Abstract
The Wnt/β-catenin pathway is involved in diverse cell functions governing development and disease. β-Catenin, a central mediator of this pathway, binds to members of the TCF/LEF family of transcription factors to modulate hundreds of genes. Active Wnt/β-catenin/TCF-4 signaling plays a significant role in repression of HIV-1 replication in multiple cell targets, including astrocytes. To determine the mechanism by which active β-catenin/TCF-4 leads to inhibition of HIV replication, we knocked down β-catenin or TCF/LEF members in primary astrocytes and astrocytomas transiently transfected with an HIV long terminal repeat (LTR)-luciferase reporter that contained an integrated copy of the HIV LTR-luciferase construct. Knockdown of either β-catenin or TCF-4 induced LTR activity by 2- to 3-fold under both the episomal and integrated conditions. This knockdown also increased presence of serine 2-phosphorylated RNA polymerase II (Pol II) on the HIV LTR as well as enhanced its processivity. Knockdown of β-catenin/TCF-4 also impacted tethering of other transcription factors on the HIV promoter. Specifically, knockdown of TCF-4 enhanced binding of C/EBPβ, C/EBPδ, and NF-κB to the HIV LTR, while β-catenin knockdown increased binding of C/EBPβ and C/EBPδ but had no effect on NF-κB. Approximately 150 genes in astrocytes were impacted by β-catenin knockdown, including genes involved in inflammation/immunity, uptake/transport, vesicular transport/exocytosis, apoptosis/cellular stress, and cytoskeleton/trafficking. These findings indicate that modulation of the β-catenin/TCF-4 axis impacts the basal level of HIV transcription in astrocytes, which may drive low level/persistent HIV in astrocytes that can contribute to ongoing neuroinflammation, and this axis also has profound effects on astrocyte biology.
INTRODUCTION
HIV invades the central nervous system (CNS) early in the course of natural infection and leads to HIV-associated dementia (HAD) in up to 30% of infected/untreated individuals. Advances in current antiretroviral therapy remarkably decreased the incidence of frank dementia, but a spectrum of HIV-associated neurocognitive disorders (HAND) persists in approximately 50% of infected individuals (18, 19). HAND is manifested by a decline in memory, learning, and executive function that confers impairment in day-to-day activity. HIV-mediated neuropathogenesis, depending on severity of disease, includes reactive astrocytosis, myelin pallor, and perturbations in synaptic and dendritic density that may also include selective neuronal loss. The mechanisms underlying HAND are still not entirely clear but are believed to be driven by neuroinflammation mediated by inflammatory cytokines as well as by HIV virotoxins such as Tat and gp120 expressed within or released from infected cells, causing dysregulation and/or apoptosis of neurons and glial cells.
Although the primary cell targets for productive HIV replication in the brain are monocyte/macrophages and microglia, emerging data point to a contributing role of astrocytes in driving lower levels of HIV replication and even constituting a sanctuary site for HIV in the CNS. Astrocytes constitute 40 to 70% of brain cells and perform vital functions critical for maintenance of blood-brain barrier integrity, release of neurotrophic factors, metabolism of toxic neurotransmitters, and immune surveillance by secretion of cytokines/chemokines. Historically, the role of astrocytes as permissive hosts for productive HIV replication has been unclear. Recent data suggest that astrocytes are an important target for HIV latency and under certain conditions support productive HIV replication (9, 10). Postmortem tissue from patients with various degrees of HAND demonstrates that up to 19% of astrocytes are positive for HIV Env DNA, which correlates with their close proximity to perivascular macrophages (10). Most importantly, without HAND or close proximity to perivascular macrophages, up to 3% of astrocytes are HIV Env+, a number that is reminiscent of the size of the latent HIV pool within CD4+ memory T cells (10, 39). Despite detection of considerable HIV DNA within astrocytes, the numbers of HIV p24+ astrocytes in vivo are considerably low (11, 15, 29, 40, 41, 44). This paradox suggests that, despite being CD4 negative, astrocytes do support HIV entry and are largely latently infected in vivo but that, under the appropriate microenvironment signals, they can lead to low levels of HIV replication. A low level of HIV replication is thought to be a driving force in residual neuroinflammatory processes in the CNS and in continued seeding of HIV in the CNS. In vitro, HIV infection of resting/nonprimed astrocytes is nonproductive, with evidence for HIV integration but not release (8). When pretreated with gamma interferon (IFN-γ), a cytokine associated with severity of HAD (25, 37), astrocytes become productively infected with HIV.
HIV infection of astrocytes, albeit less efficient than that in macrophages and microglia, is still highly relevant to the overall pathogenesis of HIV in the CNS. Because astrocytes are abundant in the CNS, by sheer number alone, any level of HIV replication within these cells may have dramatic effects on HIV-mediated pathogenesis. Particularly, astrocytes may contribute to HIV neuropathogenesis by releasing virotoxins (e.g., gp120, Tat, Nef, and Vpr), increasing the overall viral load within the CNS and/or inducing host inflammatory responses. These collective mechanisms are detrimental to neuronal and glial health. Persistent HIV replication, even at low levels, is especially detrimental in a largely non-self-renewal compartment such as the CNS. We have been investigating the conditions and signaling pathways that drive HIV replication in astrocytes. We demonstrated that astrocytes have robust Wnt/β-catenin signaling that is associated with suppression of HIV. When this pathway is down-modulated by a biologic signal such as IFN-γ (8, 26) or by loss-of-function molecular studies, HIV replication is enhanced.
The Wnt family is comprised of 19 soluble secreted cysteine-rich glycoproteins that regulate signaling pathways that control transcriptional activity of hundreds of genes involved in cell differentiation, communication, apoptosis/survival, and proliferation. Wnt signaling includes β-catenin-dependent (canonical) and β-catenin-independent (noncanonical) pathways (20). The most extensively studied Wnt pathway is the Wnt/β-catenin canonical pathway. Dysregulation of Wnt/β-catenin has been linked to a number of disease conditions, including neurodegenerative diseases, psychiatric diseases, cancers, asthma, and even wound healing (20).
Canonical Wnt/β-catenin signaling is initiated by binding of a Wnt ligand to a member of the Frizzled (Fz) family of seven transmembrane receptors and a coreceptor such as low-density lipoprotein receptor-related proteins 5 and 6 (LRP5/6). The central mediator of the canonical pathway is β-catenin, a multifunctional protein that either can associate with cadherins at the cell membrane to regulate cellular adhesion or can translocate to the nucleus, where it functions as a transcriptional coactivator. Shuttling of β-catenin between the membrane, cytoplasm, and nucleus is largely controlled by its phosphorylation state, which is regulated by a wide variety of tyrosine and serine/threonine kinases. When Wnt signaling is inactive, a multiprotein destruction complex composed of axin, adenomatous polyposis coli (APC), casein kinase 1α (Ck1α), and glycogen synthase kinase 3β (GSK3β) binds to cytosolic β-catenin. Ck1-mediated phosphorylation on Ser45, followed by GSK3β-mediated phosphorylation on Thr 41, Ser33, and Ser37, targets β-catenin for ubiquitination by βTrcp and degradation through the proteasomal pathway. Binding of a Wnt ligand initiates a cascade of events that results in destabilization of the destruction complex and accumulation of a stable, hypophosphorylated β-catenin that is able to translocate to the nucleus and associate with a member of the T cell factor/lymphoid enhancer factor (TCF/LEF) family of transcription factors. Within the nucleus, β-catenin displaces negative regulatory elements from TCF/LEF, such as transducin-like enhancer (TLE) protein and histone deacetylases (HDACs), and recruits cofactors such as BCL9, Pygopus (Pygo), and CBP/p300 to activate transcription of Wnt target genes. Members of the TCF/LEF family of transcriptional factors are critical in canonical β-catenin signaling. They are downstream effectors of this pathway and include LEF1, TCF-1, TCF-3, and TCF-4. They all contain a high mobility group (HMG) domain, allowing them to induce a sharp bend in the DNA helix, and without a coactivator such as β-catenin, they are associated with gene repression.
