Abstract
There are 80 trimeric, glycoprotein spikes that cover the surface of an alphavirus particle. The spikes, which are composed of three E2 and E1 glycoprotein heterodimers, are responsible for receptor binding and mediating fusion between the viral and host-cell membranes during entry. In addition, the cytoplasmic domain of E2 interacts with the nucleocapsid core during the last stages of particle assembly, possibly to aid in particle stability. During assembly, the spikes are nonfusogenic until the E3 glycoprotein is cleaved from E2 in the trans-Golgi network. Thus, a mutation in E2 potentially has effects on virus entry, spike assembly, or spike maturation. E2 is a highly conserved, cysteine-rich transmembrane glycoprotein. We made single cysteine-to-serine mutations within two distinct regions of the E2 ectodomain in both Sindbis virus and Ross River virus. Each of the E2 Cys mutants produced fewer infectious particles than wild-type virus. Further characterization of the mutant viruses revealed differences in particle morphology, fusion activity, and polyprotein cleavage between Sindbis and Ross River virus mutants, despite the mutations being made at corresponding positions in E2. The nonconserved assembly defects suggest that E2 folding and function is species dependent, possibly due to interactions with a virus-specific chaperone.
INTRODUCTION
The alphavirus particle is comprised of two concentric protein shells, the glycoprotein spikes and the nucleocapsid core, separated by a lipid membrane (7, 66). The outer protein shell consists of 80 trimeric, glycoprotein spikes embedded in a host-derived lipid membrane and arranged with icosahedral symmetry. The spikes are trimers of E2 and E1 protein heterodimers (7, 26, 66). The spike proteins are required to initiate infection; the E2 glycoprotein is responsible for receptor binding (5, 24, 53), and the E1 glycoprotein mediates fusion between the viral and host-cell membranes during entry (14, 45, 59). The nucleocapsid core is located in the center of the particle and is composed of 240 copies of the capsid protein arranged with icosahedral symmetry around the viral RNA genome (7, 66). The endodomain of E2 interacts with the nucleocapsid core, presumably to aid in particle stability (11, 15, 32, 48).
Alphavirus assembly is a highly regulated process comprised of multiple cleavage and oligomerization events. The alphavirus genome encodes five structural proteins that are translated as a single polyprotein, capsid-E3-E2-6K-E1 (where 6K indicates a 6,000-molecular-weight protein), from a subgenomic RNA (54). Capsid is cleaved from the rest of the polyprotein and remains in the cytoplasm, where nucleocapsid core formation occurs (38). The E3 glycoprotein contains a signal sequence that translocates the remaining polyprotein, E3-E2-6K-E1 (also referred to as the spike proteins), into the endoplasmic reticulum (ER) (1, 2, 38). Once inserted into the ER, the cellular enzyme signalase cleaves the polyprotein into E3-E2 (also referred to as pE2 or p62), 6K, and E1 (28, 68). In the ER, pE2 and E1 associate into heterodimers, three of which oligomerize to form nonfusogenic spikes (30, 31, 41). The nonfusogenic spikes are then transported through the Golgi network to the plasma membrane via the host's secretory system (9). At the plasma membrane, the spikes are arranged in hexagonal arrays (56). The nucleocapsid cores interact with the E2 endodomain to initiate budding from the host cell (63, 69). When E1 and E2 are expressed individually, E1 is retained in the ER and a small amount of E2 is transported to the cell surface. This observation suggests that heterodimerization of E1 and pE2 is required for efficient spike oligomerization and transport to the cell surface (29, 31, 58).
E1 and E2 are highly conserved, cysteine-rich transmembrane glycoproteins that require ER chaperones to fold properly. The cellular proteins BiP and protein disulfide isomerase mediate E1 folding, whereas calnexin/calreticulum and Erp57 mediate E2 folding (39–43). Atomic structures show that E1 contains eight disulfide bonds (26) and E2 contains six disulfide bonds (27, 57). E1 undergoes at least three conformational changes in the ER, and each E1 intermediate differs in either the number or arrangement of disulfide bonds (40, 42). The E3 glycoprotein does not directly participate in E1 folding. In contrast, it is unknown if E2 folds through a series of disulfide bond rearrangements or how many E2 intermediates are formed. E2 does require chaperone activity from E3 to fold properly (6, 40, 43). In the absence of E3 or if E3 is replaced with an ER signal sequence, E2 does not fold properly and E2-E1 heterodimers do not form (31). E3 also plays a regulatory role in alphavirus assembly. Cleavage of E3 from pE2 by the cellular enzyme furin is necessary for rendering the nonfusogenic spikes fusion competent, a process called spike maturation (21, 49, 51, 58, 68). Inappropriate or untimely pE2 cleavage could cause the spikes to prematurely convert from the stable conformation to the meta-stable, fusion-competent conformation (17, 57).
Using the atomic structures of Chikungunya virus (CHIK) E2 and Sindbis virus (SINV) E2 as templates (27, 57), we mutated cysteines within two distinct regions of the E2 ectodomain in both SINV and Ross River virus (RRV). The E2 Cys mutants were attenuated in virus growth compared to the parental viruses. Furthermore, the mutants had defects in polyprotein cleavage, particle morphology, and fusion activity. Interestingly, mutations in SINV and RRV at corresponding Cys residues did not yield similar phenotypes. Our results suggest that the E2 glycoprotein of SINV folds differently than the E2 glycoprotein of RRV, perhaps due to interactions with a virus-specific chaperone.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells (American Type Culture Collection, Manassas, VA) were grown in minimal essential medium (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA), nonessential amino acids, l-glutamine, and penicillin-streptomycin. BHK-21 cells were grown at 37°C in the presence of 5% CO2. C6/36 cells (American Type Culture Collection, Manassas, VA) were grown in the same medium but at 28°C in the presence of 5% CO2. The virus strains used in this work were the TE12 strain of SINV (33) and the T48 strain of RRV (25).
Generation of mutant viruses.
All mutations in the TE12 cDNA clone and the T48 cDNA clone were introduced using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA). For each construct, the region corresponding to the E2 glycoprotein was sequenced. Wild-type and mutant cDNA clones were linearized with SacI and in vitro transcribed with SP6 polymerase (46).
Electroporation of BHK-21 cells was performed as previously described (47). Briefly, approximately 107 BHK-21 cells were trypsinized, washed three times with phosphate-buffered saline (PBS), and resuspended with PBS to a final volume of 500 μl. The cells were mixed with in vitro-transcribed RNA in a 2-mm-gap cuvette and pulsed once at 1.5 kV, 25 μF, and 200 Ω using a Bio-Rad Gene Pulser Xcell electroporation system (Bio-Rad Laboratories, Hercules, CA). Following a 10-min recovery at room temperature, the cells were diluted 1:10 in BHK cell medium and incubated at 37°C in the presence of 5% CO2. At the indicated time points, virus was harvested and titer was determined using standard plaque assay procedure (46).
For electroporation of C6/36 cells, approximately 107 cells were washed two times with PBS and resuspended in PBS to a final volume of 500 μl. The cells were mixed with in vitro-transcribed RNA in a 2-mm-gap cuvette and pulsed twice at 0.4 kV, 25 μF, and 800 Ω using the Bio-Rad Gene Pulser Xcell electroporation system (Bio-Rad Laboratories, Hercules, CA). Following a 1-min recovery at room temperature, the cells were diluted 1:10 in C6/36 cell medium and incubated at 28°C in the presence of 5% CO2. At 25 h postelectroporation, virus was harvested and titer was determined on BHK-21 cells using standard plaque assay procedure (46).
