Abstract
Background
A large proportion of basic research into disease mechanisms has leveraged genetic findings to model and understand etiology. There has been broad success in finding disease linked mutations using traditional positional cloning approaches, however, because of the requirements of the method, these successes have been limited by the availability of large, well characterized families. Because of these and other limitations the genetic basis of many diseases, and in many families remains unknown across myriad diseases.
Recent Developments
Exome sequencing uses DNA enrichment methods and massively parallel nucleotide sequencing to comprehensively discover and type protein-coding variants throughout the genome. Coupled with growing databases that contain known variants, exome sequencing affords the ability to find genetic mutations and risk factors in families and samples that were deemed insufficiently informative for previous genetic studies. Not only does this method afford discovery in families that linkage and positional cloning methods were unable to use, but compared to this method, it is much quicker and cheaper. Exome sequencing has had initial success in many rare diseases.
Where next?
Exome sequencing is being adopted widely and we can expect a landslide of mutation discovery, similar to the deluge of genome wide association findings reported over the past 5 years. It is to be expected that exome sequencing will enable not only the discovery of rare causal variants, but also protein coding risk variants. This method will have application in both the research and clinical arena and sets the scene for the use of whole genome sequencing.
At this moment, progress toward a full resolution of the genetic basis of disease is being significantly aided by a fast moving technology, exome sequencing. This method promises to speed up discovery of the genetic causes of disease, in both the research and the clinical setting.
The method of exome sequencing has been covered elsewhere1, although there are several methods they all use a similar principle; reducing a genomic DNA sample to one that is enriched for the protein coding regions of the genome (exons), followed by very high throughput sequencing of that exon-enriched sample. In short, a method to rapidly identify protein-coding mutations, including, missense, non-sense, splice site and small deletion/insertion mutations.
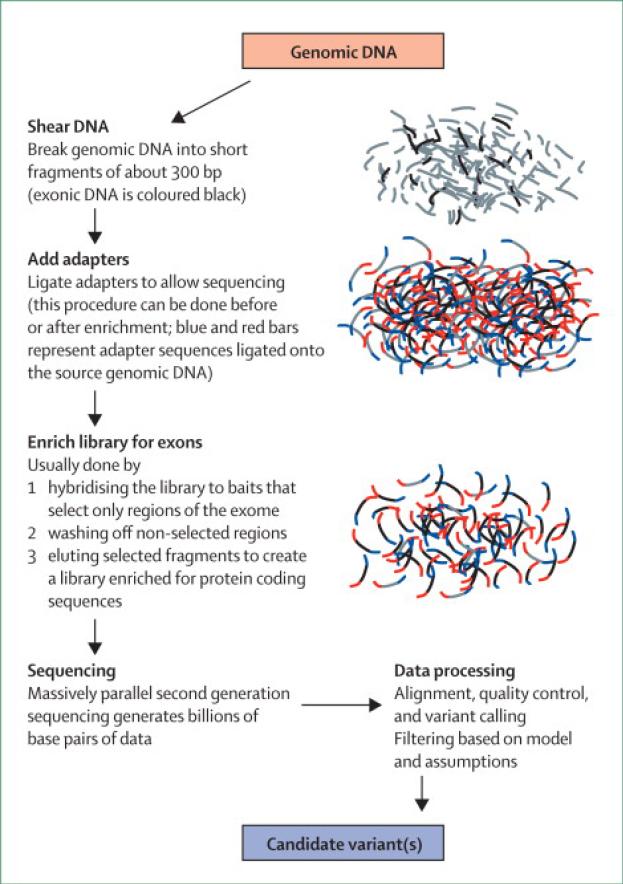
Exome sequencing uses second-generation sequencing, which generates sequence data from hundreds of millions of short DNA fragments in parallel. The sequencing of input libraries is, to all intents and purposes, random; each of the fragments that happen to be in the DNA library applied to the sequencer has a fairly even chance of being sequenced. Thus, directive sequencing of specific DNA fragments is determined by creating a DNA library solely consisting of, or enriched for, the DNA regions of interest. In the context of exome sequencing, this target selection is performed using one of several enrichment products; each of which aim to produce a DNA sample where the content is made up of the protein coding and regulatory regions of the genome. There are some limitations to this method. First, capture is not complete, in early experiments ~8% of the regions of interest resisted the enrichment strategy2 and although this has improved, it will likely never reach 100%; second, this method is not currently useful for identifying repeat mutations (such as triplet repeats in spinocerebellar ataxia); and third, copy number variants are difficult to detect with exome sequencing. However, given the distribution of variants, exome-sequencing remains an efficient way to identify the majority of mutations altering protein sequence in any single DNA sample. Although exome-sequencing is relatively new to the market, it has been rapidly adopted by the research community (figure).