Given that β-catenin/TCF-4 inhibits HIV replication (8, 23, 36), we investigated here whether the inhibition of HIV replication mediated by β-catenin/TCF-4 is at the level of HIV transcriptional activity in astrocytes and determined the impact of β-catenin signaling on activity of transcriptional factors prominent in regulation of HIV promoter activity. Studying astrocyte biology is complex because of the caveats associated with the in vitro model systems used. Primary human progenitor-derived astrocytes and human fetal astrocytes, which are the norm in the literature, are still fetal astrocytes in nature and may not mimic the behavior of adult astrocytes in the context of HIV comorbidity. Several cell lines have been used to model adult astrocytes, but these cells are transformed; and their behavior may also be distinct from nontransformed cells. We approached this challenge by using primary human progenitor-derived astrocytes as well as two astrocytoma cell lines so that our findings would not be limited to a particular cell line.
MATERIALS AND METHODS
Cell culture.
U87MG and U251MG astrocytoma cell lines were obtained from the NIH AIDS Research and Reference Reagent Program (Frederick, MD) and the American Type Culture Collection (ATCC, Manassas, VA), respectively. They were propagated in Dulbecco's modified eagle's medium (DMEM; Gibco Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS; Sigma, St. Louis, MO) and 1% penicillin-streptomycin (Gibco Invitrogen) in a 5% CO2 humidified atmosphere at 37°C. Progenitor-derived astrocytes (PDAs) were generated from neural progenitor cells as previously described (24). Briefly, progenitor cells (provided by Eugene Major, NINDS, NIH, MD) were seeded on poly-d-lysine coated T-75 tissue culture flasks at 2 × 106 cells/flask and maintained in progenitor medium consisting of neurobasal medium (Gibco Invitrogen) supplemented with 0.5% bovine albumin (Sigma), neurosurvival factor (NSF) (Lonza, Walkersville, MD), N2 components (Gibco Invitrogen), 25 ng/ml basic fibroblast growth factor (bFGF), 20 ng/ml epidermal growth factor (EGF) (R & D Systems, Minneapolis MN), 50 μg/ml gentamicin (Lonza), and 2 mM l-glutamine (Gibco Invitrogen). To induce differentiation, progenitor medium was replaced with PDA medium containing DMEM supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, and 50 μg/ml gentamicin. Cultures were >90% positive for glial fibrillary acidic protein (GFAP) after 30 days of differentiation. Both adherent primary cells and cell lines were removed by treatment with 1 mM EDTA for 5 min, with gentle scraping or pipetting multiple times.
Plasmid construction.
Genomic DNA was obtained from HIV-1 Bal-infected peripheral blood mononuclear cells ([PBMCs] after 3 days of infection) using a DNeasy blood and tissue kit (Qiagen, Germantown, MD). The long terminal repeat (LTR) was amplified using the primers Sn25 (5′-TCGACTCGAGGACAAGATATCCTTGATTTGT) and Sn26 (5′-TCGACTCGAGTTTGGCGTACTCACCAGTCG) cut with XhoI (underlined sequences) and cloned into XhoI-predigested pGL4.19 plasmid (Promega, Madison, WI). Recombinant plasmid was subjected to restriction digestion with SFI1 and EcoRV to confirm the presence of a single copy of the LTR insert and further sequenced to confirm its proper orientation, yielding a wild-type LTR (WT-LTR) plasmid.
Plasmid and siRNA transfections.
An NF-κB PathDetect cis-reporting system plasmid was purchased from Agilent Technologies. Expression of the Photinus pyralis (firefly) luciferase gene in this reporter plasmid is controlled by a synthetic promoter that contains direct repeats of the transcription recognition sequences for nuclear factor κB (NF-κB). At 48 h of small interfering RNA (siRNA) knockdown, the reporter plasmid or WT-LTR plasmid at 0.5 μg was transfected into designated cells using TransIT-LT1 reagent, per recommendations of the manufacturer (Mirus Bio LLC, Madison, WI), and a luciferase assay was performed after 24 h of transfection. On-Targetplus SMARTpool siRNAs specific for TCF-1, TCF-3, TCF-4, LEF1, CTNNB (β-catenin), and scrambled siRNA were obtained from Thermo Scientific (Waltham, MA) and transfected into cell lines and PDAs using Lipofectamine RNAiMAX (Invitrogen) according to the reagent protocol. Cells were approximately 60 to 70% confluent at the time of transfections.
Generation of LTR-Luc cell lines.
To prepare stable cell lines expressing the LTR-driven luciferase (LTR-Luc), we transfected U87MG and U251MG cells with wild-type (WT)-LTR plasmid. The plasmid is of pGL4.19 background with neomycin as marker gene. After 48 to 72 h of transfection, the cells were removed, washed with phosphate-buffered saline (PBS), and split into half and cultured in the presence of G418 (25 μg/ml for U87MG and 50 μg/ml for U251MG) at a low cell density (20 to 40% confluence) until they reached full confluence, which was typically within 4 to 6 days. G418 was added at day 2 after seeding. Subsequently (4 to 6 days after seeding), the cells were detached. These were considered generation 1 cells. Those cells were then replated as indicated above in the presence of G418 treatment, and this process was repeated for several generations until episomal plasmid was diluted out and chromosomally integrated HIV LTR reporter that can drive neomycin resistance remained. The presence of HIV LTR and neomycin genes was confirmed by PCR while luciferase was confirmed by the luciferase reporter assay.
Luciferase reporter assays.
Approximately 24 h after transfection of the indicated reporter plasmids, the cells were harvested and washed with PBS once; then 100 μl of passive lysis buffer was added, and the cells were incubated at 37°C for 10 to 12 min. The cells were then spun at 5,000 rpm for 4 min to remove debris, and 10 to 20 μl was used to assay for luciferase activity using a dual luciferase reporter assay (Promega) in a single-injector luminometer. Total protein concentration was measured using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific), and relative light units were normalized to μg/ml of protein. Graphs were plotted from data obtained as a mean of three independent experiments, with standard deviations shown as error bars.
Quantitative real-time RT-PCR.