Quantitation of genome copy by RT and qRT-PCR.
BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA, resuspended in BHK cell medium, and incubated at 37°C in the presence of 5% CO2. At 25 h postelectroporation, the medium was harvested and spun at 3,500 × g for 15 min at 4°C to remove cells and cell debris. The cleared medium containing either wild-type or mutant virus (5 μl) was mixed with 500 ng of both nsP1 and E2 reverse transcription (RT) primers (SINV nsP1 [5′-AACATGAACTGGGTGGTG-3′] and SINV E2 [5′-ATTGACCTTCGCGGTCGGATTCAT-3′] or RRV nsP1 [5′-GCTCTGGCATTAGCATGG-3′] and RRV E2 [5′-GAACATCATGACCAGCCATA-3′]). The samples were incubated at 94°C for 5 min and 70°C for 5 min and transferred to ice. Once on ice, an RT mixture containing 50 mM Tris-HCl, pH 8.3, 75 mM KCl, 10 mM dithiothreitol, 3 mM MgCl2, 0.5 mM deoxynucleoside triphosphate, RNase inhibitor (GenScript, Piscataway, NJ), and ImProm-II reverse transcriptase (Promega, Madison, WI) was added to each sample. Following addition of the RT mixture, the samples were incubated at 25°C for 5 min, 42°C for 45 min, and 75°C for 15 min.
The cDNA prepared as described above (2 μl) was mixed with 1× Brilliant SYBR green reagent (Stratagene, La Jolla, CA) and 250 nM forward and reverse detection primers to a final volume of 25 μl per well in a 96-well plate. Multiple quantitative RT-PCR (qRT-PCR) measurements were made for each sample. Standard curves were generated by serially diluting wild-type SINV or RRV cDNA clones. All data sets included paired standard curves, which were generated by plotting the observed threshold cycle (CT) value using an exponential regression. The number of genome copies present within each qRT-PCR mixture was calculated using the observed CT values for each sample and the equation obtained from the standard curve.
Examination of polyprotein cleavage by Western blot analysis of cell lysates.
BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA, resuspended in BHK cell medium, and incubated at 37°C in the presence of 5% CO2. At the indicated time points, the cells were lysed with BHK cell lysis buffer (10 mM Tris, pH 7.4, 20 mM NaCl, 0.4% deoxycholic acid, 1% NP-40, and 1 mM EDTA). The cell lysates were solubilized in reducing SDS sample buffer. Solubilized cell lysates were analyzed by SDS-PAGE and probed with polyclonal antibodies recognizing pE2/E2 and capsid (Cocalico, Reamstown, PA).
Immunofluorescence analysis of glycoprotein cell surface expression.
BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA. At 18 h postelectroporation, the cells were fixed with freshly prepared 1.5% paraformaldehyde (Electron Microscopy Science, Hatfield, PA) for 15 min at room temperature. After fixation, the cells were washed with PBS and probed for cell surface glycoprotein expression using polyclonal antibodies recognizing SINV pE2/E2 and E1 (Antibodies Inc., Davis, CA) or RRV pE2/E2 (Cocalico, Reamstown, PA) and the Alexa Fluor 488 (Invitrogen, Carlsbad, CA) secondary antibody. The cells were imaged using an Olympus 1X71 fluorescence microscope (Olympus, Center Valley, PA).
Biotinylation analysis of glycoprotein cell surface expression.
BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA. At 18 h postelectroporation, the medium was replaced with l-methionine- and l-cysteine-free Dulbecco's minimal essential medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 5% dialyzed fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). The cells were incubated at 37°C in the presence of 5% CO2 for 15 min. Following the 15-min incubation, 50 μCi/ml [35S]cysteine-methionine (PerkinElmer, Waltham, MA) was added to the medium, and the cells were incubated for an additional 2 h. After radiolabeling, the cells were washed with PBS and cell surface proteins were biotinylated for 30 min at room temperature with 2.5 mM succinimidyl-6-(biotinamido)hexanoate (NHS-LC-biotin; Pierce, Rockford, IL). The biotin solution was prepared in PBS immediately before use. The biotinylated cells were lysed with BHK cell lysis buffer, and cell lysates were incubated with streptavidin resin (Pierce, Rockford, IL) for 1 h at room temperature with rocking. Following incubation, the resin was pelleted and washed three times with PBS. Biotinylated proteins were eluted from the resin by adding reducing SDS sample buffer and boiling at 95°C for 10 min. The eluted proteins were analyzed by SDS-PAGE.
Western blot analysis of released virus particles purified by pelleting.
Medium containing either wild-type or mutant virus was spun at 3,500 × g for 15 min at 4°C to remove cells and cell debris. The cleared medium was overlaid onto a 27% sucrose cushion and spun at 185,000 × g for 2.5 h at 4°C. Pelleted virus particles were resuspended in HNE buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, and 0.1 mM EDTA).
The resuspended particles were solubilized in reducing SDS sample buffer. Solubilized particles were analyzed by SDS-PAGE and probed with two polyclonal antibodies recognizing pE2/E2 and capsid (Cocalico, Reamstown, PA). Band intensities were quantitated using ImageQuant (version 5.2) software (Molecular Dynamics, Sunnyvale, CA).
Transmission electron microscopy (TEM).
Virus samples purified by pelleting through a 27% sucrose cushion (3 μl) were applied to a Formvar- and carbon-coated 400-mesh copper grid (Electron Microscopy Sciences, Hatfield, PA) and stained with 1% uranyl acetate. The stained grids were analyzed using a JEOL 1010 transmission electron microscope (Tokyo, Japan) operating at 80 kV. Images were recorded using a Gatan UltraScan 4000 charge-coupled-device camera (Pleasanton, CA).
Production of radiolabeled virus and purification using a step gradient.
BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA, resuspended in BHK cell medium, and incubated at 37°C in the presence of 5% CO2. At 18 h postelectroporation, the medium was replaced with l-methionine- and l-cysteine-free DMEM (Invitrogen, Carlsbad, CA) supplemented with 5% dialyzed fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). The cells were incubated at 37°C in the presence of 5% CO2 for 15 min. Following the 15-min incubation, 50 μCi/ml [35S]cysteine-methionine (PerkinElmer, Waltham, MA) was added to the medium and the cells were incubated for an additional 24 h. Following virus production, medium containing either radiolabeled wild-type or radiolabeled mutant virus was spun at 3,500 × g for 15 min at 4°C to remove cells and cell debris. The cleared medium was then overlaid onto a 27% and 60% sucrose step gradient and spun at 250,000 × g for 2.5 h at 4°C. Following ultracentrifugation, virus present at the 27% and 60% sucrose step interface was extracted. Radiolabeled virus was solubilized in reducing SDS sample buffer and analyzed by SDS-PAGE. Band intensities were quantitated using ImageQuant (version 5.2) software (Molecular Dynamics, Sunnyvale, CA).
Liposome-mediated E1 trimerization assay.
Radiolabeled virus was mixed with 0.8 mM liposomes (1-palmitoyl-2-oleoyl-sn-glycero-3-choline [POPC], 1-palmitoyl-2-oleoyl-sn-glycero-3-ethanolamine [POPE], sphingomyelin [SM], and cholesterol [Chl] in a ratio of 1:1:1:1.5). Liposomes (large, unilamellar vesicles) were prepared in HNE buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, and 0.1 mM EDTA) using an Avanti miniextruder with a 0.1-μm polycarbonate membrane pore size (Avanti Polar Lipids, Alabaster, AL). The virus-liposome mixture was adjusted to the indicated pH with 0.1 M morpholineethanesulfonic acid, 0.2 M acetic acid buffer and incubated at 20°C for 5 min. After the 5-min incubation, the virus-liposome mixture was adjusted to pH 7.4 with a pretitrated volume of 1 M NaOH. The neutralized samples were solubilized in nonreducing SDS sample buffer, and the formation of SDS-resistant E1 trimers was assayed using SDS-PAGE.
