Abstract
A rodent or other small animal model for HIV-1 has not been forthcoming, with the principal obstacles being species-specific restriction mechanisms and deficits in HIV-1 dependency factors. Some Carnivorans may harbor comparatively fewer impediments. For example, in contrast to mice, the domestic cat genome encodes essential nonreceptor HIV-1 dependency factors. All Feliformia species and at least one Caniformia species also lack a major lentiviral restriction mechanism (TRIM5α/TRIMCyp proteins). Here we investigated cells from two species in another carnivore family, the Mustelidae, for permissiveness to the HIV-1 life cycle. Mustela putorius furo (domesticated ferret) primary cells and cell lines did not restrict HIV-1, feline immunodeficiency virus (FIV), equine infectious anemia virus (EIAV), or N-tropic murine leukemia virus (MLV) postentry and supported late HIV-1 life cycle steps comparably to human cells. The ferret TRIM5α gene exon 8, which encodes the B30.2 domain, was found to be pseudogenized. Strikingly, ferret (but not mink) cells engineered to express human HIV-1 entry receptors supported productive spreading replication, amplification, and serial passage of wild-type HIV-1. Nevertheless, produced virions had relatively reduced infectivity and the virus accrued G→A hypermutations, consistent with APOBEC3 protein pressure. Ferret cell-passaged HIV-1 also evolved amino acid changes in the capsid cyclophilin A binding loop. We conclude that the genome of this carnivore can provide essential nonreceptor HIV-1 dependency factors and that ferret APOBEC3 proteins with activity against HIV-1 are likely. Even so, unlike in cat cells, HIV-1 can replicate in ferret cells without vif substitution. The virus evolves in this novel nonprimate cell adaptive landscape. We suggest that further characterization of HIV-1 adaptation in ferret cells and delineation of Mustelidae restriction factor repertoires are warranted, with a view to the potential for an HIV-1 animal model.
INTRODUCTION
Exogenous lentiviruses infect species in four mammalian orders: Primates, Perissodactyla, Artiodactyla, and Carnivora. Endogenous and now apparently extinct lentiviruses have been identified in several Lagomorpha and lemur genomes (22, 33, 34). Extant lentiviruses exhibit narrow tropisms with no cross-order and highly limited cross-species infection. HIV-1, for example, cannot replicate in a sustained fashion or cause disease in any species besides Homo sapiens (3). These impediments have been central considerations for animal model development, and they reflect two complementary issues: viral requirements for specific cellular cofactors and the antiviral activities of species-specific restriction factors such as APOBEC3 proteins, TRIM5 proteins, and tetherin (52, 59, 60, 63). Lentiviruses have evolved counterdefenses to restriction. Impressively, it is now believed that the primate lentiviral accessory genes (vif, vpu, vpr, vpx, and nef) are largely devoted to this role (43). Central plus-strand initiation provides an additional defense against APOBEC3G editing of the unduplexed minus strand (28).
Recently, HIV-1 clones that contain only SIVmac vif or vif and capsid sequences were shown to evade macaque TRIM5-alpha and APOBEC3 restrictions (24, 26, 29, 32), and a vif-only chimera replicated for up to 6 months in pig-tailed macaques (24). Chronic replication and disease have not yet been observed, but this approach is promising for achieving an HIV-1 animal model, and it highlights the centrality of the known restrictions. In contrast, progress toward transgenic rodent and other common small laboratory animal models for HIV-1 has been confounded not only by multiple specific restrictions but also by complex viral life cycle blocks, particularly to proper particle assembly (5, 9, 17, 45, 64). Such a model would be valuable and informative whether or not macaque HIV-1 models become more fully realized, because of practical limitations intrinsic to research on these nonhuman primates and because of insights that could be gained from observing how the host responds and how HIV-1 evolves as it transitions into a different mammalian order.
Carnivorans comprise over 260 species of placental mammals. They group phylogenetically into two suborders, the Feliformia (Felidae, Hyaenidae, Herpestidae, and others) and the Caniformia (Canidae, Ursidae, Pinnepedia, Mustelidae, etc.). Variants of feline immunodeficiency virus (FIV) currently infect approximately half of Felidae, and FIV-ancestral lentiviruses have been endemic in the Panthera (lion) lineage since at least the late Pleistocene and perhaps earlier (4, 53, 68). AIDS very similar to the human syndrome results in one feline species, the domestic cat, in which the virus is pandemic and acquisition occurred relatively recently. Differences in the respective host-lentiviral equilibria of Primates and Carnivora are also informative and potentially exploitable. For example, considering restriction factors, Feliformia lack functioning antiviral Trim5α or TRIMCyp genes (48), as does at least one Caniformia species, the dog (57). The domestic cat does have an effective APOBEC3 repertoire that restricts HIV-1 (19, 49, 50, 62). However, when FIV Vif was stably expressed in trans in a feline cell line (CrFK) that also expressed HIV-1 entry receptors, productive spreading replication was enabled (62). vif-chimeric HIV-1 clones that encode FIV Vif in cis replicated in such cells, too (62, 73). The most important implication of these results is that, except for entry receptors, the domestic cat genome can supply the dependency factors needed for HIV replication, which is a fundamental difference from the mouse (9). SIVmac Vif was also effective in mediating feline APOBEC3 evasion, showing for the first time that a Vif could function effectively in a different mammalian order (62). Since corroborated for SIVmac Vif and extended to visna virus Vif as well (38), this is an exception to the general theme of narrow species specificity in evolved retroviral evasions.
Based on these results, we here examined cells of a different carnivore family, Mustelidae (suborder Caniformia). There are good precedents for effective Mustelidae models of human viral diseases. One species, the domesticated ferret, is a favored experimental host for studies of important human RNA virus pathogens (influenza virus, severe acute respiratory syndrome [SARS] coronavirus, and Nipah virus). No Mustelidae antiretroviral restriction factors have been cloned or characterized.
MATERIALS AND METHODS
HIV-1 entry receptor-expressing stable Mustelidae cell lines.