RNA was isolated using TRIzol reagent (Invitrogen), according to the manufacturer's recommendations. Subsequently, cDNA was synthesized using a Quantitect reverse transcription kit (Qiagen). Real-time reverse transcription-PCR (RT-PCR) was performed using a Quantitect SYBR green PCR kit (Qiagen) in a 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) using 7500 software, version 2.0.1. Melting curve analysis was performed to ensure the amplification of a single product. Primers used were the following: TCF1-F, 5′-AGGCCAAGAAGCCAACCATCAAGA, and TCF1-R,5′-ACTCTGCAATGACCTTGGCTCTCA; TCF3-F, 5′-TGCAGTGAGCGTGAAATCACCAGT, and TCF3-R, 5′-AATGGCTGCACTTTCCTTCAGGGT; TCF4-F, 5′-TCGGCAGAGAGGGATTTAGCTGATGT, and TCF4-R, 5′-CTTTCCCGGGATTTGTCTCGGAAACT; LEF1-F, 5′-AAGCATCCAGATGGAGGCCTCTACAA, and LEF1-R, 5′ TGATGTTCTCGGGATGGGTGGAGAAA; β-cat-F, 5′-TCTTGCCCTTTGTCCCGCAAATCA, and β-cat-R,5′-TCCACAAATTGCTGCGTCCCA; c-Myc-F, 5′ TTCGGGTAGTGGAAAACCAG, and c-Myc-R, 5′-AGTAGAAATACGGCTGCACC; CCDN1-F, 5′-TCTGGCATTTTGGAGAGGAAG, and CCDN1-R, 5′-CATCTACACCGACAACTCCATC; LCN2-F, 5′-GAAGACAAAGACCCGCAAAAG, and LCN2-R, 5′-CTGGCAACCTGGAACAAAAG; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-CTTCAACGACCACTTTGT, and GAPDH-R, 5′-TGGTCCAGGGGTCTTACT. Fold change in mRNA expression was calculated by relative quantification using the comparative threshold cycle (CT) method with GAPDH as an endogenous control.
ChIP.
Chromatin immunoprecipitation assay (ChIP) assays were performed from cells using antibodies (Abs) as described below. Cells were seeded to 60 to 70% confluence, treated as described in the figure legends, and then processed for ChIP beginning with cross-linking proteins to DNA by 1.0% formaldehyde. Chromatin was sonicated five times for 20 s each, generating DNA fragments of about 500 to 1,000 bp. The sonicated supernatants containing the DNA were diluted with ChIP dilution buffer (0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, and 167 mM NaCl) to a total volume of 5.5 ml and precleared by rotation for 1 h at 4°C with ChIP-prepared protein A/G beads (beads were washed twice with 1 ml of TNE50 buffer [100 mM Tris-HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA] plus NP-40, resuspended in 650 μl, and 40 μl of single-stranded DNA [ssDNA; 10 mg/ml] and 75 μl of bovine serum albumin [BSA] 10 mg/ml] were added). No proteases or RNases were used for the extraction. The extract was centrifuged at 3,000 rpm for 10 min at 4°C, and the lysate was transferred to a fresh tube. Supernatant (500 μl) was reserved for input, and then 10 μg of each antibody (anti-polymerase II [Pol II] large subunit N-20; Santa Cruz) (data not shown), or the C-terminal domain of phosphoserine-2 (pSer2 CTD) (H5; Covance) was added to the reaction mixture. After overnight rotation at 4°C, the immune complexes were collected by addition of ChIP-prepared protein A/G beads. After extensive washes, the immune complexes were eluted twice with 1% SDS-NaHCO3 solution for 30 min at room temperature. The eluted complexes were treated with NaCl solution and reverse cross-linked overnight. DNA was extracted using 1:1 phenol-chloroform (500 μl), followed by the addition of 1 ml of absolute ethanol and 3 M sodium acetate (50 μl), and samples were incubated at −20°C for at least 20 to 30 min. The solution was spun for 20 min at 14,000 rpm at 4°C, followed by a 70% ethanol wash and 5-min spin. DNA pellet was resuspended in 1× Tris-EDTA (TE) buffer and stored at 4°C. Afterwards, DNA was purified by a PCR purification Kit (BiONEER) and amplified by PCR. Primer pairs for PCR analysis of chromatin immunoprecipitation-amplified sequences flanked the HIV-1 LTR Nuc-1 (+10 to +165) and Env (+8990 to +9120). Readings were normalized to either Tat-deficient samples or input, as specified in the figure legends. ChIP to detect NF-κB and C/EBP (CCAAT/enhancer binding protein) on the LTR was performed as described above except that the pulldown was performed using 5 μg of the immunoprecipitating antibody (Abcam, Cambridge, MA), and elution of DNA was performed using a SimpleChIP Enzymatic IP Kit (Cell Signaling, Danvers, MA).
Microarray gene expression assays.
Total RNA was isolated as described earlier and then purified using an RNeasy Mini Kit (Qiagen). The integrity of the RNA samples was analyzed on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Using an Applause RNA Amplification System (NuGEN, San Carlos, CA), 50 ng of total RNA was used for each sample for amplification following the manufacturer's protocol. The resulting cDNA was fragmented and labeled using a NuGEN Encore Biotin Module. A Human GeneChip Gene 1.0 ST array (Affymetrix), which contains 28,869 genes, each represented on the array by approximately 26 probes spread across the gene, was used for hybridization. The hybridization was performed at 45°C with 60-rpm rotation in an Affymetrix Hybridization Oven 640 for 17 h. The hybridized arrays were then scanned on an Affymetrix GeneChip Scanner 3000 7G, after washing and staining on an Affymetrix Fluidics Station 450.
Microarray data analysis.
The array image data were preprocessed and saved as cel files using the Command Console (Affymetrix). Data were further analyzed by normalization and statistical analysis using an unpaired t test, with a P value cutoff of 0.05, using GeneSpring GX, version 10.5, software (Agilent Technologies). The P value computations were done asymptotically along with a Benjamin Hochberg false-discovery rate (FDR) multiple testing correction. Further analyses were performed for differentially expressed genes using a fold change cutoff of 1.5. Hierarchical clustering using the similarity measure of Euclidean distance and linkage rule of the centroid and gene ontology (GO) analysis were performed using GeneSpring GX, version 10.5.
Western blotting.
Ten to 20 μg of total cell lysate or nuclear extract was separated by 10% SDS-PAGE, transferred onto a nitrocellulose membrane, blocked with Superblock (Thermo Scientific) containing 0.1% Tween 20 (T20) for 1 h, incubated with primary antibody for 1 h at room temperature (RT) for β-catenin (1: 8,000, rabbit; Sigma) or overnight at 4°C for other TCF/LEF proteins (1:1,000, TCF/LEF1 antibody sampler kit; Cell Signaling) at indicated dilutions in Superblock–0.1% T20. Membranes were washed extensively with Tris-buffered saline–Tween 20 (TBST) and incubated with secondary antibody conjugated to horseradish peroxidase (HRP) (1:50,000 [Sigma] in Superblock–0.1% T20) for 45 min at RT. Membranes were again washed extensively in TBST and developed with SuperSignal West Femto maximum-sensitivity substrate (Thermo Scientific) according to instructions.
Flow cytometry.
Intracellular staining of cells for detection of β-catenin by flow cytometry was performed as previously described (26). For cellular proliferation studies, cells were preloaded with 5 μM 5,6-carboxy fluorescein diacetate succinimidyl ester (CFSE), treated with indicated siRNAs for 3 days, detached from the surface using PBS containing EDTA, washed once with PBS, and then fluorescence acquired. Cell viability was measured using an annexin V staining kit according to the protocol provided by the manufacturer (BD Biosciences, Franklin Lakes, NJ). Data were acquired in a FACSCalibur flow cytometer (BD Biosciences) and analyzed with FlowJo (Tree Star, Ashland, OR).
Statistical analysis.
