Abstract
The replication of many viruses involves the formation of higher-order structures or replication “factories.” We show that the key replication enzyme of foot-and-mouth disease virus (FMDV), the RNA-dependent RNA polymerase, forms fibrils in vitro. Although there are similarities with previously characterized poliovirus polymerase fibrils, FMDV fibrils are narrower, are composed of both protein and RNA, and, importantly, are seen only when all components of an elongation assay are present. Furthermore, an inhibitory RNA aptamer prevents fibril formation.
TEXT
Foot-and-mouth disease virus (FMDV) is one of the most important animal pathogens of agricultural significance, yet key aspects of its replication are still unclear. The virus occurs as seven distinct serotypes and is a member of the Picornaviridae, which also includes important human pathogens, such as poliovirus (PV) and rhinovirus. Replication of the positive-stranded RNA genome occurs via a negative-stranded intermediate and involves several viral proteins as well as specific sequences within the untranslated regions (UTRs) at the 5′ and 3′ ends. The 5′ UTR is exceptionally long (∼1,200 nucleotides), constituting one-seventh of the viral genome. It includes a 400-nucleotide region predicted to be double stranded in structure (3), a poly(C) tract of ∼200 nucleotides (20), a series of pseudoknots (3, 6), a cis-acting replication element (cre) necessary for the uridylation of the peptide primer of replication (VPg) (13, 15), and an internal ribosome entry site (IRES) necessary for the initiation of protein synthesis (18). The 3′ UTR is shorter but includes two stem-loop structures and a poly(A) tract. Several of these RNA structural elements are known or thought to be important in genome replication. The engine of genome replication is an RNA-dependent RNA polymerase enzyme (RdRp), termed 3Dpol, with the involvement of its precursor, 3CD. RNA synthesis is primed by a uridylated form of VPg, which is the product of the 3B gene. Uniquely, the FMDV genome encodes three tandem copies of 3B (VPg), and it is tempting to speculate that this feature is causally linked to the exceptionally fast replication rate characteristic of the virus. Some strains of FMDV complete the entire life cycle in only 3 h in vitro (20). Uridylation of VPg to form VPgpUpU is carried out by 3Dpol but requires the presence of 3CD in an ancillary catalytic role to work efficiently (15). Although synthetic versions of VPg peptide can be uridylated in vitro, the true precursor in vivo (3AB or 3BCD) is unknown and controversial. Replication of the viral genome is complex and difficult to study in vivo, but aspects of the process can be replicated in vitro, making them more amenable to investigation.
Replication of many positive-stranded RNA viruses is thought to occur in “factories” (4), allowing compartmentalization, concentration, and protection of the components and products. Consistent with this, FMDV 3Dpol has been shown to concentrate on membranes of small, smooth vacuoles (19). The Kirkegaard group has identified fibrous structures (approximately 120 Å in diameter) associated with membranes within PV-infected cells. Furthermore, they have demonstrated that recombinant PV 3Dpol alone can form fibers/fibrils, sheets, and tubes of 400 to 1,000 Å in diameter in vitro (11). In this communication, we show that FMDV 3Dpol also forms fibrils in vitro, but only when all of the components necessary for an active elongation assay are present. Furthermore, these structures do not form in the presence of a previously characterized RNA aptamer inhibitory to 3Dpol (5).
