Abstract
The formation of replication compartments, the subnuclear structures in which the viral DNA genome is replicated, is a hallmark of herpesvirus infections. The localization of proteins and viral DNA within human cytomegalovirus replication compartments is not well characterized. Immunofluorescence analysis demonstrated the accumulation of the viral DNA polymerase subunit UL44 at the periphery of replication compartments and the presence of different populations of UL44 in infected cells. In contrast, the viral single-stranded-DNA binding protein UL57 was distributed throughout replication compartments. Using “click chemistry” to detect 5-ethynyl-2′-deoxyuridine (EdU) incorporation into replicating viral DNA and pulse-chase protocols, we found that viral DNA synthesis occurs at the periphery of replication compartments and that replicated viral DNA subsequently localizes to the interior of replication compartments. The interiors of replication compartments also contain regions in which UL44 and EdU-labeled DNA are absent. The treatment of cells with a viral DNA polymerase inhibitor reversibly caused the dispersal of both UL44 and EdU-labeled viral DNA from replication compartments, indicating that ongoing viral DNA synthesis is necessary to maintain the organization of replication compartments. Our results reveal a previously unappreciated complexity of the organization of human cytomegalovirus replication compartments.
INTRODUCTION
The replication of viral genomes takes place in discrete sites within the cell, which enables viruses to concentrate and organize factors required for genome replication. During herpesvirus infection, a drastic and dynamic reorganization of the nucleus is observed, including the partitioning of host cell chromatin and the rearrangement of cellular nuclear proteins due primarily to the development of viral replication compartments (20, 23, 26).
The formation of human cytomegalovirus (HCMV) replication compartments in infected cells has been observed, as has the localization of several viral proteins within them (2, 10, 21). It is unclear how viral proteins are organized within replication compartments, and it is unknown where viral DNA synthesis occurs within compartments.
In a previous report from our laboratory, we assayed the localization of the presumptive viral DNA polymerase processivity subunit UL44 (also known as ICP36) in infected cells as a marker for infected-cell nuclei (10). Although we did not comment upon it at the time, we observed that UL44 accumulates at the periphery of replication compartments. To our knowledge, no viral protein in any herpesvirus replication compartment had shown this distribution, so we wished to investigate this observation further, hypothesizing that it might signal how DNA synthesis is organized within replication compartments. We therefore examined the localization of UL44, another viral DNA replication protein, and viral DNA synthesis within replication compartments.
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblast (HFF) cells (ATCC CRL-1684; American Type Culture Collection) were used in all experiments. HCMV laboratory strain AD169 was used. Virus expressing FLAG-tagged UL44 (HCMV-FLAG44) was described elsewhere previously (28).
Immunofluorescence (IF).
HFF cells (5 × 104) were plated onto glass coverslips. Cells were mock infected or infected with AD169 or HCMV-FLAG44 (28) (multiplicity of infection [MOI] of 3) in the presence or absence of phosphonoformic acid (PFA) (520 μM). Cells were fixed at room temperature (RT) with 4% formaldehyde in Dulbecco's phosphate-buffered saline (DPBS) at the time points indicated in the text. Where indicated, cells were incubated prior to fixation with 10 μM 5-ethynyl-2′-deoxyuridine (EdU) (Invitrogen) or 200 μM thymidine. Also, where indicated, 520 μM PFA (Sigma) was added. When necessary, EdU and PFA were washed out of cells by rinsing cells 3 times with tissue culture medium that did not contain either molecule. Following fixation, cells were washed with DPBS and permeabilized at RT for 10 min with 0.5% Triton X-100 dissolved in DPBS. Where indicated, EdU incorporated into DNA was detected by using “click chemistry” (25) with a fluorescent azide (Alexa Fluor 488; Invitrogen) according to the manufacturer's instructions (Invitrogen). Once rinsed again with DPBS, cells were incubated in 0.5% bovine serum albumin (BSA) dissolved in DPBS for 20 min at RT. Primary antibodies (Abs) in 0.5% BSA dissolved in DPBS were applied and incubated for 1 h at 37°C. Antiserum was removed by rinsing cells once with 0.5% Tween dissolved in DPBS and twice with DPBS, each time for 5 min with rocking. This procedure was repeated for the secondary antibodies. Where indicated, coverslips were incubated in DPBS containing 10 μg/ml Hoechst 33342 for 5 min before mounting. Coverslips were mounted onto microscope slides with ProLong Antifade (Invitrogen-Molecular Probes) and imaged by using either deconvolution microscopy or spinning-disk confocal microscopy.