Statistical analyses were performed using Prism software (GraphPad Prism, San Diego, CA). The variables were compared using the Student t test when the data were normally distributed. When the data were not normally distributed, the two groups were compared using a nonparametric Mann-Whitney test. All tests were two-tailed, and a P value of <0.05 was considered significant.
Nucleotide sequence accession number.
The sequence of the LTR was deposited in GenBank under accession number HQ882192.
RESULTS
Human astrocytes express downstream transcriptional effectors of the canonical Wnt/β-catenin signaling pathway.
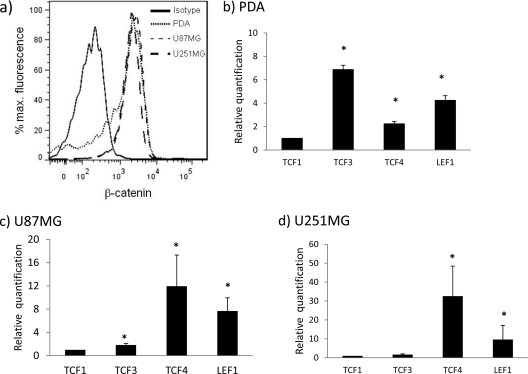
Active β-catenin is robustly expressed in primary human progenitor-derived astrocytes (PDAs) (Fig. 1a). The level of expression was also comparable in two astrocytoma cell lines (U87MG and U251MG) (Fig. 1a). β-Catenin, via canonical signaling, regulates the expression of hundreds of target genes by interacting with downstream transcription factor TCF-1, TCF-3, TCF-4, or LEF1. Using quantitative real-time RT-PCR (qRT-PCR) for TCF/LEF mRNAs, we show that PDAs express all members of the TCF/LEF family (Fig. 1b). Because the level of TCF-1 mRNA was the least abundant, all values were compared to TCF-1 expression. In PDAs, we show that the levels of TCF-3 and LEF1 mRNA were the most abundant, followed by lower levels of TCF-4 (Fig. 1b). TCF-4 mRNA was the most abundant in the two astrocytoma cell lines (U87MG and U251MG), followed by LEF1 (Fig. 1c and d). These data indicate that although PDAs and astrocytoma cells express all TCF/LEF family members, their mRNA expression patterns vary depending on the cell types.
Fig 1.
Human astrocytes have high levels of endogenous β-catenin and express the downstream transcription factors of the Wnt pathway, TCF-1, TCF-3, TCF-4, and LEF1. (a) β-Catenin protein was measured by intracellular flow cytometry using an antibody specific for the active, hypo-phosphorylated form of β-catenin from U87MG and U251MG cells and PDAs. (b to d) PDAs and U87MG and U251MG cells were cultured for 24 h, and then mRNA expression of TCF-1, TCF-3, TCF-4, and LEF1 was measured by real-time RT-PCR and normalized to the housekeeping gene GAPDH. Results are expressed relative to the least abundant TCF/LEF member, TCF-1. All results represent at least three independent experiments performed in duplicate. *, P < 0.05 in comparison with TCF-1.
Knockdown of β-catenin and its downstream transcription factors.
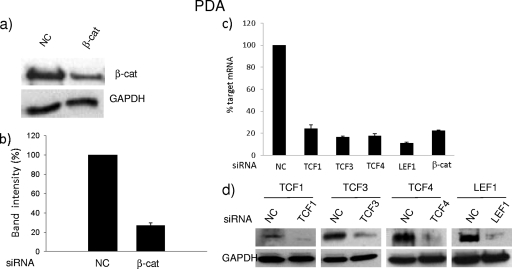
To assess the role of endogenous β-catenin and TCF/LEF on HIV transcriptional activity, we transfected PDAs and astrocytic cell lines with a SMARTpool siRNA for β-catenin. Initially the knockdown was optimized in U87MG cells, whereby 20 nM siRNA was sufficient to knock down β-catenin protein by >90% (see Fig. S1a in the supplemental material). Using this concentration of siRNA for β-catenin, we observed a >80% reduction in its mRNA and protein levels at 24 h and 72 h posttransfection, respectively, in U87MG (see Fig. S1b to d) and U251MG (see Fig. S1e and f) cells and PDAs (Fig. 2a and b). This knockdown of β-catenin had no effect on cell viability, as measured by annexin V staining (see Fig. S2a), or on cell proliferation, as measured by a CFSE dye tracking assay at 72 h posttransfection (see Fig. S2b). Knockdown of β-catenin was also consistent with reduction in β-catenin-dependent target gene expression, such as downregulation of c-Myc and cyclin D1 and induction of lipocalin 2 (see Fig. S2c). Collectively, these data demonstrate the efficiency of 20 nM SMARTpool siRNA in downregulating β-catenin mRNA and protein levels and in impacting its downstream target genes. A similar strategy was employed to knock down TCF/LEF family members. It also resulted in efficient knockdown of TCF/LEF family members, without notable toxicity, as shown for PDAs (Fig. 2c and d), U87MG, and U251MG cell lines (see Fig. S3 in the supplemental material).
Fig 2.
Efficacy of knockdown of β-catenin and TCFs/LEF1 in PDAs. PDAs were transfected with scrambled or β-catenin siRNAs (20 nM) (a and b) or siRNA targeting TCF/LEF members (c and d). At 72 h posttransfection, immunoblotting was performed to detect the level of β-catenin knockdown (a) and its respective densitometry (b) or TCF/LEF knockdown (d). Some cells were also harvested at 24 h posttransfection, and RT-PCR was performed to detect mRNA levels of TCF/LEF knockdown (c). Data were normalized to GAPDH and are represented as percent target mRNA compared to negative control (NC) siRNA. All results represent at least three independent experiments performed in duplicate.
To assess the impact of β-catenin knockdown on the transcriptome of PDAs, we evaluated differentially expressed genes in cells that were transfected with scrambled siRNA versus those that had been transfected with β-catenin siRNA. We show that β-catenin knockdown had profound effects on the expression of 150 genes for PDAs, with a fold change of at least 1.5 (see Table S1 in the supplemental material). The genes impacted fall under five broad categories: (i) inflammation/immunity, (ii) uptake/transport, (iii) vesicular transport/exocytosis, (iv) apoptosis/cellular stress genes, and (v) cytoskeleton/trafficking (see Table S2).
Knockdown of β-catenin and TCF-4 induces HIV transcription in transiently transfected and stably integrated HIV LTR reporter constructs.
To directly assess the role of β-catenin and its downstream transcriptional cofactors in regulating HIV LTR activity, astrocytic cell lines or primary astrocytes were transfected with a full-length HIV LTR linked to a firefly luciferase reporter gene with siRNA for β-catenin, siRNA for LEF1/TCF family members, or a scrambled siRNA. Transfection with green fluorescent protein (GFP) was used to monitor transfection efficiency, which was equivalent between cell types (see Fig. S4 in the supplemental material). Knocking down β-catenin or TCF-4 consistently induced HIV LTR activity by 1.5- to 4-fold in PDAs and astrocytic cell lines (Fig. 3). TCF-1 and TCF-3 knockdown had no impact on HIV LTR activity in all cells evaluated (Fig. 3). While β-catenin and TCF-4 knockdown consistently induced HIV LTR activity, the LEF1 knockdown effect was cell type specific. Knockdown of LEF1 had no effect on HIV promoter activity in PDAs and U87MG cells but showed a significant induction in LTR activity in U251MG cells by 8-fold (Fig. 3). These studies demonstrate that β-catenin and TCF-4 are negative regulators of HIV LTR transcription and that this response is consistent between human progenitor-derived-primary astrocytes and the astrocytoma cell lines evaluated.