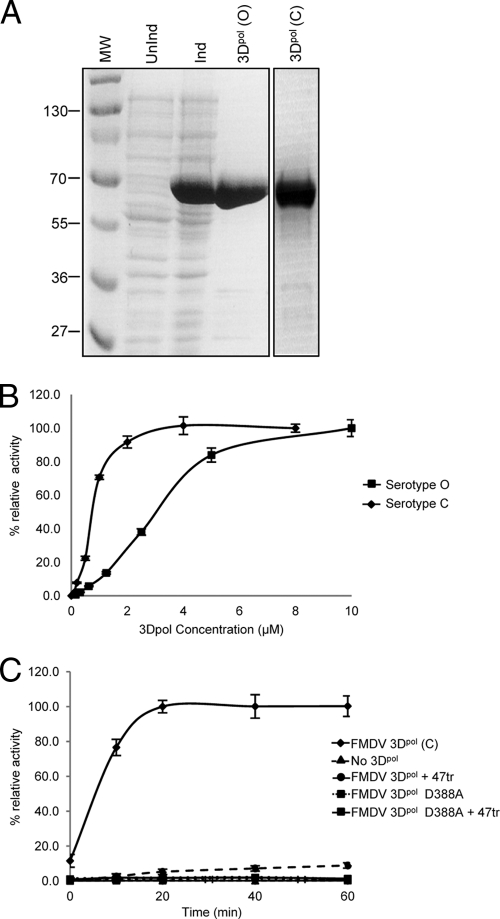
Both serotype O and C enzymes were used in this study, together with catalytically inactive D388A serotype C mutant protein (a gift from Esteban Domingo, Madrid [1]). Serotype C 3Dpol [3Dpol(C)] was purified as described previously (7). The 3Dpol (serotype O) gene was subcloned from pUb-3Dpol (15) into pET22b-3Dpol. The protein was expressed in Escherichia coli as a C-terminal hexahistidine fusion and was affinity purified by adsorption to a nickel-Sepharose column (in 50 mM phosphate [pH 7.8], 300 mM NaCl, and elution with a gradient of up to 500 mM imidazole). The 3Dpol samples were more than 95% pure, as shown in Fig. 1A. The wild-type (WT) proteins were shown to be active in elongation assays in which [32P]UTP incorporation was measured using a poly(A) template and an oligo(U)15 primer (5). However, under these conditions the relationship between enzyme concentration and nucleotide incorporation was not linear, indicative of a degree of cooperativity (Fig. 1B). This is particularly evident with the less-active serotype O enzyme. The addition of ZnCl2 (60 μM) to enhance the activity according to previously reported PV protocols (2, 8, 9, 12, 21) had little effect. Using a similar assay, we showed that the D388A mutant was unable to incorporate [32P]UTP, as expected (Fig. 1C). Interestingly, cooperativity had been demonstrated previously for other viral polymerases, including PV 3Dpol (11, 17).
Fig 1.
Purification and activity of FMDV 3Dpol. (A) Samples of BL21(DE3) pLysS E. coli transformed with pET22-3Dpol(O) were analyzed by 15% SDS-PAGE either prior to IPTG (isopropyl-β-D-thiogalactopyranoside) induction (UnInd) or 2 h postinduction (Ind). A sample of 3Dpol(C) is also shown. MW, molecular weights in thousands. (B) Samples of 3Dpol(C) and 3Dpol(O) were assayed for elongation activity using the incorporation of [32P]UTP in a MOPS (morpholinepropanesulfonic acid)/NaCl buffer (with 5 mM MgCl2) with a poly(A) template (0.1 μg/μl) and an oligo(U)15 primer (1 μM) as described previously (5). Samples were assayed after 20 or 25 min, respectively. (C) The activities of wild-type 3Dpol(C) and the active-site mutant D388A (1 μM) were investigated over time in a similar assay. The effects of preincubation with aptamer 47tr (4 μM; 20 min) are also shown.
Previously, we selected RNA aptamers to 3Dpol of the serotype C virus. RNA aptamers are derived from the process of in vitro selection and bind to their target molecules with high affinity and specificity, making them useful as molecular tools and potentially as inhibitors (5). We characterized aptamers that inhibited the activity of 3Dpol in an elongation assay. One of the aptamers was truncated to 32 mer (F47tr, 5′-GGGUUAACAGAAAACCUCAGUUGCUGGGUUGU-3′), maintaining both its inhibitory capacity and its affinity for 3Dpol. Here, we also show that preincubation of 3Dpol with 47tr resulted in no incorporation of [32P]UTP (Fig. 1C).
