vol] surfactant P20) for 600 s. The experiment was repeated twice. The data were acquired and analyzed using Biacore T100 evaluation software (version 3.2).
Characterization of antibody 65C6-HA complexes by negative-stain electron microscopy.
Hemagglutinin (HA) from strain A/Shenzhen/406H/06 (H5N1) was prepared by disrupting purified virus in 1% N,N-dimethyl dodecylamine-N-oxide (LDAO) (Fluka)–10 mM Tris (pH 8.0) for 30 min at 4°C. HA and NA glycoproteins were recovered from the 55,000 rpm (10 min) supernatant, and HA was purified by sucrose gradient centrifugation and ion-exchange chromatography as described before (11a). The generation of HA-antibody immune complexes was performed as described before (46). Briefly, HA was diluted in PBS (pH 7.2) to 50 μg/ml for optimum spreading on carbon support films. Antibody 65C6 was added in increasing amounts until only a few molecules remained unbound to HA. HA-antibody complex suspensions were absorbed to thin carbon films freshly stripped from mica, floated on a 1% (wt/vol) solution of sodium silicotungstate (pH 7.0), and air dried. Micrographs were taken under minimum-dose conditions known from analyses of periodic specimens to preserve detail.
Epitope mapping of 65C6 using yeast display.
Epitope mapping of 65C6 was carried out at two levels, the domain level and the fine-epitope level. To do the domain-level epitope mapping of 65C6, a previously generated combinatorial library of H5 HA fragments displayed on the surface of yeast (Saccharomyces cerevisiae) cells was used as described before (52). Briefly induced yeast cells (106 to 107) were collected by centrifugation (12,000 rpm/s, 1 min), washed once with PBS, and incubated with 500 ng of antibody 65C6 on ice for 1 h. Cells were washed twice with cold PBS and then incubated with phycoerythrin (PE)-labeled anti-human IgG (1:200 dilution) on ice for 45 min. Cells were then washed twice with cold PBS and analyzed by fluorescence-activated cell sorting (FACS) using an Aria II instrument (BD). PE-positive yeast clones were sorted and sequenced.
To do the fine-epitope mapping of 65C6, a random mutagenesis library was constructed using error-prone PCR. The template used for library construction was the HA fragment from position 51 to 260 defined by the above-described domain-level epitope mapping. The PCR products were gel purified and extracted using a QIAquick gel extraction kit (Qiagen). The construction, growth, and expression of the library using yeast surface display were carried out as described before (52). Briefly, induced yeast cells (106 to 107) were collected by centrifugation, washed once with PBS, and incubated with 500 ng of antibody 65C6 and anti-c-myc chicken IgY (1:200 dilution) on ice for about 1 h. Cells were washed twice with cold PBS and then incubated with PE-labeled anti-human IgG (1:200 dilution) and Alexa Fluor 488-labeled goat anti-chicken IgG (1:400 dilution) on ice for 45 min. Cells were then washed twice with cold PBS and analyzed by FACS using an Aria II instrument (BD). PE-negative yeast clones were sorted and sequenced.
Prophylaxis and therapy in mice with antibody 65C6.
To test the prophylactic effect of 65C6, four groups of female BALB/c mice (12 mice per group; 6 to 8 weeks old; average weight, 20 g) were inoculated i.p. with 1 ml PBS containing 15 mg/kg, 5 mg/kg, and 1 mg/kg of 65C6 or 15 mg/kg of control antibody TG15. After 4 h, 6 mice in each group were i.n. inoculated with 50 μl PBS containing 5 MLD50 of HPAI H5N1 A/Shenzhen/406H/06 virus and the other 6 mice in each group were i.n. inoculated with 50 μl PBS containing 5 MLD50 of HPAI H5N1 A/Cambodia/P0322095/05 virus. Mice were monitored daily for survival and weight loss until day 14 postinfection. Mice that lost more than 35% of their initial body weight were euthanized according to the study protocol. To measure the therapeutic effect of human monoclonal antibodies, two groups of female BALB/c mice (24 mice per group; 6 to 8 weeks old; average, weight 20 g) were i.n. inoculated with 50 μl PBS containing 5 MLD50 of HPAI H5N1 A/Shenzhen/406H/06 or A/Cambodia/P0322095/05 virus, respectively. After 24, 48, or 72 h, 6 mice from each group were injected i.p. with 1 ml of control antibody TG15 or antibody 65C6 at 40 mg/kg. Mice were monitored daily for survival and weight loss until day 14 postinfection. Mice that lost more than 35% of their preinfection body weight were euthanized.
Statistical analysis.
The weight and the survival of each individual animal were counted as an individual value for statistical analysis. Kaplan-Meier survival curves were used to measure differences between the different prophylactic and therapeutic groups.
Nucleotide sequence accession numbers.
Antibody sequences have been deposited in GenBank (accession no. JF274048 to JF274053).
RESULTS
Development of human monoclonal antibodies 65C6, 100F4, and 3C11.