Adherent cell lines and T cell lines were maintained in Dulbecco modified Eagle medium (DMEM) and RPMI 1640 medium, respectively, with 10% heat-inactivated fetal calf serum (FCS), penicillin-streptomycin, and l-glutamine. Mpf, a ferret (Mustela putorius furo) brain-derived cell line, and Mv.1.Lu, an American mink (Neovison vison, formerly Mustela vison) fetal lung-derived cell line, were obtained from ATCC. The M. putorius furo lung cell line, FtAEpC, was recently derived as described previously (37). Mpf.CD4.X41 cells were derived in two selection steps, with FIV-based lentiviral vectors (55) used consecutively as described previously (62). One vector encoded hCD4 plus neo (G418 resistance), and the second encoded hCXCR4 plus pac (puromycin resistance), with each receptor and resistance gene linked by an intervening internal ribosome entry site (IRES). Cells were selected and maintained in 1 mg/ml G418 and 1 μg/ml puromycin. To establish Mpf.CD4.X42, FtAEpC.CD4.X4, and Mv.1.Lu.CD4.X4 cell lines, a single HIV-1-based lentiviral vector derived from TSINcherry (40) was used; the transfer vector has the following elements in series: hCD4-porcine teschovirus 2A (P2A) peptide-hCXCR4-IRES-pac. Cell surface expression was verified by flow cytometry using mouse anti-hCXCR4 (RD Systems) and anti-hCD4 (BD Biosciences Pharmingen; phycoerythrin and fluorescein isothiocyanate [FITC] conjugated, respectively). Competence for gp120-mediated entry was assayed by infecting with an HIV-1 LAI luciferase reporter virus kindly provided by M. Emerman (54). Luciferase activities were determined by lysing cells with cold phosphate-buffered saline (PBS; 1%) and Tween 20 followed by assay with SteadyGlo or BrightGlo (Promega) in a TopCount NXT microplate scintillation and luminescence counter (Perkin-Elmer). Activities were normalized for total protein (Bio-Rad) or for cell number counted at the time of cell lysate collection. Mean luciferase activity ± standard deviation (SD) from duplicate measurements was calculated.
Primary mononuclear cells.
Spleen, bone marrow, and lymph nodes from 3 different ferret donors were used. Organs were finely minced in PBS supplemented with 2% FCS to allow extravasation of cells. After passage through a strainer, mononuclear cells were purified by Ficoll centrifugation and maintained in RPMI supplemented with 10% FCS, 0.05 mM β-mercaptoethanol, phytohemagglutinin E (PHA-E; 2 μg/ml), and human interleukin-2 (IL-2) (in conditioned medium from murine L2.23 feeder cells, a gift of T. Miyazawa). PHA-E was discontinued 48 h after isolation. Human peripheral blood mononuclear cells (PBMC) were purified by Ficoll centrifugation of cells eluted from Mayo Clinic Blood Bank apheresis machine leukoreduction system chambers. Transduction with challenge vector HIV-1luc+ was performed with six serial 1:3 dilutions in a 24-well plate (200,000 cells/well). At day 5 after transduction, cells were counted and lysed to assay for luciferase activity as described above. Five million cells were then plated in a 10-cm tissue dish and transduced with equal amounts of HIV-1luc+. At day 5, cells were collected, centrifuged, and lysed for immunoblotting as described below. Supernatants were collected and concentrated by ultracentrifugation over a sucrose cushion in an SW32Ti swinging bucket rotor at 25,000 rpm for 2 h and then resuspended in 300 μl of PBS, a portion of which was set aside for p24 measurements and the rest of which was directly lysed in Laemmli buffer with β-mercaptoethanol and then boiled for 10 min before being loaded into polyacrylamide gels for immunoblotting. Control supernatants from uninfected cells were collected and processed the same way.
Vectors and viruses.
HIV-1luc− and HIV-1luc+, the vesicular stomatitis virus G protein (VSV-G)-pseudotyped NL4-3R−E−Δ426 and NL4-3R+E−Δ426 luciferase reporter viruses, have been described previously (40). Replication-competent NL4-3 clones that express the Vif proteins of HIV-1 NL4-3 or SIVmac239 (HIV-1VH and HIV-1VS) from a vif frame engineered to not overlap integrase are those of Stern et al. (62). TRIP-luc was constructed by exchanging firefly luciferase for gfp in TRIP-green fluorescent protein (GFP), a gift of Pierre Charneau. Replication-competent viruses were produced by transfection of 293T cells with 10 μg plasmid DNA in 75-cm2 flasks. Particle normalization utilized reverse transcriptase (RT) activity or p24 antigen. RT activity was determined using a 32P-based RT assay as described previously (40). p24 antigen was measured using the Zeptometrix enzyme-linked immunosorbent assay (ELISA) kit. Mean p24 ± SD from duplicate measurements for each sample was calculated. Infectivity per ng of p24 was determined by titration on GHOST cells according to the NIH AIDS Research and Reference Reagent Program protocol. For infections with p24-normalized viruses, 3 × 105 entry receptor-complemented cells were infected in six-well plates. The cells were washed 24 to 36 h later with DMEM five times to remove input virus, and a time zero p24 sample was collected. Cultures were maintained by splitting them 1:5 or 1:10 when confluent, and supernatants were sampled every 2 to 4 days for p24 measurements. Supernatants were filtered (0.45 μm) before passage to uninfected cells.
Hypermutation analysis.
Virus particles were pelleted by ultracentrifugation over a sucrose cushion for 2 h at 25,000 rpm. Viral RNA was isolated (RNeasy; Qiagen), and reverse transcribed with a Transcriptor first-strand cDNA synthesis kit (Roche). Genomic segments spanning gag-vpr and the 5′ and 3′ long terminal repeat (LTR) and leader were amplified with Phusion Hot Start DNA polymerase. Products were gel purified and cloned (StrataClone Ultra Blunt PCR cloning kit; Stratagene). Eight to 10 independent clones for each virus were sequenced.
Cloning of an Mpf cell cyclophilin A (CypA) cDNA and ferret TRIM5α exon 8 sequences.