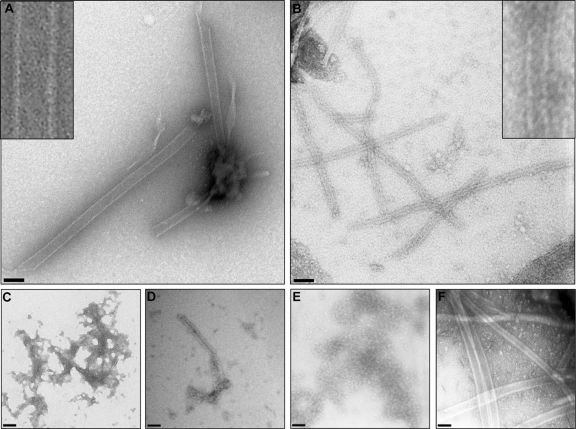
To investigate the products of elongation assays (as described in Fig. 1C), samples were negatively stained prior to examination by transmission electron microscopy (TEM) according to a previously described protocol (22). PV 3Dpol was included as a control and shown to form fibrils similar to those described previously (11, 23) (Fig. 2A). In contrast, FMDV 3Dpol alone was disordered and aggregated, showing no evidence of defined higher-order structures (Fig. 2C). This result is in agreement with crystallographic data in which differences in crystal packing between the PV and FMDV enzymes suggest that FMDV 3Dpol alone is unlikely to form higher-order structures (7). Products of the FMDV elongation assay, however, appeared as ordered fibrils by TEM (Fig. 2B). The fibrils were 250 ± 27 Å (n = 30) in width and were narrower and more regular than the fibrils observed with isolated PV 3Dpol (460 ± 32Å; n = 30). Fibrils formed by both PV 3Dpol and FMDV 3Dpol were variable in length. Both C and O 3Dpol were capable of fibril formation (Fig. 2B and D, respectively); however, the fibrils formed with the more active serotype C enzyme were more regular in morphology. There was no evidence for the formation of defined higher-order structures other than fibrils, unlike PV 3Dpol, which also formed sheets and tubes. Also in contrast with PV 3Dpol, all components of an elongation assay were necessary for the formation of the FMDV 3Dpol structures (i.e., primer, template, magnesium ions, and NTPs). The presence of dithiothreitol (DTT) or Zn2+ had no effect; however, EDTA inhibited both [α-32P]UTP incorporation in the elongation assay and the formation of fibrils as seen by TEM (Table 1), indicating that fibril formation was not a spontaneous event, but rather requires active elongation to occur.
Fig 2.
Formation of fibrils during the polymerase elongation assay. (A) PV 3Dpol alone (3 μM). (B) FMDV 3Dpol(C) (1 μM) in the context of a full elongation assay after 30 min. The FMDV fibrils have a regular width averaging 250 Å but exhibit considerable variations in length. (C) FMDV 3Dpol(C) alone. (D) Same as panel B but with the substitution of 3Dpol(O) for 3Dpol(C). (E) Same as panel B but with the addition of equimolar aptamer 47tr. (F) Same as panel A but with the addition of equimolar aptamer 47tr. The insets in panels A and B are higher-magnification views. Negative-stain TEM; scale bars, 500 Å.
Table 1.
