Abstract
We previously established that at 3 years postseroconversion, ∼30% of HIV-infected individuals have cross-reactive neutralizing activity (CrNA) in their sera. Here we studied the kinetics with which CrNA develops and how these relate to the development of autologous neutralizing activity as well as viral escape and diversification. For this purpose, sera from five individuals with CrNA and one elite neutralizer that were obtained at three monthly intervals in the first year after seroconversion and at multiple intervals over the disease course were tested for neutralizing activity against an established multiclade panel of six viruses. The same serum samples, as well as sera from three individuals who lacked CrNA, were tested for their neutralizing activities against autologous clonal HIV-1 variants from multiple time points covering the disease course from seroconversion onward. The elite neutralizer already had CrNA at 9.8 months postseroconversion, in contrast with the findings for the other five patients, in whom CrNA was first detected at 20 to 35 months postseroconversion and peaked around 35 months postseroconversion. In all patients, CrNA coincided with neutralizing activity against autologous viruses that were isolated <12 months postseroconversion, while viruses from later time points had already escaped autologous neutralizing activity. Also, the peak in gp160 sequence diversity coincided with the peak of CrNA titers. Individuals who lacked CrNA had lower peak autologous neutralizing titers, viral escape, and sequence diversity than individuals with CrNA. A better understanding of the underlying factors that determine the presence of CrNA or even an elite neutralizer phenotype may aid in the design of an HIV-1 vaccine.
INTRODUCTION
Antibodies with the ability to neutralize autologous human immunodeficiency virus type 1 (HIV-1) are formed within the first 3 months of HIV-1 infection (29, 41). Most of these neutralizing antibodies are strain specific, yet there are HIV-1-infected individuals in whom cross-reactive neutralizing activity (CrNA) that can neutralize different subtypes of HIV-1 is elicited (2, 10, 12, 31, 34). A vaccine should ideally be capable of eliciting this type of neutralizing activity, since it may be able to provide protection against infection with different HIV-1 subtypes. The prevalence of CrNA among HIV-1-infected individuals is about 10 to 30%, as described in different cohorts (2, 10, 12, 31, 34). Only about 1% of HIV-1-infected individuals fulfill the definition of an “elite neutralizer,” that is, an HIV-1-infected individual with unusually potent CrNA against a majority of HIV-1 subtypes (34). Based on their neutralizing activity against multiple unrelated HIV-1 variants, cross-reactive neutralizing antibodies are considered to be directed against conserved regions of the virus. Epitopes on autologous viruses early in infection may trigger the development of CrNA, and it remains to be established whether the development of CrNA is due to the presence of unique epitopes, a unique B cell repertoire or other unique host characteristics, or a completely random event.
CrNA is not associated with a prolonged asymptomatic course of HIV-1 infection (10, 12, 24a, 26, 38). Indeed, the prevalences of CrNA were similar in long-term nonprogressors and progressors, and in both groups of individuals, serum neutralizing activity against autologous HIV-1 variants faded over time due to viral escape, explaining at least in part the lack of effect of potent CrNA on the clinical course of infection (37). Interestingly, CrNA against heterologous virus variants was preserved over the course of infection, despite the escape of autologous virus.
The virus seems to employ various mechanisms to escape from neutralizing antibodies, such as amino acid substitutions, insertions, or deletions, especially in the variable loops of Env, thereby changing or occluding the neutralizing epitopes, after which the neutralizing antibody can no longer bind (6, 22, 29, 30, 41). Although CrNA does not protect from disease progression, in several nonhuman primate studies, passive transfer of known cross-reactive neutralizing antibodies could completely block infection by a chimeric simian-human immunodeficiency virus (SHIV) (7, 15, 16, 19). A vaccine should therefore be capable of eliciting this type of neutralizing activity (20).
It has been shown that CrNA increases with time of infection (13, 21, 31, 38), and most broadly neutralizing antibodies (BrNAbs) seem to have gone through several rounds of somatic hypermutation before potent CrNA was achieved (32). In other infections, effective neutralizing antibodies arise much earlier than in HIV-1 infection. The questions remains how and when broadly neutralizing antibodies develop, and if and how type-specific neutralizing responses contribute to this development. Although the envelope of the infecting HIV-1 is likely to play a role in this process (18, 28), we focused here on the kinetics with which CrNA develops. For this purpose, we retrospectively studied six individuals whom we previously identified to have CrNA in serum at ∼35 months after seroconversion (SC) (12). We compared the kinetics with which neutralizing activity in serum against autologous and heterologous viruses (CrNA) developed in the first months and years after infection and examined how, in turn, the virus adapted to the humoral response.
MATERIALS AND METHODS
Participants and viruses.
All individuals studied here were selected from the Amsterdam Cohort Studies on HIV and AIDS in homosexual men (ACS) (9) (Table 1). Participants were selected based on their CrNA as previously established (12). The top six participants, C1 (cohort identification number [ID] ACH18877), C2 (ACH19308), C3 (ACH11668), C4 (ACH18814), C5 (ACH18818), and C6 (ACH11694), were selected on the basis of the highest geometric mean 50% inhibitory concentration (IC50) titers against a panel of 23 heterologous viruses from different subtypes and the highest total number of viruses neutralized; they included one elite neutralizer (C1) as defined by Simek et al. (34). Three other participants with no CrNA were also included: N1 (ACH19961), N2 (ACH18766), and N3 (ACH19489). All participants were infected with HIV-1 subtype B and seroconverted during active follow-up. None of the individuals received combination antiretroviral therapy during the follow-up period for the present study.
Table 1.
Patient characteristics
| Patient ID | Neutralizing activity | Yr of SC | Yrs to CD4 count of <200/AIDS | Yrs to HAART | X4 switch | CD4 count at set point | Viral (log) load at set point (copies/ml) |
|---|---|---|---|---|---|---|---|
| 18877 (C1) | Elite neutralizer | 1986 | 7.5 | NAa | No | 380 | 4 |
| 19308 (C2) | CrNA | 1991 | 4.7 | NA | Yes | 380 | 4.4 |
| 11668 (C3) | CrNA | 1986 | 5.1 | NA | Yes | 380 | 4.5 |
| 18814 (C4) | CrNA | 1987 | NA | 8.6 | No | 500 | 5.1 |
| 18818 (C5) | CrNA | 1986 | 7.1 | NA | No | 510 | 5.4 |
| 11694 (C6) | CrNA | 1985 | 3.2 | NA | Yes | 240 | 4.8 |
| 19961 (N1) | Non-CrNA | 1987 | 3.2 | NA | Yes | 530 | 4.5 |
| 18766 (N2) | Non-CrNA | 1988 | 5.9 | NA | Yes | 760 | 4.3 |
| 19489 (N3) | Non-CrNA | 1985 | NA | 6.6 | No | 550 | 3.1 |
NA, not applicable.