Degenerate primers were designed from the canine and feline sequences. The forward primer, which contained a hemagglutinin (HA) epitope tag, was FelHuFerCypA (5′-ATATGGATCCACCATGTACCCATACGACGTCCCAGACTACGCTATGGTCAACCCCAYCRTGTT-3′), and the reverse primer was KpnIFelFerCypA (5′-ATATGGTACCTTAGATYTGTCCACAGTCAGCAATGG-3′). Mpf, FtAEpC, and Mv.1.Lu TRIM5α exon 8 sequences were isolated using the primers described by McEwan et al. (47, 48), i.e., gex8 feT5f, ATCCCTYTYACAGKGTCACA, and gex8 feT5r, MATGAARAGAAYKTATAGATGAGAAACC, where M = A/C, K = G/T, R = G/A, and Y = C/T.
Capsid mutants H87Q and A92T.
The H87Q and A92T capsid mutants were constructed in the HIV-1–GFP or NL4-3 backbone by site-directed mutagenesis using the QuikChange Lightning site-directed mutagenesis kit (Agilent). For virus-like particle (VLP) saturation assays, a fixed dose of GFP-encoding vector was coinfected with 4-fold serial dilutions of VSV-G-pseudotyped HIV-1 vector encoding pac. Cyclosporine (CsA; Paddock Laboratories) was obtained from the Mayo Clinic pharmacy and used at 5 μM. GFP-positive cells were counted by fluorescence-activated cell sorting (FACS) 48 after transduction.
Immunoblotting.
Cells were lysed in RIPA buffer (150 mM NaCl, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, 1% NP-40, 150 mM Tris-HCl, pH 8.0) with protease inhibitors (Complete Mini; Boehringer). Protein was quantified with the Bradford assay. Twenty micrograms of lysate was boiled in Laemmli buffer with β-mercaptoethanol for 10 min and then electrophoresed in 12% Tris-HCl gels (Bio-Rad) and transferred over 1 h to Immobilon P membranes (Millipore). The blocked membranes were incubated for 2 h with the primary antibody (Ab) anti-CypA (Santa Cruz rabbit polyclonal 133494) at 1:250 and then washed with Tris-buffered saline–Tween 20 (TBST) three times for 7 min each. Afterward, membranes were incubated for 1 h at room temperature with the secondary Ab, goat anti-rabbit–horseradish peroxidase (HRP) (Calbiochem), at 1:4,000. After being washed with TBST 3 times for 10 min each, membranes were incubated in SuperSignal West Pico chemiluminescent substrate (Pierce) for 1 to 2 min and exposed to film. Human and ferret primary mononuclear cell lysates and supernatants were electrophoresed in 10% Tris-HCl gels (Bio-Rad) and transferred over 1 h to Immobilon P membranes (Millipore). Blocked membranes were incubated overnight with primary anti-p24 (mouse monoclonal, Abcam 9071) at 1:2,000 and then washed with TBST three times for 7 min each. Afterward, membranes were incubated for 2 h at room temperature with the secondary Ab, goat anti-mouse–HRP (Calbiochem). After being washed with TBST 3 times for 10 min each, membranes were incubated in Lumi-lightplus Western blot substrate (Roche) for 1 to 2 min and exposed to film.
Nucleotide sequence accession number.
The sequence of exon 8 of the ferret TRIM5α gene from ferret (Mpf and FtAEpC) cells was deposited in GenBank under accession no. JQ048543.
RESULTS
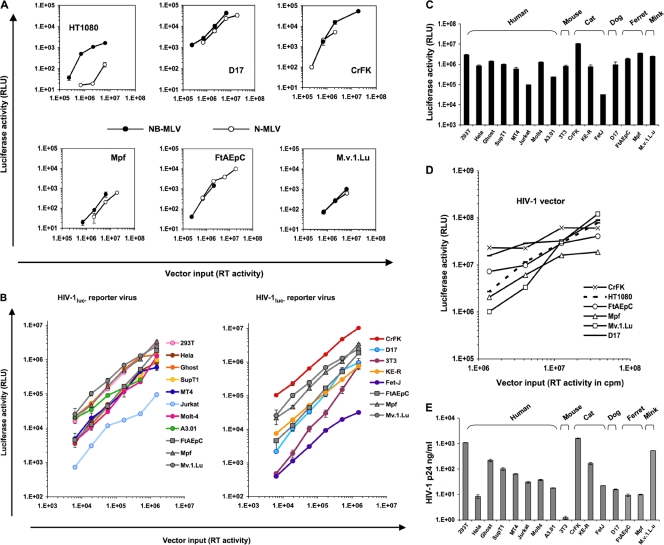
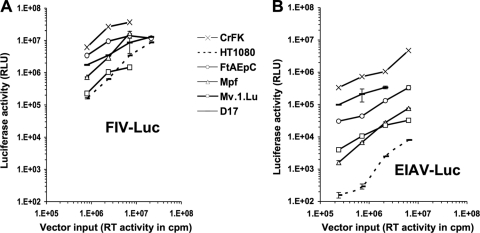
Pseudotyped luciferase (luc) reporter viruses and vectors were used initially to compare human, rodent, and carnivore cell lines (Fig. 1). These included three lines from two Mustelidae species: a recently established ferret (Mustela putorius furo) lung cell line (FtAEpC cells [37]), an M. putorius furo brain cell line (Mpf cells [67]), and an American mink (Neovison vison, formerly Mustela vison) fetal lung cell line (Mv.1.Lu cells [27]). In agreement with a previous study that included the latter two (65), we found that all three Mustelidae cell lines as well as dog and cat cells supported equivalent N- and NB-murine leukemia virus (MLV) infection, whereas N-MLV-restricting human HT1080 cells were much less efficiently infected with N-MLV (Fig. 1A). The Mustelidae and two other carnivore lines were also readily susceptible to HIV-1luc reporter virus infection, yielding luciferase activities that equaled or exceeded those of human cells infected with the same inputs (Fig. 1B and C). Since luc is expressed from the nef open reading frame in HIV-1luc (40), this virus demonstrates competence for the following postentry stages: reverse transcription, integration, and Tat/U3-promoted early (Rev-independent) viral gene expression (40). Similar results were observed when primary human PBMC and primary ferret mononuclear cells obtained from spleen, bone marrow, and lymph nodes were compared (Fig. 2A). Furthermore, similarly equivalent transduction was observed in ferret cell lines with a genome-minimized trans-packaged HIV-1 vector in which an internal human cytomegalovirus (CMV) promoter drives expression (Fig. 1D) and with analogously organized single-cycle FIV and equine infectious anemia virus (EIAV) vectors (Fig. 3A and B).