Formation of 3Dpol(C) fibrils under different experimental conditions
| Assay or component | Presence of fibrils |
|---|---|
| Full elongation assay | + |
| Full elongation assay | |
| With catalytically inactive 3Dpol | − |
| Without 3Dpol | − |
| Without template | − |
| Without primer | − |
| With aptamer 47tr (1–5 μM) | − |
| With RNase (25 μg/ml) | − |
| With proteinase K (500 μg/ml) | − |
| With EDTA (10 mM) | − |
| With DTT (5 mM) | + |
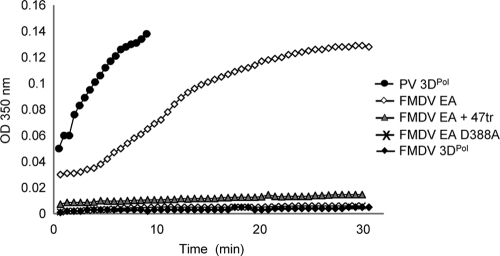
The TEM data correlated with light-scattering measurements of the assay undertaken over the same time scale (Fig. 3). A rapid increase in absorbance at 350 nm was observed with samples of isolated PV 3Dpol. A similar, but slower, increase was seen with FMDV 3Dpol (saturating after 25 to 30 min) when all components of an elongation assay were present. No increase in light scattering was detected when FMDV 3Dpol alone was incubated over time. In addition, substitution of WT 3Dpol for the D388A mutant also resulted in no increase in light scattering over time and no fibril formation visible by TEM (Fig. 3 and Table 1). Furthermore, fibrils were not observed in samples treated with RNase A (0.5 μg per 20-μl reaction mixture) or proteinase K (10 μg per 20-μl reaction mixture) at the endpoint of the assay (i.e., after 30 min) (Table 1). This indicates that the fibrils are composed of both 3Dpol and single-stranded RNA, rather than an RNA duplex. The walls of the fibrils were measured to be 60 to 70 Å in thickness, consistent with the presence of a single polymerase molecule (7) and suggestive of the structures being tubular in nature. However, further structural studies will be required to confirm this hypothesis. Interestingly, fibrils were also observed when part of the 5′ UTR (negative sense) was used as a template in an elongation assay, although these were less uniform, probably due to the heterogeneity of the RNA used (16). However, this does indicate that the fibrils are not an artifact resulting from the use of a poly(A) template and that 3Dpol is able to unfold structured RNA in order to produce a product.
Fig 3.
Light-scattering measurements of fibril formation over a time course of up to 30 min at room temperature. Symbols: solid circles, PV 3Dpol alone (3 μM); open diamonds, FMDV 3Dpol elongation assay (EA) with 3 μM 3Dpol(C) and the method described for Fig. 1C; solid diamonds, FMDV 3Dpol alone; shaded triangles, FMDV 3Dpol EA (as above) in the presence of equimolar aptamer 47tr; cross, EA using FMDV 3Dpol active-site mutant D388A. OD350 nm, optical density at 350 nanometers.
In order to determine the effects of aptamer 47tr on fibril formation, this molecule was included in the assays. Preincubation of 3Dpol(C) with 47tr (at an equimolar concentration for 30 min prior to the addition of other components of the elongation assay) inhibited the formation of fibrils observable by TEM (Fig. 2E). This result correlated with no increase in light scattering detectable over time in the presence of the aptamer (Fig. 3). However, addition of the aptamer at the endpoint of the assay had no effect. The inhibitory effect of the aptamer on enzyme activity therefore correlates with the absence of fibrils, again demonstrating that these formed only when the polymerase was active. We had demonstrated previously that the aptamer had no inhibitory effect on the ability of PV 3Dpol to incorporate [32P]UTP in an elongation assay (5). In agreement with this, the aptamer had no effect on the formation of PV 3Dpol fibrils by TEM (Fig. 2F).
The formation of a fibrillar replication lattice could have several benefits in viral replication. It could serve to increase the local concentration of components, and a clustering of binding sites could increase the efficiency of replication. It is also tempting to speculate that the higher-order structures have a role in preventing the formation of double-stranded RNA. During replication, a negative-stranded cRNA is first produced, which then acts as a template for the synthesis of positive strands. If the RNA is “coated” in polymerase molecules during this process, forming fibril-like structures, then the formation of a double-stranded negative/positive RNA complex could be prevented. This would allow replication to proceed without the necessity to melt an RNA duplex, which would be energetically demanding and therefore inefficient. It must be noted that in vivo, a number of other viral and host proteins (e.g., RNA helicase [10]) are also involved in replication. Our studies are therefore continuing with the use of an FMDV replicon (14) in order to probe the formation of higher-order structures in cells where all replication components are present.
ACKNOWLEDGMENTS
We thank Armando Arias, Graham Belsham, Ian Goodfellow, and Esteban Domingo for 3Dpol serotype C and O constructs, samples of PV 3Dpol, and the D388A mutant proteins, respectively.
We gratefully acknowledge financial support from BBSRC.