Clonal virus variants were obtained as described previously (33, 33, 39, 40). The quasispecies of clonal virus isolated from peripheral blood mononuclear cells (PBMC) are highly identical to the quasispecies in the serum of the same patient (11). We selected a maximum of five virus variants per individual per time point to be tested for autologous neutralization sensitivity. To prevent a potential change in the neutralization sensitivity of the virus variants during in vitro culture, the number of virus passages in PBMC was kept to a minimum.
The Amsterdam Cohort Studies are conducted in accordance with the ethical principles set out in the Declaration of Helsinki, and written consent was obtained prior to data collection. The study was approved by the Academic Medical Center Institutional Medical Ethics Committee.
U87/pseudovirus assay for testing of HIV-1 cross-reactive neutralizing activity in serum.
Where available, sera from the 6 cohort participants who developed CrNA were obtained every 3 months post-SC until 1 year, then every 6 months until 2 years post-SC, and then every 2 years until death, initiation of highly active antiretroviral therapy (HAART), or lost to follow-up. Sera were tested for cross-reactive neutralizing activity in a pseudovirus assay involving tier 2 viruses in a single round of viral infection, developed by Monogram Biosciences (25, 29). The viral panel consisted of 6 pseudoviruses with envelope sequences from primary isolates of HIV-1 subtypes A, B, C, and CRF01_AE that were resistant (n = 1), moderately resistant (n = 3), or moderately sensitive (n = 2) based on previously determined neutralization sensitivities to sera from subtype B-infected individuals and monoclonal antibodies (MAbs) b12, 2G12, and 4E10. This 6-virus panel covered 93% of the variation in neutralization of a larger pseudovirus panel (n = 15) (34). Previously we have shown that the CrNA of patient serum samples as determined with an independent 23-virus panel correlated strongly with the CrNA determined with the smaller 6-virus panel used in this study (Spearman's r, 0.91; P, <0.0001) (12). Pseudotyped viral particles were produced by cotransfection of HEK293 cells with an expression vector carrying the HIV-1-derived gp160 gene (eETV) and an HIV-1 genomic vector carrying a luciferase reporter gene (pRTV1.F-lucP.CNDO-ΔU3). Forty-eight hours after transfection, pseudovirus stocks were harvested, and small aliquots were tested for infectivity using U87 target cells expressing CD4, CCR5, and CXCR4. Pseudovirus stocks were tested and normalized for infectivity prior to testing in the neutralization assay. Diluted pseudoviruses were incubated for 1 h at 37°C with serial dilutions of the patient sera, after which the U87 target cells were added. The ability of patient sera to neutralize viral infection was assessed by measuring luciferase activity 72 h after viral inoculation in comparison with that for a control infection with a virus pseudotyped with the murine leukemia virus envelope (aMLV). Neutralization titers are expressed as the reciprocal of the plasma dilution that inhibited virus infection by 50% (IC50). Neutralization titers were considered positive if they were 3 times greater than that for the negative aMLV control. The lowest serum dilution used in the assay was 1:40. For the calculation of IC50s for viruses that were not inhibited by the 1:40 serum dilution, we assumed that 50% inhibition would have occurred at a 1:20 serum dilution.
PBMC-based assay for testing autologous HIV-1-neutralizing activity in serum.
Clonal virus variants of participants were tested for their relative neutralization sensitivities to autologous serum and pooled sera from healthy, uninfected individuals. Participant C6 (ACH11694) was excluded, since clonal viral variants could not be obtained from his PMBC. PBMC were obtained from buffy coats from 10 healthy seronegative blood donors and were pooled prior to use. Cells were isolated by Ficoll-Isopaque density gradient centrifugation and were then stimulated for 3 days in Iscove modified Dulbecco medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 U/ml), Ciproxin (5 μg/ml), and phytohemagglutinin (PHA; 5 μg/ml) at a cell concentration of 5 × 106/ml. After inoculation, the cells (106/ml) were grown in the absence of PHA in a medium supplemented with recombinant interleukin-2 (20 U/ml; Chiron Benelux, Amsterdam, The Netherlands) and Polybrene (hexadimethrine bromide) (5 μg/ml; Sigma, Zwijndrecht, The Netherlands). To prevent possible complement-mediated antibody inhibition of virus infection, complement in human sera and fetal bovine serum was inactivated by a 30-min incubation at 56°C.
From each virus isolate, an inoculum of 30 50% tissue culture infective doses in a total volume of 50 μl was incubated for 1 h at 37°C with decreasing concentrations of the serum (starting concentration, 1:50) in 96-well microtiter plates. Subsequently, 105 PHA-stimulated PBMC were added to the mixtures of virus with serum. After 4 h of incubation, PBMC were washed once in 150 μl of phosphate-buffered saline, after which 150 μl fresh medium was added. On day 7 or 11, depending on viral growth, virus production in culture supernatants was analyzed by an in-house p24 antigen capture enzyme-linked immunosorbent assay (35). Background measurements were performed with pooled sera from uninfected individuals, and neutralization titers were expressed as the reciprocal serum dilutions that established the IC50s for virus infection. Experiments were performed in triplicate. When possible, the IC50s were determined by linear regression. To calculate IC50s for viruses that were not inhibited by the 1:50 serum dilution, we assumed that 50% inhibition would have occurred at a 1:25 serum dilution.
Sequence analysis.
The HIV envelope gp160 gene was PCR amplified from DNA isolated from PBMC that were infected in vitro with a single clonal HIV-1 variant, and the PCR product was subsequently sequenced as described previously (1, 4, 27). Nucleotide sequences were aligned using ClustalW in the BioEdit software package (14) and were edited manually. Potential N-linked glycosylation sites (PNGS) were identified using N-GlycoSite (43) at the HIV database website (http://www.hiv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html). Pairwise nucleotide distances and divergences were calculated with the Kimura 2-parameter model of evolution included in the MEGA 4 software package.