For deconvolution microscopy, cells were imaged on an Axioplan 2 microscope (Carl Zeiss, Inc., Thornwood, NY) with a 63× objective and a Hamamatsu charge-coupled-device (CCD) camera (model C4742-95). Images were deconvolved by using the inverse filter algorithm with Axiovision (Rel.4.5) software.
For spinning-disk confocal microscopy, images were acquired by using an inverted spinning-disk confocal microscope based on an Axiovert 200 M inverted microscope (Zeiss), a CSU-X1 spinning-disk confocal unit (Yokogawa Electric Corporation, Tokyo, Japan), a spherical aberration correction (SAC) device (Infinity Photo-Optical, Boulder, CO), and a 63× objective lens (Plan-Apochromat with a 1.4 numerical aperture [NA]; Zeiss). Images shown were obtained by acquiring sequential optical planes of the entire cell spaced by 0.15 μm in the z axis and projecting those planes by using Slidebook 4.2 (Intelligent Imaging).
Primary antibodies recognizing UL44 (αICP36 antibody [CA006; Virusys], CH16 [22] [Virusys], and 28-21 [3] [a kind gift from Bill Britt, University of Alabama]), a murine monoclonal Ab (MAb) recognizing UL57 (Virusys), and rabbit polyclonal antibody F7425 recognizing FLAG (Sigma) were used at a dilution of 1:100. All fluorescently labeled secondary antibodies (Alexa Fluor 488 or Alexa Fluor 594) were obtained from Molecular Probes and used at a dilution of 1:1,000.
For each experiment, 3 to 5 cells representative of the phenotypes observed for each coverslip (5 × 104 cells) were imaged and analyzed in detail. Each experiment was performed at least twice.
RESULTS
Localization of UL44 in the infected-cell nucleus.
Our laboratory has previously used UL44 as a marker for infected-cell nuclei in studies of the nuclear lamina in HCMV-infected cells (10). Unexpectedly, in those studies, UL44 appeared to localize to the periphery of replication compartments (10). To explore this observation further, we mock infected or infected HFF cells with HCMV strain AD169 and stained the cells with a monoclonal antibody (MAb) recognizing UL44 (αICP36 antibody) for analysis by immunofluorescence (IF). We also stained cells with Hoechst stain to visualize host cell chromatin. Cells were imaged by using deconvolution microscopy. Single deconvolved sections are shown in Fig. 1A. In mock-infected cells, host cell chromatin could be observed throughout the nucleus, and no staining of UL44 was observed (Fig. 1Ai to iii). In infected cells at 48 h postinfection (p.i.) and 96 h p.i. (Fig. 1Aiv to ix), UL44 appeared to accumulate in a ringlike distribution at the periphery of viral replication compartments. Only a faint staining of UL44 was observed within the interior of viral replication compartments, and host cell chromatin was excluded from these compartments. We observed some marginalization of host cell chromatin in the infected-cell nucleus, reminiscent of the host cell chromatin partitioning seen in herpes simplex virus (HSV)-infected cells (20) (Fig. 1A, arrows). The number of UL44-positive compartments at each time point was calculated (Fig. 1B). Typically, infected cells contained 2 to 3 compartments at 48 h p.i. By 96 h p.i., most cells contained only one large compartment. The change in the number of replication compartments over time may represent the coalescence of replication compartments as infection progresses, as was reported previously for HSV (4, 27, 30).
Fig 1.
Localization of UL44 and host cell chromatin in infected cells. (A) HFF cells were infected at an MOI of 3, fixed at the indicated time points, and stained for IF analysis by deconvolution microscopy with Hoechst reagent (shown in green) and αICP36 MAb recognizing UL44. The anti-ICP36 antibody was detected by a secondary antibody conjugated to a red fluorophore. Images were obtained by acquiring sequential optical planes in the z axis and deconvolved to remove out-of-focus light from individual planes. A single deconvolved focal plane is shown in each panel. Hoechst staining (green) is shown in the left column, and UL44 staining (red) is shown in the middle column. Panels in the right column show the images in the right and middle columns merged. Arrows indicate areas of chromatin marginalization. (B) Number of compartments containing UL44 in infected cells. At each time point, the numbers of compartments in 100 cells were determined.