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Arias A, et al. 2005. Mutant viral polymerase in the transition of virus to error catastrophe identifies a critical site for RNA binding. J. Mol. Biol. 353:1021–1032 [DOI] [PubMed] [Google Scholar]
- 2. Cho MW, Richards OC, Dmitrieva TM, Agol V, Ehrenfeld E. 1993. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J. Virol. 67:3010–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clarke BE, et al. 1987. Potential secondary and tertiary structure in the genomic RNA of foot and mouth disease virus. Nucleic Acids Res. 15:7067–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cook PR. 1999. The organization of replication and transcription. Science 284:1790–1795 [DOI] [PubMed] [Google Scholar]
- 5. Ellingham M, Bunka DHJ, Rowlands DJ, Stonehouse NJ. 2006. Selection and characterization of RNA aptamers to the RNA-dependent RNA polymerase from foot-and-mouth disease virus. RNA 12:1970–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Escarmís C, Dopazo J, Dávila M, Palma EL, Domingo E. 1995. Large deletions in the 5′-untranslated region of foot-and-mouth disease virus of serotype C. Virus Res. 35:155–167 [DOI] [PubMed] [Google Scholar]
- 7. Ferrer-Orta C, et al. 2004. Structure of foot-and-mouth disease virus RNA-dependent RNA polymerase and its complex with a template-primer RNA. J. Biol. Chem. 279:47212–47221 [DOI] [PubMed] [Google Scholar]
- 8. Gohara DW, et al. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275:25523–25532 [DOI] [PubMed] [Google Scholar]
- 9. Hobson SD, et al. 2001. Oligomeric structures of poliovirus polymerase are important for function. EMBO J. 20:1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lawrence P, Rieder E. 2009. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J. Virol. 83:11356–11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lyle JM, Bullitt E, Bienz K, Kirkegaard K. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218–22122 [DOI] [PubMed] [Google Scholar]
- 12. Lyle JM, et al. 2002. Similar structural basis for membrane localization and protein priming by an RNA-dependent RNA polymerase. J. Biol. Chem. 277:16324–16331 [DOI] [PubMed] [Google Scholar]
- 13. Mason PW, Bezborodova SV, Henry TM. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McInerney GM, King AM, Ross-Smith N, Belsham GJ. 2000. Replication-competent foot-and-mouth disease virus RNAs lacking capsid coding sequences. J. Gen. Virol. 81:1699–1702 [DOI] [PubMed] [Google Scholar]
- 15. Nayak A, Goodfellow IG, Belsham GJ. 2005. Factors required for the uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro. J. Virol. 79:7698–7706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nayak A, et al. 2006. Role of RNA structure and RNA binding activity of foot-and-mouth disease virus 3C protein in VPg uridylylation and virus replication. J. Virol. 80:9865–9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pata JD, Schultz SC, Kirkegaard K. 1995. Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1:466–477 [PMC free article] [PubMed] [Google Scholar]
- 18. Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325 [DOI] [PubMed] [Google Scholar]
- 19. Polatnick J, Wool SH. 1983. Correlation of surface and internal ultrastructural changes in cells infected with foot-and-mouth disease virus. Can. J. Comp. Med. 47:440–444 [PMC free article] [PubMed] [Google Scholar]
- 20. Rowlands DJ, Harris TJ, Brown F. 1978. More precise location of the polycytidylic acid tract in foot and mouth disease virus RNA. J. Virol. 26:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Semler BL, Hanecak R, Dorner LF, Anderson CW, Wimmer E. 1983. Poliovirus RNA synthesis in vitro: structural elements and antibody inhibition. Virology 126:624–635 [DOI] [PubMed] [Google Scholar]
- 22. Stonehouse NJ, Stockley PG. 1993. Effects of amino acid substitution on the thermal stability of MS2 capsids lacking genomic RNA. FEBS Lett. 334:355–359 [DOI] [PubMed] [Google Scholar]
- 23. Tellez AB, et al. 2011. Interstitial contacts in an RNA-dependent RNA polymerase lattice. J. Mol. Biol. 412:737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]