Abstract
Mason-Pfizer monkey virus (M-PMV), like some other betaretroviruses, encodes a G-patch domain (GPD). This glycine-rich domain, which has been predicted to be an RNA binding module, is invariably localized at the 3′ end of the pro gene upstream of the pro-pol ribosomal frameshift sequence of genomic RNAs of betaretroviruses. Following two ribosomal frameshift events and the translation of viral mRNA, the GPD is present in both Gag-Pro and Gag-Pro-Pol polyproteins. During the maturation of the Gag-Pro polyprotein, the GPD transiently remains a C-terminal part of the protease (PR), from which it is then detached by PR itself. The destiny of the Gag-Pro-Pol-encoded GPD remains to be determined. The function of the GPD in the retroviral life cycle is unknown. To elucidate the role of the GPD in the M-PMV replication cycle, alanine-scanning mutational analysis of its most highly conserved residues was performed. A series of individual mutations as well as the deletion of the entire GPD had no effect on M-PMV assembly, polyprotein processing, and RNA incorporation. However, a reduction of the reverse transcriptase (RT) activity, resulting in a drop in M-PMV infectivity, was determined for all GPD mutants. Immunoprecipitation experiments suggested that the GPD is a part of RT and participates in its function. These data indicate that the M-PMV GPD functions as a part of reverse transcriptase rather than protease.
INTRODUCTION
Retroviral structural proteins and enzymes are synthesized as the polyprotein precursors Gag and Gag-Pol or Gag, Gag-Pro, and Gag-Pro-Pol, depending on the number of frameshifting or readthrough of termination codon events. Envelope glycoproteins are translated from a spliced env mRNA as the precursors of surface and transmembrane domains. The Gag-containing polyprotein precursors form immature particles either at a distinct site within the cytoplasm or at the plasma membrane of infected cells. The Gag polyprotein mediates the specific packaging of two copies of unspliced genomic RNA. During or shortly after budding, retroviral protease (PR) is activated and liberates individual proteins from their polyprotein precursors. This step leads to the reorganization of the immature particle into a mature, fully infectious virus.
Mason-Pfizer monkey virus (M-PMV) is a member of the family Betaretroviridae, and although it was first isolated from rhesus monkey breast carcinoma, it is not oncogenic (5, 10, 17). Instead, infected macaques suffer a severe immunodeficiency syndrome with a pathology similar to that of infection with lentiviruses such as human immunodeficiency virus (HIV) (6). M-PMV is a simple retrovirus that utilizes a cis-acting viral RNA segment for the transport of incompletely spliced genomic RNA. This segment, termed the constitutive transport element (CTE) (3, 9), is recognized by a cellular factor called Tip-associated protein (TAP), which specifically binds the CTE and stimulates the nuclear export of unspliced mRNA (13).
Bioinformatic analyses of several eukaryotic proteins associated with mRNA processing led to the identification of a short, glycine-rich region called the G-patch domain (GPD) (1). Interestingly, this GPD was found in several betaretroviruses (M-PMV, simian retrovirus type 1 [SRV-1], SRV-2, SRV-4, Jaagsiekte sheep retrovirus [JSRV], and squirrel monkey retrovirus [SMRV]) and in some class II-related endogenous retroviruses (human endogenous retrovirus K [HERV-K]) (1, 12) (Fig. 1D). The GPD is an approximately 40-amino-acid-long region that is characterized by the presence of six highly conserved glycine residues in the sequence hhxxxGaxxGxGhGxxxxG(x)nG, where “h” stands for bulky, hydrophobic residues (I, L, V, or M); “a” stands for aromatic residues (F, Y, or W); and “x” stands for any residue. Various G-patch-containing proteins have been found to be involved in mRNA splicing (14, 24, 28, 32) and DNA repair (7, 11) and to be overexpressed in the great majority of breast cancer cases (19). In most G-patch-containing proteins, this conserved domain is usually combined with other well-defined RNA binding domains, such as the RRM, dsRED, SWAP, R3H, C4, and C2H2 fingers (1).
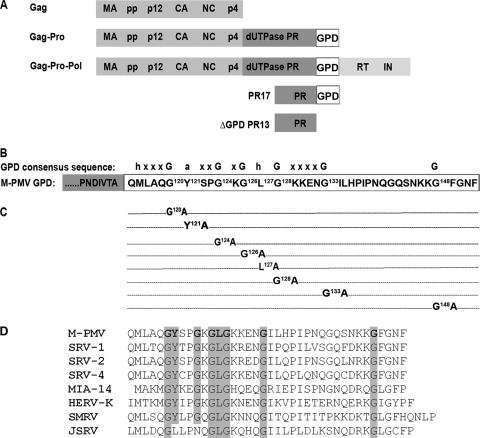
Fig 1.
Schematic diagram of the M-PMV GPD domain. (A) Schematic organization of M-PMV polyprotein precursors showing the location of the G-patch domain (GPD). (B) Consensus sequence of GPD, where “h” stands for I, L,V, or M; “a” stands for F, Y, or W; “x” stands for any residue; and “G” stands for glycine. (C) Schematic diagram of the mutations introduced within the M-PMV GPD used in this study. (D) Amino acid alignment of the GPDs of several betaretroviruses: M-PMV (Mason-Pfizer monkey virus), SRV-1 (simian retrovirus type 1), SRV-2 (simian retrovirus type 2), SRV-4 (simian retrovirus type 4), MIA-14 (mouse intracisternal A particles), HERV-K (human endogenous retrovirus K), SMRV (squirrel monkey retrovirus), and JSRV (Jaagsiekte sheep retrovirus).
The M-PMV GPD is synthesized as part of the Gag-Pro and also the Gag-Pro-Pol polyprotein precursors. Within these polyproteins, the GPD is localized between PR and reverse transcriptase (RT). Whereas a GPD at the C terminus of mature PR has been identified and studied (2, 27), the possible presence of a GPD at the N terminus of mature RT has not yet been determined. The mature PR is a 17-kDa, proteolytically active protein (PR17) containing the GPD at its C terminus. This PR17 protein undergoes further C-terminal autoprocessing into a 13-kDa form without a GPD (PR13; ΔGPDPR). Both PR17 and PR13 are present in released virions and display similar proteolytic activities and substrate specificities (33, 34). Previously reported in vitro experiments showed that the M-PMV GPD within PR17 binds single-stranded DNA or RNA oligonucleotides (27). In contrast to PR17, the PR13 protease, which lacks the GPD, does not bind nucleic acids. Moreover, a single-point mutation of the highly conserved tyrosine residue (Y121) of the GPD abolished the binding of nucleic acid to M-PMV PR17 (27) and significantly decreased virus infectivity (2).
The precise function of GPDs in cellular proteins is not clear. The GPD has been reported to mediate interactions with both nucleic acids and protein. A direct interaction with nucleic acid was described previously, e.g., for GPDs of the DNA repair enzyme of Toxoplasma gondii (TgDRE) (11). The GPD of tuftelin-interacting protein 11 (TFIP11), which is a protein component of the spliceosome complex, is necessary for interactions with the putative pre-mRNA-splicing factor ATP-dependent RNA helicase (DHX15) (28, 32). The GPD of Prp2, an RNA-dependent ATPase that activates the spliceosome, plays a cooperative role in splicing upon its interaction with the Saccharomyces cerevisiae protein Spp2 (24). The G-patch protein Pfa1p interacts directly with Prp43p through its GPD to stimulate PrP43p ATPase and helicase activities, which are required for efficient 20S pre-rRNA processing and thus for ribosome biogenesis (18, 31).