Fig 1.
Assessment of gammaretroviral and HIV-1 life cycle stages in Mustelidae and other mammalian cell lines. (A) Lack of restriction to N-tropic MLV luciferase vectors in Mustelidae cell lines. HT1080 cells were used as the positive-control line. The same viral preparation was used for all lines. (B to E) Early and late HIV-1 viral gene expression in Mustelidae and other mammalian cell lines. (B and C) Indicated cells were transduced with increasing doses of VSV-G-pseudotyped reporter virus HIV-1luc. Curves for the three Mustelidae are colored gray. Error bars represent standard deviations of duplicate measurements. (D) Cells were transduced with an HIV-1 vector in which transfer vector luciferase expression is driven by an internal CMV promoter. Cell lysates from equal numbers of cells were assayed for luciferase activity 5 days later. (E) p24 antigen, measured at day 5 postransduction, in supernatants of the respective cells transduced in panel C.
Fig 2.
Early and late HIV-1 viral gene expression in primary human and ferret cells. Primary mononuclear cells from human peripheral blood and ferret spleen, bone marrow, and lymph nodes were transduced with increasing doses of VSV-G pseudotyped reporter virus HIV-1luc+. (A) Luciferase activity was measured at 5 days postransduction in cell lysates. (B) Immunoblotting for HIV-1 capsid protein. (C) p24 antigen measured at day 5 postransduction, in supernatants of the respective cells transduced in panel B.
Fig 3.
FIV and EIAV infection. The indicated cells were transduced with increasing doses of luciferase-encoding single-cycle vectors derived from FIV (A) or EIAV (B). Cell lysates from equal numbers of cells were assayed for activity 72 h later. Error bars represent standard deviations of duplicate measurements.
Using primers validated by McEwan et al. to amplify TRIM5α exon 8 from mink, dog, and various feline species genomic DNA (47, 48), we amplified and sequenced exon 8 of the ferret TRIM5α gene from ferret (Mpf and FtAEpC) cells (see above) and found that the Feliformia-specific premature stop codon (48) is lacking, as was previously reported for the dog and mink exons (48, 57). However, multiple other stop codons were present in all reading frames, indicating pseudogenization as in the dog (57). No ferret TRIMCyp transcript could be identified by PCR using primers anchored in the ferret CypA sequence (determined in the present study; see below) and sets of degenerate primers homologous to Carnivora and human exon 2 (data not shown). Late HIV-1 life cycle events were assessed initially by measuring HIV-1 p24 production. Mouse (3T3) cells displayed the previously well-established (9, 17, 46, 64) assembly block to HIV-1, whereas Mustelidae cell lines and primary cells yielded robust HIV-1 p24 production similar to that of human cells (Fig. 1E and Fig. 2B and C). Taken together, these data indicate that major pre- and postintegration portions of the HIV-1 life cycle are grossly unimpaired in each of the three Mustelidae cell lines tested as well as in primary ferret mononuclear cells derived from spleen, bone marrow, and lymph nodes. There is an absence of TRIM5α/TRIMCyp/Fv1-type postentry blocks to lentiviral and gammaretroviral life cycles, substantial Tat transactivation function, and substantial Rev-mediated protein production, assembly, and particle release in these cells.
Based on these experiments, we proceeded to test directly whether mink or ferret cells could support productive, spreading replication of HIV-1. We derived stable cell lines that express human CD4 and CXCR4 and verified cell surface expression of these proteins by flow cytometry (Fig. 4A). Entry receptor competence was verified by infecting the receptor-complemented cells with HIV-1 LAI-luc (69) (Fig. 4B). The Mustelidae cells were then challenged with HIV-1 NL4-3 (2) and two variants of NL4-3, HIV-1VS and HIV-1VH (62). HIV-1VS utilizes a separation of the normally overlapping integrase and vif reading frames to encode the Vif protein of SIVmac (62), and HIV-1VH is a matched control virus that encodes HIV-1 Vif in the same manner. HIV-1VS but not HIV-1VH or wild type HIV-1 NL4-3 replicates in feline CrFK.CD4.X4 cells (62).
Fig 4.
Mustelidae cell lines with functional HIV-1 entry receptors. FIV vectors that coencode the neo or pac resistance markers (62) were used along with G418 and puromycin selection, respectively, to introduce the two receptors serially into Mpf cells, yielding the Mpf.CD4.X41 cell line. An HIV-1 TSIN series (40) lentiviral vector was also constructed to encode the receptor and coreceptor transgenes linked by a porcine teschovirus 2A (P2A) peptide, with pac coencoded by a downstream IRES. Using this vector, a second Mpf line (Mpf.CD4.X42) as well as FtAEpC.CD4.X4 and Mv.1.Lu.CD4.X4 cell lines was derived and maintained with puromycin selection. (A) Flow cytometry was performed to determine the expression of human CD4 and CxCR4 on the surface of the derived cells. All cells were labeled with the antibodies except for the SupT1 cells in the top left panel. The parental Mustelidae species cells not complemented with receptors were used as negative controls (left, bottom three panels), and SupT1 was used as a positive control (top right panel). (B) gp120-mediated entry. Cells with or without receptors were infected with increasing doses of native enveloped HIV-1 LAI-luc reporter virus (54). Cell lysates from equal numbers of cells were assayed for luciferase activity 72 h later.