Statistical analysis.
Differences in length or PNGS in gp160 between different time points for the same individual were evaluated by analysis of variance (ANOVA). Where differences were present (P, <0.05), specific differences between two time points were tested with the Bonferroni multiple-comparison test. Spearman's rank correlation coefficient was used to assess the association of the geometric mean titers of each patient serum that were obtained on the two viral panels, as well as the development of potent CrNA and the amount of virus neutralized over calendar time. Differences between individuals with and without CrNA in the mean diversity of gp160 sequences isolated from clonal HIV-1 variants within the first 5 years postseroconversion were calculated and were compared by an unpaired t test. Analyses were performed in GraphPad Prism 4 (GraphPad Software, La Jolla, CA).
RESULTS
Development of cross-reactive neutralizing activity over the course of HIV-1 infection.
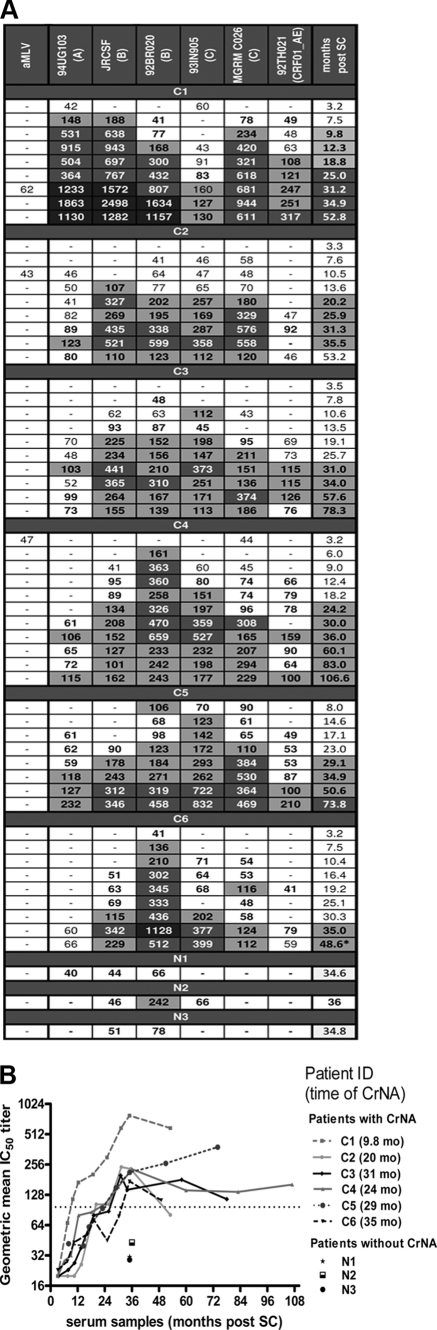
Previously we determined that 33% of individuals from the ACS had CrNA in serum at ∼35 months post-SC (12). Here we wanted to see how soon after seroconversion CrNA developed in the sera of six individuals with the most potent CrNA at 35 months post-SC, including one elite neutralizer. Sera were screened for neutralizing activity on a virus panel that represents several clades and that was created by an analytical selection algorithm to identify both the potency and the breadth of neutralizing activity in serum (34). Sera from 82 individuals were previously screened on this virus panel as well as on a larger panel of 23 viruses belonging to subtypes A, B, C, and D, and the classifications of sera with respect to neutralization potency against these two virus panels were strongly correlated (Spearman's r, 0.91; P, <0.0001). Across the six viruses of the smaller panel, the upper 25% quartile geometric mean IC50 titer in sera from the initial 82 patients was 97; we subsequently used this titer as the threshold for CrNA, together with the condition that at least four out of six viruses be neutralized.
Sera were selected at three monthly time points, starting at ∼3 months post-SC and continuing until 12 months post-SC; then at six monthly time points until 35 months post-SC; and subsequently at two yearly time points until death, the initiation of HAART, or lost to follow-up (Fig. 1A). In the sera of all participants, some low-titer heterologous neutralizing activity could already be detected at 6 to 8 months post-SC, while sera from three participants already had some detectable heterologous neutralizing reactivity as early as 3 months post-SC (C1, C4, and C6), which could have been present prior to this first time point tested. Five of the six participants developed CrNA between 20.2 and 35 months post-SC, while in the elite neutralizer (C1), CrNA had developed as early as 9.8 months post-SC (Fig. 1B). For all participants except C5, CrNA peaked between 31 and 36 months post-SC, after which the titer decreased slightly and subsequently remained stable throughout infection. In only one participant (C5) did the neutralization titer in serum still increase after 35 months of infection.
Fig 1.
Development of cross-reactive neutralizing activity over the course of HIV-1 infection. (A) Breadth and potency of neutralizing activity in longitudinally obtained serum samples from six individuals who had cross-reactive neutralizing activity (CrNA) and three individuals who lacked CrNA in serum at 35 months postseroconversion (post-SC). Serum samples were obtained at different time points post-SC (rightmost column) from one elite neutralizer (C1) and five individuals with CrNA (C2 to C6) and at one time point post-SC from individuals who lacked CrNA (N1 to N3). The virus panel (top) included 6 viruses from subtypes A, B, C, and CRF_AG. Amphotropic murine leukemia virus (aMLV) served as a negative control. Values represent the reciprocal serum dilution at which 50% inhibition of virus infection was achieved. IC50 titers are indicated as follows: −, <1:40; white background, ≥3 times the value for aMLV; light gray background, ≥1:100; dark gray background, ≥1:300; black background, ≥1:1,000. Months post-SC are color coded in the same way based on the geometric mean IC50 across the 6 viruses. (B) Geometric mean IC50s for each patient across the 6 viruses over the course of the infection. The time point at which CrNA was reached is given in parentheses to the right of the patient ID. To fulfill the definition of CrNA, sera had to neutralize ≥4 viruses above background with a geometric mean IC50 of ≥1:97 (dotted line) as indicated on the y axis.
The development of potent CrNA (Spearman's r, 0.77; P, <0.0001) and the number of viruses neutralized (Spearman's r, 0.83; P, <0.0001) were strongly correlated over calendar time, as described previously (37).