Different populations of UL44 in infected cells.
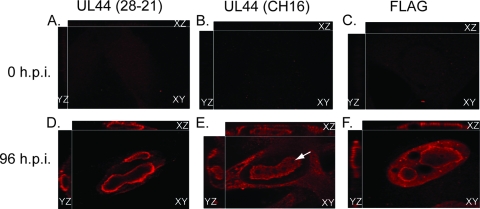
The staining of HCMV-infected cells with αICP36 MAb (Fig. 1) indicated that UL44 is found predominantly at the periphery of replication compartments, with little protein inside or outside replication compartments in infected-cell nuclei. This distribution was similar to the staining of UL44 at the periphery of replication compartments described in our previous report using MAb 28-21 (10). In contrast, Penfold and Mocarski previously observed high levels of UL44 within the interior of replication compartments when infected cells were stained with MAb CH16 (21). To better understand the UL44 distribution, cells were infected with AD169 or a virus expressing FLAG-tagged UL44 (HCMV-FLAG44) (28) and stained with a panel of UL44 MAbs or a rabbit polyclonal antibody recognizing FLAG (Fig. 2). Cells were imaged by using a spinning-disk confocal microscope.
Fig 2.
Detection by IF analysis of UL44 and FLAG-tagged UL44 in HCMV-infected cells. Uninfected HFF cells (A, B, and C) or HFF cells infected at an MOI of 3 with either HCMV strain AD169 (D and E) or HCMV-FLAG44 (28) (F) (fixed at 96 h p.i.) were prepared for IF analysis by spinning-disk confocal microscopy by staining with MAbs recognizing UL44 (MAbs 28-21 [A and D] and CH16 [B and E]) or FLAG (anti-FLAG rabbit polyclonal Ab) (C and F). Abs were detected by using secondary antibodies conjugated to a red fluorophore. Three-dimensional (3D) images were acquired by acquiring sequential optical planes in the z axis using spinning-disk confocal microscopy. Each panel shows the merged xz and yz images from each focal plane in the 3D stack acquired during microscopy and a single confocal plane in xy. The antibody used in each case is noted above each column. The time points analyzed are indicated at the left. The arrow in panel E indicates an area where UL44 staining by MAb CH16 could not be observed within the interior of the replication compartment.
AD169-infected cells stained with αICP36 MAb showed phenotypes similar to those shown in Fig. 1 (results not shown). In AD169-infected cells stained with MAb 28-21 (3) (Fig. 2D), UL44 accumulated at the periphery of replication compartments, and faint UL44 staining was detected inside and outside compartments, similar to the distribution of UL44 in AD169-infected cells stained with αICP36 MAb (Fig. 1). In HCMV-FLAG44-infected cells, using a rabbit polyclonal antibody against the FLAG epitope, we observed a distribution of staining similar to that found by using MAb 28-21 (Fig. 2F). For AD169-infected cells stained with MAb CH16 (22), we again found UL44 concentrated at the periphery of replication compartments; however, notable staining was also observed within the interior of replication compartments and throughout the cytoplasm (Fig. 2E). This pattern was similar to that observed previously by Penfold and Mocarski (21) and further confirms that UL44 accumulates at the periphery of replication compartments. The data suggested that a population of UL44 recognized by MAb CH16 is present within the interior of compartments. Also, for cells stained with MAb CH16, we found regions in the interior of the replication compartment where UL44 could not be observed (an example is indicated with an arrow in Fig. 2E). With all of the antibodies, we observed similar results whether cells were analyzed at 48 h p.i. (data not shown) or at 96 h p.i. No obvious staining was observed at 0 h p.i. in each case (Fig. 2A to C).
In summary, our results raise the possibility of different populations of UL44 being present within the nucleus: at least one at the periphery of replication compartments and at least one that is within the interior of replication compartments. We do not know the molecular basis for the differences in staining patterns of the different antibodies. Regardless, these findings and our inability to detect UL44 in certain regions of the interior of the replication compartments suggest the presence of multiple subcompartments within HCMV replication compartments.