RESULTS
Rationale for site-directed mutations in the E2 glycoprotein.
The atomic structures of two alphavirus E2 glycoproteins have been determined and are structurally very similar (26, 56). From the CHIK E2 structure, three ordered domains, A, B, and C (57) (Fig. 1A and B), are seen. Domain A is located in the central portion of E2, domain B corresponds to the distal end of E2, which covers the E1 fusion peptide and contains the receptor-binding domain (5, 24, 34, 53, 57, 62), and domain C is adjacent to the viral membrane. There is a β ribbon which forms two polypeptide strands and connects domain A to domain B (Fig. 1B). Two sets of two-stacked disulfide bonds, one in domain A and one in domain B, are formed between conserved Cys residues in the E2 ectodomain (27, 57). Disulfide bonds 1 and 2 are stacked in domain A, and disulfide bonds 5 and 6 are stacked in domain B (Fig. 1B). The atomic structure of SINV E2 is similar to that of CHIK E2, except for domain B, which is disordered in SINV E2 (27) (Fig. 1C).
Fig 1.
Structure and sequence of the alphavirus E2 glycoprotein. (A) Atomic structure of the Chikungunya virus E2-E1 heterodimer (57) (Protein Data Bank accession number 3N42). The E2 glycoprotein is colored in blue. The E1 glycoprotein is colored in red. (B) Atomic structure of the Chikungunya virus E2 glycoprotein (57) (Protein Data Bank accession number 3N42); (C) atomic structure of the Sindbis virus E2 glycoprotein (27) (Protein Data Bank accession number 3MUU). In both panel B and panel C, domain A is colored in green, domain C is colored in magenta, and the β ribbon is colored in orange. In panel B, domain B is colored in gray. In panel C, the presumed location of domain B is indicated with a gray oval. Cys residues, which form stacked disulfide bonds, are colored in blue. Stacked disulfide bonds are numbered, consistent with the numbering in Table 1, and indicated with arrows. (D) Amino acid sequence alignments of a representative group of alphavirus E2 glycoproteins. Conserved Cys residues in the E2 ectodomain are in blue bold. Disulfide bonds in the E2 ectodomain are numbered above the corresponding Cys residues. The disulfide bond numbering is consistent with that in Table 1. The E2 transmembrane region is underlined. CHIK, Chikungunya virus; RRV, Ross River virus; SFV, Semliki Forest virus; SINV, Sindbis virus.
It has been demonstrated that E1 undergoes disulfide bond reshuffling during spike assembly (40, 42). We hypothesize that E2 also undergoes disulfide bond reshuffling, possibly by interacting with the E3 glycoprotein (47). Amino acid sequence alignments of E2 from a representative group of alphaviruses (Fig. 1D) show that the Cys residues in the E2 ectodomain are strictly conserved, both in number and in position (57). Further analysis shows multiple repeats of C-(X)1-5-C, which is a motif that is shared by many redox proteins, including protein disulfide isomerase (65, 67). We chose to mutate Cys residues that were part of these C-(X)1-5-C motifs to determine how virus assembly was affected. To understand how folding intermediates impact the assembly and function of alphavirus particles, we made single Cys-to-Ser mutations in the following disulfide bonds: disulfide bond 1 in domain A (residues SINV C16 and RRV C19), disulfide bond 2 in domain A (residues SINV C19 and RRV C22), disulfide bond 5 in domain B (residues SINV C201 and RRV C201), and disulfide bond 6 in domain B (residues SINV C220 and RRV C220) (Fig. 1B and C and Table 1). There are 4 conserved Cys residues in the E2 ectodomain that are not part of a C-(X)1-5-C motif (Fig. 1D). These residues make up disulfide bonds 3 and 4 and are not part of this study. In our mutants, we have one free, unpaired Cys residue. As a result, different transient states during protein folding may occur, but it is unlikely that the folded protein will have a novel fold.
Table 1.
Cysteine mutations made in the E2 ectodomain of Sindbis and Ross River viruses
| Disulfide bond | Cysteine mutated in: |
Location in E2 | |
|---|---|---|---|
| SINV | RRV | ||
| 1 | 16 | 19 | Domain A |
| 2 | 19 | 22 | Domain A |
| 3 | No mutations | No mutations | Domain A |
| 4 | No mutations | No mutations | β ribbon |
| 5 | 201 | 201 | Domain B |
| 6 | 220 | 220 | Domain B |
The SINV and RRV E2 Cys mutants have decreased levels of infectivity compared to wild-type virus.
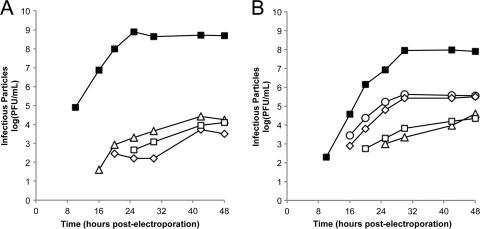
Each of the SINV and RRV E2 Cys mutants was reduced in titer compared to the parental virus, with SINV C16S producing no detectable infectious particles. SINV C19S and SINV C220S were reduced in titer by 5 log units compared to wild-type SINV (Fig. 2A). In contrast, growth of the RRV E2 Cys mutants was divided into two groups. In one group, RRV C22S and RRV C220S were reduced in titer by 4 log units compared to wild-type RRV. The second group contained viruses, RRV C19S and RRV C201S, that were reduced in titer by 2 log units compared to wild-type RRV (Fig. 2B). The experiments to obtain growth curves were performed twice for each E2 Cys mutant. Since the growth curves were initiated by electroporation, the number of infectious virus particles produced by each mutant varied between experiments. However, the ratios of infectious mutant particles produced to infectious wild-type particles produced were the same in each experiment. Similar patterns of decreased infectivity were also observed when the Aedes albopictus cell line C6/36 was used to propagate the E2 Cys mutants (data not shown).
Fig 2.
Production of infectious virus particles by the SINV and RRV E2 Cys mutants. BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA. At the indicated time points, the medium was harvested and titers were determined by standard plaque assay procedure. The experiments to obtain growth curves were performed twice for each E2 Cys mutant. The results from one representative experiment are shown. (A) Growth curves of the SINV E2 Cys mutants. ■, wild-type SINV; △, SINV E2 C19S; ♢, SINV E2 C201S; □, SINV E2 C220S. SINV E2 C16S produced no detectable infectious particles. (B) Growth curves of the RRV E2 Cys mutants. ■, wild-type RRV; ○, RRV E2 C19S; △, RRV E2 C22S; ♢, RRV E2 C201S; □, RRV E2 C220S.
Plaque sizes were smaller for each of the E2 Cys mutants than the parental virus. Wild-type SINV and RRV had plaque sizes that were 3 to 4 mm in diameter at 36 h postinfection. Viruses with Cys mutations in domain A of E2 (SINV C19S, RRV C19S, and RRV C22S) had plaque sizes that were 1 to 3 mm in diameter. Viruses with Cys mutations in domain B of E2 (SINV C201S, RRV C201S, SINV C220S, and RRV C220S) had plaque sizes that were less than 1 mm in diameter.
