Abstract
The small mRNA (SmRNA) of all Bunyaviridae encodes the nucleocapsid (N) protein. In 4 out of 5 genera in the Bunyaviridae, the smRNA encodes an additional nonstructural protein denominated NSs. In this study, we show that Andes hantavirus (ANDV) SmRNA encodes an NSs protein. Data show that the NSs protein is expressed in the context of an ANDV infection. Additionally, our results suggest that translation initiation from the NSs initiation codon is mediated by ribosomal subunits that have bypassed the upstream N protein initiation codon through a leaky scanning mechanism.
INTRODUCTION
The Andes virus (ANDV), a rodent-borne hantavirus member of the Bunyaviridae family of viruses, is the major etiological agent of hantavirus cardiopulmonary syndrome (HCPS) in South America (34). Like all Bunyaviridae members, ANDV features a tripartite genome consisting of three different negative-polarity single-stranded RNA segments, designated large (L), medium (M), and small (S), packed into helical nucleocapsids (18, 36). The L and M segments encode the RNA polymerase and a glycoprotein precursor that is cotranslationally processed to yield two envelope glycoproteins (Gc and Gn). The small mRNA (SmRNA) of all hantaviruses, including ANDV, encodes the nucleocapsid (N) protein, yet 4 out of 5 genera in the Bunyaviridae family encode an additional nonstructural protein, designated NSs (16, 35, 51).
In some, but not all, members of the Bunyaviridae, the SmRNA encompasses an overlapping (+1) open reading frame (ORF) for NSs (9, 32, 35, 41). The NSs protein is variable in size, ranging from 83 to 109 residues, among viruses known to encode it (8). NSs is described as a nonessential protein, but when expressed it contributes to viral pathogenesis, acting mainly as an interferon antagonist through diverse mechanisms depending upon the virus in question (2, 4, 16, 28, 29, 46, 52). Several additional functions have been ascribed to this viral protein, for example, the NSs protein of San Angelo virus and Bunyamwera virus (BUNV) have been shown to play a role in shutting off host cell protein synthesis (4, 6, 12). Among the hantaviruses, the NSs protein recently has been identified in Puumala virus (PUUV)- and Tula virus (TULV)-infected cells (16, 51). The SmRNAs of other hantaviruses, such as Prospect Hill virus (PHV), Sin Nombre virus (SNV), and ANDV, have a putative NSs ORF (31, 35), and the expression of the protein has been predicted (17, 31, 35, 47) yet remains to be shown.
In this study, we present evidence suggesting that the overlapping (+1) open reading frame (ORF) found in the ANDV SmRNA encodes an NSs protein. The expression of the NSs protein in the context of a viral infection is shown. Using virus-like mRNAs designed to mimic the ANDV SmRNA, we determined that the translation of the NSs ORF relies on a leaky scanning mechanism from the upstream N initiation codon.
MATERIALS AND METHODS
Monoclonal antibodies against ANDV NSs.
A peptide corresponding to the C-terminal end of the ANDV NSs (residues 32 to 63 of ANDV strain CHI-7913; GenBank accession no. 30313863 [47]), SGNSRDNLQIWWQLKNWLQNQLIQQGLSLMTI, was synthesized and purified by high-performance liquid chromatography (HPLC) over a QC column from New England Peptide Inc. (Gardner, MA). The purity of the peptide reached 87%. Monoclonal antibodies were prepared as previously described (20). Positive hybridoma cell lines were identified by an enzyme-linked immunosorbent assay (ELISA) in which their respective supernatants showed reactivity against the NSs peptide but not against negative-control peptides. Anti-NSs antibodies were further classified based on their ability to recognizing native or denatured recombinant NSs protein using a strip immunoblot assay (SIA) vacuum blot test. SIA was conducted as previously described (11) but using native or heat-denatured affinity-purified recombinant glutathione S-transferase (GST)-NSs protein. Native or heat-denatured GST or recombinant GST-N protein was used as a negative control. SIAs were developed using a chemiluminescent reaction as described below. The protocol used to express the GST recombinant proteins is described below.
Cells and virus.
HeLa (CCL-2), 293T (CRL-11268), NIH 3T3 (CRL-1658), and Vero E6 (Vero C1008; ATCC CRL 156) cells were grown in Dulbecco's modified Eagle medium (DMEM; HyClone, Logan, UT) containing 10% bovine fetal serum (HyClone, Logan, UT), 1% amphotericin B, and 1% penicillin-streptomycin (Gibco BRL, Life Technologies Corporation, Carlsbad, CA) at 37°C in a 5% CO2 atmosphere. ANDV strain CHI-7913 was propagated in Vero E6 cells as previously described (10, 11, 30). Supernatants of Vero E6 cells were used to infect naïve cells at a multiplicity of infection (MOI) of 1 for 2 h, followed by an extensive wash, as previously described (11, 30).
Plasmids.