A blood sample from a Chinese adult who had recovered from subclade 2.3.4 H5N1 infection (50) was collected at 6 months postinfection. The plasma derived from the blood sample exhibited high neutralizing-antibody titers against both clade 1 and subclade 2.3.4 H5N1 viruses (P. Zhou et al., data not shown). Memory B cells isolated from patient PBMCs were immortalized with EBV in the presence of CpG as described by Traggiai et al. (41). Culture supernatants were screened by HA and NA pseudotype-based neutralization (PN) assay as described before (43). As stated in Materials and Methods, we did two independent EBV transformation experiments. In our first experiment we observed that neutralizing-antibody secretion by EBV-transformed cells was not stable. After more than two rounds of subcloning, neutralization activity in culture supernatants dramatically decreased. Therefore, in the second experiment, after one round of subcloning, total RNA was isolated and VH, Vκ, and Vλ gene segments were RT-PCR amplified with a panel of forward and reverse primers (see Table S2 in the supplemental material) and inserted into Drosophila S2 cell expression vectors containing constant regions of human γ1, κ1, and λ1 gene segments (Fig. 1A). We then performed a pairwise, across-board cotransfection with heavy- and light-chain gene expression vectors into Drosophila S2 cells to identify the heavy- and light-chain pairs required for the generation of neutralizing monoclonal antibodies. From approximately 16,000 EBV-transformed B cell supernatant samples, 6 supernatants exhibited at least 2-log10 neutralization activity. From these positive wells, we isolated RNA, constructed heavy- and light-chain gene libraries, did across-board cotransfection into Drosophila S2 cells, and eventually generated 3 stable Drosophila S2 cell transfectant clones that secrete fully human monoclonal antibodies 65C6, 100F4, and 3C11 (see Materials and Methods for details). Among them, antibody 65C6 was generated from the first experiment, in which two rounds of subcloning were carried out, whereas antibodies 100F4 and 3C11 were generated from the second experiment, in which only one round of subcloning was carried out (see Materials and Methods). Stable Drosophila S2 transfectants that secrete human monoclonal antibody TG15 specific for HIV-1 gp41 were also generated and used as a control. Using the Wave bioreactor system, stable Drosophila S2 cell transfectant clones that secrete these human monoclonal antibodies were grown in perfusion culture. Culture supernatants were harvested, and the human monoclonal antibodies in these culture supernatants were purified. The amino acid sequences of the VH and the VL chains of antibodies 65C6, 100F4, and 3C11 are shown in Fig. S1 in the supplemental material. The VH chains of 65C6, 3C11, and 100F4 belong to 5-a*03, 5-a*03, and 4-61*03, respectively, and the VL chains of 65C6, 3C11, and 100F4 belong to Vκ3D-15*01, Vκ2D-28*01, and Vλ1-40*01, respectively. Compared to germ line sequences, the VH and the Vκ chains of antibody 65C6 contain 11 and 5 mutations, respectively, the VH and the Vκ chains of antibody 100F4 contain 12 and 7 mutations, respectively, and the VH and the Vλ chains of antibody 3C11 contain 14 and 4 mutations, respectively.
Binding specificity and affinity of antibodies 65C6, 100F4, and 3C11.
To test the specific binding of human antibodies, HIV-1, HA, and NA virus-like particles (VLPs) were electrophoresed on a 10% SDS-polyacrylamide gel and transferred onto a PVDF membrane, and the binding of antibodies 65C6, 100F4, 3C11, and TG15 was measured by Western blot analysis. Mouse immune sera elicited with H5 HA plasmid DNA were used as a positive control. Figure 1B shows that the TG15 control specifically binds to envelope protein derived from HIV-1 VLPs but not to those from influenza virus HA and NA VLPs, whereas mouse immune sera specifically bind to HA0, HA1, and HA2 of HA and NA VLPs but not to envelope protein from HIV-1 VLPs. Antibodies 65C6, 100F4, and 3C11 specifically bind HA0 and HA1 but not H5 HA2 or HIV-1 envelope proteins. Thus, all three antibodies 65C6, 100F4, and 3C11 recognize epitopes in the HA1 domain of HA.
To measure the affinity of the antibodies, surface plasmon resonance using recombinant H5 HA (A/Anhui/05/01, subclade 2.3.4) was performed. Figure 1C shows the association and dissociation curves of antibodies 100F4, 65C6, and 3C11 at the indicated concentrations of HA. Based on these curves, the dissociation constant (KD) values of antibodies 100F4, 65C6, and 3C11 were calculated to be 2.42 × 10−9, 4.14 × 10−8, and 7.02 × 10−8 M, respectively (Fig. 1D).
Neutralization breadths and potencies of antibodies 65C6, 100F4, and 3C11.