The total number of particles produced, both infectious and noninfectious, was determined using qRT-PCR (70). Briefly, qRT-PCR primers corresponding to regions of nsP1 and E2 were used to quantify the number of viral RNA genome copies, or total number of particles, present in the medium. For each of the SINV and RRV E2 Cys mutants, we observed at most a 1.5-log-unit decrease in the total number of particles produced, indicating a slight to moderate reduction in particle assembly compared to wild-type virus (data not shown). However, the ratio of total number of particles/total number of infectious particles (particle-to-PFU ratio) was 2 to 4 log units higher for each of the E2 Cys mutants than wild-type virus. Our approach for determining particle-to-PFU ratios assumes that there is one genome copy per particle. If multicored particles are formed, there could hypothetically be five nucleocapsid cores per virion, resulting in a particle-to-PFU ratio of 5 for a single infectious particle. Nonetheless, the several-log-unit increase in particle-to-PFU ratio observed for each of the E2 Cys mutants suggests that only a small percentage of the released virions were competent to infect a new cell.
The Cys mutations in E2 affect pE2 cleavage differently in SINV and RRV.
One of the last steps in spike assembly is spike maturation when the cellular enzyme furin cleaves pE2 (E3-E2) to yield the individual E3 and E2 glycoproteins (21, 49, 68). This cleavage is necessary to make the viral spikes fusion competent for entry into a new cell. The presence of large quantities of pE2 relative to E2 could indicate either improper folding or improper heterodimerization with E1 in virus-infected cells. To determine if the SINV and RRV E2 Cys mutants had pE2 cleavage kinetics similar to that of wild-type virus, we monitored the presence of pE2 and E2 in cell lysates over time (Fig. 3).
Fig 3.
pE2 processing characterization of the SINV and RRV E2 Cys mutants. BHK-21 cells were electroporated with in vitro-transcribed wild-type or mutant viral RNA. At the indicated time points, cells were lysed with BHK cell lysis buffer. Cell lysates were solubilized in reducing SDS sample buffer. The solubilized cell lysates were analyzed by SDS-PAGE and probed with polyclonal antibodies recognizing pE2/E2 (top blot for each construct) and capsid (bottom blot for each construct) by Western blot. Migration of pE2, E2, and capsid bands is indicated to the right of each blot. Migration of molecular mass standards is indicated to the left of each blot. (A) Wild-type SINV and RRV; (B) pE2 furin cleavage mutants of SINV and RRV; (C and D) domain A mutants; (E and F) domain B mutants.
For wild-type SINV and RRV, approximately equal amounts of pE2 and E2 were observed (Fig. 3A). For SINV and RRV pE2 mutants (viruses with altered furin cleavage sites [21]), the majority of the cellular protein was present in the pE2 form (Fig. 3B). Levels of capsid are shown in all samples as a loading control. We observed two distinct cleavage patterns for the E2 Cys mutants: those that had similar levels of pE2 and E2 in cell lysates (SINV C19S, RRV C201S, and RRV C220S) and those that had predominantly pE2 in cell lysates (RRV C19S, RRV C22S, SINV C201S, and SINV C220S). Interestingly, the corresponding Cys mutations in E2 of SINV and RRV did not yield similar pE2 cleavage patterns (compare the SINV and RRV panels in Fig. 3D to F). The domain A mutation of SINV C19S displayed a pE2 processing pattern similar to that of wild-type SINV. In contrast, the domain A mutants in RRV, RRV C19S and C22S, showed predominantly pE2 in cell lysates. Differences in pE2 and E2 glycoprotein quantities were also seen in the domain B mutants. The pE2 glycoprotein is predominant for SINV C201S and SINV C220S. For RRV C201S and RRV C220S, similar amounts of pE2 and E2 were present in cell lysates.
Although pE2/E2 were detected in all of the E2 Cys mutants except SINV C16S, there were delays in the timing of detection. In the SINV mutants (C19S, C201S, and C220S), pE2 and E2 were detected 5 to 10 h later than in wild-type SINV, 25 h postelectroporation versus 20 h postelectroporation. We observed a similar delay with RRV C22S but not with RRV C201S or RRV C220S. These delays suggest that the detectable amount of protein was observed at later time points due to a reduction in the accumulation of pE2/E2 in the cell.
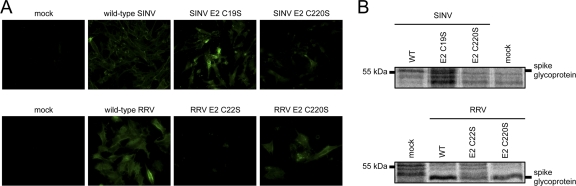
The decreased levels of virus production and the abundance of pE2 compared to E2 in cell lysates are consistent with overall E2 misfolding resulting in decreased surface expression of E2. Immunofluorescence and cell surface biotinylation experiments showed that E1/E2 were present at the plasma membrane, albeit at reduced amounts, for SINV C19S, SINV C220S, RRV C22S, and RRV C220S (Fig. 4). Thus, while pE2 processing is misregulated in some of our mutants, there are fusogenic spikes at the plasma membrane because we detected E1/E2 at the plasma membrane (Fig. 4) and we observed the release of infectious particles (Fig. 2). The reduced amounts of E1/E2 at the plasma membrane may impact virus assembly in several ways. Particles that are not fully covered by viral glycoprotein spikes, multicored, and/or nonicosahedral may be released from the host cell. The released particles may also be less stable than wild-type particles, possibly due to improper E2-nucleocapsid core interactions.
Fig 4.
Cell surface pE2/E2 and E1 expression by the SINV and RRV E2 Cys mutants. BHK-21 cells were electroporated with in vitro-transcribed wild-type (WT) or mutant viral RNA. (A) Immunofluorescence probing for cell surface glycoprotein expression. At 18 h postelectroporation, the cells were fixed with freshly prepared 1.5% paraformaldehyde. The fixed cells were probed with polyclonal antibodies recognizing SINV pE2/E2 or E1 and RRV pE2/E2, and the secondary antibody was Alexa Fluor 488. The cells were imaged at ×20 magnification using an Olympus 1X71 fluorescence microscope. Cell surface glycoprotein expression is false colored in green. (B) Biotinylation probing for cell surface glycoprotein expression. At 18 h postelectroporation, the cells were incubated in l-methionine- and l-cysteine-free medium for 15 min. After 15 min, [35S]cysteine-methionine was added to the medium and the cells were incubated for an additional 2 h. Radiolabeled cells were washed with PBS, and cell surface proteins were biotinylated for 30 min at room temperature with 2.5 mM NHS-LC-biotin. Following biotinylation, the cells were lysed with BHK cell lysis buffer. Biotinylated proteins were purified from cell lysates using streptavidin resin and eluted by adding reducing SDS sample buffer and boiling at 95°C for 10 min. The eluted proteins were analyzed by SDS-PAGE. Migration of spike glycoprotein bands is indicated to the right of each image. Migration of molecular mass standards is indicated to the left of each image.
For the remainder of our studies, we characterized SINV C19S and RRV C22S to represent viruses with mutations in domain A of E2. We characterized SINV C220S and RRV C220S to represent viruses with mutations in domain B of E2.
The SINV E2 Cys mutants form large, misassembled particles.
The E2 Cys mutants show reduced infectivity and, in some cases, atypical pE2 cleavage patterns compared to wild-type SINV and RRV. If the E2 Cys mutations inhibit furin cleavage of E3 from E2, then uncleaved pE2 may be incorporated into the released particles. The presence of pE2 in the released particle reduces virus infectivity (21). Alternatively, the Cys mutations in E2 may prevent proper lateral spike interactions at the plasma membrane or interactions between E2 and the nucleocapsid core during particle budding. Consequently, the glycoprotein spikes would not envelop a single nucleocapsid core and virions with multiple cores could form. Thus, it is necessary to determine the composition and morphology of the released E2 Cys mutant particles.