Autologous neutralizing activity during primary infection in individuals with and without cross-reactive neutralizing activity.
We and others have demonstrated previously that the presence of CrNA in serum is not associated with prolonged asymptomatic survival of HIV-1 infection (10, 12, 24a, 26, 38), due to the rapid selection of escape variants of the virus (37). CrNA is thought to be directed against conserved regions of the virus, which makes escape through amino acid changes in the epitope less likely, although not entirely impossible. An alternative mechanism of escape is the shielding of epitopes, which would be in line with the increased length of variable loops in the envelope and the increased number of potential N-linked glycosylation sites that have been associated with a neutralization resistance phenotype. Escape through either mechanism, driven by autologous neutralizing activity, which generally occurs around 3 months after infection, may create the sequential virus variation that can drive affinity maturation of the antibody response, ultimately resulting in CrNA. Here we wanted to compare the kinetics with which autologous neutralizing activity developed in five individuals with CrNA and three without CrNA. Participant C6 was excluded, because no clonal virus variants could be isolated from his PBMCs. Moreover, we performed analyses to determine whether CrNA and non-CrNA patients differed with respect to the potency of the autologous neutralizing immune response and the time after infection at which viral escape from autologous neutralization occurred.
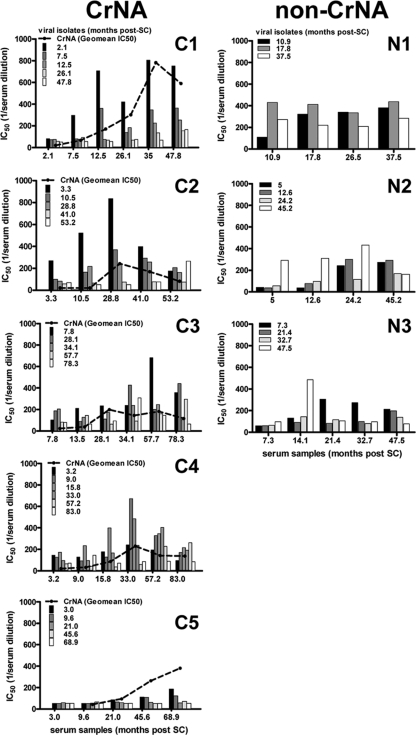
Autologous viruses were obtained during the acute phase of infection and 6 and 12 months thereafter. Subsequently, virus variants were obtained every 2 years until death, initiation of HAART, or loss to follow up. For each individual, all viruses from all time points were tested for sensitivity to neutralization using autologous serum from the same time points. For the elite neutralizer (C1), there was no neutralizing activity in serum at 2.1 months post-SC, while serum from 7.5 months post-SC did neutralize viruses that were obtained 2.1 months post-SC. Neutralizing titers increased over time, with a dip at 26 months post-SC (Fig. 2). Contemporaneous viruses were minimally neutralized at the last time point measured (47.8 months post-SC), but not at all at any other time point. Most viruses were neutralized only at the subsequent time point, and viral escape was present from 7 months post-SC, since neutralizing titers were lower for viruses isolated from 7 months post-SC than for viruses isolated from 2.1 months post-SC.
Fig 2.
Cross-reactive and autologous neutralizing activity developed with similar kinetics. The height of individual bars indicates the average IC50s in the serum (y axis) at the indicated month post-SC (x axis), as determined by linear regression of IC50s achieved on ≤5 autologous virus variants per time point. C1 to C5, individuals with CrNA; N1 to N3, individuals who lacked CrNA. The geometric mean IC50s across the heterologous 6-virus panel are shown as dashed lines for individuals with CrNA. Patient C6 is absent from this analysis, because no viruses could be isolated.
For the remaining four patients with CrNA as well, contemporaneous viruses were less susceptible to neutralization than viruses from earlier time points (Fig. 2). Furthermore, in each individual, de novo responses against newly emerging virus variants were elicited. Participants C3 and C4 had a less typical profile of autologous neutralization, with virus variants from later time points inducing stronger neutralizing responses than earlier virus variants; however, escape of later viruses from neutralization occurred at 34 months post-SC for participant C3 and at 15.8 months post-SC for participant C4. Participant C5 had very low autologous neutralizing titers compared to those of the other participants, and only viral isolates from the first year of infection were neutralized by autologous serum from 46 months post-SC onward.
We next explored whether three participants with no CrNA in their sera had neutralizing activity in serum against autologous viruses, even though they were unable to neutralize heterologous HIV-1 strains. Moreover, we compared the emergence of autologous neutralizing activity in serum and the emergence of viral escape between participants with and without CrNA (Fig. 2). The three participants lacking CrNA who were tested here did not have a typical autologous neutralizing profile. Two participants had higher neutralizing titers against later viruses with serum from earlier time points. For participant N1, neutralizing titers remained more or less the same until 37.5 months post-SC, whereas for participant N2, neutralizing titers for viral isolates from the first three time points increased over time. Overall, viruses from participants with CrNA seemed to escape from autologous neutralizing activity more rapidly than HIV-1 variants from patients who lacked CrNA. Indeed, in all patients who lacked CrNA, later-stage viruses were still sensitive to neutralization by autologous sera from early time points. The slower emergence of HIV-1 escape variants possibly reflects the limited immune pressure in these individuals, which was also evident from the lower peak neutralization titers. With the exception of C5, all individuals with CrNA had peak IC50 neutralizing titers above 650, while the three individuals who lacked CrNA had peak autologous neutralizing titers no higher than 480. Although only three individuals in this study lacked CrNA, this observation is in agreement with our earlier findings that individuals with the lowest CrNA also had the lowest autologous neutralizing titers (6, 38).
Development of cross-reactive neutralizing activity coincided with neutralization of early virus variants by autologous serum.
The contribution of autologous neutralization in the development of CrNA remains unclear. Here we compared the autologous and heterologous neutralizing activities in serum samples from the same patients. CrNA was measured at more time points than the autologous neutralizing activity. Therefore, the geometric mean IC50 titers in serum on the 6-virus panel closest to the time points at which the autologous neutralizing titers were established were plotted (Fig. 2). In the elite neutralizer (C1), the geometric mean IC50 titer against heterologous HIV-1 variants increased with the same kinetics as the neutralizing activity against autologous viruses from the early time points (2.1 and 7.5 months post-SC). At 26 months post-SC, the neutralizing titers against both autologous and heterologous viruses had remained stable; they subsequently increased again to peak at 35 months post-SC and decreased again thereafter. A similar pattern was observed for participants C2, C4, and C5. In participant C3, neutralizing titers against heterologous viruses peaked earlier than the neutralizing activity against autologous virus but were still high at 57.7 months post-SC, when neutralizing titers against viruses from the first time point (7.8 months post-SC) were highest. In all participants, the decrease in neutralizing titers against heterologous viruses corresponded with a waning of the autologous neutralizing responses over time.