FIGURE.
Schematic of the current method applied to allow exome sequencing
Mendelian disorders: a gold rush
The primary successes for exome sequencing have been in finding mutations that cause rare, familial forms of disease. The strength of this approach lies in comprehensive discovery of protein coding variants throughout the genome. This effectively means that DNA samples collected from small families, and isolated affected individuals, which could not be used for mutation identification through traditional linkage and positional cloning, can now be used to discover mutations causing disease. Exome sequencing of a DNA sample from a single individual will typically reveal ~25,000 variants; the challenge then lies not in finding variants, but in identifying the particular mutation responsible for disease. A common next step, when looking for extremely rare causal mutations involves filtering against variants known to exist in the general population, as these are unlikely to be disease causing. Genome wide association (GWA) studies have shown us that generation of large, accessible, reference data sets is an economical way to generate control data3 and this is a model that is being emulated in exome sequencing. At present the most broadly used reference panel for exome sequencing is the data derived from the 1000 genomes project, an initiative that will ultimately sequence the genomes of approximately 2500 individuals at low coverage (http://www.1000genomes.org/)4. The aim of this project is to provide a comprehensive resource on human genetic variation across a large series of individuals. Filtering the 25,000 or so variants identified in a single sample against known 1000 genomes and dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) variants typically reduces the list of candidate mutations down to ~700. Further filtering steps often include removing variants that don't appreciably alter amino acid sequence, do not fit with the mode of inheritance of disease, do not segregate with disease (in families), and those that are predicted computationally to have minimal consequences in terms of protein structure. This formulaic approach to winnowing down a candidate list of variants to define the causal variants has proven successful in a variety of diseases, including most recently, the successful identification of new Parkinson's disease causing mutations. 2, 5-24
As more sequence data accumulates in the public domain, the list of candidate variants after filtering will decrease, but with this the likelihood of erroneously filtering out a disease causing mutation increases. This is perhaps less likely in very young onset severe diseases, but probably quite a likely occurrence in later onset diseases, those with quite benign phenotypes, or with mutations that have a low penetrance. Mutation carriers who are too young to express disease, do not exhibit symptoms because of decreased penetrance, or have a phenotype that may not be noticed, may be sequenced and their data deposited in reference panels. Further, some (perhaps all) reference data is associated with very limited phenotypic data, so patients with, or who will get, common diseases are likely to be included but not identified.
Exome sequencing, beyond Mendelian disorders
There is increasing interest in leveraging the power of exome sequencing in diseases that do not exhibit a Mendelian mode of transmission. Exome sequencing has particular potential in one such case, diseases caused by non-inherited or de novo mutations. Typically this can be achieved by complementary approaches; first, and most simply, sequencing a group of cases suspected to have disease causing de novo mutations, and looking for a gene that is commonly mutated; second, and perhaps more compelling, sequencing parent-affected child trios, to establish the variant(s) that are present only in the affected children. De novo mutations were particularly resistant to previous genetic efforts aimed at gene localization and identification. In this context exome sequencing will not only help identify novel genetic causes of disease, but may also show that de novo mutations are a more common cause of disease than previously expected. There has already been some success in identifying these mutations using exome sequencing for varied diseases, including multiple malformation and mental retardation disorders.13, 19, 25, 26
Fittingly, in diseases with a complex genetic component, the situation is more complex. In this scenario, exome sequencing is being used to identify variability in protein coding regions that alters risk for, rather than causes, disease. There are numerous examples of this type of coding risk variant, although in general those that have been discovered to date are quite common variants and thus detectable by genome wide association studies, or were found through positional and functional candidate based screening strategies. Classic examples include APOE in Alzheimer's disease, CFH in age related macular degeneration, and GBA/LRRK2 in Parkinson's disease.27-31 The power of exome sequencing in this regard is that it will allow the identification of common protein coding risk alleles, and, given suitable power and study design, the identification of rare risk alleles. This latter approach centers on a somewhat complicated analytical design that involves assessing the collective burden of multiple risk alleles at a locus. While this approach remains largely unproven, initial work in epilepsy suggests that this method can reveal quite complex relationships between established disease associated variants, and enable risk prediction modeling32.