By introducing alanine-scanning mutations at the most highly conserved GPD amino acid positions, we attempted to elucidate the function of the GPD in the M-PMV life cycle. Neither the point mutations nor the deletion of the entire GPD affected protease activity and specificity. Assembly, morphogenesis of assembled immature particles, viral genomic RNA incorporation, and envelope glycoprotein incorporation were also unaffected. However, changes were observed in the infectivity of the GPD mutants as a consequence of a significantly lowered reverse transcriptase activity. Based on these results, we conclude that the GPD is required for the function of reverse transcriptase rather than protease.
MATERIALS AND METHODS
Viral constructs.
All DNA manipulations were carried out by using standard subcloning techniques, and plasmids were propagated in Escherichia coli DH5α cells. All newly created constructs were verified by DNA sequencing. To create point mutations within the GPD of a proviral DNA vector, we used a helper vector prepared by the ligation of a SacI-Eco72I fragment corresponding to nucleotides 1165 to 3275 of M-PMV into pUC19 (MHelppUC19). Point mutations within the GPD were created by two-step PCR mutagenesis using primers carrying the appropriate mutation and suitable restriction site (X) and MHelppUC19 as a template. The obtained fragments were digested with SacI-X and X-Eco72I (where X is the enzyme that cuts at the restriction site introduced by the mutagenic primers), and both fragments were ligated into MHelppUC19. Following sequence verification, the SacI-Eco72I fragment of MHelppUC19, carrying the appropriate mutation, was inserted into pSARM4 (25) (kindly provided by E. Hunter), i.e., a plasmid that contains the entire infectious M-PMV proviral genome. For the single-round infectivity assay, the M-PMV Env expression vector pTMO (4) and the pSARM-EGFP vector, in which EGFP (enhanced green fluorescent protein [GFP]) replaces the env gene (20) (both kindly provided by E. Hunter), were used.
Further details of the cloning strategy and full sequences of all PCR primers can be obtained from the authors upon request.
Cell growth and virus production.
HEK 293T cells were grown in Dulbecco's modified Eagle medium (DMEM; PAA Laboratories, Linz, Austria) supplemented with 10% fetal bovine serum (PAA Laboratories) and 1% l-glutamine (PAA Laboratories). Typically, 1 day before transfection, HEK 293T cells were plated at a density of 3 × 105 cells/ml and then transfected with the wild-type (wt) or mutant proviral DNAs using Fugene HD transfection reagent (Roche Molecular Biochemicals) according to the manufacturer's instructions. Supernatants were harvested at 48 h posttransfection, filtered through a 0.45-μm filter, and centrifuged through a 20% sucrose cushion for 1 h at 200,000 × g in a Beckman SW41Ti rotor.
Protein expression, radioactive labeling, and quantification of particle release.
HEK 293T cells transfected with the appropriate DNA were grown for 48 h posttransfection, starved for 30 min in methionine- and cysteine-deficient DMEM (Sigma), and then pulse-labeled for 30 min with 125 μCi/ml of Tran35Slabel (MGP, Czech Republic). The labeled cells were then chased in complete DMEM for 16 h. The cells from pulse and pulse-chase experiments were washed with phosphate-buffered saline (PBS), lysed in 1 ml of lysis buffer A (1% Triton X-100, 1% sodium deoxycholate, 0.05 M NaCl, 25 mM Tris [pH 8.0]) on ice for 30 min, and clarified by centrifugation at 13,000 × g for 1 min. The culture medium of the chased cells was filtered through a 0.45-μm filter, and SDS was added to a final concentration of 0.1%. Viral proteins were immunoprecipitated from the cells and culture media with a polyclonal rabbit anti-M-PMV CA or anti-GPD antibody (1:1,000 dilution) and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Radiolabeled proteins were visualized on a Typhoon PhosphorImager.
env expression was detected by pulse-labeling with [3H]leucine. HEK 293T cells transfected with mutant and wild-type DNAs were grown for 48 h, starved for 1 h in leucine-free DMEM, and then pulse-labeled with [3H]leucine (1.0 mCi/ml; 75 μl/35-mm well) for 30 min. The chase experiment was carried out for another 16 h. A goat anti-M-PMV antibody (22) was added to the cleared lysates of the cell- or medium-associated viral proteins prepared as described above. The immunoprecipitated viral proteins were separated by SDS-PAGE, dried, and analyzed by use of a Typhoon PhosphorImager.
To quantify the particles released, the radiolabeled protein bands of 35S-pulse-labeled Gag (Pr78) and pulse-chase-labeled virion-associated CA(p27) were quantified by using ImageQuant TL (Amersham Biosciences). Values of released viral proteins are shown as a relative concentration of CA correlated to the level of intracellular Gag in individual samples.
Western blotting.
HEK 293T cells were transfected with the wild-type or mutant proviral constructs. Virions from the culture supernatants were harvested at 48 h posttransfection as described above. The cells were washed with PBS and resuspended in 2× SDS protein loading buffer (PLB). Cell- and particle-associated viral proteins were separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and detected by using polyclonal antibodies against various M-PMV proteins. Polyclonal anti-M-PMV CA, MA, and PR antibodies were prepared by immunizing a rabbit with the appropriate purified protein or, in the case of the anti-GPD antibody, with a synthetic peptide (CYSPGKGLGKKENGILHPIPNQGQ) corresponding to M-PMV PR amino acid residues 121 to 143. The rabbit anti-M-PMV RT and IN antibodies were kindly provided by J. Snášel. The goat anti-M-PMV antibody was kindly provided by E. Hunter (22).
Quantitative Western blotting.
The quantification of virions was carried out by using quantitative Western blotting using West Femto chemiluminescent substrate (Thermo Scientific) followed by ImageQuant TL (Amersham Biosciences) quantification. The amount of the CA protein in individual viral samples was determined by comparison with a standard curve prepared with each Western blot. The standard curve was prepared from recombinantly expressed and purified M-PMV CA (23). The samples of the purified CA protein (1 to 2,000 ng) were separated on 15% SDS-PAGE gels and transferred onto a nitrocellulose membrane. After overnight blocking in blocking buffer (Blocker Casein in Tris-buffered saline [TBS]; Thermo Scientific), the rabbit anti-M-PMV CA antibody (at a 1:1,000 dilution in blocking buffer) was added to the membrane, and the membrane was incubated for 2 h at 4°C. The membrane was washed 3 times for 15 min with TBS. Blots were then incubated with goat anti-rabbit antibody (sc-2030 at a 1:1,000 dilution in blocking buffer; Santa Cruz) for 60 min. After three 15-min washes with TBS, the membrane was developed by using West Femto chemiluminescent substrate and an LAS-2000 imager. Protein band densities were quantified by use of ImageQuant TL (Amersham Biosciences). The linear range for purified CA(p27) from 20 to 600 ng was determined. The same Western blot procedure described as that above was performed with the samples of released virions. The amounts of CA(p27) in viral samples were determined by a comparison of their ImageQuant TL signals with those of recombinant CA(p27) in a calibration curve.
Single-round infectivity assay.