Visualization of UL44 and UL57 within viral replication compartments.
We next wished to compare the localization of UL44 with that of another viral DNA replication protein. We therefore assayed the localization of the HCMV single-stranded-DNA (ssDNA) binding protein UL57 relative to that of FLAG-tagged UL44. We infected cells with a virus expressing FLAG-tagged UL44 (HCMV-FLAG44) (28), stained the cells with a murine MAb recognizing UL57 and a rabbit polyclonal antibody recognizing FLAG (Fig. 3), and analyzed the IF image using spinning-disk confocal microscopy. At 96 h p.i., FLAG-tagged UL44 was concentrated at the periphery of replication compartments and was more diffusely distributed throughout the nucleus (Fig. 3iv). In contrast, UL57 was observed in punctate structures throughout replication compartments (Fig. 3v), reminiscent of the staining for ICP8 observed for HSV-infected cells (5, 23). No obvious colocalization of FLAG-tagged UL44 and UL57 was observed in the interior of the replication compartments; however, a few punctate areas of colocalization could be seen at the periphery (Fig. 3vi). Similar results were obtained when cells were analyzed at 48 h p.i. (data not shown). Because UL57 can bind to the viral genome, these results suggested that viral DNA is present both at the periphery of the replication compartment, where at least one form of UL44 concentrates, and in the interior of the replication compartment.
Fig 3.
Analysis of UL44 and UL57 localization in infected cells. HFF cells were infected at an MOI of 3 with HCMV-FLAG, fixed at different time points, stained for IF analysis using a polyclonal antibody recognizing FLAG and a MAb recognizing UL57, and analyzed by spinning-disk confocal microscopy. Panels in the left column show cells stained with an Ab recognizing FLAG and a secondary antibody conjugated to a red fluorophore. Panels in the middle column show cells stained with an Ab recognizing UL57 and a secondary antibody conjugated to a green fluorophore. Panels in the right column show the images in the right and middle columns merged. The white box in panel vi is a magnified view of the indicated area.
Visualization of viral DNA within viral replication compartments.
We next sought to understand the localization of viral DNA and UL44 in viral replication compartments by labeling viral DNA. A previous report (21) indicated that bromodeoxyuridine (BrdU)-labeled DNA is present within replication compartments and throughout the nucleus of HCMV-infected cells. In our preliminary experiments, the labeling of HCMV-infected HFF cells with BrdU was very inefficient (data not shown). Instead, we assayed the localization of viral DNA by IF using 5-ethynyl-2′-deoxyuridine (EdU) (Fig. 4A). As described elsewhere previously (25), EdU incorporated into replicating DNA is recognized by a fluorescent azide via a Cu(I)-catalyzed [3 + 2] cycloaddition reaction (“click chemistry”).
Fig 4.
Visualization of EdU-labeled viral DNA in infected cells. (A) HFF cells were infected at an MOI of 3 with AD169, fixed at the indicated time points after 60 min of incubation with EdU, stained for IF analysis using an αICP36 MAb recognizing UL44, and treated with a green fluorophore to detect EdU incorporated into DNA. Cells were analyzed by spinning-disk confocal microscopy. Images were obtained by acquiring and merging sequential optical planes in the z axis. Panels in the left column show cells stained with a MAb recognizing UL44 and a secondary antibody conjugated to a red fluorophore. Panels in the middle column show cells treated with a green fluorescent azide to detect EdU. Panels in the right column show the images in the right and middle columns merged. Each panel shows the merged xz and yz images from each focal plane in the 3D stack acquired during microscopy and a single confocal plane in xy. An area where neither UL44 nor EdU-labeled DNA could be detected is indicated with an arrow in panel vi. The white box in panel vi is a magnified view of the indicated area. (B) Cells were pulsed with EdU for 30 min (i) and then incubated with thymidine for 30 min in the absence of EdU (ii). Infected cells were prepared as described above for panel A. Each panel shows the merged signals from the red (UL44) and green (EdU) fluorophores. Each image shows a single focal plane from the z axis of the cells analyzed.