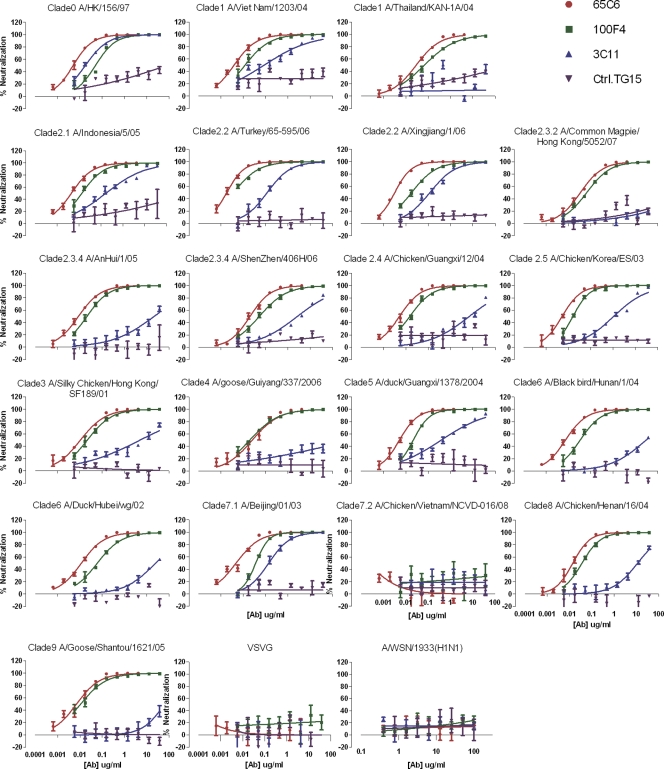
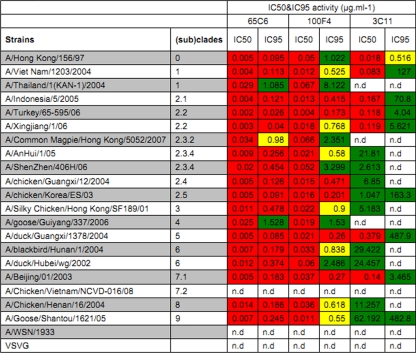
To determine the breadths and potencies of human antibodies 65C6, 100F4, and 3C11, they were titrated in a PN assay against a panel of 20 H5N1 and 1 H1N1 pseudotypes as well as a pseudotype expressing VSV-G (see Table S1 in the supplemental material). As shown in Fig. 2, the control antibody TG15 did not exhibit any neutralization activity against any of these H5N1 and H1N1 pseudotypes. Antibody 3C11 neutralized 16 H5N1 pseudotypes, with IC50s ranging from 0.018 μg/ml against A/Hong Kong/156/97 to 62.192 μg/ml against A/Goose/Shantou/1621/05, but neutralized only H5N1 pseudotypes derived from A/Hong Kong/156/97, A/Turkey/65-595/2006, A/Xingjiang/1/2006, and A/Beijing/01/2003, with IC95 values at 0.516, 4.04, 5.612, and 3.465 μg/ml, respectively (Fig. 2 and Table 1). In contrast, except for A/Chicken/Vietnam/NCVD-016/08 (subclade 7.2), both antibodies 65C6 and 100F4 neutralized all clades and subclades of H5N1 pseudotypes tested with a higher degree of potency. Antibody 100F4 at <0.5 μg/ml neutralized 6 H5N1 pseudotypes with the IC95 and at <1 μg/ml neutralized 13 H5N1 pseudotypes with the IC95, while for the remaining 6 H5N1 pseudotypes between 1.022 and 8.122 μg/ml was required to reach the IC95 (Table 1). Most strikingly, although it had lower binding affinity to soluble HA than antibody 100F4 (Fig. 1D), antibody 65C6 exhibited higher neutralization activity than antibody 100F4. Antibody 65C6 at <0.5 μg/ml neutralized 16 H5N1 pseudotypes with the IC95 and at <1 μg/ml neutralized 17 H5N1 pseudotypes with the IC95, while for 2 H5N1 pseudotypes, derived from A/Thailand/KAN-1A/04 and A/Goose/Guiyang/337/06, 1.085 and 1.528 μg/ml were needed to reach the IC95 (Table 1). None of these three antibodies exhibited any neutralization activity against H1N1 and VSV-G pseudotypes (Fig. 2 and Table 1).
Fig 2.
Cross clade neutralization by antibodies 65C6, 3C11, and 100F4. A VSV-G pseudotype was used as a negative control.
Table 1.
IC50s and IC95s of antibodies 65C6, 100F4, and 3C11 against a panel of H5N1 pseudotypesa
The doses of antibodies 65C6, 100F4, and 3C11 that result in the IC50 and IC95 to a given H5N1 pseudotype are shown. n.d., not detected. Green, >1 μg/ml required to reach IC50 and IC95; yellow, between 0.5 and 1 μg/ml required to reach IC50 and IC95; red, <0.5μg/ml required to reach IC50 and IC95.
To further test the breadth and potency of neutralization activity of 65C6, we performed hemagglutination inhibition (HI) assays against a panel of influenza A viruses, including H1N1, H2N2, H3N2, and H5N1. Again depending on the H5N1 strains, between 0.3 and 2.7 μg/ml of antibody 65C6 completely inhibited all 6 H5N1 viruses tested, while >170 μg/ml of antibody 65C6 still did not inhibit H1N1, H2N2, and H3N2 viruses, indicating that antibody 65C6 recognizes a conserved epitope on HAs from almost all clades and subclades of the H5 subtype and that this epitope is not shared by HAs of H1, H2, and H3 subtypes (Table 2).
Table 2.
The concentrations of antibody 65C6 required to completely inhibit hemagglutination of 8 HA units of virus measured by HI assay
| Subtype | Virus | Clade or subclade | IgG (μg ml−1) |
|---|---|---|---|
| H5N1 | A/Viet Nam/1194/2004 | 1 | 0.66 |
| A/Indonesia/5/2005 | 2.1 | 2.7 | |
| A/Bar headed goose/Qinghai/1A/2005 | 2.2 | 1.3 | |
| A/Whooper swan/Mongolia/244/2005 | 2.2 | 0.66 | |
| A/Turkey/1/2005 | 2.2 | 0.33 | |
| A/Anhui/1/2005 | 2.3 | 0.33 | |
| H1N1 | A/California/7/2009 | >170 | |
| H2N2 | A/Singapore/1/1957 | >170 | |
| H3N2 | A/Aichi/2/1968 | >170 |
Binding site of antibody 65C6.