Fig 3.
Knockdown of β-catenin and TCF-4 induces basal transcription of HIV LTR in astrocytes. PDAs, U87MG cells, or U251MG cells were transfected with the indicated siRNAs. After 48 h, cells were transfected with LTR-Luc, incubated for another 24 h, lysed, and assayed for luciferase activity. Data represent at least three independent experiments performed in duplicate. *, P < 0.05 in comparison with scrambled siRNA negative control (NC). RLU, relative light units.
As a retrovirus, HIV DNA integrates into the host genome, and its expression is likely impacted by the overall chromatin structure. To ensure that the impact of β-catenin and TCF-4 on HIV promoter activity is not a consequence of transient transfection, we generated stably transfected U87MG and U251MG cell lines with HIV LTR linked to a firefly luciferase and the neomycin gene (U87MG-HIV-LTR-LUC and U251MG-HIV-LTR-LUC, respectively). These cells stably expressed HIV LTR reporter activity with occasional spikes of enhanced HIV reporter activity (Fig. 4a). Typically, these spikes in reporter activity occurred intermittently in later (9th and 10th) passages postintegration of the LTR reporter into the genome (e.g., cells from passages 9 and 10 were grown to confluence and split in the presence of neomycin). We assessed the effect of β-catenin and TCF-4 knockdown using 5th- to 7th-generation LTR-Luc cells, for which reporter activity was consistently low. Knocking down β-catenin or TCF-4 in stably transfected U87MG-HIV-LTR-LUC or U251MG-HIV-LTR-LUC induced HIV promoter activity by 2-fold relative to scrambled siRNA (Fig. 4b and c). These data demonstrate that β-catenin and TCF-4 negatively regulate HIV LTR promoter activity and are capable of doing so within or outside the chromatin architecture.
Fig 4.
Knockdown of β-catenin and TCF-4 induces transcription of stably integrated LTR in astrocytes. Stably integrated LTR-Luc was developed in U87MG and U251MG cells as described in Materials and Methods. (a) Stably transfected U87MG cells were grown for the indicated number of passages, and firefly luciferase activity was assayed in each passage. Data points represent luciferase readings from at least two independently generated LTR-Luc lines, with luciferase readings taken in duplicate and normalized to total protein content. (b and c) U87MG-HIV-LTR-LUC and U251MG-HIV-LTR-LUC cells were transfected with the indicated siRNAs. After 72 h, cells were lysed and assayed for luciferase activity. Experiments were performed in duplicate at least three times. *, P < 0.05 in comparison with a scrambled siRNA negative control (NC).
Knockdown of TCF-4 significantly enhanced docking and processivity of Pol II on HIV DNA.
β-Catenin does not directly bind to DNA but rather partners with LEF/TCF transcriptional factors to regulate gene expression. To directly assess the impact of TCF-4 knockdown on HIV transcription, we evaluated tethering of a transcription-competent Pol II on HIV LTR promoter and Env regions. U87MG cells were transfected with pNL4-3 provirus in the presence of TCF-4 siRNA or scrambled siRNA, and then a chromatin immunoprecipitation (ChIP) assay was performed utilizing anti-Pol II Abs or an isotype control, and subsequently the HIV LTR R/U5 region or Env region was amplified by real-time PCR. Knockdown of TCF-4 dramatically enhanced detection of Pol II on both the HIV LTR and Env regions (Fig. 5a and b). The anti-Pol II antibody (pSer2 CTD [H5]) used here recognizes the active form of the polymerase as serine 2 phosphorylation is associated with processive and elongating RNA polymerase. There was only minimal Pol II association with these regions in control siRNA-treated cells (Fig. 5a and b, lanes 2) despite ample amounts of chromatin in these samples (see Fig. S5a in the supplemental material). A similar observation was made using 293T cells, where by presence of pSer2-Pol II in the HIV LTR and Env was enhanced by approximately 10- and 6-fold, respectively (see Fig. S5b). As expected, knocking down Sp1 had no effect on detection of pSer2-Pol II (Fig. 5a and b). ChIP performed using the U87MG-HIV-LTR-LUC astrocytoma cell line with TCF-4 knockdown also demonstrated enhanced presence of pSer2-Pol II on the HIV LTR. As a control for PolII Ser2 antibody in the ChIP assay, cytomegalovirus (CMV)-Env was transfected and subjected to ChIP assay simultaneously for the presence of processive Pol II on the Env region (Fig. 5c). Together, these data demonstrate that TCF-4 is a repressor of HIV transcriptional activity, independent of cell type, whereby its inhibition leads to enhanced pSer2-Pol II docking on HIV LTR and processivity.
Fig 5.
Knockdown of TCF-4 enhances docking and processivity of pSer2-Pol II on HIV DNA. (a and b) U87MG cells were knocked down for TCF-4 or transfected with a scrambled control. After 48 h, cells were transfected with pNL4-3 and rested for an additional 24 h. ChIP was performed using antibody against Pol II pSer2 (H5) or an IgG isotype control, followed by DNA purification and amplification of the LTR and Env regions by PCR. DNA products were run on an agarose gel and analyzed using a PhosphorImager to quantify band intensity, reported in panel b as arbitrary light units. (c) U87MG cells containing integrated copies of the LTR-Luc and episomal CMV-ENV constructs were transfected with TCF-4 siRNA or a scrambled control. After 48 h, ChIP was performed using either antibody against pSer2-Pol II or an IgG isotype control. Data represent at least three independent experiments. *, P < 0.05.
β-Catenin and TCF-4 knockdown enhance C/EBPβ/δ tethering on the HIV LTR while TCF-4 knockdown, independent of β-catenin, negatively regulates NF-κB tethering on the HIV LTR and NF-κB reporter activity.
We evaluated the impact of β-catenin and TCF-4 on tethering of key transcriptional factors on the HIV LTR. We elected to focus on C/EBP and NF-κB because they are positive regulators of HIV transcription. C/EBPα, -β, and -δ are expressed in brain tissue (1, 7). We evaluated the activity of C/EBPβ and -δ because C/EBPα is only weakly expressed in astrocytes (14) and is downregulated under inflammatory conditions, whereas C/EBPβ and -δ are upregulated by inflammation (7). U87MG cells were transiently transfected with an LTR-luciferase construct, and ChIP was performed for these transcriptional factors accordingly. β-Catenin knockdown enhanced tethering of C/EBPβ and C/EBPδ to the HIV LTR, while TCF-4 knockdown increased binding of the p65 subunit of NF-κB, C/EBPβ, and C/EBPδ (Fig. 6a to c). Knockdown of TCF-4 and not β-catenin significantly enhanced NF-κB reporter activity by 2.5-fold in comparison to scrambled siRNA (Fig. 6d). These data demonstrate that β-catenin and TCF-4 negatively regulate tethering of C/EBP on the HIV LTR and that TCF-4, independent of β-catenin, negatively regulates NF-κB binding to the HIV LTR as well as its reporter activity. This finding is especially intriguing because it demonstrates that TCF-4, independent of β-catenin, can differentially impact the expression of transcriptional factors important in HIV LTR promoter activity. The net outcome of these interactions is likely to regulate the extent of HIV promoter activity.