We incubated AD169-infected HFF cells with EdU for 1 h at 0 h p.i. and 96 h p.i. and then prepared the cells for IF assays by performing the cycloaddition reaction to detect incorporated EdU and staining with αICP36 MAb to recognize UL44. Images were obtained at 96 h p.i. using spinning-disk confocal microscopy. UL44 again accumulated at the periphery of replication compartments, with faint UL44 staining in the interior of the compartment and throughout the nucleus (Fig. 4Aiv). Punctate EdU staining was seen mainly in the interior of the viral replication compartment (Fig. 4Av). We also observed regions within viral replication compartments where neither UL44 nor EdU-labeled viral DNA was found (Fig. 4Avi, white arrow), which may represent subcompartments in the interior of replication compartments. Nevertheless, the colocalization of EdU and UL44 was evident at the periphery of replication compartments (Fig. 4Avi). Similar results were obtained when cells were analyzed at 48 h p.i. (results not shown and see below).
To investigate where viral DNA synthesis occurred within replication compartments, we performed a pulse-chase experiment at 48 h p.i. (Fig. 4B). We pulse-labeled HCMV-infected HFF cells at 48 h p.i. with EdU for 30 min (the minimum period of labeling that consistently gave a signal [results not shown]) and then chased cells with thymidine for 30 min to stop EdU labeling. After the 30-min pulse with EdU, we observed the incorporation of EdU into DNA predominantly at the periphery of replication compartments, colocalizing with UL44 detected by αICP36 MAb (Fig. 4Bi). When EdU was chased with thymidine, DNA containing EdU accumulated within the interior of the compartments, and punctate staining of EdU at the periphery of replication compartments was less evident (Fig. 4Bii). Similar results were obtained when cells were analyzed at 96 h p.i. (results not shown).
In sum, we found that viral DNA synthesis occurs at the periphery of viral replication compartments and that replicated viral DNA is then found in the interior of the compartments.
Localization of UL44 and EdU-labeled DNA in the presence of an inhibitor of viral DNA synthesis.
We next sought to determine if active viral DNA synthesis plays a role in the organization of viral replication compartments once the compartment is formed. We therefore labeled infected cells with EdU at 48 h p.i. for 1 h. After incubation with EdU, infected cells were left untreated or were treated with an inhibitor of viral DNA synthesis (phosphonoformic acid [PFA]) in the presence of EdU. Cells were collected at different times after treatment with PFA and EdU (Fig. 5iv to xii). After 90 min, the PFA was removed and washed out, and cells were collected at different times in the presence of EdU (Fig. 5xiii to xx).
Fig 5.
Localization of UL44 and EdU-labeled DNA in the presence of PFA. HFF cells were infected at an MOI of 3. At 48 h p.i., cells were incubated with EdU for 60 min and fixed (i to iii) or then treated with phosphonoformic acid (PFA) in the presence of EdU. Cells were fixed at the indicated time points after PFA treatment (iv to xii), PFA was then washed out of cells, and the cells incubated further in the presence of EdU (xiii to xxi). Cells were stained for IF analysis by using an anti-ICP36 MAb recognizing UL44 and treated with a green fluorophore to detect EdU incorporated into DNA. Cells were analyzed by spinning-disk confocal microscopy. Panels in the left column show cells stained with a MAb recognizing UL44 and a secondary antibody conjugated to a red fluorophore. Panels in the middle column show cells treated with a green florescent azide to detect EdU. Panels in the right column show the images in the right and middle columns merged. All cells were imaged using the same magnification, but because they are imaged using different focal planes, they may appear to be different sizes.
In the absence of PFA, we observed a distribution of UL44 and EdU similar to what we had already observed (Fig. 5i to iii). Treatment with PFA caused a dispersal of UL44 and EdU-labeled viral DNA from replication compartments, which was quite pronounced by 90 min (Fig. 5iv to xii). Also, by 90 min, EdU staining was notably less bright, most likely due to the lack of incorporation of EdU in the absence of viral DNA replication. When PFA was washed from the cells (Fig. 5xiii to xx), EdU-labeled DNA colocalized with punctate UL44 staining within 30 min (Fig. 5xiii to xv). Over time, the integrity of replication compartments was reestablished, and UL44 was again concentrated at the periphery of large replication compartments that contained EdU-labeled DNA (Fig. 5xvi to xxi).