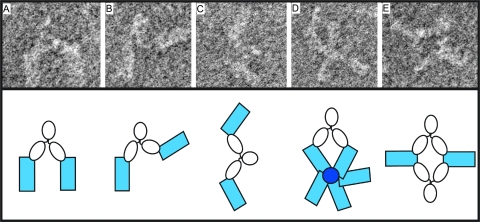
Figure 3 shows five representative images of HA-antibody 65C6 complexes obtained by negative-staining electron microscopy. HA protein was derived from autologous strain A/Shenzhen/406H/06 (subclade 2.3.4). Figure 3A to C show that one antibody binds two individual HA molecules. Each Fab of the antibody binds near an end of HA and forms a constant angle of about 110° with HA. Figure 3D shows that one antibody binds to two individual HA molecules in a five-HA aggregate “rosette.” In this complex, each Fab also binds near an end of HA and forms a constant angle of about 110° with HA. Since it is known that the “membrane anchor” regions of HA associate with each other in the absence of detergent to form “rosettes,” the binding site on the HA for the Fab is at the membrane-distal tip of HA.
Fig 3.
Binding site of antibody 65C6. Five representative complexes (A to E) of H5 HA and antibody 65C6 determined by negative-staining electron microscopy are shown. (A, B, and C) Complexes of 2 HA molecules linked to 1 65C6 molecule; (D) complex of 1 65C6 molecule linked to 2 HA molecules of a “rosette” of 5 HA molecules associated through their transmembrane domains; (E) is complex of 2 HA molecules plus 2 65C6 molecules. To aid interpretation, a schematic diagram for each complex is shown underneath. Blue rectangles, HA protein; white ovals, Fab and Fc domains of antibody 65C6.
Epitope mapping of antibody 65C6.
To map the neutralization epitope of antibody 65C6, yeast display analysis was carried out at both the domain level and the fine-epitope level as described before (52). To perform the domain-level epitope mapping, yeast (Saccharomyces cerevisiae) cells that display a combinatorial library of H5 HA fragments were incubated with antibody 65C6, followed by FACS analysis and cell sorting. Sequencing analysis of HA fragments isolated from PE-positive yeast clones indicated that the 65C6 epitope resides within the HA fragment comprising amino acid residues 51 to 260 (T. Zuo et al., data not shown).
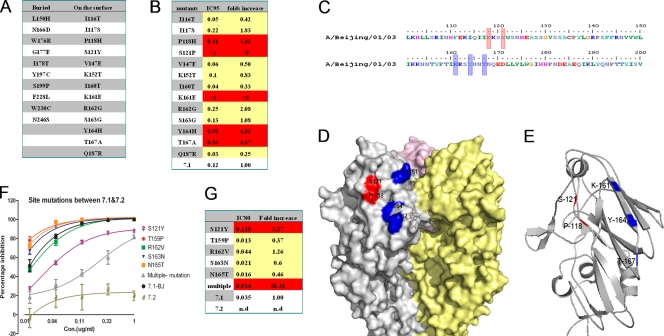
To perform the fine-epitope mapping, Saccharomyces cerevisiae cells that display a random mutagenesis library of the HA fragment comprising amino acid residues 51 to 260 were constructed and incubated with antibody 65C6, followed by FACS analysis and cell sorting. Sequencing analysis of HA fragments isolated from PE-negative (65C6-resistant) yeast clones identified 23 amino acid mutations that abolish the binding of 65C6 (Fig. 4A). Among them, 13 amino acid residues, at positions 116, 117, 118, 121, 147, 152, 160, 161, 162, 163, 164, 167, and 187 were on the surface of the HA molecule, while the remaining 10 residues were underneath the surface, suggesting that they are not directly contacted by antibody 65C6.
Fig 4.
Amino acid residues involved in neutralization epitopes. (A) List of 23 single-amino-acid mutants to which antibody 65C6 can no longer bind, determined using Saccharomyces cerevisiae cells that display a random mutagenesis library of the HA fragment comprising amino acid residues 51 to 260 in fine-epitope mapping. Among them, 10 amino acid mutations are underneath the HA surface and the other 13 are on the surface of HA. (B) Concentrations of antibody 65C6 required to reach 95% neutralization (IC95) in PN assay and relative fold increases against 13 single surface amino acid mutants compared to the parental HA. Red indicates single-amino-acid mutants that are more resistant to neutralization by antibody 65C6. (C) Five amino acid residues, at positions 118, 121, 161, 164, and 167 (highlighted in red), that are involved in HA derived from H5N1 strain A/Beijing/01/03 (subclade 7.1) as determined by the combined yeast display screen and PN assay. (D) The 3D structure shows that the 5 amino acid residues 118, 121, 161, 164, and 167 (highlighted in red and blue) are adjacent to each other on the surface of HA. (E) The 3D structure shows that the 5 amino acid residues 118, 121, 161, 164, and 167 are adjacent to each other as depicted in ribbon representation. (F) Titration of antibody 65C6 against 5 single HA mutants and one HA mutant with all 5 amino acid substitutions for subclade 7.1 compared to parental subclades 7.1 and 7.2. (G) Concentrations of antibody 65C6 required to reach 80% neutralization (IC80) in PN assay and relative fold increases against 5 single HA mutants and one HA mutant with all 5 amino acid substitutions for subclade 7.1 compared to parental subclade 7.1. Red indicates single- or multiple-amino-acid mutants that are more resistant to neutralization by antibody 65C6.