Fig 6.
Knockdown of β-catenin and TCF-4 modulates C/EBP and NF-κB tethering on HIV LTR. PDAs at 60 to 70% confluence were transfected with β-catenin siRNA, TCF-4 siRNA, or a scrambled siRNA negative control (NC). After 48 h, cells were transiently transfected with an LTR-Luc construct. At 24 h post-LTR transfection, cells were cross-linked with 1.0% formaldehyde and sonicated to generate chromatin preparations (5 × 106 cells/sample). ChIP was performed using antibodies against C/EBPβ, C/EBPδ, or NF-κB p65 subunit (5 μg antibody/IP). Data shown are normalized to a nontargeting IgG control. *, P < 0.05 in comparison with control siRNA. (d) PDAs at 60 to 70% confluence were transfected with β-catenin siRNA, TCF-4 siRNA, or a scrambled siRNA negative control (NC). After 48 h, cells were transfected with an NF-κB luciferase reporter plasmid, and a luciferase assay was performed after an additional 24 h. Results are reported as fold change in relative light units (RLU) compared to the negative control and are normalized to the amount (micrograms) of cellular protein. All data represent at least three independent experiments performed in duplicate. *, P < 0.05 in comparison with the negative control.
DISCUSSION
HIV enters astrocytes in a nonclassical way, either via endocytosis and escape of HIV from the endosome (27, 30, 42), utilization of alternative receptors for HIV entry including the promiscuous CC chemokine receptor D6 (28), or a decreased requirement of neurotropic HIV strains for CD4 (16). Despite lack of a clear understanding of how HIV enters astrocytes, a number of groups of investigators have demonstrated the presence of HIV protein and/or DNA within astrocytes in HIV-positive (HIV+) postmortem tissue (11, 15, 29, 40, 41, 44). By and large, without a diagnosis of HIV-associated dementia, the degree of HIV infection within astrocytes is relatively low in comparison to that in perivascular macrophages and microglia. Nonetheless, the few infected astrocytes have detrimental effects in the CNS, such as promoting disruption of blood-brain barrier integrity (15). More recent sophisticated assessment of HIV infection in astrocytes demonstrated a considerable level of HIV Env in astrocytes from HAD patients and up to 3% in the absence of HAND (10), which is similar to the size of the HIV pool in the major latent reservoir of HIV, the CD4+ memory T cell compartment (39). Evaluating the true size of HIV productive replication in astrocytes from postmortem tissue is complicated by several factors such as presence of a large population of astrocytes that are not GFAP positive, making detection of these cells in postmortem tissue difficult. Further, infected astrocytes may preferentially undergo apoptosis (40). Certain environmental signals may trigger astrocytes to be more supportive of productive HIV replication. We previously demonstrated that IFN-γ, which is elevated in cerebral spinal fluid (CSF) of HIV+ patients with neurological complications of AIDS (neuroAIDS) and is secreted by infiltrating lymphocytes as well as resident macrophages/microglia and astrocytes, induces productive HIV replication in astrocytes (8, 9). The mechanism by which IFN-γ enhances HIV replication in astrocytes is dependent on the β-catenin signaling pathway. Specifically, IFN-γ downregulates β-catenin signaling and leads to enhanced HIV replication by inducing an antagonist of the β-catenin pathway (DKK1) in a Stat 3-dependent manner (26). In this study, we directly assessed the impact of β-catenin and its downstream transcriptional cofactors on HIV transcription in astrocytes.
Using primary fetal progenitor astrocytes as well as two astrocytoma cell lines (U87MG and U251MG), we determined that the β-catenin/TCF-4 axis regulates HIV transcriptional activity. We detected all four TCF/LEF family members in our astrocyte model systems; however, when all of these factors were knocked down, only TCF-4 demonstrated an effect on HIV LTR activity. Interestingly, LEF1 is described as an inducer of HIV transcription in Jurkat T cells (38). However, in astrocytes, the effect of LEF1 on HIV transcription is cell type specific. LEF1 knockdown had no effect on HIV LTR activity in PDAs and in U87MG cells, but it enhanced HIV transcription in U251MG cells. This may reflect the ability of LEF1 to function as a dominant negative factor for TCF-4 and perhaps potentiates its effects in a cell-type-specific manner. Because TCF-4 consistently demonstrated an inhibitory effect on HIV promoter activity in all cell types evaluated, including primary astrocytes, we focused on TCF-4. Although Wortman et al. transfected an astrocytic cell line with TCF-4 and demonstrated that TCF-4 leads to repression of HIV LTR activity, they ruled out a role for β-catenin (45). In their studies, a TCF-4 mutant lacking binding sites for β-catenin still downregulated LTR transcription, and they concluded that these effects were β-catenin independent (45). β-Catenin expression and its mediated signaling events are robust in astrocytes (8). Therefore, there may still have been endogenous β-catenin expression that can partner with endogenous TCF-4 to mediate the repressed effects. In our study, by knocking down endogenous β-catenin expression, we demonstrated a role for β-catenin in repressing HIV promoter activity. To our knowledge, this is the first report to indicate that β-catenin in association with TCF-4 leads to repressed LTR activity and that it can have effects on the extent of C/EBP and NF-κB tethering on the HIV LTR.
The mechanism by which β-catenin/TCF-4 suppress HIV replication is likely to be multifaceted, involving both direct and indirect interactions between β-catenin/TCF-4 and the HIV LTR. TCF-4 contains an HMG domain and is a DNA bender. It is conceivable that TCF-4 docks on unique sites within the HIV LTR and bends them in a way that makes them inaccessible to general transcription machinery and/or transcriptional activators. Indeed, we show here that knocking down TCF-4 enhances RNA Pol II processivity in both the LTR region and the Env region. Although β-catenin/TCF-4 largely turn on gene expression, few studies have now pointed to this same complex as turning off gene expression by recruiting HDACs (21). Efforts in the lab are ongoing to establish the direct interaction between TCF-4 and the HIV LTR. Indirect effects of β-catenin/TCF-4-mediated inhibition of HIV transcription may include impacting transcriptional factors important for HIV transcription. We demonstrated here that β-catenin inhibited the binding of C/EBPβ and -δ to the HIV LTR. The HIV promoter has binding sites for both C/EBP (CCAAT/enhancer binding protein) and NF-κB which induce HIV transcription (17, 32, 33). The association between β-catenin signaling and C/EBP activity is particularly interesting, given recent evidence from Burdo et al. showing that different regions of the brain contain LTRs with distinct C/EBP binding sites (5). Furthermore, C/EBP sequence variants that bind Vpr with high affinity occur at a higher frequency in brain tissue from patients with HAD (6). In addition to enhancing HIV transcription, excessive C/EBPβ and C/EBPδ activity can increase neuroinflammation that contributes to HAND by enhancing production of proinflammatory cytokines such as interleukin-6 (IL-6), IL-1β, and tumor necrosis factor alpha (TNF-α) by astrocytes (31). Similarly, we have shown that TCF-4 negatively regulates NF-κB, which is important for robust HIV replication. This finding may be particularly relevant in HIV-infected astrocytes, given that NF-κB binding is required for TAR-independent LTR transactivation in these cells (2). Deng et al. previously reported that β-catenin suppresses NF-κB activity in several cancer cell models by a mechanism that requires the presence of other cellular factors (12, 13). Intriguingly, in our study knockdown of β-catenin had no effect on NF-κB transcriptional activity or binding to the HIV LTR, which suggests that β-catenin is not involved in TCF-4-mediated regulation of NF-κB in astrocytes. The mechanism by which TCF-4 suppresses NF-κB activity is unclear. Inhibition may involve direct interaction between TCF-4 and NF-κB and/or indirect effects on upstream regulators. One mechanism by which TCF-4 inhibits HIV transcription is by preventing phosphorylation of another important transcription factor, Sp1, by DNA-dependent protein kinase (DNA-PK) (34). Several components of the NF-κB pathway, including IKKα, IKKβ, and p50, are also targets of DNA-PK (3, 4, 22). It is possible that TCF-4 suppresses NF-κB DNA binding by antagonizing DNA-PK activity. To our knowledge, this is the first report of TCF-4 inhibiting NF-κB activity independent of β-catenin.