Molecular basis of neutralizing antibody escape.
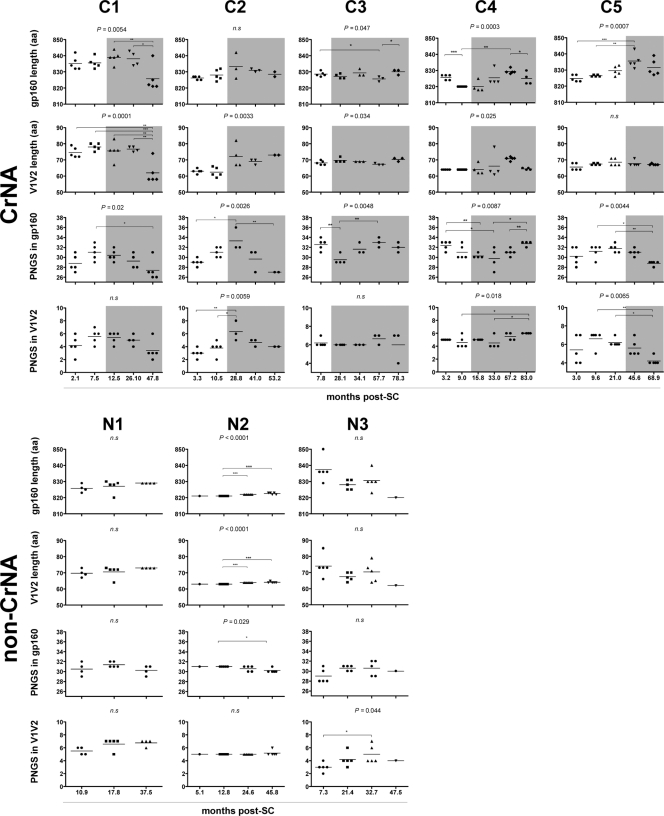
To determine the molecular basis of neutralization escape or possibly the induction of CrNA, the gp160 envelopes of all the autologous viral clones used in this study were sequenced. Overall, changes in the total length of gp160 were observed for 4 of the 5 individuals with CrNA (Fig. 3). In the elite neutralizer (C1), the length of gp160 remained the same until 26.1 months post-SC, whereas viruses that were obtained at the last time point before loss to follow-up had significantly shorter gp160 than the viruses from earlier time points. In participant C3, after an initial decrease, a small increase in the length of gp160 was observed during the last time point before AIDS. The same initial decline in the length of gp160 was seen in participant C4 but took place immediately after the first time point post-SC; the length of gp160 started to increase at 33 months post-SC, ending lower again prior to HAART initiation. In participant C5, there was an increase in the length of gp160 from the first time point until 45.6 months post-SC. There was no association between changes in the length of the total gp160 and the emergence of CrNA in sera. For participants who lacked CrNA in sera, gp160 length variation was less evident than it was in participants with CrNA; only one non-CrNA participant showed a slight change in gp160 length over the course of the infection.
Fig 3.
Longitudinal analysis of changes in the length of gp160 and the number of PNGS. Data for individuals with CrNA (C1 to C5) are grouped at the top, and data for individuals lacking CrNA (N1 to N3) are grouped at the bottom. Each dot represents one clonal virus variant. The horizontal bars indicate mean values per time point. P values above graphs were calculated by ANOVA across all time points, and individual differences were calculated using Bonferroni's multiple-comparison test for independent samples. Shaded areas indicate the presence of CrNA. The lengths of gp160 and V1V2 and the numbers of PNGS in gp160 and V1V2 are shown for both groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s, nonsignificant. Patient C6 is absent from this analysis because no viruses could be isolated.
Total PNGS in gp160 also differed between participants (Fig. 3). HIV-1 variants from three participants with CrNA (C1, C2, and C5), including the elite neutralizer (C1), showed an initial increase in the number of PNGS; however, the peak in the number of PNGS occurred earlier after SC in the elite neutralizer, and subsequently, the total number of PNGS declined, coinciding with the emergence of CrNA. Participants C3 and C4 had an inverse pattern, with a declining number of PNGS until 33 and 28.1 months post-SC, respectively, whereupon CrNA developed in both participants, coinciding with an increase in the amount of PNGS. These two participants also had an autologous neutralization pattern different from those of the other three participants with CrNA: in some instances, the later viruses were more sensitive to neutralization than viruses from earlier time points. Among individuals without CrNA, one (N3) had a slight increase in the number of PNGS from primary to chronic infection, after which the number remained stable. The remaining two participants showed no decrease, or only a slight decrease, in the number of PNGS in gp160. The presence of PNGS in the footprint of glycan-dependent antibodies, such as amino acid positions N160 and N332 for antibodies PG9, PG16, and 2G12 and the recently identified PGT antibodies, was checked in all sequences in order to investigate potential antibody-specific escape. A PNGS on N332 was present at all time points in all viruses analyzed from all patients (data not shown). The loss of the PNGS on position N160 was seen in all clones from the last time point for the elite neutralizer (C1). However, viral escape occurred earlier in this patient and therefore cannot be attributed solely to the loss of this PNGS. For all other patients with CrNA, individual viral clones showed the absence of a PNGS on N160, but its absence was not associated with viral escape, whereas all viruses from the individuals who lacked CrNA had a PNGS at position N160.