Virus particles in the medium were pelleted through a 27% sucrose cushion. The pelleted particles were resuspended in HNE buffer, analyzed by SDS-PAGE, and probed for E2 and capsid by Western blotting (Fig. 5A). Wild-type and pE2 virus are shown as controls for the migration of E2 and pE2 (E3-E2) bands, respectively. SINV C19S incorporated E2 into the released particles, while SINV C201S particles contained an aberrant species of E2, which migrated more slowly than E2 but faster than pE2. The nature of this protein is unknown. In contrast to wild-type SINV, each of the pelleted SINV E2 Cys mutants contained an abundance of capsid over E2, suggesting the presence of multicored particles. Individual wild-type SINV capsid and E2 values were set to 1, which made the ratio of capsid/E2 equal to 1. Both SINV C19S and SINV C201S showed ∼5 times higher levels of capsid and 10-fold less E2 glycoprotein relative to wild-type SINV. As a result, the capsid/E2 ratios for these mutants were approximately 80-fold higher than the ratio observed in wild-type SINV. For SINV C220S, capsid levels were also ∼5-fold higher than those for the wild type, but very little E2 glycoprotein was detected. In contrast to the SINV E2 Cys mutants, each of the RRV E2 Cys mutants incorporated cleaved E2 into the released particles (Fig. 5A). The relative levels of capsid to E2 were higher in the RRV E2 Cys mutants, ranging from 1.2 to 6, than wild-type RRV but were not as high as those observed for the SINV E2 Cys mutants. The individual levels of capsid and E2 in the RRV E2 Cys mutants were also much closer to wild-type RRV levels. The increased ratios of capsid/E2 in the E2 Cys mutants compared to wild-type particles could be due to single-cored or multicored particles that have reduced amounts of spike proteins on the surface of the released virus particles.
Fig 5.
Composition and morphology of the released SINV and RRV E2 Cys mutant particles. Wild-type (WT) and mutant viruses were purified from medium by pelleting through a 27% sucrose cushion. The pelleted particles were resuspended in HNE buffer. (A) Composition of the pelleted E2 Cys mutant particles. Purified particles were solubilized in reducing SDS sample buffer. The solubilized particles were analyzed by SDS-PAGE and probed with polyclonal antibodies recognizing pE2/E2 (top blot for each construct) and capsid (bottom blot for each construct) by Western blotting. For wild-type SINV, SINV pE2, and the SINV E2 Cys mutants, 1 × 106 PFU was loaded onto the gel. For wild-type RRV, RRV pE2, and the RRV E2 Cys mutants, 1 × 104 to 8 × 105 PFU was loaded onto the gel. For each of the E2 Cys mutants, the total number of particles loaded onto the gel is much greater than the number of PFU due to the high particle/PFU ratio. Migration of pE2, E2, and capsid bands is indicated to the right of each blot. Migration of molecular mass standards is indicated to the left of each blot. (B) TEM images of pelleted E2 Cys mutant particles. Purified particles were applied to a Formvar- and carbon-coated 400-mesh copper grid and stained with 1% uranyl acetate. The stained grids were imaged at ×40,000 magnification.
To examine virion morphology, specifically looking for single-cored versus multicored particles, the pelleted particles were analyzed by negative-stain TEM (Fig. 5B). Wild-type SINV and RRV produced spherical particles that were 70 nm in diameter, which is consistent with previous results (7, 66). Although SINV C19S assembled a small number of particles that were similar in size and morphology to wild-type SINV, both SINV C19S and SINV C220S predominantly produced large, pleomorphic particles. The high particle-to-PFU ratios for the SINV E2 Cys mutants and the abundance of capsid over E2 in the pelleted particles (Fig. 5A) suggest that the aberrantly formed virions contain multiple nucleocapsid cores. The morphologies of the SINV C19S and SINV C220S particles were very similar to those of multicored particles observed by Ivanova et al. (19).
TEM analysis revealed that the majority of the particles produced by RRV C22S and RRV C220S were similar in size to those produced by wild-type RRV. The RRV E2 Cys mutants produced a small number of large particles that had irregular morphologies. These findings were consistent with the high particle-to-PFU ratios and the relative amounts of E2 and capsid in the pelleted RRV E2 Cys mutant particles (Fig. 5A).
Host-cell proteins are potentially incorporated into the SINV and RRV E2 Cys mutant particles.
During alphavirus assembly, the spikes form hexagonal arrays at the plasma membrane (56). These arrays are thought to serve as the sites for E2 and nucleocapsid core interactions during particle budding. In contrast to other enveloped viruses, alphaviruses do not overtly incorporate host-cell proteins into the released particles (4, 16, 44). It is hypothesized that the lateral spike interactions at the cell surface prevent the incorporation of host-cell proteins into the released particles (56). The Cys mutations in E2 may cause improper spike organization at the plasma membrane. Thus, interactions between E2 and the nucleocapsid core could be disrupted and host-cell proteins may be incorporated into the mutant virions, leading to particles with atypical morphologies (Fig. 5B).
To determine if host protein was present in the E2 Cys mutant particles, we purified [35S]cysteine-methionine-labeled virus using a two-step (27% and 60%) sucrose gradient. Virus was radiolabeled during infection to ensure that only host-cell and viral proteins and not contaminating proteins from the medium or FBS were labeled. Step gradients were used to purify fragile particles that may have been damaged during pelleting through a sucrose cushion. Virus was isolated from the 27% and 60% sucrose interface, the particle number was determined by qRT-PCR, and roughly equal numbers of virions (within 1 log unit) were analyzed by SDS-PAGE (Fig. 6). As expected for wild-type SINV and RRV, only E1/E2 and capsid and no additional proteins were observed in the purified particles. Each of the E2 Cys mutant particles contained proteins in addition to E1/E2 and capsid. These additional proteins may be derived from the host cell, from viral structural polyproteins (E3-E2-6K-E1) that were not properly cleaved or modified during assembly, or from improperly oligomerized viral proteins.
Fig 6.
Composition of radiolabeled SINV and RRV E2 Cys mutant particles. BHK-21 cells were electroporated with in vitro-transcribed wild-type (WT) or mutant viral RNA. At 18 h postelectroporation, the cells were incubated in l-methionine- and l-cysteine-free medium for 15 min. After 15 min, [35S]cysteine-methionine was added to the medium and the cells were incubated for an additional 24 h. Following production, radiolabeled virus was purified from the medium through a 27% and 60% sucrose step gradient. Particles present at the 27% and 60% sucrose step interface were extracted and solubilized in reducing SDS sample buffer. The solubilized particles were analyzed by SDS-PAGE. For wild-type SINV and the SINV E2 Cys mutants, 5 × 107 to 4 × 108 particles were loaded onto the gel. For wild-type RRV and the RRV E2 Cys mutants, 1 × 109 to 3 × 1010 particles were loaded onto the gel. Particle number was quantitated using qRT-PCR. Migration of E1/E2 and capsid bands is indicated to the left of each set of images.