In the present experiments in Mustelidae cells, the pattern was different. Productive, spreading replication of wild-type HIV-1 NL4-3, HIV-1VH, and HIV-1VS was observed in ferret but not mink cells (Fig. 5A to C). This was the case in the FtAEpC.CD4.X4 cell line and in two independently derived stable Mpf.CD4.X4 cell lines (Mpf.CD4.X41 and Mpf.CD4.X42, which were made with different receptor-transducing systems). The viruses could be multiply passaged and amplified (Fig. 5D and E). In contrast, the receptor-complemented mink cell line Mv.1.Lu.CD4.X4, though clearly enabled for efficient viral entry (Fig. 4B), did not support productive HIV-1 replication with any of the above viruses, including HIV-1 first passaged through the ferret cell lines (data not shown).
Fig 5.
Productive HIV-1 replication in ferret cells. (A and B) Mpf.CD4.X41 cells were infected with HIV-1VH and HIV-1VS, and virus produced was serially passaged 5 additional times. Input inocula were 1 ng p24. (C) Mpf.CD4.X42 cells were infected with 10 ng p24 of HIV-1 NL4-3, and virus produced was serially passaged 5 additional times. (D and E) Third-passage HIV-1 HIV-1VH(mpfP3) and HIV-1VS(mpfP3) viruses from Mpf.CD4.X41 replication experiments were serially passaged 4 times on FtAEpC.CD4.X4 cells. The passage numbers above the curves reflect the total passages on both ferret lines, while the numbers in the symbol keys refer to the number of passages on FtAEpC.CD4.X4 cells. (F) Infectivity determined by titration on GHOST cells.
These results showed that wild-type HIV-1 will replicate productively and spread in ferret cells if entry receptors are provided. Nevertheless, we found that virions produced in these cells were still less infectious per unit of p24 antigen than were virions produced in human cell lines. Figure 4F shows GHOST cell titrations of third-passage viruses from the ferret cell line Mpf.CD4.X41, which we refer to as HIV-1VH(mpfP3) and HIV-1VS(mpfP3). Comparison is made to virus produced in maximally permissive human 293T cells. This producer cell-dependent loss of infectivity in ferret cells suggested that APOBEC3 proteins may be targeting HIV-1. Therefore, we amplified and sequenced long terminal repeat (LTR) and gag/pol-vpr segments of HIV-1VH(mpfP3) and HIV-1VS(mpfP3) genomes as well as from later FtAEpC cell passages (Fig. 6A). A signature of APOBEC3 protein activity, G→A hypermutation, was observed. This genome editing is nevertheless manifestly not lethal since HIV-1 was still amplified exponentially and passaged robustly in ferret cell lines. Another finding was a premature stop codon in vpr, a frequent occurrence when HIV-1 is passaged in any cultured cells (51). In HIV-1VH(mpfP3), a vif reading frame-preserving closure of the artificial integrase-vif separation arose through deletion of a 62-nucleotide (nt) segment (Fig. 6B and its legend). This change, effectively a reversion to wild type, was found at the 5′ end of vif in 8/10 clones and was likely consequent to the duplication of 5′-GGAAAACAG-3′ (56, 62) at the 5′ end of vif in the input virus, which facilitated recombination during reverse transcription (72) to produce the virus illustrated in Fig. 6B. While G-to-A changes predominated, other kinds of mutations were also seen (Fig. 6A). For example, as previously observed to happen with rat and rabbit APOBEC1 (10, 30), a significant number of plus-strand C-to-T mutations were observed (45, versus 161 G-to-A changes). This outcome might reflect RNA deamination as well (10, 30). The dinucleotide contexts observed for cytidine deamination can vary substantially with different APOBEC proteins (6). Here in ferret cells, it was different from the pattern typically seen with human A3G, where CC and TC (edited minus-strand cytidine underlined) contexts predominate (23, 44). Instead, a broader dinucleotide context pattern was observed in the 161 G-to-A mutations that occurred with passage in ferret cells: CC, 46 (29%); TC, 32 (20%); GC, 27 (17%); and AC, 56 (35%). This is reminiscent of more diverse dinucleotide patterns observed previously for different feline A3 proteins (50).
Fig 6.
Sequencing of HIV-1VH(mpfP3) and HIV-1VS(mpfP3) viruses isolated from passage 3 of HIV-1VH and HIV-1VS on the Mpf.CD4.X41 cell line. (A) Sequencing of 290,607 nt (8 to 10 clones per segment) was performed, revealing 161 G→A changes. (B) Recombinant HIV-1VH found in 8/10 clones. Capital letters indicate original mutations used to separate the integrase and vif frames. (C) Capsid mutations selected in the unstructured CypA binding loop of HIV-1 CA. H87Q and A92T were present in 10 of 10 and 9 of 9 sequenced clones, respectively.
Vif did not accrue consistent amino acid changes in any of the viruses. However, capsid did. Two coding changes arose in the cyclophilin A (CypA) binding loop, which is complexly involved in the viral life cycle, including in TRIM5 protein restriction (41). For HIV-1VH(mpfP3), a T→A change at nt 261 in capsid and, for HIV-1VS(mpfP3), a G→A mutation at nt 274 produced, respectively, H87Q and A92T mutations (Fig. 6C). As the selection of two different mutations in this functionally significant region of capsid after passage in ferret cells was intriguing, we introduced them prospectively, alone and in combination, into HIV-1 NL4-3 reporter viruses and full-length clones. Examined in this genetically defined context, the HIV-1 capsid mutants were found to have moderately increased infectivity in ferret cells compared to wild type (WT) (Fig. 7A).
Fig 7.
Infection of ferret cells with WT, H87Q, A92T, and H87Q/A92T NL4-3 clones. (A) HIV-1GFP reporter viruses. The number of GFP-positive cells was determined by FACS at 48 h. The upper and lower graphs show the results of two independent experiments with different vector preparations. Student's 2-tailed t test was used to determine P values for comparisons of the titers of WT virus with each of the capsid mutants. All calculated P values were <0.05 except for the comparison of WT and H87Q/A92T mutants in FtAEpC cells (asterisk, upper right plot; P = 0.07). (B) Full-length WT and mutant viral clone challenges of Mpf.CD4.X41 cells.