To test whether these 13 surface mutations of H5 HA would affect the neutralization activity of antibody 65C6, genes encoding 13 full-length H5 HA mutants derived from H5N1 strain A/Beijing/01/03 (subclade 7.1) were constructed and used to generate H5N1 pseudotypes. The susceptibility of H5N1 pseudotypes to neutralization by antibody 65C6 was determined by PN assay (see Fig. S3 in the supplemental material). Compared to the wild-type H5N1 pseudotype, H5N1 pseudotypes expressing H5 HA mutants with a mutation at position 116, 117, 147, 152, 160, 162, 163, or 187 were similarly or even more susceptible to neutralization by antibody 65C6, whereas H5N1 pseudotypes expressing H5 HA mutants with a mutation at position 118, 121, 161, 164, or 167 were more resistant to neutralization by antibody 65C6 (Fig. 4B). Interestingly all these resistant amino acid residues are adjacent to each other on the surface according to the three-dimensional (3D) structure of the HA model (Fig. 4C, D, and E).
Within amino acid stretches of residues 117 to 121 and 159 to 167 of H5 HA, there are 5 amino acid differences at positions 121, 159, 162, 163, and 165 between 65C6-susceptible strain A/Beijing/01/2003 (subclade 7.1) and 65C6-resistant strain A/Chicken/Vietnam/NCVD-016/08 (subclade 7.2). To determine the involvement of these residues in the neutralization epitope of antibody 65C6, we constructed 5 HA single mutants and one HA mutant with mutations in all 5 amino acid by replacing these amino acids from subclade 7.1 with those from subclade 7.2 and used them to generate H5N1 pseudotypes. Figure 4F and G show the neutralization activities of antibody 65C6 against these H5N1 mutant pseudotypes. Compared to the subclade 7.1 wild-type H5N1 pseudotype, H5N1 mutant pseudotypes with single mutations at positions 159, 163, and 165 exhibited higher neutralization sensitivity. In contrast, the H5N1 mutant pseudotype with a single mutation at position of 121 was resistant (3.37-fold increases at the IC80) to 65C6 neutralization. Most strikingly, the H5N1 mutant pseudotype with all 5 amino acid mutations was much more resistant (26-fold increase at the IC80) to neutralization by antibody 65C6 than the H5N1 mutant pseudotype with a single mutation at position 121 (Fig. 4G).
Prophylactic efficacy of antibody 65C6.
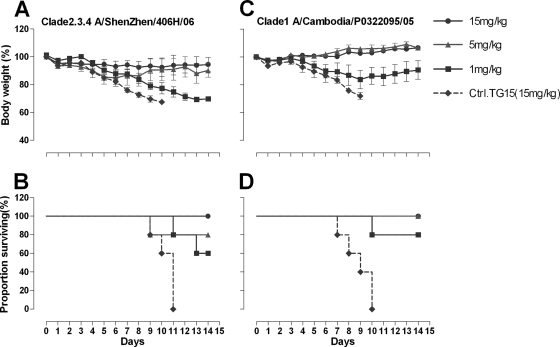
To determine whether the potent in vitro neutralization activity displayed by antibody 65C6 would be predictive of its prophylactic efficacy in vivo, female BALB/c mice were passively administrated (i.p.) 15, 5, and 1 mg/kg of antibody 65C6 or 15 mg/kg of control antibody TG15 and then challenged i.n. with 5 MLD50 of HPAI H5N1 A/Shenzhen/406H/06 and A/Cambodia/P0322095/05 viruses. A dose of 5 MLD50 was chosen to ensure 100% mortality in the control group. Figure 5A shows the time course of body weight changes and Fig. 5B shows the survival rate of each group during 14 days after challenge with HPAI H5N1 A/Shenzhen/406H/06 virus. Figure 5C shows the time course of body weight changes and Fig. 5D shows the survival rate of each group during 14 days after challenge with HPAI H5N1 A/Cambodia/P0322095/05 virus. In the group of mice that were inoculated with 15 mg/kg of control antibody TG15, severe sickness of mice became evident on day 3 after the challenge with H5N1 A/Shenzhen/406H/06, and all mice died. In contrast, in the group of mice that were inoculated with 1 mg/kg of antibody 65C6, all 5 mice got sick at 4 to 6 days postchallenge and 2 mice died, while the remaining 3 mice survived. In the group of mice that were inoculated with 5 mg/kg of antibody 65C6, mice got sick at 5 to 7 days postchallenge and 1 mouse died, while the remaining 4 mice survived. In the group of mice that were inoculated with 15 mg/kg of antibody 65C6, no mice became sick or lost weight, and all survived.
Fig 5.
Prophylactic efficacy of antibody 65C6 in mice. (A) Time course of body weight changes. (B) Survival rate of each group challenged with HPAI H5N1 A/Shenzhen/406H/06 virus. (C) Time course of body weight changes. (D) Survival rate of each group challenged with HPAI H5N1 A/Cambodia/P0322095/05 virus. Survival rate was calculated as percent survival within each experimental group (n = 5 mice per experimental group).
In the group of mice that were inoculated with 15 mg/kg of control antibody TG15, after the challenge with A/Cambodia/P0322095/05 virus, rates of sickness, weight loss, and death similar to those for mice infected with A/Shenzhen/406H/06 were observed. In the group of mice that were inoculated with 1 mg/kg of antibody 65C6, all 5 mice got sick and one mouse died, while the remaining 4 mice survived. In the groups of mice that were inoculated with 5 or 15 mg/kg of antibody 65C6, no mice became sick and all survived.