It is worth noting that C/EBPβ, C/EBPδ, and NF-κB have a synergistic relationship in inflammation. C/EBP family members can form heterodimers with NF-κB subunits that can potently activate the HIV-1 promoter by tethering on NF-κB binding sequences (35). Additionally, cooperativity between these transcription factors has been observed with many promoters that contain adjacent C/EBP and NF-κB binding sites, including genes that encode the proinflammatory cytokine IL-6 and the chemokine IL-8 (31). It is possible that induction of C/EBP activity by TCF-4 knockdown contributes to enhanced NF-κB binding and reporter activity via cooperative binding. In a broader context, synergistic activation of C/EBPβ, C/EBPδ, and NF-κB in astrocytes may contribute to robust production of proinflammatory cytokines and chemokines leading to enhanced HIV replication and recruitment of immune cells into the CNS and contributing to glial activation and neuronal injury.
Although we demonstrated that β-catenin and TCF-4 suppress HIV LTR promoter activity, there may be additional pathways by which β-catenin suppresses HIV replication at steps beyond HIV transcription. β-Catenin functions as a transcriptional coactivator for TCF-4, or it can bind to cadherins through α-catenin to regulate cell adhesion and communication. In lymphocytes, Vpu enhances HIV release by dissociating β-catenin from E-cadherin (36). Increased association of E-cadherin with β-catenin leads to depressed HIV release from cells that express E-cadherin such as macrophages. These data suggest that multiple mechanisms may be involved by which β-catenin inhibits HIV replication, especially in cells that are negative for E-cadherin such as T cells. In T cells overexpression of β-catenin leads to a dramatic inhibition of HIV replication (data not shown). Conversely, β-catenin inhibition by dominant negative β-catenin construct enhances HIV replication in astrocytes as well as peripheral blood lymphocyte (PBLs) (8, 23). These data demonstrate the inverse relationship between β-catenin expression and HIV replication in multiple cell types.
In the U87MG astrocytoma LTR-Luc cell line that we generated, HIV LTR activity was modest; however, under certain passage numbers, we observed a significant spike in promoter activity. These spikes are quite interesting as possible phenomena to describe virologic blips in astrocytes. Typically, astrocytes do not support a high level of productive HIV replication. In U87MG-HIV-LTR-LUC cells these spikes may mimic incidences driving higher levels of HIV basal promoter activity; this could be a consequence of chromatin remodeling of integrated DNA between early and later passages of cells, the milieu of passaged astrocytes, their relative confluence over time, or some other unknown mediator driving these spikes. It is difficult to determine exactly the reason for this phenomenon, but it is an interesting tool to understand factors that can drive virologic blips or higher levels of promoter activity within astrocytes which, consequently, may promote higher expression of early transcripts (Tat) that can mediate neuropathogenesis of surrounding cells or within the secreting cell itself, or that can reach a threshold level of Tat to enhance full transcriptional activity and virion release. Because the spikes are transient, it is likely that there astrocytes may have inherent mechanisms that downregulate HIV transcriptional activity. Based on our data it is conceivable that intact/robust β-catenin signaling within astrocytes suppresses any significant HIV replication within these cells and that HIV resides as a latent virus within these cells. However, environmental factors, such as IFN-γ or others that lead to downregulation of β-catenin signaling, can lead to enhanced HIV replication in astrocytes. Consequently, astrocytes under permissive conditions for HIV replication may continue to seed the CNS with HIV, leading to inflammatory responses within the CNS and biologic effects promoting HIV neuropathogenesis. Understanding the interplay between β-catenin signaling and HIV can inform novel approaches to manipulate β-catenin signaling to potentially be used as a therapeutic strategy to purge the latent reservoir of HIV in the CNS and/or or other targets where β-catenin functions to repress HIV replication.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the following grants from the National Institutes of Health: R01 NS060632 to L.A.-H., F31 NS071999 to L.J.H., and AI043894, AI074410-01 and AI078859 to F.K. The studies were also supported by the Chicago Developmental Center for AIDS Research (D-CFAR, P30 AI 082151), an NIH-funded program supported by NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NCCAM.
We thank Eugene O. Major (National Institute of Neurological Disease and Stroke, National Institutes of Health, Bethesda, MD) for providing us with the progenitor-derived astrocytes.