In the case of replicating virus, capsid mutants replicated to higher peak levels than did wild-type virus in ferret cells (Fig. 7B). We then tested effects of cyclosporine (Fig. 8A through C). In these experiments, the increase in infectivity conferred by the CypA binding loop mutations was again observed in ferret cells and also in owl monkey kidney (OMK) cells (Fig. 8A through C). CsA, which disrupts TRIMCyp restriction (59), produced the well-known dramatic augmenting effect in OMK cells for wild-type NL4-3 and each of the mutants (Fig. 8A). The moderately increased infectivity of H87Q in the absence of CsA in these experiments (Fig. 8A) is consistent with that observed previously in OMK cells (31). In clear contrast, CsA did not boost infectivity in ferret cells for any of the viruses (Fig. 8B and C); rather, a slight inhibitory effect was discernible, particularly for A92T. Since levels of CypA have been reported to play a role in determining HIV-1 infectivity in certain contexts (70), we performed immunoblotting for this protein. CypA was clearly present in the ferret cells, and its levels were also similar to those in human cells (Fig. 8D). A ferret CypA cDNA was isolated by reverse transcriptase PCR (RT-PCR) (Fig. 8E); sequencing and determination of the predicted amino acid sequence did not reveal significant differences in the known HIV-1 capsid-interacting regions (13, 20, 71).
Fig 8.
Cyclosporine experiments in ferret cells and CypA alignments. (A to C) Infectious titers of HIV-1 GFP vector were determined in the presence (black columns) and absence (gray columns) of 5 μM CsA for owl monkey kidney (A), Mpf (B), and FtAEpC (C) cells. (D) Immunoblotting for CypA in human and Mustelidae cell lines. Jurkat ppia−/− cells (14) were used as a negative control. (E) Alignment of the ferret and human CypA amino acid sequences reveals high conservation overall and complete conservation in the central hydrophobic pocket involved in HIV-1 capsid binding. The amino acids involved in HIV-1 capsid binding, which are known from several mutational and high-resolution structural studies (13, 20, 71), are highlighted with boxes. Comparison is made between the human protein reference sequence (top line, accession no. NP_066953) and the predicted amino acid sequence determined from the ferret CypA cDNA isolated from Mpf cell mRNA in the present study (middle line) and a sequence predicted from an M. putorius furo whole-genome shotgun (WGS) sequence contig (bottom line, accession no. AEYP01032892). The dashed arrows at each end indicate the span of the degenerate primers used to obtain the Mpf cell cDNA sequence. Stringent selection pressure for amino acid level conservation is apparent. For example, at the nucleotide sequence level (not shown), there were 49 differences between Mpf cell CypA and human CypA in the coding region of the mRNA (10.1% nonidentity), but virtually all were synonymous. Only three amino acid differences were present, and these were located at both termini (arrowheads indicating the fifth and the final two C-terminal amino acids); the T5I difference (black arrowhead) is uncertain since both threonine and isoleucine were encoded by the degenerate primer, the domestic cat sequence has an isoleucine at this position, and the ferret WGS contig predicts a threonine. Polymporphism is evident in the two ferret sequences; they differed at 10 nt (not shown), but besides T5I there were only two other predicted amino acid differences, A26S and R37H.
To complete the analysis with respect to the capsid mutants, VLP saturation experiments were performed. A dose-dependent, clear VLP saturation effect was observed in rhesus FrHK4 cells as anticipated (8, 16), but no such effects occurred in ferret cells (Fig. 9A to D).
Fig 9.
VLP saturation experiments in ferret cells. FRhK4, Mpf, and FtAEpC cells were infected with a fixed dose of wild-type HIV-1 GFP (A) and capsid mutants H87Q (B), A92T (C), and H87Q/A92T (D), in the presence of increasing amounts of HIV-1 VLPs. While the VLPs clearly released Lv1 restriction in the rhesus macaque cells as anticipated, they had no effect in ferret cells.
DISCUSSION
The results of this study show that, unlike the mouse genome (9), the ferret genome can supply the nonreceptor dependency factors needed for productive, spreading HIV-1 replication. Moreover, in contrast to cells of another carnivore that share this property (62, 73), vif gene substitution was not needed and wild-type HIV-1 was capable of replication and serial passage. As in feline cells, G→A hypermutation and producer cell-dependent infectivity reductions were observed, but in both ferret cell lines they did not prevent productive viral replication. The selection of capsid mutations is also strong corroborative evidence that continuous viral replication occurred. Whether the HIV-1 or SIVmac Vif protein produces partial APOBEC3 mitigation in ferret cells, or might be evolved by repeated passage to acquire it, deserves further specific analysis. The absence of postentry capsid-targeting defenses against N-MLV and lentiviruses is consistent with the apparent lack of an intact Trim5α or TRIMCyp gene in this species. We observed similarly robust completion of early and late events in primary ferret cells (Fig. 2). We were not able to complement primary ferret mononuclear cells with HIV-1 receptors, and lymphoid-lineage cell lines are not yet available. Therefore, the extent to which specific relevant primary cell types (CD4+ T cells) in ferrets in vivo express all needed nonreceptor dependency factors and/or might manifest additional restrictions will be a worthy subject for further study.