Therapeutic efficacy of antibody 65C6 in vivo.
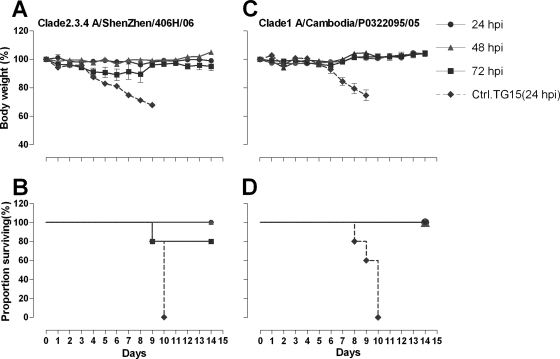
Having demonstrated potent prophylactic efficacy of antibody 65C6 in vivo, we next determined whether the potent in vitro neutralization activity displayed by 65C6 would be predictive of its therapeutic efficacy in vivo. To accomplish this, female BALB/c mice were inoculated i.n. with 5 MLD50 of HPAI H5N1 viruses A/Shenzhen/406H/06 and A/Cambodia/P0322095/05 and then injected i.p. with 40 mg/kg of antibody 65C6 or control antibody TG15 at 24, 48, and 72 h postinfection. Figure 6A shows the time course of body weight changes and Fig. 6B shows the survival rate of each group during 14 days after challenge with HPAI H5N1 A/Shenzhen/406H/06 virus. Figure 6C shows the time course of body weight changes and Fig. 6D shows the survival rate of each group during 14 days after challenge with HPAI H5N1 A/Cambodia/P0322095/05 virus. In the group of mice that were injected with 40 mg/kg of control antibody TG15 at 24 h after the infection, severe sickness, weight loss, and death similar to what was found in prophylactic studies were observed. In contrast, after injection with 40 mg/kg of antibody 65C6 at 24, 48, and 72 h postinfection, all mice infected with A/Cambodia/P0322095/05 survived with no weight loss (Fig. 6C and D), and except for one mouse that was injected with antibody at 72 h postinfection, all mice infected with A/Shenzhen/406H/06 also survived with no significant weight loss (Fig. 6A and B).
Fig 6.
Therapeutic efficacy of antibody 65C6 in mice. (A) Time course of body weight changes. (B) Survival rate of each group infected with HPAI H5N1 A/Shenzhen/406H/06 virus and treated with 40 mg/kg of antibody 65C6 at 24, 48, and 72 h postinfection. (C) Time course of body weight changes. (D) Survival rate of each group infected with HPAI H5N1 A/Cambodia/P0322095/05 virus and treated with 40 mg/kg of antibody 65C6 at 24, 48, and 72 h postinfection. Survival rate was calculated as percent survival within each experimental group (n = 5 mice per experimental group).
DISCUSSION
Continuous zoonotic transmission of HPAI H5N1 viruses to humans, with 60% mortality and for which there are few specific treatment options, remains a major public health threat. In this study, we report the development of three fully human monoclonal antibodies, 65C6, 100F4, and 3C11, from immortalized memory B cells of an individual who had recovered from infection with a subclade 2.3.4 HPAI H5N1 virus in China (50).
Perhaps, the most important findings of this study are that one such antibody, 65C6, neutralizes H5N1 strains derived from all clades and subclades of H5 HA with a high degree of potency, except for subclade 7.2 (Fig. 2 and Table 1). These findings indicate that the neutralization epitope of 65C6 is conserved among diverse H5N1 strains. The fact that antibody 65C6 was established from memory B cells of an individual who had recovered from HPAI H5N1 virus infection implies that the 65C6 epitope elicits a neutralizing antibody response during the natural infection. Similarly, Whittle et al. (43a) recently described another subtype-specific broadly neutralizing antibody, CH65, isolated from single plasma cells of an individual vaccinated against seasonal influenza, that neutralizes 30 out of 36 H1N1 strains from the past 30 years. However, the epitope recognized by antibody CH65 is in the receptor binding site and the antigenic site of Sb.
Another important finding is that antibody 65C6 exhibits potent prophylactic and therapeutic efficacy in vivo. Intraperitoneal injection of 65C6 at 1 mg/kg or higher significantly protects mice from infection with lethal doses of HPAI H5N1 viruses (Fig. 5), and i.p. injection of 40 mg/kg of 65C6 at 72 h after lethal-dose infection with HPAI H5N1 viruses significantly protects mice from weight loss and the requirement for euthanasia (Fig. 6). Thus, antibody 65C6 could have potential for use in treatment of human pandemic or zoonotic H5N1 cases.