Virions purified via a step gradient (Fig. 6) showed ratios of capsid protein to E1/E2 proteins that were similar to those for wild-type SINV and RRV. This is in contrast to virions purified through a sucrose cushion (Fig. 5A), where the SINV E2 Cys mutants showed an abundance of capsid protein. This result is consistent with the observation that SINV C19S, SINV C220S, and RRV C22S formed large, misassembled particles that contain multiple nucleocapsid cores (Fig. 5B). Multicored particles would likely sediment through the 27% and 60% sucrose step interface, whereas particles with a single nucleocapsid core would remain at the step gradient interface. Interestingly, RRV C220S particles purified via a step gradient contained an abundance of capsid over E2 with a ratio of ∼90 (Fig. 6). Nevertheless, the majority of the particles observed by TEM for this mutant were single cored. One explanation is that there may be fewer spikes present on the surface of RRV C220S particles.
The SINV E2 Cys mutants contain E1 trimers in the released virus particles.
In order to assess the presence and function of the spike proteins on the particle surface, we looked at the ability of the mutant particles to undergo E1 trimerization, the first step in the fusion process during entry. Alphavirus particles enter the host cell through receptor-mediated endocytosis (10, 18, 34). In the endosome, the E2 and E1 heterodimers dissociate in response to a decrease in pH. The spikes then undergo a major conformational change as E1 trimers form and fusion between the viral and host-cell membrane occurs. If the spikes are misassembled, the pH requirements to form E1 trimers may be altered compared to those for wild-type virus (23, 50).
E1 trimerization can be recapitulated in vitro by mixing purified virus with liposomes at low pH (52). We incubated the E2 Cys mutants with liposomes (POPC, POPE, SM, and Chl at a 1:1:1:1.5 molar ratio) under three different pH environments, pH 7.4, 5.5, or 4.8. The virus-liposome mixtures were then pH neutralized and analyzed by SDS-PAGE under nonreducing conditions (Fig. 7A to F). Consistent with previous reports (52), both wild-type SINV and RRV displayed E1 trimerization activity when treated at pH 5.5 and 4.8 in the presence of liposomes, but not at pH 7.4 (Fig. 7A and B, respectively). Trimers dissociated into E1 monomers when the virus-liposome mixtures were heated to 95°C (lanes labeled boiled +) or treated with a reducing agent prior to SDS-PAGE analysis (data not shown).
Fig 7.
E1 trimerization activity in SINV and RRV E2 Cys mutant particles. Radiolabeled wild-type (A and B) or mutant (C to F) virus was mixed with 0.8 mM liposomes (PC, PE, SM, and Chl at 1:1:1:1.5). The virus-liposome mixture was adjusted to the indicated pH for 5 min. After 5 min, the virus-liposome mixture was pH neutralized and solubilized with nonreducing SDS sample buffer. The solubilized virus-liposome mixtures were analyzed by SDS-PAGE. At each pH tested, the neutralized samples were split into equal aliquots and either boiled at 95°C (+ lanes) or not boiled (− lanes) prior to loading onto the gel. For wild-type SINV and the SINV E2 Cys mutants, 2 × 105 to 1 × 106 particles were loaded into each lane. For wild-type RRV and the RRV E2 Cys mutants, 4 × 106 to 9 × 107 particles were loaded into each lane. Particle number was quantitated using qRT-PCR. Migration of E1 trimer and E1/E2 bands is indicated to the right of each set of images. (G and H) Radiolabeled wild-type or mutant virus, in the absence of liposomes, was solubilized in nonreducing SDS sample buffer and analyzed by SDS-PAGE. For each virus, samples were either boiled at 95°C or not boiled prior to loading onto the gel. Migration of E1 trimer and E1/E2 bands is indicated to the right of each set of images.
The SINV E2 Cys mutants formed E1 trimers at each pH tested, indicating that trimerization occurred in a pH-independent manner (Fig. 7C and E). Furthermore, in the absence of liposomes (Fig. 7G), E1 trimers and monomers were present in the purified SINV E2 Cys mutant particles; trimers of E1 did not require the presence of liposomes to form. However, in the presence of liposomes, there was a complete loss of the E1/E2 monomer band, indicating that additional trimerization had occurred. These results suggest that the Cys mutations in E2 disrupted spike assembly such that a portion of the E1 glycoprotein present on the particle surface was in the trimeric state. In the absence of a host-cell membrane, premature trimer formation would render the spikes fusion incompetent and the virus noninfectious (37). We speculate that the E2 Cys mutations interfere with E2 folding and cause misregulation of spike formation and pE2 cleavage. As a result, E1 trimers were already formed when the spikes reached the plasma membrane and were present on the surface of the SINV E2 Cys mutant particles.
In contrast to the domain A SINV C19S mutant, RRV C22S demonstrated E1 trimerization activity in a pH-dependent manner, similar to wild-type RRV (Fig. 7D). For RRV C220S, a domain B mutant, the E1 trimer band intensities were too weak to make a definitive conclusion about E1 trimerization activity (Fig. 7F). The lack of E1 trimerization is consistent with RRV C220S having reduced infectivity, possibly due to small amounts of viral spikes on the particle surface (Fig. 6). In purified particles, E1 trimers were not observed on the surface of the RRV E2 Cys mutant particles (Fig. 7H). Thus, in RRV, we speculate that the E2 Cys mutations did not impact E2 folding and/or pE2 cleavage in a manner similar to that in SINV.
DISCUSSION
The E2 glycoprotein is a major component of the viral spikes and has numerous functions both during assembly and in the mature particle. First, E2 is responsible for mediating receptor binding during a viral infection (5, 24, 53). Second, E2 modulates pE2-E1 heterodimerization and spike oligomerization during assembly (30, 31, 41). Third, cleavage of E3 from E2 is required to render the spikes fusion competent (21, 49, 51, 58, 68). Fourth, E2 interacts with the nucleocapsid core during budding. Fifth, E2 must dissociate from E1 for fusion to occur during entry (23, 50). Therefore, a mutation in E2 could affect virus binding to a host cell, spike assembly, spike maturation, and spike disassembly during entry. Mutations that alter particle formation early in assembly may also have one or more downstream effects. This work examined conserved cysteines within two distinct regions, domains A and B, of the E2 ectodomain through site-directed mutagenesis. As predicted, these mutants were defective at several stages of the virus life cycle. Surprisingly, we observed nonconserved assembly defects when corresponding mutations were made in the E2 glycoprotein of SINV and RRV (Table 2). Thus, although the Cys residues and disulfide bonds are conserved, functional differences exist between SINV and RRV E2.
Table 2.
Summary of E2 Cys mutant phenotypes in Sindbis and Ross River viruses
| Virus | Domain site of mutation | Relative levels of pE2 | Capsid and E2 levels in particles | Morphology of particles | Potential host-cell proteins | E1 trimer formation |
|---|---|---|---|---|---|---|
| SINV WTa | pE2 similar to E2 | Capsid similar to E2 | Spherical, 70-nm diam | No | pH dependent | |
| SINV C19S | A | pE2 similar to E2 | Capsid >>> E2 | Large, multicored | Yes | pH independent, trimers in particles |
| SINV C220S | B | Predominantly pE2 | Capsid >>> E2 | Large, multicored | Yes | pH independent, trimers in particles |
| RRV WT | pE2 similar to E2 | Capsid similar to E2 | Spherical, 70-nm diam | No | pH dependent | |
| RRV C22S | A | Predominantly pE2 | Capsid > E2 | Single cored | Yes | pH dependent |
| RRV C220S | B | pE2 similar to E2 | Capsid > E2 | Single cored | Yes | Unclear |
WT, wild type.