The capability to repeatedly passage a primate lentivirus through the novel adaptive environment of a nonprimate cell allows experimental selection for continued viral evolution. So far, after three passages, we have observed selection of two capsid CypA binding loop mutations, H87Q and A92T. We found that these confer moderately increased infectivity in ferret cells and that disruption of CypA interaction with CsA only slightly affects this (Fig. 7 and 8). The reasons that these mutations arose are not clear because of the ambiguities that persist about the roles that CypA plays in retroviral life cycles, but they may represent optimal fitness of the CypA binding loop in the presence of CypA but in the absence of any functionally antiviral TRIM5 protein. CypA, which is highly conserved between mammals, is a peptidyl-prolyl isomerase that binds lentiviral capsids. It catalyzes the cis/trans isomerization of the G89-P90 peptide bond in the HIV-1 capsid protein (11). CypA has distinctive and at times opposite context-dependent effects (18, 20, 42). The protein promotes infectivity in human cells (25, 61, 66) but is in contrast necessary for Trim5α restriction in rhesus macaque and African green monkey cells (7). While multiple possibilities have been proposed for the role of CypA in the viral life cycle, a unifying mechanistic explanation remains elusive. It has been hypothesized that this peptidyl-prolyl isomerase protects HIV-1 from human cell restriction by competing with Trim5α binding (31) or that it shields HIV-1 from an unknown antiviral factor in human cells since the stimulatory effect of CypA on HIV-1 infectivity is independent of human Trim5α (58, 59). Some CypA binding loop mutations alleviate restriction in Old and New World monkey cells; similar effects are seen with CsA (15, 31). Of interest, H87Q decreases the binding affinity of HIV-1 CA for CypA by 4.8-fold (71) and was reported to confer a replication advantage to HIV-1 in CypA-rich human cells (21). The mutation is present in 19.7% of natural isolates in the Los Alamos database (15), and in human cells it can confer HIV-1 resistance to the effects of CsA (i.e., CypA independence) (15, 31). H87Q has also been reported to mediate escape from cytotoxic T lymphocyte responses, emerging in the late phase of infection in 13.7% of patients infected with HIV-1 (36, 39).
A92T has not been previously reported, but a charge-adding mutation at this residue, A92E, is known to arise when HIV-1 is passaged in HeLa cells in the presence of CsA, thus conferring CsA resistance and in some cell lines actually conferring CsA dependence on the virus (1, 12); this phenomenon was later shown to reflect high levels of CypA in HeLa cells (70). Our experiments make it clear that CypA is present at substantial levels in the Mustelidae cell lines that we used (Fig. 8D) and also that its amino acid sequence is conserved versus human CypA in the hydrophobic regions known to form the retroviral capsid-interacting domain (13) (Fig. 8E). Thus, these mutations that developed in ferret cells may represent HIV-1 adaptation to a situation where CypA is present but there is no Trim5 protein pressure.
In contrast to ferret cells, we did not observe spreading HIV-1 replication in the mink (Mv.1.Lu.CD4.X4) cell line. This result, which was verified in repeated experiments, is at variance with a prior report (35). As was discussed previously (62), it may be possible to reconcile these differences by considering that a single round of provirus generation and p24 production occurred in the experiments of Koito et al. (35).
Our results add to emerging evidence that cells of carnivore species appear in general to harbor relatively few restrictions to HIV-1 replication, and prominent among these are APOBEC3-mediated restrictions. Reference 19 provides a recent review of carnivore cell restrictions. We also conclude that, like the domestic cat and unlike the mouse, the ferret genome encodes the major nonreceptor dependency factors for this primate lentivirus. There are robust precedents for modeling human RNA pathogens in the ferret. A ferret genome sequencing project is nearing completion (http://www.broadinstitute.org/scientific-community/science/projects/mammals-models/ferret-genome-project). We suggest that further characterization of HIV-1 adaptation in ferret cells and delineation of Mustelidae restriction factor gene repertoires are warranted, with a view to several possible benefits. These include potentials for developing an eventual HIV-1 animal model, for gaining basic insights into mechanisms of species-specific retroviral restriction, and for exploring the extent to which HIV-1 will evolve when confronted with the novel adaptive landscape of a nonprimate cell.
ACKNOWLEDGMENTS
We thank M. Stern for assistance with initial experiments in Mpf cells, J. Luban for Jurkat CypA-knockout cells, and M. Emerman, J. Olsen, and P. Charneau for plasmids.
We are grateful for funding from NIH AI47536 and AI77344 and the Tietze Foundation.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Aberham C, Weber S, Phares W. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Adachi A, et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ambrose Z, KewalRamani VN, Bieniasz PD, Hatziioannou T. 2007. HIV/AIDS: in search of an animal model. Trends Biotechnol. 25:333–337 [DOI] [PubMed] [Google Scholar]
- 4. Antunes A, et al. 2008. The evolutionary dynamics of the lion Panthera leo revealed by host and viral population genomics. PLoS Genet. 4:e1000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baumann JG, et al. 2004. Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 78:12537–12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beale RC, et al. 2004. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J. Mol. Biol. 337:585–596 [DOI] [PubMed] [Google Scholar]
- 7. Berthoux L, Sebastian S, Sokolskaja E, Luban J. 2005. Cyclophilin A is required for TRIM5alpha-mediated resistance to HIV-1 in Old World monkey cells. Proc. Natl. Acad. Sci. U. S. A. 102:14849–14853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Besnier C, Takeuchi Y, Towers G. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. U. S. A. 99:11920–11925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bieniasz PD, Cullen BR. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868–9877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bishop KN, Holmes RK, Sheehy AM, Malim MH. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. [DOI] [PubMed] [Google Scholar]
- 11. Bosco DA, Eisenmesser EZ, Pochapsky S, Sundquist WI, Kern D. 2002. Catalysis of cis/trans isomerization in native HIV-1 capsid by human cyclophilin A. Proc. Natl. Acad. Sci. U. S. A. 99:5247–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Braaten D, et al. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170–5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Braaten D, Ansari H, Luban J. 1997. The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 71:2107–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Braaten D, Luban J. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chatterji U, et al. 2005. Naturally occurring capsid substitutions render HIV-1 cyclophilin A independent in human cells and TRIM-cyclophilin-resistant in owl monkey cells. J. Biol. Chem. 280:40293–40300 [DOI] [PubMed] [Google Scholar]
- 16. Cowan S, et al. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. U. S. A. 99:11914–11919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diaz-Griffero F, Taube R, Muehlbauer SM, Brojatsch J. 2008. Efficient production of HIV-1 viral-like particles in mouse cells. Biochem. Biophys. Res. Commun. 368:463–469 [DOI] [PubMed] [Google Scholar]
- 18. Dorfman T, Weimann A, Borsetti A, Walsh CT, Gottlinger HG. 1997. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J. Virol. 71:7110–7113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fadel H, Poeschla E. 2011. Retroviral restriction and dependency factors in primates and carnivores. Vet. Immunol. Immunopathol. 143:179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gamble TR, et al. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285–1294 [DOI] [PubMed] [Google Scholar]