Challenges and Opportunities
It is extremely likely that exome sequencing, and if not genome sequencing, will also have significant impact in the clinical setting, outside of identifying genes that were previously unknown to contain disease causing mutations. In the context of genetic testing, rather than screening an inherently limited panel of genes for a particular set of diseases, why not just sequence the whole exome? This opens the door to a lot of possibilities, first, rapid genetic diagnosis and screening, when known disease-causing mutations are detected; second an inevitable expansion of the phenotypic range of disease associated with particular mutations or mutations in particular genes.
Exome sequencing has already proven its worth in the former; for many neurological diseases, where there is a long list of candidate genes and loci, it is often cheaper and certainly quicker to find mutations by exome sequencing. For many diseases with a high degree of genetic heterogeneity, screenings are often only designed to catch the most common mutations, or alterations in the most frequently mutated genes. Montenegro and colleagues demonstrated the power of exome sequencing in exactly this situation, with the analysis of a family with Charcot-Marie-Tooth (CMT) disease.33 Rather than screen the 35 genes known to contain mutations causing this disease, the authors used exome sequencing in 2 affected family members; with these data they were able to identify GJB1 mutations as a cause of CMT in this family. This case is also illustrative, because GJB1 mutation would have been ruled out for screening because of a reported male-to-male inheritance, incompatible with mutation of this gene, which lies on chromosome X. The use of exome sequencing by these authors likely saved time and money in reaching a genetic diagnosis. The increasing adoption of exome sequencing in the research setting, means that these data are becoming easier to process and analyze, in addition to becoming much cheaper to generate; many laboratories are now able to generate an exome for <$1500 a sample. When compared to the costs and time required for conventional screening, such a comprehensive approach represents value for money. Despite this still being a new technology, exome sequencing in genetically heterogeneous neurological diseases has already identified TECR mutations in non-syndromic mental retardation,10 WDR62 mutations in severe brain development malformations,34TGM6 mutations in ataxia,23 and a WRN mutation in atypical Werner's syndrome.35
As mentioned above, exome sequencing is likely to find mutations in genes previously linked to disease, but associated with a phenotype distinct from the one being tested. This has been elegantly demonstrated with the identification of VCP mutations as a cause of amyotrophic lateral sclerosis (ALS).15 In this article Johnson and colleagues had used exome sequencing to identify the genetic lesion responsible for an autosomal dominant form of ALS in a large pedigree from Italy. Surprisingly VCP mutations, including the same amino-acid change identified in this ALS kindred, had previously been linked to Paget's disease, inclusion body myopathy and frontotemporal dementia. Therefore this finding broadened the clinical and pathological phenotype of VCP mutations to include ALS and TDP-43 inclusions. Notably, further work by this group showed that VCP mutations are an appreciable cause of familial ALS, responsible for ~2% of cases in this group. As this work shows, broadening the phenotype associated with mutations has the potential to inform on the etiologic basis of disorders by uniting what is known about the biological underpinnings of apparently unrelated disorders into a single model.
One might also predict that as exome data accumulates we will get greater resolution on the role of mutations in disease. Exome data will not only help to identify pathogenic variants, including those of previously unknown or equivocal pathogenicity, but it will also help in determining penetrance, expressivity, and prevalence of mutations in particular populations. Related to the notion of penetrance, in the sequencing of normal individuals we might expect to find variants that were previously thought to cause fully penetrant disease, and these data will call into question pathogenicity of some published variants. As recent work describing the high phenocopy rate in families with apparently monogenic PD shows, in even the most ‘simple’ families, confounding genetic, environmental, or stochastic factors likely effect presentation and penetrance.36 In the beginning exome sequencing will raise many questions and often reveal an apparently confusing relationship between genetic variants and disease, but with time, accumulating data will help bring resolution to many outstanding clinico-genetic questions.
One clinical challenge that is particular to more comprehensive genetic approaches comes with the inevitable discovery of mutations unrelated to the condition in question. These secondary, or collateral, findings will be common in exome sequencing, and indeed one of the first exome publications, which sought to identify mutations for Miller syndrome, also described the identification of mutations causing ciliary dyskinesia within the same family.20, 37 The question then arises, what does a clinician do when they identify a mutation of proven or even potential clinical relevance; how should the disclosure of these mutations be handled. These are not necessarily simple issues, particularly when considering the confounds and problems such as disclosing carrier status, non-paternity, reduced penetrance, lack of viable treatment options and interpretation of risk factors. As has been elegantly argued previously, these issues in particular will require thoughtfully constructed research37 and continuing education of both health care providers and the general public.