The infectivities of wt M-PMV and the GPD mutants were determined as described previously by Stansell et al. (26). HEK 293T cells were cotransfected with the pSARM-EGFP expression vector (20), containing either wt or GPD mutations, and the glycoprotein expression vector pTMO (4). At 48 h posttransfection, the culture supernatants were collected and filtered through a 0.45-μm filter, and each sample was normalized for capsid protein content by quantitative Western blotting. The volume of the culture supernatant used to infect HEK 293T cells was adjusted such that equivalent amounts of virus were added to each sample. The medium volume was adjusted to 4 ml with complete DMEM, and the cells were incubated for an additional 48 h. The cells were fixed with 4% formaldehyde, and the number of GFP-positive cells was determined by using flow cytometry (BD FACSaria).
Quantitative RT-PCR (qRT-PCR).
HEK 293T cells were transfected with wild-type or mutant proviral constructs. At 48 h posttransfection, the virus-containing medium was filtered through a 0.45-μm filter and centrifuged for 1 h at 200,000 × g in a Beckman SW41Ti rotor. The viral pellet was resuspended in 100 μl of PBS. An aliquot of 30 μl of the sample was added to an equal volume of PLB and analyzed by quantitative Western blotting as described above. Three units of DNase I (RNase free; Qiagen) was added to 70 μl of the sample, the sample was incubated for 1 h at 37°C, the volume was adjusted to 140 μl with PBS, and the sample was used to isolate viral RNA using the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's instructions. An aliquot of 5 μl of isolated RNA from each sample was used for reverse transcription by mixing with 0.5 μg oligo(dT)18 in a final volume of 12.5 μl. After incubation at 65°C for 5 min, 200 U of RevertAid H Minus Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Fermentas), 20 U of RiboLock RNase inhibitor (Fermentas), deoxynucleoside triphosphate (dNTP) mix (to a final concentration of 1 mM), and 4 μl of reaction buffer (5×) were added to each sample to a final volume of 20 μl, and the samples were incubated for 60 min at 42°C. Subsequently, 2 μl of each reverse transcription product was used for quantitative real-time PCR (qRT-PCR) using a LightCycler 480 real-time PCR system (Roche). Each quantitative PCR sample of a total volume of 20 μl contained 10 μl DyNAmo Hot Start SYBR green mix (Finnzymes) and each primer (Generi Biotech) to a final concentration of 50 nM. M-PMV CA-specific primers CA1ss (5′-GTG GAA TCT GTA GCG GAC AA-3′) and CA1as (5′-ATT ACC GGC TTG TTG GTT TC-3′) were designed by using GenScript Real-Time PCR Primer Design. The temperature profile was 95°C for 15 min, followed by 45 cycles of amplification as follows: a denaturation step at 94°C for 30 s, an annealing step at 60°C for 30 s, and an elongation step at 72°C for 1 min. Melting-curve analysis was performed at between 55°C and 90°C in 0.5°C increments. Six replicates were analyzed for each sample, including control samples containing water or isolated RNA in place of cDNA. The specific amplification of each cDNA was confirmed by melting-curve analysis. The data were analyzed by use of GenEx software (MultiD). The viral genomic RNA contents in individual samples were determined in two independent experiments, and the obtained values were corrected for different quantities of CA(p27) in released particles, determined by quantitative Western blot analysis.
RT activity assay.
For analyses of particle-associated reverse transcriptase activity, M-PMV particles were isolated by ultracentrifugation from the supernatant of HEK 293T cells at 48 posttransfection with wt and GPD mutant proviral DNAs. The viral pellets were resuspended in 100 μl buffer L (50 mM Tris [pH 8] containing 100 mM KCl, 0.05% Nonidet P-40, and 2 mM dithiothreitol [DTT]). An aliquot of 50 μl was used for quantitative Western blot analysis to normalize the CA(p27) content in the released M-PMV particles. Aliquots of normalized viral sample (up to 10 μl) were mixed with 40 μl of a reaction mixture with a final composition of 50 mM Tris (pH 8), 90 mM KCl, 8 mM MgCl2, 0.05% Nonidet P-40, 0.3 μg dA oligonucleotide, 0.6 μg poly(rU), 20 mM DTT, and 10 μCi [α-33P]dATP, and the mixture was incubated at 37°C for 1 h. Five-microliter aliquots of the samples were spotted onto a Whatman DE-81 paper, washed with SSC buffer (1.5 mM sodium citrate buffer [pH 7] and 150 mM NaCl), and analyzed with a Typhoon PhosphorImager.
Electron microscopy (EM) of tissue culture cells.
HEK 293T cells transiently expressing the wild-type or mutant M-PMV proviral constructs grown in 100-mm plastic dishes were washed with PBS, scraped into a microtube, and prefixed with freshly prepared 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.5). After washing with 0.1 M cacodylate buffer (pH 7.5), the cells were postfixed in 1% osmium tetroxide, dehydrated in an ethanol series (30, 50, 70, 80, 90, and 100%), and embedded in fresh Embed 812 or Agar 100 epoxy resin in a labeled Beem capsule. Ultrathin sections (70 nm) of cells were cut with a diamond knife on an RMC MT 7000 ultramicrotome and placed onto 200-mesh copper grids. The sections were contrasted with uranyl acetate and lead citrate. A Jeol JEM-1200EX analytical transmission electron microscope operating at 60 kV was used for analysis.
Immunoprecipitation.
Virions from 30 ml of each culture medium from HEK 293T cells expressing either wt M-PMV or the ΔGPD mutant were pelleted by centrifugation through a 20% sucrose cushion and resuspended in 200 μl of buffer L (50 mM Tris [pH 8] containing 100 mM KCl, 0.05% Nonidet P-40, and 2 mM DTT). A 10-μl aliquot was used for RT activity assays, and 100 μl was used for Western blot analyses. To remove GPD-containing proteins from the viral lysate, 10 μl anti-GPD antibody and 20 μl protein A-agarose (Invitrogen) were added to 90 μl of viral lysate. After overnight incubation, 14 μl of viral lysate containing the unbound material was used for the RT activity assay. Immunoprecipitated proteins were washed three times with 1 ml buffer L and then washed with 1 ml PBS, resuspended in PLB, and analyzed by Western blotting.
RESULTS
Production, release, and processing of the wild-type and GPD mutant polyproteins.
The sequence encoding the M-PMV G-patch domain (GPD), like those of other GPD-containing betaretroviruses, is located at the 3′ end of the pro reading frame directly prior to the pro-pol frameshift sequence (Fig. 1A and B). In order to investigate the role of the GPD in the M-PMV life cycle, a series of mutations replacing the most highly conserved amino acids in the GPD was prepared. The conserved glycine residues (G120, G124, G126, G128, G133, and G148), Y121, and L127 were replaced with alanine residues (Fig. 1C). The mutant lacking the entire G-patch domain (ΔGPDPR13), but possessing an intact frameshift sequence, allowing the production of the Gag-Pro-Pol polyprotein precursor, was prepared as described previously (2).