These results indicated that viral DNA synthesis is required for maintaining the organization of HCMV replication compartments. Also, the appearance of large replication compartments from small punctate bodies may represent the coalescence of smaller replication compartments to form larger replication compartments, as also suggested by the data shown in Fig. 1.
DISCUSSION
Here we provide insights into the organization of HCMV replication compartments, demonstrating that viral DNA synthesis occurs at the periphery of each compartment within a layer in which the viral DNA polymerase subunit UL44 concentrates. The viral single-stranded-DNA binding protein UL57 distributes both at the periphery of the replication compartment and in its interior. The interior of the compartment also contains a population of UL44, areas where replicated viral DNA can be found, and regions where neither UL44 nor viral DNA was detected. Thus, there are subcompartments present within replication compartments. Viral DNA synthesis appears to be required to maintain the distribution of UL44 and viral DNA within replication compartments.
As viral DNA synthesis takes place at the periphery of replication compartments, UL44 and UL57 in the interior of the compartments are most likely not involved directly in viral DNA synthesis. HSV replication compartments are the sites of transcription of late HSV genes (13, 24). The HSV counterpart of UL57, ICP8, is important for the regulation of late HSV gene transcription (7, 8, 18). Additionally, it was reported previously that UL44 has a role in the transcription of late HCMV genes (11, 12). Therefore, UL57 and UL44 within the interior of replication compartments may be involved in late viral transcription.
Our findings that EdU-labeled viral DNA is found first at the periphery of replication compartments and then in the interior of compartments in pulse-chase experiments suggest that there is a mechanism facilitating the movement of replicated viral genomes within the compartment and/or the movement of the periphery of the compartment away from replicated genomes. What this mechanism is remains unknown. However, the packaging of HSV viral DNA into virus capsids occurs within replication compartments (19). Thus, we speculate that the packaging of viral DNA may facilitate the movement of viral DNA into the interior of replication compartments. It is possible that the regions within replication compartments where no UL44 or EdU-labeled DNA could be observed may be sites of capsid assembly. The encapsidation of EdU-labeled viral DNA at these sites may have prevented the detection of EdU in our analysis. The factors determining the efficient packaging of viral DNA are still under investigation but could involve UL44. TRS1, a protein that we have previously found to associate with UL44 in infected cells (29), is required for efficient viral DNA packaging (1).
The formation of HSV replication compartments has been extensively studied (14–17, 20, 23, 31). Several of the observations made in this report are consistent with features of HSV replication compartments, including the localization of the viral single-stranded-DNA binding protein within replication compartments and the dispersal of viral proteins from replication compartments in the presence of an inhibitor of viral DNA synthesis (13, 23). However, important features of HSV compartments differ from the features of the HCMV replication compartments that we describe here. BrdU staining of HSV DNA and the immunolocalization of several HSV proteins involved in viral DNA synthesis indicate that viral DNA synthesis takes place throughout HSV replication compartments (5, 6, 9, 14, 23) and not at the periphery of compartments. Also, the HSV counterpart of HCMV UL44, UL42, has yet to be observed to be concentrated at the periphery of replication compartments. To date, UL42 has been observed only diffusely distributed throughout HSV replication compartments (6, 9), although the form of UL42 recognized by the antibodies used in those studies is unclear. Thus, the structures of HSV and HCMV replication compartments differ. Recently, it was demonstrated that HSV replication compartments coalesce at and reorganize nuclear speckles to enhance late viral mRNA export (4). The differences between HSV and HCMV replication compartment organizations that we observed here may reflect differences in the requirements of each virus to utilize subnuclear structures during virus replication.
ACKNOWLEDGMENTS
We thank Martha Simpson-Holley for helpful discussions. We gratefully acknowledge Bill Britt (University of Alabama) for providing reagents and all members of the Coen laboratory for their support.
This work was supported by NIH grants AI19838 and AI26077 to D.M.C., NIH grant AI06106 to D.M.K., and grants GM 075252 (NIH) and U54 AI057159 (New England Regional Center of Excellence in Biodefense and Emerging Infectious Disease, Core Imaging Facility) to T.K.