More mutations, quicker, cheaper. So what?
There exists a point of view that there is little point in finding new genetic causes of disease, because we have made little progress in understanding the consequences of the mutations we know of. Not only does this remind me of the Luddites resistance to the technological advances of the Industrial Revolution (http://en.wikipedia.org/wiki/Luddite), but also to my mind this argument falls down. First, it is simply not true, an easy example is provided by the understanding of the role of amyloid processing in Alzheimer's disease that was imparted by the discovery of APP, PS1 and PS2 mutations. Second, this argument ignores non-research aspects of mutation identification, in particular our responsibility to find a cause of disease for patients, most of whom desire a diagnosis even if it doesn't come with a treatment. Lastly, this logic centers on the idea that etiologic based therapies are not likely, or not important, or both, because etiologic-based therapy will need to be grounded on a complete understanding of the disease process. Such an understanding requires the foundation of knowledge provided by defined disease-initiating events. There exists here a philosophical difference between those who believe in something of a systems based approach, where the more information (genes) we have to put in a model, the more reliable and complete our understanding of the network involved, to those who believe in a more reductionist approach to science, where the consequences of a single perturbation to a system should be studied one by one. However, even a truly reductionist approach would allow the separate study of perturbations at several genes and a subsequent search for overlap in the network of downstream effects. In either case, the more genes we can link to a disease entity, the more complete our understanding of the affected systems, and, the greater our understanding of the likely consequences of therapeutic interventions directed at those systems.
A second argument against applying exome sequencing appears to revolve around the notion that something else better will come along soon. Many genomic technologies are indeed transient in nature, and exome sequencing is likely no exception. The next logical step, taking us to whole genome sequencing, offers the advantages of comprehensive genomic coverage, an easier route to finding structural genomic mutations, and no intervening library enrichment steps. Inhibiting this transition are the high price point and large analytical burden (~100x the data of an exome). Critics have suggested that there is just too much uncertainty to deal with in non coding sequences and at too high a price to ever make whole genome sequencing viable; however, per base pair costs for genomic technologies continue to drop precipitously and scientists have never stayed away from large datasets for much time. Clearly, as both the cost and analytical limitations decrease whole genome sequencing will become the dominant technology. Indeed, this method has already been successful in identifying genetic lesions underlying disease, including the identification of a novel cause of CMT.38-41 Given this, one question frequently asked is “why not wait?” My view on this is simple: we cannot afford to wait. We are in a race to cure diseases. We know exome-sequencing works, and it is relatively affordable. The earlier we make genetic findings, the earlier we can begin integrating these discoveries into an understanding of the disease process. This surely places us on the road toward etiologic based therapies sooner and with a more complete set of tools. We should make hay while the sun shines; even if we know tomorrow's weather is pretty good too.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Department of Health and Human Services; project Z01 AG000958-08.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19(R2):R145–51. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Via M, Gignoux C, Burchard EG. The 1000 Genomes Project: new opportunities for research and social challenges. Genome Med. 2010;2(1):3. doi: 10.1186/gm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vilarino-Guell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, et al. VPS35 Mutations in Parkinson Disease. Am J Hum Genet. 2011;89(1):162–7. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimprich A, Benet-Pages A, Struhal W, Graf E, Eck SH, Offman MN, et al. A Mutation in VPS35, Encoding a Subunit of the Retromer Complex, Causes Late-Onset Parkinson Disease. Am J Hum Genet. 2011;89(1):168–75. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanna-Cherchi S, Burgess KE, Nees SN, Caridi G, Weng PL, Dagnino M, et al. Exome sequencing identified MYO1E and NEIL1 as candidate genes for human autosomal recessive steroid-resistant nephrotic syndrome. Kidney Int. 2011 doi: 10.1038/ki.2011.148. [DOI] [PubMed] [Google Scholar]