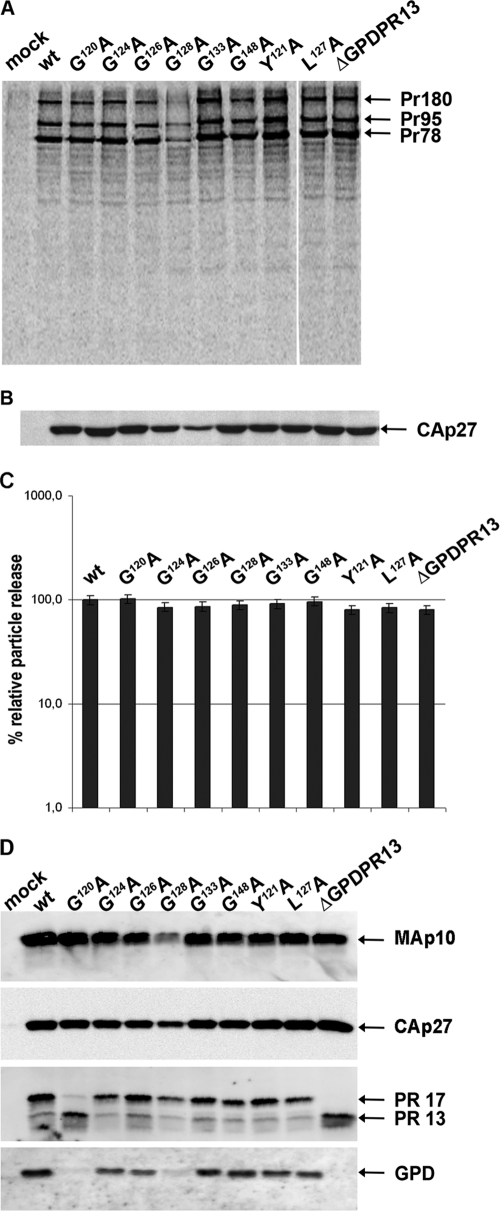
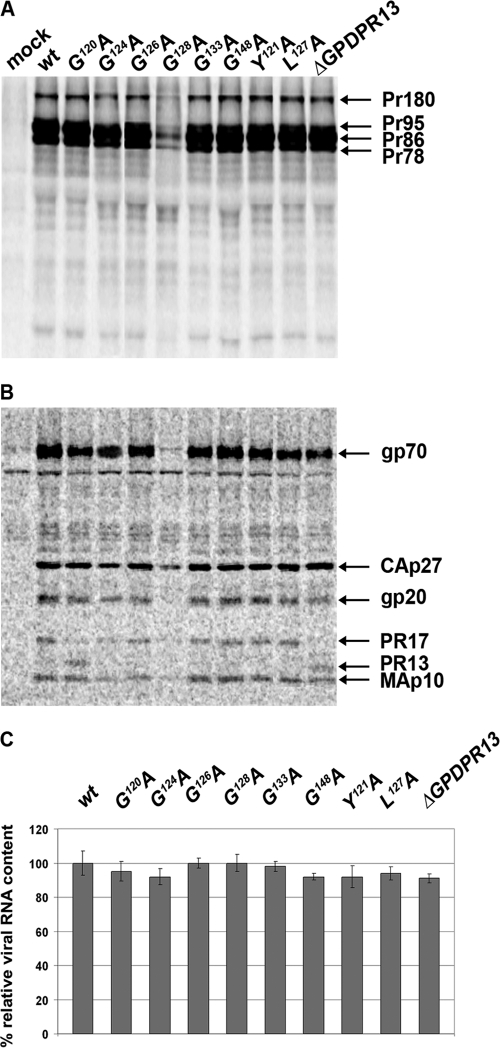
The expression of M-PMV polyproteins and virus release were evaluated by using a pulse-chase experiment with HEK 293T cells transfected with the wild-type and the mutant proviral constructs (pSARM4). At 48 h posttransfection, the proteins were metabolically labeled with 35S and chased for 16 h. Both cell- and virus-associated proteins were immunoprecipitated with rabbit anti-M-PMV capsid protein serum (Fig. 2A and B). Correct molecular masses (Pr78, Pr95, and Pr180) were found for polyprotein precursors (Gag, Gag-Pro, and Gag-Pro-Pol, respectively) of mutants and the wild type (Fig. 2A). A mature capsid protein [CA(p27)] of the proper molecular mass, i.e., 27 kDa, was detected in the wild-type and the mutant viral particles released into the culture medium (Fig. 2B). The relative ratios of cell- and virion-associated protein levels varied negligibly among the wild type and the GPD mutants (Fig. 2C), suggesting that particle release was not affected by the GPD mutations. A lower level of expression of the G128A mutant (Fig. 2A) than those of the wild type and the other mutants was consistently observed for several independent vector DNA preparations. This effect, which we think is caused by mRNA instability, was reflected in all phenotypic traits of the G128A mutant with respect to the wild type and all the other mutants.
Fig 2.
Synthesis, release, and processing of wild-type M-PMV and GPD mutants. HEK 293T cells were transfected with wild-type or mutant M-PMV proviral DNAs. Viral proteins were metabolically labeled with [35S]cysteine-methionine mix for 30 min and then chased for 16 h. M-PMV CA(p27)-related proteins were then immunoprecipitated from the cells and culture media and analyzed by SDS-PAGE. (A) Intracellular M-PMV proteins Gag Pr78, Gag-Pro Pr95, and Gag-Pro-Pol Pr180 immunoprecipitated from the cell lysate after a 30-min pulse. (B) Released M-PMV CA(p27) immunoprecipitated from the culture medium 16 h after the chase. (C) Quantification of the release of wt M-PMV and GPD mutants. Band intensities of 35S-pulse-labeled Gag (Pr78) and released CA(p27) were calculated. The relative percentage of CA released into the culture medium was corrected for the intracellular expression of individual samples. (D) Western blot analysis of released wild-type and GPD-related mutant proteins. At 48 h after transfection of HEK 293T cells with wild-type and GPD mutant proviral DNAs, the VLPs from the culture medium were collected by centrifugation through a 20% sucrose cushion. The viral proteins were analyzed by SDS-PAGE, blotted onto a nitrocellulose membrane, and detected by use of rabbit antibodies raised against (from top to bottom) MAp10, CA(p27), PR13, and the GPD.
None of the GPD mutants exhibited an alteration of Gag polyprotein processing compared to the wild type, as processed mature matrix (MAp10) and capsid proteins of the correct molecular masses were observed for all viruses (Fig. 2D). It is known that the 17-kDa M-PMV protease splits off its GPD, yielding the shorter, 13-kDa form (ΔGPDPR13) (34) shortly after virus release (Fig. 1A). Such a proteolytic cleavage of PR17 to ΔGPDPR13 was confirmed for all GPD mutant viruses released from the HEK 293T cells by using anti-MPMV PR and anti-GPD antibodies (Fig. 2D). An acceleration of the proteolytic cleavage of PR17 (yielding ΔGPDPR13) was observed for the G120A mutant (Fig. 2D).
The split-off 4-kDa fragment generated by the autocatalytic cleavage of PR17 was detected neither in the wt nor in the GPD mutants. This suggests that the inability to detect the fragment was not caused by a putative altered antigenicity of the mutants and the wt. Based on the fact that the rabbit anti-GPD antibody efficiently recognized both the wt and mutant GPDs at the C terminus of PR17 (Fig. 2D), we conclude that the split-off 4-kDa fragment was further degraded.