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Adamo JE, Schroer J, Shenk T. 2004. Human cytomegalovirus TRS1 protein is required for efficient assembly of DNA-containing capsids. J. Virol. 78:10221–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahn JH, Jang WJ, Hayward GS. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Britt WJ, Vugler L. 1987. Structural and immunological characterization of the intracellular forms of an abundant 68,000 Mr human cytomegalovirus protein. J. Gen. Virol. 68:1897–1907 [DOI] [PubMed] [Google Scholar]
- 4. Chang L, et al. 2011. Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. U. S. A. 108:E136–E144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Bruyn Kops A, Knipe DM. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857–868 [DOI] [PubMed] [Google Scholar]
- 6. de Bruyn Kops A, Uprichard SL, Chen M, Knipe DM. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252:162–178 [DOI] [PubMed] [Google Scholar]
- 7. Gao M, Knipe DM. 1991. Potential role for herpes simplex virus ICP8 DNA replication protein in stimulation of late gene expression. J. Virol. 65:2666–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Godowski PJ, Knipe DM. 1985. Identification of a herpes simplex virus function that represses late gene expression from parental viral genomes. J. Virol. 55:357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goodrich LD, Schaffer PA, Dorsky DI, Crumpacker CS, Parris DS. 1990. Localization of the herpes simplex virus type 1 65-kilodalton DNA-binding protein and DNA polymerase in the presence and absence of viral DNA synthesis. J. Virol. 64:5738–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hamirally S, et al. 2009. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5:e1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Isomura H, et al. 2008. Noncanonical TATA sequence in the UL44 late promoter of human cytomegalovirus is required for the accumulation of late viral transcripts. J. Virol. 82:1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Isomura H, et al. 2007. The late promoter of the human cytomegalovirus viral DNA polymerase processivity factor has an impact on delayed early and late viral gene products but not on viral DNA synthesis. J. Virol. 81:6197–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Knipe DM, Senechek D, Rice SA, Smith JL. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liptak LM, Uprichard SL, Knipe DM. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lukonis CJ, Burkham J, Weller SK. 1997. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J. Virol. 71:4771–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lukonis CJ, Weller SK. 1996. Characterization of nuclear structures in cells infected with herpes simplex virus type 1 in the absence of viral DNA replication. J. Virol. 70:1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lukonis CJ, Weller SK. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McNamee EE, Taylor TJ, Knipe DM. 2000. A dominant-negative herpesvirus protein inhibits intranuclear targeting of viral proteins: effects on DNA replication and late gene expression. J. Virol. 74:10122–10131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mettenleiter TC. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661–665 [DOI] [PubMed] [Google Scholar]
- 21. Penfold ME, Mocarski ES. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46–61 [DOI] [PubMed] [Google Scholar]
- 22. Pereira L, Hoffman M, Gallo D, Cremer N. 1982. Monoclonal antibodies to human cytomegalovirus: three surface membrane proteins with unique immunological and electrophoretic properties specify cross-reactive determinants. Infect. Immun. 36:924–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quinlan MP, Chen LB, Knipe DM. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857–868 [DOI] [PubMed] [Google Scholar]
- 24. Randall RE, Dinwoodie N. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67(Pt 10):2163–2177 [DOI] [PubMed] [Google Scholar]
- 25. Salic A, Mitchison TJ. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simpson-Holley M, Colgrove RC, Nalepa G, Harper JW, Knipe DM. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840–12851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sourvinos G, Everett RD. 2002. Visualization of parental HSV-1 genomes and replication compartments in association with ND10 in live infected cells. EMBO J. 21:4989–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Strang BL, Boulant S, Coen DM. 2010. Nucleolin can associate with the human cytomegalovirus DNA polymerase accessory subunit UL44 and is necessary for viral replication J. Virol. 84:1771–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strang BL, Geballe AP, Coen DM. 2010. Association of the human cytomegalovirus DNA polymerase subunit UL44 with the viral proteins IRS1 and TRS1. J. Gen. Virol. 91:2167–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taylor TJ, McNamee EE, Day C, Knipe DM. 2003. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology 309:232–247 [DOI] [PubMed] [Google Scholar]
- 31. Zhong L, Hayward GS. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]