The length and number of PNGS in the V1V2 region of env have been shown to play a crucial role in both autologous and heterologous viral escape (5, 6, 8, 36). In contrast to the normal increase in V1V2 length over the course of infection, the viral variants from the elite neutralizer already had exceptionally long V1V2 regions at the start of infection (72 to 79 amino acids) compared with normal V1V2 length (∼65 amino acids) (8) and the length of V1V2 in the other individuals with CrNA (63 to 68 amino acids); these remained long until the last time point before loss to follow-up (47.8 months post-SC), when they declined to an average of 62 amino acids (Fig. 3). For two of the individuals with CrNA (C2 and C4), an increase in V1V2 length was observed, while participant C3 showed a decrease in V1 length and an increase in V2 length after the appearance of CrNA. Participant C5 showed no variation in V1V2 length, which could be due to low autologous neutralizing titers, despite the presence of high heterologous neutralizing titers. Although HIV-1 variants from patients who lacked CrNA did show some variation in V1V2 length, there were no changes in the length of the V1V2 domain over the course of infection, which could be due to the absence of neutralizing pressure.
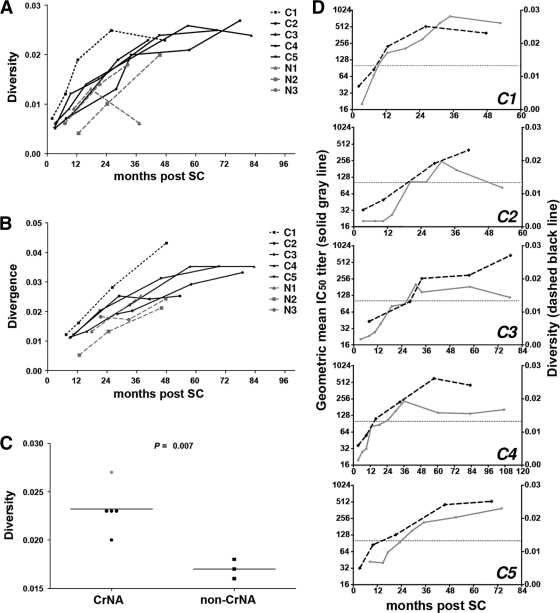
In order to look at the overall immune pressure on the virus per individual, the mean diversity within the gp160 sequences from the viral isolates per time point was calculated over the course of infection (Fig. 4A), as well as the sequence divergence between consecutive time points (Fig. 4B). These analyses revealed that HIV-1 variants from the elite neutralizer (C1) at each time point had a higher Env sequence diversity than those from any of the other individuals, as well as a higher sequence divergence between consecutive time points. The overall mean diversity across all clones per patient within the first 5 years of infection was significantly higher (P = 0.007) for individuals with CrNA than for patients who lacked CrNA (Fig. 4C), indicating stronger immune pressure on the virus in patients with CrNA. The average number of viral clones and amount of follow-up time per patient were similar for the two groups. Also, the kinetics with which the sequence diversity increased and peaked over the course of infection correlated with the increase and peak in the geometric mean IC50 titer across the 6-virus panel (Fig. 4D), further supporting the role of CrNA in viral escape through viral diversification.
Fig 4.
Diversity and divergence of HIV-1 gp160 sequences. (A) Sequence diversity across all clones of gp160 per time point is shown for individuals with (black lines) and without (gray dashed lines) CrNA, including one elite neutralizer (black dotted line) (C1), following seroconversion. (B) Divergence between subsequent time points during follow-up. Each dot depicts the latest of the subsequent time points for the same individuals for whom results are shown in panel A. (C) The mean diversity across all viral clones isolated within 5 years postseroconversion per individual is shown for individuals with and without CrNA. The elite neutralizer is represented by the gray dot. (D) Sequence diversity (gp160) (right y axis) and geometric mean IC50s across the 6-virus panel (left y axis) per time point depicted across the period of infection for individuals with CrNA. Patient C6 is absent from this analysis because no viruses could be isolated.
DISCUSSION
The prevalence of CrNA in HIV-1-infected individuals in different cohorts has been established at 10 to 30% (2, 10, 12, 31, 34). Because a vaccine should likely be able to elicit this type of neutralizing activity to protect against infection, the question of how CrNA develops in vivo remains to be answered. Here we compared two patient groups, those with and those without CrNA, within the Amsterdam cohort studies. All patients were Caucasian homosexual men infected with HIV-1 subtype B and had similar disease courses.
The mechanism that leads to the development of CrNA is not well defined. The ability to mount a cross-reactive neutralizing response could likely be partially dependent on the transmitted autologous virus, or viral escape from the immune system could lead to slight changes in the envelope that could enhance the affinity maturation of these antibodies, ultimately resulting in better binding of the viral envelope. The relationship between autologous and heterologous neutralizing activities could offer insight into the underlying mechanism by which CrNA develops. Here we showed that individuals with CrNA differed in their autologous neutralizing activity. Two individuals had potent neutralizing responses against autologous viruses that were isolated during primary infection, while two other individuals showed stronger neutralization of viruses that were isolated later in infection. However, the presence of CrNA does not ensure good autologous neutralization, as was seen for participant C5. The possible virus that induced CrNA could have escaped the autologous response, while the heterologous viruses were not placed under the same immune pressure from this particular patient. Sera from all individuals showed limited neutralizing activity against contemporaneous viruses. The peak of autologous neutralizing responses in the individuals with CrNA ranged from 28 to 69 months post-SC, while no clear peak could be observed in individuals who lacked CrNA. Viral escape was evident in all individuals with CrNA, although in two individuals escape was delayed. In the elite neutralizer, escape by autologous viruses occurred simultaneously with the development of CrNA. Additionally, we have shown that individuals who lacked CrNA in serum at 35 months post-SC could still mount an autologous neutralizing response, albeit with neutralizing titers lower than those of the majority of patients with CrNA. However, our study groups are too small to enable us to draw clear conclusions on the difference in autologous responses between patients who have or lack CrNA. Surprisingly, viruses isolated at later stages of infection in some cases were more sensitive to neutralization than viruses isolated early in infection. This may reflect the lack of immune pressure by CrNA driving the escape from neutralization, as opposed to the situation in individuals with CrNA. This was also evident from the differences in molecular changes in the envelope that have been associated with escape from humoral immune pressure. Indeed, changes in envelope length and in the number of potential N-linked glycosylation sites (PNGS) were more evident in individuals with CrNA than in those without CrNA. This was also evident from the higher genetic diversity of gp160 across all viral clones isolated within the first 5 years of infection for patients with CrNA than for those without CrNA. Also, the marked genetic diversity at all time points in the elite neutralizer suggests an immune pressure higher even than that of other individuals with CrNA and much higher than that of patients who lacked CrNA. Furthermore, the kinetics with which CrNA developed in these individuals coincided with the kinetics of emergence of a genetically diverse viral population and peaked simultaneously, suggesting that an increase in the potency of the neutralizing antibodies may have driven viral diversity. In turn, increased viral diversity may have driven the affinity maturation of the neutralizing antibodies, since the peak of viral diversity was not necessarily beyond the peak of the heterologous neutralization response.