The detection of Pol-derived enzymes, i.e., RT and IN, was unsuccessful due to a combination of two factors: the high level of cross-reactivity of anti-RT and anti-IN antibodies and the very small amount of these Pol-derived enzymes (100-fold lower molar concentration than that of CA). The N terminus of M-PMV RT is still unknown. Therefore, it is possible that the GPD remains part of the RT after Gag-Pro-Pol polyprotein processing. However, using an anti-GPD antibody, we failed to detect any GPD-related protein other than PR17, suggesting that either the RT concentration is below the immunodetection limit or the GPD is not present at the N terminus of RT. Nevertheless, reverse transcriptase activity was verified in all released viral particles (see below).
Morphogenesis of immature virus particles.
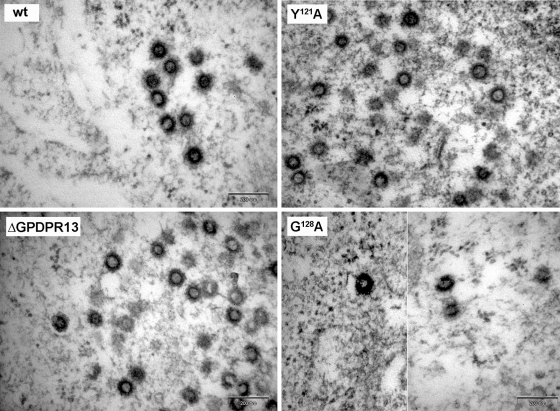
To analyze the effect of the GPD mutations on immature particle assembly, HEK 293T cells were transiently transfected with the wild-type and mutant proviral vectors. The cells were fixed at 48 h posttransfection, and ultrathin sections were prepared. Similar to the wild type, all the mutants assembled into regular immature particles within the cytoplasm of the transfected cells. A panel of EM images of selected mutants (ΔGPDPR13, Y121A, and G128A) is shown in Fig. 3. We did not observe the previously described effect of the Y121S mutation within the M-PMV GPD, i.e., the switch of the assembly mode from cytoplasmic assembly (typical for M-PMV) to that at the plasma membrane (2). The G128A mutant also assembled immature particles within the infected cells despite its lower level of production than the wild type (Fig. 3).
Fig 3.
Transmission EM images of HEK 293T cells expressing wild-type and mutant M-PMV proviruses. At 48 h after transfection of HEK 293T cells with wild-type or mutant proviral vectors, the cells were fixed in glutaraldehyde and postfixed in 1% osmium tetroxide. The sections were contrasted with uranyl acetate and lead citrate and analyzed by using a Jeol JEM-1200EX analytical transmission electron microscope. Bars, 200 nm.
The GPD is important for infectivity of the virus.
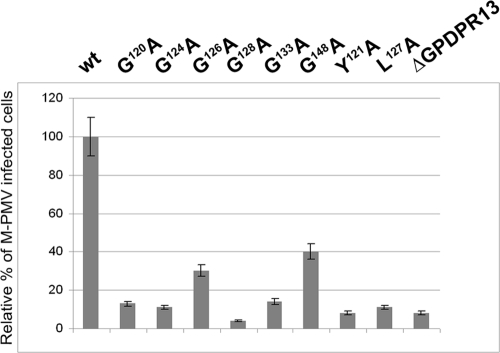
We further analyzed the effect of the GPD mutations on M-PMV infectivity in a single-round assay (26). For this purpose, the mutations were engineered into pSARM-EGFP, which expresses EGFP instead of Env (20). Wild-type and mutant EGFP proviruses were used for the cotransfection of HEK 293T cells simultaneously with an Env expression vector (pTMO) (4), and the culture medium supernatants containing released viruses were normalized for CA(p27) content based on results from quantitative Western blotting and were used for infections of fresh HEK 293T cells. At 48 h postinfection, the cells were analyzed by flow cytometry. The results from three independent experiments are summarized in Fig. 4. The percentages of EGFP-positive cells, i.e., those infected with the recombinant viruses, were around 40% for the wild type and below 0.5% for the negative control. The relative infectivity of the wild type was regarded as 100%. The deletion of the entire GPD as well as any mutation introduced within the GPD caused a drop in infectivity. Except for the G148A and G126A mutants, which exhibited about 40% and 30% of the wild-type infectivity, respectively, the level of infectivity of the mutants ranged from around 10 to 15% of the wild-type value.
Fig 4.
Relative infectivity of M-PMV GPD mutants, determined by a single-round assay. HEK 293T cells were cotransfected with wild-type or GPD mutant pSARM-EGFP and pTMO vectors. At 48 h posttransfection, the virus from the culture medium was filtered and normalized for CA(p27) by quantitative Western blotting. Equivalent amounts of virions were used to infect fresh HEK 293T cells. At 48 h postinfection, the cells were harvested, and the number of GFP-positive cells was determined by flow cytometry (BD FACSaria). The mean percentages of five independent infectivity measurements (with calculated standard deviations) for each mutant relative to the wild type are shown.
Due to the decreased infectivity of the GPD mutants, we explored additional possible roles of the M-PMV GPD. Based on the previously reported functions of other cellular GPD-containing proteins, we examined whether the M-PMV GPD (i) is involved in the splicing of env mRNA, (ii) contributes to the export of unspliced genomic RNA from the nucleus or its incorporation into viral particles, and (iii) directly affects the RT activity or incorporation of the Gag-Pro-Pol precursor into the virus particle.
Expression and incorporation of envelope glycoproteins are GPD independent.
Several cellular GPD-containing proteins have been reported to be involved in mRNA splicing (14, 24, 28, 32). Therefore, we analyzed the possibility that the GPD might play such a role in the splicing of M-PMV env mRNA that is used for the translation of the Env polyprotein precursor Pr86. This polyprotein undergoes proteolytic cleavage, yielding the gp70 surface subunit (SU) and the gp22/20 transmembrane subunit (TM). To compare the expressions of M-PMV envelope glycoproteins in the wild-type and mutant virions, HEK 293T cells were transfected with the M-PMV proviral vectors, and the proteins were metabolically labeled with [3H]leucine and immunoprecipitated from both cell lysates and cell culture media. The immunoprecipitated samples were analyzed by autoradiography of SDS-PAGE gels. The ratios of the cell-associated M-PMV Env (Pr86), Gag (Pr78), Gag-Pro (Pr95), and Gag-Pro-Pol (Pr180) polyproteins of all the GPD mutants were similar to that of the wild type (Fig. 5A). In accordance with the results of the 35S pulse-chase experiments, we found that the level of production of the G128A mutant polyprotein was lower than that of the wild type (Fig. 5A). Similar levels and protein patterns of the mature virus-associated proteins were present in the wild type and the other GPD mutants. The mature env gene products, i.e., gp70 and gp20, as well as the gag gene products CA(p27) and MAp10 and the pro gene products PR17 and ΔGPDPR13 were detected in released virions (Fig. 5B).
Fig 5.
Expression of envelope glycoproteins and incorporation of mRNA into released particles. HEK 293T cells were metabolically labeled with [3H]leucine at 48 h after transfection with wild-type or mutant proviral vectors. The viral proteins were immunoprecipitated with goat anti-M-PMV antibody, separated by SDS-PAGE, and analyzed by use of a Typhoon PhosphorImager. (A) Cell-associated viral proteins. (B) Medium-associated viral proteins. (C) Mean relative RNA contents ± standard deviations of the wt and individual GPD mutants. Viral RNA was isolated from the purified viral particles released into the culture medium at 48 h after transfection of HEK 293T cells with wild-type or mutant proviral vectors. Following reverse transcription, qRT-PCR was used to quantify the amount of RNA incorporated into released M-PMV particles. The viral genomic RNA contents were normalized for the different particle releases of the individual samples, as determined by quantitative Western blotting.