Although antibody-based therapy is not a new therapeutic strategy, for severe influenza cases it represents a plausible intervention. For example, passive immunization by vertical acquisition of specific antibodies has been found to be associated with influenza immunity in early infancy in humans (27, 30, 37, 38). Transfusion of human blood products from patients recovered from the 1918 “Spanish flu” resulted in a 50% reduction in influenza mortality (from 37% to 16%) during the pandemic (21). Transfusion of convalescent-phase plasma from a patient recovered from H5N1 infection resulted in a dramatic reduction of viral loads and complete recovery (50). The underlying mechanism of these clinical observations is that neutralizing antibodies in these plasma samples modulate the course of viral infection and mitigate the development of acute respiratory distress syndrome and other complications (21). Thus, the human monoclonal antibodies developed in this study and in studies by other investigators (4, 5, 9, 14, 15, 18, 32, 35, 36, 39, 44, 45, 49) offer even better therapeutic options than convalescent-phase plasma samples. One of the advantages of human monoclonal antibodies is that large quantities of the antibodies can be quickly produced. Indeed, in our own study we found that over 1 g per liter per day of human monoclonal antibodies can be produced by these Drosophila S2 transfectants using Wave bioreactor and perfusion culture (Wang et al., unpublished data). Another advantage is that human monoclonal antibodies are free of adventitious agents associated with preparations of human plasma samples. Still another advantage is that since the antibodies are human in origin, when they are used to treat humans the risks of eliciting human immune responses are minimized compared with those after treatment with antibodies generated from other species.
Electron microscopy and epitope-mapping data show that antibody 65C6 binds to a conformational epitope comprising amino acid residues at positions 118, 121, 161, 164, and 167 on the tip of the membrane-distal globular domain of HA (Fig. 4). These amino acid residues are highly conserved among diverse H5 clades and subclades, except for subclade 7.2, but quite different from those of other HA subtypes (see Fig. S4 in the supplemental material), which is totally consistent with the neutralization data shown in Table 1 and Fig. 2. In the three-dimensional structure, the 65C6 epitope was located in a region similar to that for two other epitopes recognized by human antibodies 2D1 (47, 49) and FLD21.140 (17, 32). Antibody 2D1 was isolated from a survivor of the 1918 Spanish flu and neutralizes both 1918 and 2009 H1N1 viruses (47, 49). Antibody FLD21.140 was isolated from a survivor of HPAI H5N1 virus infection in Vietnam and neutralizes (sub)clades 1, 2.2, and 2.3.4. Although it does not neutralize A/Indonesia/5/05 (clade 2.1) in vitro, it protects mice from lethal challenge with A/Indonesia/5/05 (32). The fact that these three subtype-specific broadly neutralizing antibodies isolated from 3 individual patients from three independent laboratories react with epitopes in a similar region strongly indicates that neutralization epitopes that reside in this region may be immunodominant epitopes during natural infection. Thus, the unique loop(s) and antiparallel β sheet structure shown in Fig. 4E may be exploited for immunogen design to elicit a subtype-specific broad neutralizing antibody response.
In summary, using a highly sensitive pseudotype-based neutralization assay and molecular cloning techniques, we have developed three fully human monoclonal antibodies from immortalized memory B cells of an individual who had recovered from HPAI H5N1 virus infection (50). One such antibody, 65C6, that recognizes a conserved neutralization epitope on the globular membrane-distal domain of HA1 exhibits broad and potent neutralizing activity against diverse H5N1 strains from all clades and subclades (except subclade 7.2) in vitro as well as potent prophylactic and therapeutic efficacy in vivo. Thus, on the one hand antibody 65C6 may potentially be used either alone or in combination with antibodies of different binding specificities or with small-molecule inhibitors to treat human cases of various (sub)clades of H5N1 virus infection, and on the other hand immunogens based on this common H5 HA neutralization epitope may be developed to elicit broad neutralizing antibody responses against almost all (sub)clades of H5N1 viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Naldini, Turin University Medical School, Turin, Italy, for the lentiviral transfer vector and Liangzhi Xie, Medical Academy of Sciences, China, for purified H5 HA proteins used in SPR analysis. The anti-HIV-1 p24 monoclonal antibody (clone 183-H12) was provided by the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Germantown, MD.
This work was supported by research grants from the French Ministry of Health, the Chinese National Science Foundation (30671922), National Science and Technology Major Projects (2008ZX10001-010, 2009ZX10004-105, and 2009ZX10004-016), the Li Kai-Shing Foundation of Hong Kong, and the United Kingdom Medical Research Council.