Footnotes
Published ahead of print 7 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alam T, An MR, Papaconstantinou J. 1992. Differential expression of three C/EBP isoforms in multiple tissues during the acute phase response. J. Biol. Chem. 267:5021–5024 [PubMed] [Google Scholar]
- 2. Bagasra O, Khalili K, Seshamma T, Taylor JP, Pomerantz RJ. 1992. TAR-independent replication of human immunodeficiency virus type 1 in glial cells. J. Virol. 66:7522–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basu S, Rosenzweig KR, Youmell M, Price BD. 1998. The DNA-dependent protein kinase participates in the activation of NF kappa B following DNA damage. Biochem. Biophys. Res. Commun. 247:79–83 [DOI] [PubMed] [Google Scholar]
- 4. Brzoska K, Szumiel I. 2009. Signalling loops and linear pathways: NF-kappaB activation in response to genotoxic stress. Mutagenesis 24:1–8 [DOI] [PubMed] [Google Scholar]
- 5. Burdo TH, Gartner S, Mauger D, Wigdahl B. 2004. Region-specific distribution of human immunodeficiency virus type 1 long terminal repeats containing specific configurations of CCAAT/enhancer-binding protein site II in brains derived from demented and nondemented patients. J. Neurovirol 10 Suppl 1:7–14 [DOI] [PubMed] [Google Scholar]
- 6. Burdo TH, et al. 2004. High-affinity interaction between HIV-1 Vpr and specific sequences that span the C/EBP and adjacent NF-kappaB sites within the HIV-1 LTR correlate with HIV-1-associated dementia. DNA Cell Biol. 23:261–269 [DOI] [PubMed] [Google Scholar]
- 7. Cardinaux JR, Allaman I, Magistretti PJ. 2000. Pro-inflammatory cytokines induce the transcription factors C/EBPbeta and C/EBPdelta in astrocytes. Glia 29:91–97 [PubMed] [Google Scholar]
- 8. Carroll-Anzinger D, Kumar A, Adarichev V, Kashanchi F, Al-Harthi L. 2007. Human immunodeficiency virus-restricted replication in astrocytes and the ability of gamma interferon to modulate this restriction is regulated by a downstream effector of the Wnt signaling pathway. J. Virol. 81:5864–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carroll-Anzinger D, Al-Harthi L. 2006. Gamma interferon primes productive human immunodeficiency virus infection in astrocytes. J. Virol. 80:541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Churchill MJ, et al. 2009. Extensive astrocyte infection is prominent in human immunodeficiency virus-associated dementia. Ann. Neurol. 66:253–258 [DOI] [PubMed] [Google Scholar]
- 11. Conant K, et al. 1994. In vivo and in vitro infection of the astrocyte by HIV-1. Adv. Neuroimmunol. 4:287–289 [DOI] [PubMed] [Google Scholar]
- 12. Deng J, et al. 2002. beta-catenin interacts with and inhibits NF-κB in human colon and breast cancer. Cancer Cell 2:323–334 [DOI] [PubMed] [Google Scholar]
- 13. Deng J, et al. 2004. Crossregulation of NF-κB by the APC/GSK-3beta/beta-catenin pathway. Mol. Carcinog. 39:139–146 [DOI] [PubMed] [Google Scholar]
- 14. Ejarque-Ortiz A, Tusell JM, Serratosa J, Saura J. 2007. CCAAT/enhancer binding protein-alpha is down-regulated by toll-like receptor agonists in microglial cells. J. Neurosci. Res. 85:985–993 [DOI] [PubMed] [Google Scholar]
- 15. Eugenin EA, Clements JE, Zink MC, Berman JW. 2011. Human immunodeficiency virus infection of human astrocytes disrupts blood-brain barrier integrity by a gap junction-dependent mechanism. J. Neurosci. 31:9456–9465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gorry P. R., et al. 2002. June Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. 76:6277–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He G, Ylisastigui L, Margolis DM. 2002. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 21:697–705 [DOI] [PubMed] [Google Scholar]
- 18. Heaton R. K., et al. 2010. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 75:2087–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heaton R. K., et al. 2011. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J. Neurovirol. 17:3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henderson LJ, Al-Harthi L. 2011. Role of beta-catenin/TCF-4 signaling in HIV replication and pathogenesis: insights to informing novel Anti-HIV molecular therapeutics. J. Neuroimmune Pharmacol. 6:247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoverter NP, Waterman ML. 2008. A Wnt-fall for gene regulation: repression. Sci. Signal. 1:pe43. [DOI] [PubMed] [Google Scholar]
- 22. Ju J, et al. 2010. Phosphorylation of p50 NF-kappaB at a single serine residue by DNA-dependent protein kinase is critical for VCAM-1 expression upon TNF treatment. J. Biol. Chem. 285:41152–41160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar A, et al. 2008. Active β-catenin signaling is an inhibitory pathway of HIV replication in peripheral blood mononuclear cells. J. Virol. 82:2813–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lamba S, Ravichandran V, Major EO. 2009. Glial cell type-specific subcellular localization of 14-3-3 zeta: an implication for JCV tropism. Glia 57:971–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lane TE, Buchmeier MJ, Watry DD, Fox HS. 1996. Expression of inflammatory cytokines and inducible nitric oxide synthase in brains of SIV-infected rhesus monkeys: applications to HIV-induced central nervous system disease. Mol. Med. 2:27–37 [PMC free article] [PubMed] [Google Scholar]
- 26. Li W, Henderson LJ, Major EO, Al-Harthi L. 2011. IFN-γ mediates enhancement of HIV replication in astrocytes by inducing an antagonist of the β-catenin pathway (DKK1) in a STAT 3-dependent manner. J. Immunol. 186:6771–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, et al. 2004. CD4-independent infection of astrocytes by human immunodeficiency virus type 1: requirement for the human mannose receptor. J. Virol. 78:4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neil SJ, et al. 2005. The promiscuous CC chemokine receptor D6 is a functional coreceptor for primary isolates of human immunodeficiency virus type 1 (HIV-1) and HIV-2 on astrocytes. J. Virol. 79:9618–9624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nuovo GJ, Gallery F, MacConnell P, Braun A. 1994. In situ detection of polymerase chain reaction-amplified HIV-1 nucleic acids and tumor necrosis factor-alpha RNA in the central nervous system. Am. J. Pathol. 144:659–666 [PMC free article] [PubMed] [Google Scholar]
- 30. Permanyer M, Ballana E, Este JA. Endocytosis of HIV: anything goes. Trends Microbiol. 18:543–551 [DOI] [PubMed] [Google Scholar]
- 31. Poli V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273:29279–29282 [DOI] [PubMed] [Google Scholar]
- 32. Ross HL, et al. 2001. HIV-1 LTR C/EBP binding site sequence configurations preferentially encountered in brain lead to enhanced C/EBP factor binding and increased LTR-specific activity. J. Neurovirol. 7:235–249 [DOI] [PubMed] [Google Scholar]
- 33. Ross HL, et al. 2001. Interaction between CCAAT/enhancer binding protein and cyclic AMP response element binding protein 1 regulates human immunodeficiency virus type 1 transcription in cells of the monocyte/macrophage lineage. J. Virol. 75:1842–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rossi A, et al. 2006. Human immunodeficiency virus type 1 Tat prevents dephosphorylation of Sp1 by TCF-4 in astrocytes. J. Gen. Virol. 87:1613–1623 [DOI] [PubMed] [Google Scholar]
- 35. Ruocco MR, et al. 1996. Regulation of HIV-1 long terminal repeats by interaction of C/EBP(NF-IL6) and NF-κB/Rel transcription factors. J. Biol. Chem. 271:22479–22486 [DOI] [PubMed] [Google Scholar]
- 36. Salim A, Ratner L. 2008. Modulation of beta-catenin and E-cadherin interaction by Vpu increases human immunodeficiency virus type 1 particle release. J. Virol. 82:3932–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shapshak P, et al. 2004. Elevated expression of IFN-gamma in the HIV-1 infected brain. Front. Biosci. 9:1073–1081 [DOI] [PubMed] [Google Scholar]
- 38. Sheridan PL, et al. 1995. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 9:2090–2104 [DOI] [PubMed] [Google Scholar]
- 39. Siliciano JD, et al. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727–728 [DOI] [PubMed] [Google Scholar]
- 40. Thompson KA, McArthur JC, Wesselingh SL. 2001. Correlation between neurological progression and astrocyte apoptosis in HIV-associated dementia. Ann. Neurol. 49:745–752 [DOI] [PubMed] [Google Scholar]
- 41. Tornatore C, Chandra R, Berger JR, Major EO. 1994. HIV-1 infection of subcortical astrocytes in the pediatric central nervous system. Neurology 44:481–487 [DOI] [PubMed] [Google Scholar]
- 42. Vijaykumar TS, Nath A, Chauhan A. 2008. Chloroquine mediated molecular tuning of astrocytes for enhanced permissiveness to HIV infection. Virology 381:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reference deleted.
- 44. Wiley CA, Schrier RD, Nelson JA, Lampert PW, Oldstone MB. 1986. Cellular localization of human immunodeficiency virus infection within the brains of acquired immune deficiency syndrome patients. Proc. Natl. Acad. Sci. U. S. A. 83:7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wortman B, Darbinian N, Sawaya BE, Khalili K, Amini S. 2002. Evidence for regulation of long terminal repeat transcription by Wnt transcription factor TCF-4 in human astrocytic cells. J. Virol. 76:11159–11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.