Most striking from our results was the different particle morphologies observed between the SINV and RRV E2 Cys mutants. The SINV E2 Cys mutants produced large, multicored particles that appeared to contain host-cell proteins (Fig. 5B and 6). Disrupting disulfide bonds in either domain A or B in SINV has either a direct or indirect effect on lateral spike contacts and E2-capsid interactions during budding. Multicored particles or aberrantly shaped particles have also been observed when mutations were made in the 6K protein and the E2 endodomain. In these mutants, palmitoylation sites were disrupted (19, 20), possibly altering the spike organization at the plasma membrane and affecting budding. For the RRV E2 Cys domain A and B mutants, the particles contained single nucleocapsid cores but had lower levels of E2 glycoprotein in the particle, possibly due to defects in spike-capsid interactions (Fig. 5B). The RRV E2 Cys mutants also contained host-cell proteins, consistent with improper lateral spike formation at the plasma membrane (Fig. 6). We hypothesize that atypical lateral spike organization may be a direct result of the Cys mutation or an indirect, downstream effect resulting from misregulation during pE2-E1 heterodimerization or spike oligomerization. Direct assays to examine lateral spike interactions or the spacing of spike proteins at the plasma membrane have been established in some enveloped virus systems (3, 4). These approaches have not been established in the alphavirus field, but new assays are being developed. Furthermore, in the SINV E2 Cys mutants, some of the E1 glycoprotein was present in a trimeric form on the particle surface (Fig. 7G), possibly as a consequence of premature E3 cleavage from E2 due to the Cys mutation. In RRV, trimeric E1 was not detected in the virus particles (Fig. 7H), perhaps because the E2 Cys mutations did not affect the timing of E3 cleavage from E2.
The finding that a mutation in one virus has a different outcome when the corresponding mutation is made in another species has previously been observed in different virus genera. In reoviruses, the σ3 protein of strains T3A and T3D share over 95% amino acid identity (22). In T3A, the virus is sensitive to ammonium chloride treatment and protease treatment, a phenotype genetically tracked to the histidine 354 residue in the σ3 protein. However, when the corresponding Y354H mutation is made in the T3D strain, the virus is resistant to ammonium chloride treatment and displays increased protease sensitivity (8, 61, 64). In the F proteins of paramyxoviruses, which share only 40% amino acid similarity, mutations in the conserved block regions of the F1 and F2 proteins, termed CBF1 and CBF2, respectively, had different phenotypes for processing and trafficking to the cell surface (12, 13). In coronaviruses, it is the requirement of a posttranslational modification that is virus species specific. Palmitoylation of the severe acute respiratory syndrome coronavirus (SARS-CoV) S protein is not necessary for efficient interaction with the SARS-CoV M protein (36). In contrast, palmitoylation of the S protein is required for mouse hepatitis virus (MHV), also a coronavirus, to interact with the MHV M protein during assembly (55). Thus, the different phenotypes observed in different virus species may be more common than reported.
The phenotypes that we observed for the SINV and RRV E2 Cys mutants led us to propose that the chaperones that regulate E2 folding and function could be virus species specific. During spike assembly, E3 plays a chaperone role for E2 folding (6, 40, 43) and the presence of E3 is thought to prevent the spikes from prematurely converting from the stable conformation to the meta-stable conformation (17, 57). Only as the spikes pass through the trans-Golgi network is E3 cleaved, making the spikes fusion competent (21, 49, 51, 58, 68). In the atomic structure, E3 interacts with the β ribbon in E2 (57), but during assembly, E3 may be flexible and have additional interactions with domains A and B. It is known that E3 is found to be noncovalently associated with the released particle in certain alphavirus species, such as Semliki Forest virus (SFV), but in other species, E3 is released into the medium (35, 60). Interestingly, the SFV and RRV E2 glycoproteins share 69% amino acid identity and the CHIK and SFV E2 glycoproteins share 55% amino acid identity. In contrast, SINV E2, compared to SFV, CHIK, or RRV E2, has at most, 40% amino acid identity, including absolute conservation of the Cys residues. We examined regions within each domain of SINV and RRV E2 to see if the Cys residues were present in different local environments. Only the N terminus of domain A, where the domain A Cys mutations were present, has lower amino acid sequence similarity. Thus, the N terminus of domain A could potentially be a site for species-specific chaperone or host factor interactions. Therefore, association constants, covalent interactions, and cleavage requirements between E3 and E2 may be species specific.
ACKNOWLEDGMENTS
We thank members of the Indiana University virology group for helpful discussions.
This work received support from NIH Genetics, Cellular, and Molecular Sciences Training Grant T32GM007757 (to A.J.S.).
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Bonatti S, Blobel G. 1979. Absence of a cleavable signal sequence in Sindbis virus glycoprotein PE2. J. Biol. Chem. 254:12261–12264 [PubMed] [Google Scholar]
- 2. Bonatti S, Migliaccio G, Blobel G, Walter P. 1984. Role of signal recognition particle in the membrane assembly of Sindbis viral glycoproteins. Eur. J. Biochem. 140:499–502 [DOI] [PubMed] [Google Scholar]
- 3. Brown EL, Lyles DS. 2003. A novel method for analysis of membrane microdomains: vesicular stomatitis virus glycoprotein microdomains change in size during infection, and those outside of budding sites resemble sites of virus budding. Virology 310:343–358 [DOI] [PubMed] [Google Scholar]
- 4. Brown EL, Lyles DS. 2005. Pseudotypes of vesicular stomatitis virus with CD4 formed by clustering of membrane microdomains during budding. J. Virol. 79:7077–7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byrnes AP, Griffin DE. 1998. Binding of Sindbis virus to cell surface heparan sulfate. J. Virol. 72:7349–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carleton M, Lee H, Mulvey M, Brown DT. 1997. Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J. Virol. 71:1558–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng RH, et al. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark KM, et al. 2006. Reovirus variants selected for resistance to ammonium chloride have mutations in viral outer-capsid protein sigma3. J. Virol. 80:671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Curtis I, Simons K. 1988. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc. Natl. Acad. Sci. U. S. A. 85:8052–8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doxsey SJ, Brodsky FM, Blank GS, Helenius A. 1987. Inhibition of endocytosis by anti-clathrin antibodies. Cell 50:453–463 [DOI] [PubMed] [Google Scholar]
- 11. Gahmberg CG, Utermann G, Simons K. 1972. The membrane proteins of Semliki Forest virus have a hydrophobic part attached to the viral membrane. FEBS Lett. 28:179–182 [DOI] [PubMed] [Google Scholar]
- 12. Gardner AE, Dutch RE. 2007. A conserved region in the F(2) subunit of paramyxovirus fusion proteins is involved in fusion regulation. J. Virol. 81:8303–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gardner AE, Martin KL, Dutch RE. 2007. A conserved region between the heptad repeats of paramyxovirus fusion proteins is critical for proper F protein folding. Biochemistry 46:5094–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garoff H, Frischauf AM, Simons K, Lehrach H, Delius H. 1980. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature 288:236–241 [DOI] [PubMed] [Google Scholar]
- 15. Garoff H, Simons K, Renkonen O. 1974. Isolation and characterization of the membrane proteins of Semliki Forest virus. Virology 61:493–504 [DOI] [PubMed] [Google Scholar]
- 16. Hammarstedt M, Wallengren K, Pedersen KW, Roos N, Garoff H. 2000. Minimal exclusion of plasma membrane proteins during retroviral envelope formation. Proc. Natl. Acad. Sci. U. S. A. 97:7527–7532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Helenius A, Kartenbeck J, Simons K, Fries E. 1980. On the entry of Semliki forest virus into BHK-21 cells. J. Cell Biol. 84:404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ivanova L, Lustig S, Schlesinger MJ. 1995. A pseudo-revertant of a Sindbis virus 6K protein mutant, which corrects for aberrant particle formation, contains two new mutations that map to the ectodomain of the E2 glycoprotein. Virology 206:1027–1034 [DOI] [PubMed] [Google Scholar]