In 6 individuals who had CrNA in their sera at ∼35 months post-SC, we studied the kinetics with which this activity had developed. Among these 6 individuals was one so-called elite neutralizer, defined by the ability of his serum to neutralize on average more than one pseudovirus at an IC50 titer of ≥300 within a clade and across at least four clades, with an average geometric mean IC50 titer above 500 (12, 34). By frequent testing for CrNA in longitudinally obtained serum samples in the first years post-SC, we could demonstrate that CrNA required 20 to 35 months to develop and peaked between 31 and 36 months post-SC. Interestingly, all patients had neutralizing activity for at least one virus from another clade within the first year post-SC, whereas the elite neutralizer showed full CrNA at 9 months post-SC. Our observations are in line with previous studies (13, 21) showing that CrNA had generally developed at 2.5 years postinfection. While these studies show some individuals with cross-reactivity within the first year of infection, most individuals neutralize the majority of viruses only after the first year of infection.
Our study included only six patients who, from a cohort of 82 participants in the Amsterdam Cohort Studies, had the highest geometric mean neutralizing titer and the largest breadth of neutralization at approximately 35 months post-SC on a panel of 23 HIV-1 variants from four different subtypes (12). From previous studies we know that CrNA develops more slowly in some individuals (38), and we also cannot exclude the possibility that the kinetics with which CrNA develops is different in individuals with somewhat less potent CrNA. Future study may reveal a correlation between the kinetics with which CrNA develops and the degree of somatic hypermutation and/or epitope specificity.
Interestingly, the kinetics with which CrNA developed coincided with the kinetics with which the autologous neutralizing response against viruses isolated early after seroconversion developed. Also, the peak and eventual waning of the neutralization responses paralleled each other. This may suggest that the development and titer of CrNA are driven by epitopes that are exposed on primary viruses early in infection. For this study, two different neutralizing assays were used to test for autologous and heterologous neutralizing activities. For the heterologous neutralizing activity, the method and virus panels described previously by Simek et al. (34) were used. This was done in order to compare our cohort with other cohorts. However, we have previously found comparable results between the PBMC- and TZMbl-based assays (38), and the replicating clonal viruses isolated for sequencing could be used directly in the PBMC-based assay for autologous neutralizing activity.
This is the first study to show potent CrNA within the first year after infection, although it was seen in only a single elite neutralizer. The elite neutralizer (C1) neutralized 5 out of the 6 viruses at 7.5 months post-SC and did so even more potently at 9.8 months post-SC. This could indicate that the HIV-1 variant in this individual has a unique exposure of epitopes that are capable of eliciting cross-reactive neutralizing antibodies that require limited somatic mutations and affinity maturation. Interestingly, the early viruses (isolated 3.2 months post-SC) from this patient had very long variable 1 regions in gp160 compared to the early HIV-1 variants from other participants with CrNA, but also compared to global HIV-1 variants of different subtypes (8). These viral characteristics seem counterintuitive, since an increase in V1 length has been associated with viral escape (6, 38), similar to what was observed for the other individuals with CrNA in our present study. Initial epitope mapping has revealed that the neutralizing activity in the serum of our elite neutralizer is not directed against the membrane-proximal external region on gp41 or against the CD4 binding site. Also, it was insensitive to the N160 and N332 mutations, indicating an absence of PG9, PG16, 2G12, and PGT-like antibodies (data not shown). Although similarity to other known epitope specificities of broadly neutralizing antibodies remains to be tested, reactivity may potentially be directed against a novel epitope.
The broadly neutralizing antibodies known to date have highly mutated variable heavy chains compared to antibodies elicited in the secondary immune response against influenza (20). In vitro reverted unmutated ancestor antibodies from HIV-1-specific broadly neutralizing antibodies have shown little or no reactivity with the epitopes of their progeny broadly neutralizing antibodies (3, 23, 24, 44). This indicates that broadly neutralizing antibodies in general may need multiple rounds of somatic mutation and affinity maturation and that the currently known recombinant envelope glycoproteins are insufficient in promoting this process to achieve the desired mature antibodies from precursor naïve B cells. Additionally, germ line predecessors of broadly neutralizing antibodies have been shown to be different for rhesus macaques versus humans (42), which could explain the limited success of efforts at the generation of these antibodies in animal models.
We and others have shown previously that CrNA in serum at 35 months post-SC does not protect against disease progression (12, 13, 26). With the knowledge that CrNA takes 2 to 4 years to develop, one could argue that the neutralizing humoral immune response simply comes too late to have an impact on the disease course. Indeed, prior to the development of these antibodies, the immune system may have been irreversibly damaged, although obviously the ability of the patient to still develop CrNA argues against this. Moreover, in the elite neutralizer we studied here, CrNA had developed as early as 9 months post-SC and still increased until 3 years post-SC, yet this individual progressed to full-blown AIDS within 7.5 years post-SC. This result demonstrates that even early onset of CrNA may not be enough to halt disease progression and is in line with the rapid escape of HIV-1 even from CrNA (37). Our findings, taken together, emphasize that CrNA should be in place in HIV-negative individuals in order to prevent infection, since it has no impact on disease progression.
In conclusion, we have shown that CrNA can develop rapidly after HIV-1 infection is established, even within the first year after seroconversion, in an elite neutralizer, as opposed to other individuals with CrNA, where it took 20 to 35 months to develop. The kinetics with which CrNA developed paralleled the development of autologous neutralizing activity, as well gp160 sequence diversity. It is tempting to speculate that the autologous viruses that are present during primary infection in individuals who ultimately develop CrNA may have better exposure of the epitopes relevant for eliciting CrNA. Subsequent escape of these viruses, either by amino acid changes in the epitopes or by shielding of the epitope, may drive affinity maturation of the antibodies. Alternatively, individuals who develop CrNA may have a different B cell repertoire that supports the generation of antibodies with CrNA. Finally, the development of CrNA may just be random and may be dominated by the first B cell specificity that encountered the virus.