Footnotes
Published ahead of print 11 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Beigel JH, et al. 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 353:1374–1385 [DOI] [PubMed] [Google Scholar]
- 2. Buchy P, et al. 2007. Influenza A/H5N1 virus infection in humans in Cambodia. J. Clin. Virol. 39:164–168 [DOI] [PubMed] [Google Scholar]
- 3. Chen Y, et al. 2009. Broad cross-protection against H5N1 avian influenza virus infection by means of monoclonal antibodies that map to conserved viral epitopes. J. Infect. Dis. 199:49–58 [DOI] [PubMed] [Google Scholar]
- 4. Corti D, et al. 2010. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 120:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corti D, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856 [DOI] [PubMed] [Google Scholar]
- 6. de Jong MD, et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Jong MD, et al. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667–2672 [DOI] [PubMed] [Google Scholar]
- 8. Ekiert DC, et al. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ekiert DC, et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gu J, et al. 2007. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet 370:1137–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gutiérrez R, Naughtin M, Horm S, San S, Buchy P. 2009. A(H5N1) virus evolution in South East Asia. Viruses 1:335–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. 2002. H5 avian and H9 swine influenza virus haemagglutinin structures: possible origin of influenza subtypes. EMBO J. 21:865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hanson BJ, et al. 2006. Passive immunoprophylaxis and therapy with humanized monoclonal antibody specific for influenza A H5 hemagglutinin in mice. Respir. Res. 7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johansson DX, et al. 2007. Efficient expression of recombinant human monoclonal antibodies in Drosophila S2 cells. J. Immunol. Methods 318:37–46 [DOI] [PubMed] [Google Scholar]
- 14. Kashyap AK, et al. 2008. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. U. S. A. 105:5986–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kashyap AK, et al. 2010. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog. 6:e1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaverin NV, et al. 2007. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J. Virol. 81:12911–12917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khurana S, et al. 2009. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 6:e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koudstaal W, et al. 2009. Pre- and postexposure use of human monoclonal antibody against H5N1 and H1N1 influenza virus in mice: viable alternative to oseltamivir. J. Infect. Dis. 200:1870–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Le QM, et al. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 20. Lu J, et al. 2006. Passive immunotherapy for influenza A H5N1 virus infection with equine hyperimmune globulin F(ab′)2 in mice. Respir. Res. 7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luke TC, Kilbane EM, Jackson JL, Hoffman SL. 2006. Meta-analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann. Intern. Med. 145:599–609 [DOI] [PubMed] [Google Scholar]
- 22. Luo G, Chung J, Palese P. 1993. Alterations of the stalk of the influenza virus neuraminidase: deletions and insertions. Virus Res. 29:321. [DOI] [PubMed] [Google Scholar]
- 23. Oh HL, et al. 2010. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J. Virol. 84:8275–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okuno Y, Matsumoto K, Isegawa Y, Ueda S. 1994. Protection against the mouse-adapted A/FM/1/47 strain of influenza A virus in mice by a monoclonal antibody with cross-neutralizing activity among H1 and H2 strains. J. Virol. 68:517–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palladino G, Mozdzanowska K, Washko G, Gerhard W. 1995. Virus-neutralizing antibodies of immunoglobulin G (IgG) but not of IgM or IgA isotypes can cure influenza virus pneumonia in SCID mice. J. Virol. 69:2075–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peiris JS, et al. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Puck JM, Glezen WP, Frank AL, Six HR. 1980. Protection of infants from infection with influenza A virus by transplacentally acquired antibody. J. Infect. Dis. 142:844–849 [DOI] [PubMed] [Google Scholar]
- 28. Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27:493–497 [Google Scholar]
- 29. Renegar KB, Small PA, Jr, Boykins LG, Wright PF. 2004. Role of IgA versus IgG in the control of influenza viral infection in the murine respiratory tract. J. Immunol. 173:1978–1986 [DOI] [PubMed] [Google Scholar]
- 30. Reuman PD, Paganini CM, Ayoub EM, Small PA., Jr 1983. Maternal-infant transfer of influenza-specific immunity in the mouse. J. Immunol. 130:932–936 [PubMed] [Google Scholar]
- 31. Sawyer LA. 2000. Antibodies for the prevention and treatment of viral diseases. Antiviral Res. 47:57–77 [DOI] [PubMed] [Google Scholar]
- 32. Simmons CP, et al. 2007. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 4:e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smirnov YA, Lipatov AS, Gitelman AK, Claas EC, Osterhaus AD. 2000. Prevention and treatment of bronchopneumonia in mice caused by mouse-adapted variant of avian H5N2 influenza A virus using monoclonal antibody against conserved epitope in the HA stem region. Arch. Virol. 145:1733–1741 [DOI] [PubMed] [Google Scholar]
- 34. Smith GJ, et al. 2006. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl. Acad. Sci. U. S. A. 103:16936–16941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sui J, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun L, et al. 2009. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One 4:e5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sweet C, Bird RA, Jakeman K, Coates DM, Smith H. 1987. Production of passive immunity in neonatal ferrets following maternal vaccination with killed influenza A virus vaccines. Immunology 60:83–89 [PMC free article] [PubMed] [Google Scholar]
- 38. Sweet C, Jakeman KJ, Smith H. 1987. Role of milk-derived IgG in passive maternal protection of neonatal ferrets against influenza. J. Gen. Virol. 68:2681–2686 [DOI] [PubMed] [Google Scholar]
- 39. Throsby M, et al. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3:e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tiller T, et al. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329:112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Traggiai E, et al. 2004. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 10:871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tran TH, et al. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 350:1179–1188 [DOI] [PubMed] [Google Scholar]
- 43. Tsai C, et al. 2009. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine 27:6777–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a. Whittle JR, et al. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 108:14216–14221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wrammert J, et al. 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J. Exp. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wrammert J, et al. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453:667–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wrigley NG, et al. 1983. Electron microscopy of influenza haemagglutinin-monoclonal antibody complexes. Virology 131:308–314 [DOI] [PubMed] [Google Scholar]
- 47. Xu R, et al. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoshida R, et al. 2009. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 5:e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yu X, et al. 2008. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou B, Zhong N, Guan Y. 2007. Treatment with convalescent plasma for influenza A (H5N1) infection. N. Engl. J. Med. 357:1450–1451 [DOI] [PubMed] [Google Scholar]
- 51. Zhou P, Goldstein S, Devadas K, Tewari D, Notkins AL. 1998. Cells transfected with a nonneutralizing antibody gene are resistant to HIV infection: targeting the endoplasmic reticulum and trans-Golgi network. J. Immunol. 160:1489–1496 [PubMed] [Google Scholar]
- 52. Zuo T, et al. 2011. Comprehensive analysis of pathogen-specific antibody response in vivo based on an antigen library displayed on the surface of yeast. J. Biol. Chem. 286:33511–33519 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.