- 21. Gatanaga H, et al. 2006. Altered HIV-1 Gag protein interactions with cyclophilin A (CypA) on the acquisition of H219Q and H219P substitutions in the CypA binding loop. J. Biol. Chem. 281:1241–1250 [DOI] [PubMed] [Google Scholar]
- 22. Gifford RJ, et al. 2008. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U. S. A. 105:20362–20367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris RS, et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 24. Hatziioannou T, et al. 2009. A macaque model of HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 106:4425–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hatziioannou T, Perez-Caballero D, Cowan S, Bieniasz PD. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79:176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatziioannou T, et al. 2006. Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314:95. [DOI] [PubMed] [Google Scholar]
- 27. Henderson IC, Lieber MM, Todaro GJ. 1974. Mink cell line Mv 1 Lu (CCL 64). Focus formation and the generation of “nonproducer” transformed cell lines with murine and feline sarcoma viruses. Virology 60:282–287 [DOI] [PubMed] [Google Scholar]
- 28. Hu C, et al. 2010. The HIV-1 central polypurine tract functions as a second line of defense against APOBEC3G/F. J. Virol. 84:11981–11993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Igarashi T, et al. 2007. Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J. Virol. 81:11549–11552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ikeda T, et al. 2008. The antiretroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res. 36:6859–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ikeda Y, Ylinen LM, Kahar-Bador M, Towers GJ. 2004. Influence of gag on human immunodeficiency virus type 1 species-specific tropism. J. Virol. 78:11816–11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kamada K, et al. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:16959–16964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katzourakis A, Tristem M, Pybus OG, Gifford RJ. 2007. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U. S. A. 104:6261–6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Keckesova Z, Ylinen LM, Towers GJ, Gifford RJ, Katzourakis A. 2009. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology 384:7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koito A, Kameyama Y, Cheng-Mayer C, Matsushita S. 2003. Susceptibility of mink (Mustera vision)-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 77:5109–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kootstra NA, Navis M, Beugeling C, van Dort KA, Schuitemaker H. 2007. The presence of the Trim5alpha escape mutation H87Q in the capsid of late stage HIV-1 variants is preceded by a prolonged asymptomatic infection phase. AIDS 21:2015–2023 [DOI] [PubMed] [Google Scholar]
- 37. Kugel D, et al. 2009. Intranasal administration of alpha interferon reduces seasonal influenza A virus morbidity in ferrets. J. Virol. 83:3843–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. LaRue RS, Lengyel J, Jonsson SR, Andresdottir V, Harris RS. 2010. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 84:8193–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leslie AJ, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 40. Llano M, et al. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461–464 [DOI] [PubMed] [Google Scholar]
- 41. Luban J. 2007. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J. Virol. 81:1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067–1078 [DOI] [PubMed] [Google Scholar]
- 43. Malim MH, Emerman M. 2008. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 44. Mangeat B, et al. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 45. Mariani R, et al. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 75:3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mariani R, et al. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McEwan WA. 2010. Factors affecting replication and cross-species transmission of feline immunodeficiency virus. Ph.D. thesis University of Glasgow, Glasgow, United Kingdom: http://theses.gla.ac.uk/1388/ [Google Scholar]
- 48. McEwan WA, et al. 2009. Truncation of TRIM5 in Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J. Virol. 16:8270–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Münk C, et al. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 9:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Münk C, et al. 2007. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J. Virol. 81:7048–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakaya T, et al. 1996. Serial passage of human immunodeficiency virus type 1 generates misalignment deletions in non-essential accessory genes. Virus Res. 46:139–147 [DOI] [PubMed] [Google Scholar]
- 52. Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 53. Pecon-Slattery J, Troyer JL, Johnson WE, O'Brien SJ. 2008. Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology. Vet. Immunol. Immunopathol. 123:32–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peden K, Emerman M, Montagnier L. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661–672 [DOI] [PubMed] [Google Scholar]
- 55. Poeschla E, Wong-Staal F, Looney D. 1998. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354–357 [DOI] [PubMed] [Google Scholar]
- 56. Sakurai A, et al. 2004. Functional analysis of HIV-1 vif genes derived from Japanese long-term nonprogressors and progressors for AIDS. Microbes Infect. 6:799–805 [DOI] [PubMed] [Google Scholar]
- 57. Sawyer SL, Emerman M, Malik HS. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sayah DM, Luban J. 2004. Selection for loss of Ref1 activity in human cells releases human immunodeficiency virus type 1 from cyclophilin A dependence during infection. J. Virol. 78:12066–12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sayah DM, Sokolskaja E, Berthoux L, Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573 [DOI] [PubMed] [Google Scholar]
- 60. Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 61. Sokolskaja E, Sayah DM, Luban J. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800–12808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stern MA, et al. 2010. Productive replication of Vif-chimeric HIV-1 in feline cells. J. Virol. 84:7378–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stremlau M, et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 64. Swanson CM, Puffer BA, Ahmad KM, Doms RW, Malim MH. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 23:2632–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Towers G, et al. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. U. S. A. 97:12295–12299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Towers GJ, et al. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138–1143 [DOI] [PubMed] [Google Scholar]
- 67. Trowbridge RS, Lehmann J, Brophy P. 1982. Establishment and characterization of ferret cells in culture. In Vitro 18:952–960 [DOI] [PubMed] [Google Scholar]
- 68. Troyer JL, et al. 2008. FIV cross-species transmission: an evolutionary prospective. Vet. Immunol. Immunopathol. 123:159–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yamashita M, Emerman M. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 78:5670–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ylinen LM, et al. 2009. Cyclophilin A levels dictate infection efficiency of human immunodeficiency virus type 1 capsid escape mutants A92E and G94D. J. Virol. 83:2044–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yoo S, et al. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780–795 [DOI] [PubMed] [Google Scholar]
- 72. Zhang J, Temin HM. 1994. Retrovirus recombination depends on the length of sequence identity and is not error prone. J. Virol. 68:2409–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zielonka J, et al. 2010. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J. Virol. 84:7312–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]