The GPD does not affect genomic RNA incorporation.
Since the M-PMV GPD was shown previously to bind single-stranded RNA (27), we analyzed whether it might interact with the CTE to mediate the transport of full-length genomic RNA or its incorporation into viral particles. To analyze RNA packaging, viral RNA was isolated from wt and GPD mutant viral particles, and it was then reverse transcribed and quantified by use of qRT-PCR. The results shown in Fig. 5C demonstrate that all GPD mutants packaged genomic RNA with an efficiency similar to that of the wt.
The GPD enhances RT activity but not Gag-Pro-Pol incorporation.
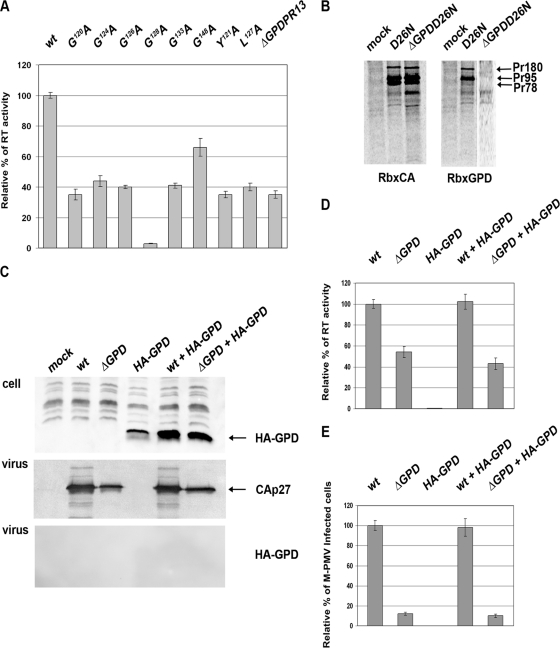
Since the GPD becomes a part of the Gag-Pro-Pol polyprotein upon the second frameshift, we tested whether the GPD might affect the activity of reverse transcriptase. For this purpose, equivalent amounts of wild-type and GPD mutant viruses normalized for CA using quantitative Western blotting were used for the RT activity assays (Fig. 6A). Indeed, a lower level of activity of the RT of all the GPD mutants than that of the wild type was observed. Interestingly, the GPD point mutations caused a decline in RT activity similar to that caused by the deletion of the entire GPD. All the mutants showed 25 to 50% of wild-type RT activity (Fig. 6A). The only exception was the G148A mutant, which exhibited about 70% of the wild-type RT activity.
Fig 6.
Reverse transcriptase activity and analysis of Gag-Pro, Gag-Pro-Pol, and hemagglutinin (HA)-GPD incorporation into M-PMV. (A) Reverse transcriptase activity of the wt and GPD mutants. HEK 293T cells were transfected with wild-type or GPD mutant proviral vectors. At 48 h posttransfection, the culture media were collected by ultracentrifugation, virus amounts were normalized for CA by using quantitative Western blotting, and equivalent amounts of virus lysates were used for RT activity assays. Mean RT activities, normalized for the different particle releases, are shown. (B) Incorporation of Gag-Pro and Gag-Pro-Pol into D26NpSARM4 and ΔGPD D26NpSARM4 particles. Viral proteins were metabolically labeled with [35S]cysteine-methionine mix, and M-PMV polyproteins were immunoprecipitated from the culture medium 16 h after a pulse using M-PMV anti-CA (Rb × CA; left) and anti-GPD (Rb × GPD; right) antibodies. (C) Incorporation of HA-GPD into ΔGPD M-PMV particles. HEK 293T cells were transfected with wild-type or ΔGPD proviral vectors with or without HA-GPD. At 48 h posttransfection, the cells and culture media were analyzed by Western blot analysis. The expression of HA-GPD in HEK 293T cell lysates was detected by using an anti-HA antibody (top). Released virions were analyzed by using an anti-CA antibody (middle) and anti-HA antibodies (bottom). The culture media of the HEK 293T cells cotransfected with wild-type or ΔGPD proviral vectors with or without HA-GPD were collected by ultracentrifugation, and virus amounts were normalized for CA by using quantitative Western blotting. (D and E) Equivalent amounts of virus lysates were used for RT activity (D) and single-round infectivity (E) assays.
Since the immunochemical detection of RT in virions, using the only available polyclonal rabbit antibodies made against the M-PMV RT peptide, failed, we could not exclude the possibility that the lower level of RT activity might be caused by an altered incorporation of Gag-Pro-Pol polyproteins into the GPD mutant virus-like particles (VLPs). To analyze the presence of the Gag-Pro-Pol polyprotein in virions, proviral vectors with inactive protease (D26ApSARM4) and with inactive protease lacking the G-patch domain (ΔGPDD26ApSARM4) were prepared. The presence of viral polyproteins within the particles released from HEK 293T cells was determined by a 35S pulse-chase experiment, followed by immunoprecipitation using anti-CA and anti-GPD antibodies (Fig. 6B, left and right, respectively). No difference in the Gag-Pro-Pol polyprotein contents in released wild-type and mutant VLPs was observed (Fig. 6B, left). Immunoprecipitation using an anti-GPD antibody confirmed the presence of the GPD within Gag-Pro and Gag-Pro-Pol in the released virus with inactive protease (Fig. 6B, right).
As the GPD is split off during protease maturation, we also inquired whether the GPD can act in trans. Similarly to data reported previously by Frenal et al. (11), we failed to detect the expression of a 40-amino-acid-long GPD. The GPD was detected only upon the expression of a construct containing two GPD copies interconnected with a GSGSGSG linker. Upon the coexpression of this double-GPD (2× GPD)-carrying vector with the ΔGPD M-PMV vector, we did not observe 2× GPD incorporation into the virions (Fig. 6C). Levels of RT activity and infectivity of ΔGPD M-PMV remained around 40 to 50% and 10%, respectively, of wt levels. This was found regardless of whether 2× GPD was coexpressed or not (Fig. 6D and E).
The GPD is a part of mature RT.
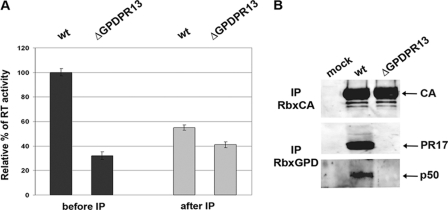
The likely reason for not detecting any GPD-related mature protein, except PR17, in a standard virus preparation (obtained from 4 ml of medium) is an insufficient amount of RT in mature virions (approximately 100-fold less than the amount of CA and 10-fold less than the amount of PR). To detect a putative transframe protein, i.e., GPD-RT, we used a highly concentrated M-PMV stock obtained by centrifugation of 2× 30-ml medium from HEK 293T cells transfected with the wild-type or ΔGPD vectors. Pelleted virions were resuspended and used for immunoprecipitation, Western blotting, and RT activity determinations. Given that the GPD would be a part of RT, we presumed that the anti-GPD antibody would immunoprecipitate the GPD-RT protein from the virus lysate. Hence, the level of activity of the RT in the unbound fraction should be lower than the level of RT activity determined before the immunoprecipitation. Indeed, a comparison of RT activities in the virus lysates before and after immunoprecipitation with anti-GPD antibody showed a decline (60%) in RT activity for the wild type. However, only a minor (10%) decrease was observed for the ΔGPD mutant (Fig. 7A). These results suggest that we immunoprecipitated the portion of RT containing the GPD. The remaining RT activity of the wild type in the supernatant fraction after immunoprecipitation was approximately 35 to 40% of the initial RT activity. Interestingly, this remaining (non-GPD-related) wild-type RT showed a level of activity similar to that detected for ΔGPD RT (40%).