- 20. Ivanova L, Schlesinger MJ. 1993. Site-directed mutations in the Sindbis virus E2 glycoprotein identify palmitoylation sites and affect virus budding. J. Virol. 67:2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain SK, DeCandido S, Kielian M. 1991. Processing of the p62 envelope precursor protein of Semliki Forest virus. J. Biol. Chem. 266:5756–5761 [PubMed] [Google Scholar]
- 22. Kedl R, Schmechel S, Schiff L. 1995. Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J. Virol. 69:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kielian M, Chanel-Vos C, Liao M. 2010. Alphavirus entry and membrane fusion. Viruses 2:796–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Klimstra WB, Ryman KD, Johnston RE. 1998. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J. Virol. 72:7357–7366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuhn RJ, Niesters HG, Hong Z, Strauss JH. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430–441 [DOI] [PubMed] [Google Scholar]
- 26. Lescar J, et al. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137–148 [DOI] [PubMed] [Google Scholar]
- 27. Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liljestrom P, Garoff H. 1991. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 65:147–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lobigs M, Garoff H. 1990. Fusion function of the Semliki Forest virus spike is activated by proteolytic cleavage of the envelope glycoprotein precursor p62. J. Virol. 64:1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lobigs M, Wahlberg JM, Garoff H. 1990. Spike protein oligomerization control of Semliki Forest virus fusion. J. Virol. 64:5214–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lobigs M, Zhao HX, Garoff H. 1990. Function of Semliki Forest virus E3 peptide in virus assembly: replacement of E3 with an artificial signal peptide abolishes spike heterodimerization and surface expression of E1. J. Virol. 64:4346–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lopez S, Yao JS, Kuhn RJ, Strauss EG, Strauss JH. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lustig S, et al. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marsh M, Bolzau E, Helenius A. 1983. Penetration of Semliki Forest virus from acidic prelysosomal vacuoles. Cell 32:931–940 [DOI] [PubMed] [Google Scholar]
- 35. Mayne JT, Rice CM, Strauss EG, Hunkapiller MW, Strauss JH. 1984. Biochemical studies of the maturation of the small Sindbis virus glycoprotein E3. Virology 134:338–357 [DOI] [PubMed] [Google Scholar]
- 36. McBride CE, Machamer CE. 2010. Palmitoylation of SARS-CoV S protein is necessary for partitioning into detergent-resistant membranes and cell-cell fusion but not interaction with M protein. Virology 405:139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McInerney GM, Smit JM, Liljestrom P, Wilschut J. 2004. Semliki Forest virus produced in the absence of the 6K protein has an altered spike structure as revealed by decreased membrane fusion capacity. Virology 325:200–206 [DOI] [PubMed] [Google Scholar]
- 38. Melancon P, Garoff H. 1987. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J. Virol. 61:1301–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Molinari M, Helenius A. 2000. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science 288:331–333 [DOI] [PubMed] [Google Scholar]
- 40. Molinari M, Helenius A. 1999. Glycoproteins form mixed disulphides with oxidoreductases during folding in living cells. Nature 402:90–93 [DOI] [PubMed] [Google Scholar]
- 41. Mulvey M, Brown DT. 1996. Assembly of the Sindbis virus spike protein complex. Virology 219:125–132 [DOI] [PubMed] [Google Scholar]
- 42. Mulvey M, Brown DT. 1994. Formation and rearrangement of disulfide bonds during maturation of the Sindbis virus E1 glycoprotein. J. Virol. 68:805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mulvey M, Brown DT. 1995. Involvement of the molecular chaperone BiP in maturation of Sindbis virus envelope glycoproteins. J. Virol. 69:1621–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. 2009. Influenza virus morphogenesis and budding. Virus Res. 143:147–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Omar A, Koblet H. 1988. Semliki Forest virus particles containing only the E1 envelope glycoprotein are infectious and can induce cell-cell fusion. Virology 166:17–23 [DOI] [PubMed] [Google Scholar]
- 46. Owen KE, Kuhn RJ. 1996. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 70:2757–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parrott MM, et al. 2009. Role of conserved cysteines in the alphavirus E3 protein. J. Virol. 83:2584–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rice CM, Bell JR, Hunkapiller MW, Strauss EG, Strauss JH. 1982. Isolation and characterization of the hydrophobic COOH-terminal domains of the Sindbis virion glycoproteins. J. Mol. Biol. 154:355–378 [DOI] [PubMed] [Google Scholar]
- 49. Salminen A, Wahlberg JM, Lobigs M, Liljestrom P, Garoff H. 1992. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 116:349–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sanchez-San Martin C, Liu CY, Kielian M. 2009. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 17:514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sjoberg M, Lindqvist B, Garoff H. 2011. Activation of the alphavirus spike protein is suppressed by bound E3. J. Virol. 85:5644–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smit JM, Bittman R, Wilschut J. 1999. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J. Virol. 73:8476–8484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith TJ, et al. 1995. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. U. S. A. 92:10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thorp EB, Boscarino JA, Logan HL, Goletz JT, Gallagher TM. 2006. Palmitoylations on murine coronavirus spike proteins are essential for virion assembly and infectivity. J. Virol. 80:1280–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. von Bonsdorff CH, Harrison SC. 1978. Hexagonal glycoprotein arrays from Sindbis virus membranes. J. Virol. 28:578–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Voss JE, et al. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712 [DOI] [PubMed] [Google Scholar]
- 58. Wahlberg JM, Boere WA, Garoff H. 1989. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J. Virol. 63:4991–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wahlberg JM, Garoff H. 1992. Membrane fusion process of Semliki Forest virus. I. Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 116:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Welch WJ, Sefton BM. 1979. Two small virus-specific polypeptides are produced during infection with Sindbis virus. J. Virol. 29:1186–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wetzel JD, et al. 1997. Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J. Virol. 71:1362–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. White J, Helenius A. 1980. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc. Natl. Acad. Sci. U. S. A. 77:3273–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wilkinson TA, Tellinghuisen TL, Kuhn RJ, Post CB. 2005. Association of Sindbis virus capsid protein with phospholipid membranes and the E2 glycoprotein: implications for alphavirus assembly. Biochemistry 44:2800–2810 [DOI] [PubMed] [Google Scholar]
- 64. Wilson GJ, et al. 2002. A single mutation in the carboxy terminus of reovirus outer-capsid protein sigma 3 confers enhanced kinetics of sigma 3 proteolysis, resistance to inhibitors of viral disassembly, and alterations in sigma 3 structure. J. Virol. 76:9832–9843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Woycechowsky KJ, Wittrup KD, Raines RT. 1999. A small-molecule catalyst of protein folding in vitro and in vivo. Chem. Biol. 6:871–879 [DOI] [PubMed] [Google Scholar]
- 66. Zhang R, et al. 2011. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 30:3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang RM, Snyder GH. 1989. Dependence of formation of small disulfide loops in two-cysteine peptides on the number and types of intervening amino acids. J. Biol. Chem. 264:18472–18479 [PubMed] [Google Scholar]
- 68. Zhang X, Fugere M, Day R, Kielian M. 2003. Furin processing and proteolytic activation of Semliki Forest virus. J. Virol. 77:2981–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljestrom P. 1994. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 13:4204–4211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zybert IA, van der Ende-Metselaar H, Wilschut J, Smit JM. 2008. Functional importance of dengue virus maturation: infectious properties of immature virions. J. Gen. Virol. 89:3047–3051 [DOI] [PubMed] [Google Scholar]