- 8.Hanson D, Murray PG, O'Sullivan J, Urquhart J, Daly S, Bhaskar SS, et al. Exome Sequencing Identifies CCDC8 Mutations in 3-M Syndrome, Suggesting that CCDC8 Contributes in a Pathway with CUL7 and OBSL1 to Control Human Growth. Am J Hum Genet. 2011 doi: 10.1016/j.ajhg.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolze A, Byun M, McDonald D, Morgan NV, Abhyankar A, Premkumar L, et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am J Hum Genet. 2010;87(6):873–81. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caliskan M, Chong JX, Uricchio L, Anderson R, Chen P, Sougnez C, et al. Exome sequencing reveals a novel mutation for autosomal recessive nonsyndromic mental retardation in the TECR gene on chromosome 19p13. Hum Mol Genet. 2011;20(7):1285–9. doi: 10.1093/hmg/ddq569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106(45):19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilissen C, Arts HH, Hoischen A, Spruijt L, Mans DA, Arts P, et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet. 2010;87(3):418–23. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42(6):483–5. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 14.Johnson JO, Gibbs JR, Van Maldergem L, Houlden H, Singleton AB. Exome sequencing in Brown-Vialetto-van Laere syndrome. Am J Hum Genet. 2010;87(4):567–9. doi: 10.1016/j.ajhg.2010.05.021. author reply 9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68(5):857–64. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krawitz PM, Schweiger MR, Rodelsperger C, Marcelis C, Kolsch U, Meisel C, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet. 2010;42(10):827–9. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- 17.Lalonde E, Albrecht S, Ha KC, Jacob K, Bolduc N, Polychronakos C, et al. Unexpected allelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum Mutat. 2010;31(8):918–23. doi: 10.1002/humu.21293. [DOI] [PubMed] [Google Scholar]
- 18.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363(23):2220–7. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42(1):30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce SB, Walsh T, Chisholm KM, Lee MK, Thornton AM, Fiumara A, et al. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am J Hum Genet. 2010;87(2):282–8. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87(1):90–4. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133(Pt 12):3510–8. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Li Y, Zhang D, Zhang H, Lu F, Liu X, et al. Exome Sequencing Identifies ZNF644 Mutations in High Myopia. PLoS Genet. 2011;7(6):e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43(6):585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vissers LE, de Ligt J, Gilissen C, Janssen I, Steehouwer M, de Vries P, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42(12):1109–12. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 27.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261(5123):921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 28.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan EK. Identification of a common genetic risk variant (LRRK2 Gly2385Arg) in Parkinson's disease. Ann Acad Med Singapore. 2006;35(11):840–2. [PubMed] [Google Scholar]
- 30.Hernandez DG, Paisan-Ruiz C, McInerney-Leo A, Jain S, Meyer-Lindenberg A, Evans EW, et al. Clinical and positron emission tomography of Parkinson's disease caused by LRRK2. Ann Neurol. 2005;57(3):453–6. doi: 10.1002/ana.20401. [DOI] [PubMed] [Google Scholar]
- 31.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med. 2009;361(17):1651–61. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klassen T, Davis C, Goldman A, Burgess D, Chen T, Wheeler D, et al. Exome sequencing of ion channel genes reveals complex profiles confounding personal risk assessment in epilepsy. Cell. 2011;145(7):1036–48. doi: 10.1016/j.cell.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montenegro G, Powell E, Huang J, Speziani F, Edwards YJ, Beecham G, et al. Exome sequencing allows for rapid gene identification in a Charcot-Marie-Tooth family. Ann Neurol. 2011;69(3):464–70. doi: 10.1002/ana.22235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467(7312):207–10. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raffan E, Hurst LA, Turki SA, Carpenter G, Scott C, Daly A, et al. Early diagnosis of Werner's syndrome using exome-wide sequencing in a single, atypical patient. Frontiers in Endocrinology. 2011;2:1–8. doi: 10.3389/fendo.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein C, Chuang R, Marras C, Lang AE. The curious case of phenocopies in families with genetic Parkinson's disease. Mov Disord. 2011 doi: 10.1002/mds.23853. [DOI] [PubMed] [Google Scholar]
- 37.Biesecker LG. Exome sequencing makes medical genomics a reality. Nat Genet. 2010;42(1):13–4. doi: 10.1038/ng0110-13. [DOI] [PubMed] [Google Scholar]
- 38.Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, et al. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6(9) doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6(6):e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N Engl J Med. 2010;362(13):1181–91. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rios J, Stein E, Shendure J, Hobbs HH, Cohen JC. Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Hum Mol Genet. 2010;19(22):4313–8. doi: 10.1093/hmg/ddq352. [DOI] [PMC free article] [PubMed] [Google Scholar]