Fig 7.
Analysis of the concentrated M-PMV stock. Virions from 30 ml of culture medium from HEK 293T cells expressing either the wild type or the ΔGPD mutant were pelleted and resuspended. Part of the viral lysate was used for immunoprecipitation using an anti-GPD antibody, and part was used for Western blot analyses. (A) The RT activities of the wild-type and ΔGPD virions were measured before and after anti-GPD antibody precipitation. (B) The presence of viral proteins was detected by Western blotting using rabbit antibodies raised against (from top to bottom) CA (Rb × CA) and GPD (Rb × GPD).
Western blot analysis of the concentrated M-PMV stock using anti-GPD antibody revealed the presence of not only PR17 but also a protein with an approximate molecular mass of 50 kDa (p50-GPD). This p50-GPD protein was detected in the wild-type but not in the ΔGPD fractions (Fig. 7B). To verify that this GPD-p50 protein is not derived from Gag or Pro polyproteins, anti-CA and anti-PR antibodies were used for immunoprecipitation. No decrease in the RT activity was observed after immunoprecipitation with anti-CA and anti-PR antibodies (data not shown), confirming that the GPD is linked to the downstream sequence. Western blot analysis confirmed that the GPD-p50 protein is recognized only with the anti-GPD antibody and not with the anti-CA and anti-PR antibodies (Fig. 7B). Thus, we hypothesize that the GPD is a part of RT and contributes to its activity.
DISCUSSION
In this study, we used alanine-scanning mutagenesis to investigate the role of the G-patch domain in the M-PMV life cycle. We did not detect any substantial changes in the processing of Gag and Gag-Pro polyproteins. The only minor difference from the wild type was an elevated level of self-processing of PR17 to PR13 in the G120A mutant. A similar acceleration of C-terminal processing was observed in vitro for the M-PMV protease Q115I mutant (mutated in the P′1 position of the PR17*PR13 cleavage site, IVTA*QMLAQ) (2). It is possible that the G120A mutation causes some local structural change in the vicinity of the PR17*PR13 cleavage site, resulting in its better accessibility. Since M-PMV PR prefers aromatic and hydrophobic residues in the P1 and P′1 positions, and the introduction of alanine at position 120 generated the sequence LAQAYSPG, we rather assume that this new sequence represents the protease cleavage site LAQA120*YSPG. This mutated site might be preferably cleaved during the processing of the protease C terminus prior to the cleavage of the original (PR17*PR13) processing site. Based on an unaltered cleavage pattern of PR17*PR13 in released virions, we can conclude that none of the other GPD mutations influenced this cleavage. The proper processing of Gag and Gag-Pro and the self-processing of the 17-kDa protease to the PR13 form suggest that the protease activity was not affected by the GPD mutations. Nevertheless, we cannot completely rule out the possibility that the processing of Gag-Pro-Pol was influenced by introduced GPD mutations.
We observed the assembly of typical D-type particles within the cytoplasm of HEK 293T cells infected with all the GPD mutants at 48 h posttransfection. It is difficult to harmonize the existence of the previously observed C-type appearance of particles for the PRΔ111-145 (i.e., ΔGPDPR13) and Y121S GPD mutants in COS cells at 20 h posttransfection (2) with our current data. Our recent research has led to an explanation of the mechanism of transport of M-PMV polyproteins. We confirmed that it is regulated by the interaction of the CTRS (cytoplasmic targeting-retention signal) of MA with a subunit of the dynein molecular motor (Tctex-1) (30). Moreover, the GPD is present only in Gag-Pro and Gag-Pro-Pol and not in Gag, suggesting that the GPD is not likely to be involved in the transport of Gag and, thus, in M-PMV morphogenesis. We hypothesize that the previously observed C-type-like immature particles of M-PMV carrying mutations in the GPD or upstream region (in PR13) were aberrant particles that accumulated at the plasma membrane in the early phase after transfection of COS cells (2).
We also examined the effect of the mutations on the incorporation of the Gag-Pro-Pol polyprotein as well as on reverse transcriptase activity. We found that the deletion of the GPD had no effect on the incorporation of the Gag-Pro-Pol polyprotein precursors. On the contrary, all mutations within the GPD influenced the activity of reverse transcriptase and subsequently led to a substantial decline in infectivity. These results clearly demonstrate the importance of the GPD for RT activity. Another piece of evidence that GPD-derived proteins exhibit RT activity came from the immunoprecipitation of wild-type and ΔGPD M-PMV proteins. The anti-GPD antibodies effectively reduced the level of RT activity of the wild-type sample to that of the ΔGPD M-PMV sample. In contrast, the immunoprecipitation did not affect the ΔGPD M-PMV sample. This finding suggests that the GPD is a part of at least some portion of mature RT.
The exact molecular mass of M-PMV RT has not yet been determined. The predicted molecular mass of mature RT is 66 kDa, and that of the RT portion without RNase H is 54 kDa; however, the precise boundary between the DNA polymerase and RNase H domains of M-PMV RT is not known. Based on our data, we speculate that M-PMV RT exists as a heterodimer consisting of GPD-p50 and 66-kDa subunits.
RTs from some other viruses have been shown to possess extended amino termini. An N-terminal extension of mouse mammary tumor virus (MMTV) RT upstream of the PR region has been observed (8, 29). Both bacterially expressed and mature viral MMTV RTs were shown previously to be active only in the presence of 27 flanking amino acid residues derived from the C terminus of the pro open reading frame protein product (8, 29). Similarly, the amino terminus of bovine leukemia virus (BLV) RT is encoded in the last 26 codons of the pro gene (21). Both MMTV and BLV RTs are transframe proteins encoded with a region spanning both the pro and pol genes. Unlike in BLV, where the pro-derived sequence of RT is absent in the protease carboxyl terminus, the C terminus of MMTV protease has a dual usage, also forming the N terminus of MMTV RT (15, 16, 21, 29). It is possible that M-PMV RT, similar to RTs from MMTV and BLV, is also a transframe protein resulting from the processing of the Gag-Pro-Pol polyprotein. The M-PMV pro-pol frameshift occurs at amino acid position 145 of GPD, and, except for G148, the entire GPD might be a part of the RT N terminus (Fig. 1C). Moreover, the RT-related GPD-p50 protein was recognized with an antibody raised against the GPD (amino acids 120 to 143 of PR upstream of the frameshift sequence CY120SPGKGLGKKENG133ILHPIPNQGQSN145*QKRFWKF-RT [the asterisk denotes the boundary between the pro and pol reading frames]). This finding, together with the results showing the importance of the first conserved glycine residue (G120) for RT activity, argues that the GPD is present at the N terminus of GPD-p50. With respect to the known ability of the GPD to bind RNA, we speculate that its physical connection with the N terminus of RT contributes to maintain RNA in a conformation advantageous for reverse transcription. Future detailed characterization and analysis of M-PMV RT are needed to elucidate the role of the GPD in RT function. Another remaining important question is, why the GPD, when dispensable for PR function, is encoded by the pro gene and not downstream of the pro-pol frameshift sequence in the pol reading frame. It remains unclear whether the GPD plays any role during PR maturation when transiently present at the C terminus of PR. In summary, our data clearly show the dispensability of the GPD for the function of mature PR and demonstrate its importance for RT activity.