Overall, the findings for our elite neutralizer prove that it is possible to achieve potent CrNA early in infection, and this individual may provide valuable clues for the design of an effective vaccine or for the creation of optimal conditions for establishing this type of humoral immune response.
ACKNOWLEDGMENTS
The Amsterdam Cohort Studies on HIV infection and AIDS, a collaboration between the Amsterdam Health Service, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, the University Medical Center Utrecht, and the Jan van Goyen Clinic, are part of The Netherlands HIV Monitoring Foundation and are financially supported by the Center for Infectious Disease Control of The Netherlands National Institute for Public Health and the Environment. This study was financially supported by The Netherlands Organization for Scientific Research (grant 918.66.628), the European Community's Sixth Framework Program, European HIV Enterprise (EUROPRISE; FP6/2007–2012), under grant agreement 037611, and the European Community's Seventh Framework Program, Next-Generation HIV-1 Immunogens Inducing Broadly Reactive Neutralizing Antibodies (NGIN) (FP7/2007–2013), under grant agreement 201433.
We also thank Kay Limoli, Sam Jauregui, and the Clinical Reference Lab at Monogram BioSciences.
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Beaumont T, et al. 2001. Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J. Virol. 75:2246–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Binley JM, et al. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651–11668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonsignori M, et al. 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boom R, et al. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bunnik EM, et al. 2010. Adaptation of HIV-1 envelope gp120 to humoral immunity at a population level. Nat. Med. 16:995–997 [DOI] [PubMed] [Google Scholar]
- 6. Bunnik EM, Pisas L, van Nuenen AC, Schuitemaker H. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burton DR, et al. 2011. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc. Natl. Acad. Sci. U. S. A. 108:11181–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Curlin ME, et al. 2010. HIV-1 envelope subregion length variation during disease progression. PLoS Pathog. 6:e1001228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Wolf F, et al. 1988. Numbers of CD4+ cells and the levels of core antigens of and antibodies to the human immunodeficiency virus as predictors of AIDS among seropositive homosexual men. J. Infect. Dis. 158:615–622 [DOI] [PubMed] [Google Scholar]
- 10. Doria-Rose NA, et al. 2010. Breadth of human immunodeficiency virus-specific neutralizing activity in sera: clustering analysis and association with clinical variables. J. Virol. 84:1631–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Edo-Matas D, et al. 2010. Genetic composition of replication competent clonal HIV-1 variants isolated from peripheral blood mononuclear cells (PBMC), HIV-1 proviral DNA from PBMC and HIV-1 RNA in serum in the course of HIV-1 infection. Virology 405:492–504 [DOI] [PubMed] [Google Scholar]
- 12. Euler Z, et al. 2010. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J. Infect. Dis. 201:1045–1053 [DOI] [PubMed] [Google Scholar]
- 13. Gray ES, et al. 2011. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J. Virol. 85:4828–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41:95–98 [Google Scholar]
- 15. Hessell AJ, et al. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hessell AJ, et al. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reference deleted. [Google Scholar]
- 18. Mahalanabis M, et al. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mascola JR, et al. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat. Med. 6:207–210 [DOI] [PubMed] [Google Scholar]
- 20. McElrath MJ, Haynes BF. 2010. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 33:542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mikell I, et al. 2011. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 7:e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore PL, et al. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mouquet H, et al. 2010. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 467:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pancera M, et al. 2010. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J. Virol. 84:8098–8110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a. Pereyra F, et al. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197:563–571 [DOI] [PubMed] [Google Scholar]
- 25. Petropoulos CJ, et al. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piantadosi A, et al. 2009. Breadth of neutralizing antibody response to human immunodeficiency virus type 1 is affected by factors early in infection but does not influence disease progression. J. Virol. 83:10269–10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quakkelaar ED, et al. 2007. Susceptibility of recently transmitted subtype B human immunodeficiency virus type 1 variants to broadly neutralizing antibodies. J. Virol. 81:8533–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rademeyer C, et al. 2007. Genetic characteristics of HIV-1 subtype C envelopes inducing cross-neutralizing antibodies. Virology 368:172–181 [DOI] [PubMed] [Google Scholar]
- 29. Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rong R, et al. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sather DN, et al. 2009. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J. Virol. 83:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scheid JF, et al. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640 [DOI] [PubMed] [Google Scholar]
- 33. Schuitemaker H, et al. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Simek MD, et al. 2009. HIV-1 elite neutralizers: individuals with broad and potent neutralizing activity identified using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 83:7337–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tersmette M, et al. 1989. Detection and subtyping of HIV-1 isolates with a panel of characterized monoclonal antibodies to HIV-p24 gag. Virology 171:149–155 [DOI] [PubMed] [Google Scholar]
- 36. van Gils MJ, et al. 2011. Longer V1V2 region with increased number of potential N-linked glycosylation sites in the HIV-1 envelope glycoprotein protects against HIV-specific neutralizing antibodies. J. Virol. 85:6986–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Gils MJ, et al. 2010. Rapid escape from preserved cross-reactive neutralizing humoral immunity without loss of viral fitness in HIV-1-infected progressors and long-term nonprogressors. J. Virol. 84:3576–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Gils MJ, Euler Z, Schweighardt B, Wrin T, Schuitemaker H. 2009. Prevalence of cross-reactive HIV-1-neutralizing activity in HIV-1-infected patients with rapid or slow disease progression. AIDS 23:2405–2414 [DOI] [PubMed] [Google Scholar]
- 39. van't Wout AB, et al. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral and vertical transmission. J. Clin. Invest. 94:2060–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van't Wout AB, Schuitemaker H, Kootstra NA. 2008. Isolation and propagation of HIV-1 on peripheral blood mononuclear cells. Nat. Protoc. 3:363–370 [DOI] [PubMed] [Google Scholar]
- 41. Wei X, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312 [DOI] [PubMed] [Google Scholar]
- 42. Yuan T, et al. 2011. Putative rhesus macaque germline predecessors of human broadly HIV-neutralizing antibodies: differences from the human counterparts and implications for HIV-1 vaccine development. Vaccine 29:6903–6910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang M, et al. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246 [DOI] [PubMed] [Google Scholar]
- 44. Zhou T, et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]