ACKNOWLEDGMENTS
We thank Romana Cubínková for excellent technical assistance and Hillary Hoffman for language correction. We thank E. Hunter for providing us with pSARM4, pTMO, and goat anti-M-PMV antibody and H. Zábranská for the ΔGPDPR13pSARM4 vector.
The work was supported by grant 204/09/1388 from the Grant Agency of the Czech Republic and grants 1M0508, Z4 0550506, ME 904, and MSM 6046137305 from the Czech Ministry of Education.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Aravind L, Koonin EV. 1999. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 24:342–344 [DOI] [PubMed] [Google Scholar]
- 2. Bauerova-Zabranska H, et al. 2005. The RNA binding G-patch domain in retroviral protease is important for infectivity and D-type morphogenesis of Mason-Pfizer monkey virus. J. Biol. Chem. 280:42106–42112 [DOI] [PubMed] [Google Scholar]
- 3. Bray M, et al. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. U. S. A. 91:1256–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brody BA, Hunter E. 1992. Mutations within the env gene of Mason-Pfizer monkey virus: effects on protein transport and SU-TM association. J. Virol. 66:3466–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chopra HC, Mason MM. 1970. A new virus in a spontaneous mammary tumor of a rhesus monkey. Cancer Res. 30:2081–2086 [PubMed] [Google Scholar]
- 6. Daniel MD, et al. 1984. A new type D retrovirus isolated from macaques with an immunodeficiency syndrome. Science 223:602–605 [DOI] [PubMed] [Google Scholar]
- 7. Dendouga N, Callebaut I, Tomavo S. 2002. A novel DNA repair enzyme containing RNA recognition, G-patch and specific splicing factor 45-like motifs in the protozoan parasite Toxoplasma gondii. Eur. J. Biochem. 269:3393–3401 [DOI] [PubMed] [Google Scholar]
- 8. Entin-Meer M, Avidan O, Hizi A. 2003. The mature reverse transcriptase molecules in virions of mouse mammary tumor virus possess protease-derived sequences. Virology 310:157–162 [DOI] [PubMed] [Google Scholar]
- 9. Ernst RK, Bray M, Rekosh D, Hammarskjold ML. 1997. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 17:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fine DL, et al. 1975. Responses of infant rhesus monkeys to inoculation with Mason-Pfizer monkey virus materials. J. Natl. Cancer Inst. 54:651–658 [PubMed] [Google Scholar]
- 11. Frenal K, et al. 2006. Structural and functional characterization of the TgDRE multidomain protein, a DNA repair enzyme from Toxoplasma gondii. Biochemistry 45:4867–4874 [DOI] [PubMed] [Google Scholar]
- 12. Gifford R, Kabat P, Martin J, Lynch C, Tristem M. 2005. Evolution and distribution of class II-related endogenous retroviruses. J. Virol. 79:6478–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gunter P, et al. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1:649–659 [DOI] [PubMed] [Google Scholar]
- 14. Herrmann G, et al. 2007. Conserved interactions of the splicing factor Ntr1/Spp382 with proteins involved in DNA double-strand break repair and telomere metabolism. Nucleic Acids Res. 35:2321–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herschhorn A, Hizi A. 2010. Retroviral reverse transcriptases. Cell. Mol. Life Sci. 67:2717–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hizi A, Herschhorn A. 2008. Retroviral reverse transcriptases (other than those of HIV-1 and murine leukemia virus): a comparison of their molecular and biochemical properties. Virus Res. 134:203–220 [DOI] [PubMed] [Google Scholar]
- 17. Jensen EM, Zelljadt I, Chopra HC, Mason MM. 1970. Isolation and propagation of a virus from a spontaneous mammary carcinoma of a rhesus monkey. Cancer Res. 30:2388–2393 [PubMed] [Google Scholar]
- 18. Lebaron S, et al. 2009. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 28:3808–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lin ML, et al. 2009. Involvement of G-patch domain containing 2 overexpression in breast carcinogenesis. Cancer Sci. 100:1443–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newman RM, et al. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc. Natl. Acad. Sci. U. S. A. 103:19134–19139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perach M, Hizi A. 1999. Catalytic features of the recombinant reverse transcriptase of bovine leukemia virus expressed in bacteria. Virology 259:176–189 [DOI] [PubMed] [Google Scholar]
- 22. Rhee SS, Hunter E. 1990. Structural role of the matrix protein of type D retroviruses in Gag polyprotein stability and capsid assembly. J. Virol. 64:4383–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rumlova M, Benedikova J, Cubinkova R, Pichova I, Ruml T. 2001. Comparison of classical and affinity purification techniques of Mason-Pfizer monkey virus capsid protein: the alteration of the product by an affinity tag. Protein Expr. Purif. 23:75–83 [DOI] [PubMed] [Google Scholar]
- 24. Silverman EJ, et al. 2004. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 24:10101–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song C, Hunter E. 2003. Variable sensitivity to substitutions in the N-terminal heptad repeat of Mason-Pfizer monkey virus transmembrane protein. J. Virol. 77:7779–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stansell E, et al. 2007. Basic residues in the Mason-Pfizer monkey virus Gag matrix domain regulate intracellular trafficking and capsid-membrane interactions. J. Virol. 81:8977–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Svec M, Bauerova H, Pichova I, Konvalinka J, Strisovsky K. 2004. Proteinases of betaretroviruses bind single-stranded nucleic acids through a novel interaction module, the G-patch. FEBS Lett. 576:271–276 [DOI] [PubMed] [Google Scholar]
- 28. Tannukit S, et al. 2009. Identification of a novel nuclear localization signal and speckle-targeting sequence of tuftelin-interacting protein 11, a splicing factor involved in spliceosome disassembly. Biochem. Biophys. Res. Commun. 390:1044–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taube R, Loya S, Avidan O, Perach M, Hizi A. 1998. Reverse transcriptase of mouse mammary tumour virus: expression in bacteria, purification and biochemical characterization. Biochem. J. 332(Pt 3):807–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vlach J, et al. 2008. D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein. Proc. Natl. Acad. Sci. U. S. A. 105:10565–10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walbott H, et al. 2010. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 29:2194–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yoshimoto R, Kataoka N, Okawa K, Ohno M. 2009. Isolation and characterization of post-splicing lariat-intron complexes. Nucleic Acids Res. 37:891–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zabranska H, et al. 2007. The role of the S-S bridge in retroviral protease function and virion maturation. J. Mol. Biol. 365:1493–1504 [DOI] [PubMed] [Google Scholar]
- 34. Zabransky A, et al. 1998. Three active forms of aspartic proteinase from Mason-Pfizer monkey virus. Virology 245:250–256 [DOI] [PubMed] [Google Scholar]