The dual-luciferase (dl) plasmids dl hepatitis C virus (HCV) and dl poliovirus (PV) have been described previously (1, 37). The dl PV plasmid was kindly provided by Nahum Sonenberg (Department of Biochemistry, McGill University, Canada). The plasmid harboring the full-length S segment of ANDV (accession number NC_003466) was kindly provided by S. C. St. Jeor (Department of Microbiology, University of Nevada) and has been described previously (31). The firefly luciferase (FLuc) reporter gene was recovered from the dual-luciferase reporter plasmid described in reference (3) by PCR using primers mFluc-1/mFluc-2 (Table 1). The amplicon was cloned into the pGemT-easy vector (Promega Corporation, Madison, WI), and the FLuc fragment was recovered by digestion with EcoRI and XbaI and cloned into the pcDNA3.1(−) myc/His B expression plasmid (Invitrogen, Carlsbad, CA), generating mFLuc plasmid. The ANDV SmRNA 5′ untranslated region (UTR; from nucleotide [nt] 1 to 127; GenBank accession no. NC_003466) was recovered by PCR and cloned upstream of FLuc in plasmid mFLuc between the NheI and EcoRI sites. Since the translation initiation codons of N (AUGN) and NSs (AUGNSs) are not in frame, primers 1AUG-W and 2AUGn-W (Table 1) were used to leave the AUGN codons in frame with the reporter, generating plasmid N-DNA (Table 2), while primers 1AUG-W and 2AUGnss-W (Table 1) were used to leave the AUGNSs codons in frame with FLuc, generating plasmid NSs-DNA (AUGNSs W/W in Table 2). The construction of plasmids for the expression of RNAs harboring translation initiation codons in different nucleotide contexts was achieved by PCR using a series of different primers described in Tables 1 and 2. To obtain the plasmid for expressing the N or NSs protein from ANDV, the N gene was amplified by PCR from the ANDV SmRNA using primers NGSTf/NGSTr or Nf/Nr (Table 1), while the NSs gene was amplified using NSsGSTf/NSsGSTr (Table 1). The amplicon generated using NGSTf/NGSTr or NSsGSTf/NSsGSTr was cloned into the pGemT-easy plasmid (Promega). The fragments were recovered by digestion with BamHI and EcoRI and cloned into the pGEX-6p-1 expression plasmid (GE Healthcare, Piscataway, NJ) used to generate the GST fusion proteins. The amplicon generated using the Nf/Nr primers was cloned directly into plasmid pcDNA4/HisMaxTopo (Invitrogen). Unexpectedly, we found that the wild-type NSs yields poor protein expression in eukaryotic cells, therefore an oligonucleotide harboring the NSs gene but with codons optimized for its expression in eukaryotic cells was designed: 5′-ATGCCCAGAAGGCAGTGGAGGTGGACCCGGATGACCCTGACAAGAGCACACTACAAAATCGACGGGCAGCTGTGTCTGCACTGGAGACCAAACAGCGAGAACCTGAGAGGCAACTTGCAGATTTGGTGGCAGCTCAAAAATTGGCTGCAAAACCAGTTGATCCAACAGGGCTTGAGCCTGATGATCATCTAA-3′. The synthetic oligonucleotide was used as the template for a PCR using primers NSsOCf/NSsOCr (Table 1). The amplicon was directly cloned into plasmid pcDNA4/HisMaxTopo (Invitrogen). The N and NSs expression constructs used the initiation codon for the expression of the His-N or His-NSs protein with pcDNA4/HisMaxTopo. The expression of a single protein from each plasmid was confirmed by Western blotting as described below. All plasmids used in this study were sequenced (Macrogen Corp., Rockville, MD). The plasmid expressing Renilla luciferase (RLuc) was constructed by introducing the RLuc gene, recovered from the pRenilla vector previously described (44), into the pCI-neo vector (Promega).
Table 1.
Primers used in this study
| Name | Orientation | Sequencea (5′ to 3′) |
|---|---|---|
| 1AUG-Kb | Sense | GCTAGCTAGTAGTAGACTCCTTGAGAAGCTACTGCTGCGAAAGCTGGAATGgGC |
| 1AUG-W | Sense | GCTAGCTAGTAGTAGACTCCTTGAGAAGC |
| 1AUG-Xc | Sense | GCTAGCTAGTAGTAGACTCCTTGAGAAGCTACTGCTGCGAAAGCTGGActcAGC |
| 2AUGnss-Kd | Antisense | GAATTCCGcCATCCTTAAGCTTTTGCCGAGC |
| 2AUGnss-W | Antisense | GAATTCCGGCATCCTTAAGCTTTTGCCGAGC |
| 2AUGn-W | Antisense | GAATTCGGCGGCATCCTTAAGCTTTTGCCGAGC |
| 2AUGn-Xe | Antisense | GAATTCGGCGGgagCCTTAAGCTTTTGCCGAGC |
| 2AUGnss-Xe | Antisense | GAATTCCGGgagCCTTAAGCTTTTGCCGAG |
| mFLuc1 | Sense | GAATTCgtgGAAGACGCCAAAAACATAAAGAAAGG |
| mFLuc2 | Antisense | TCTAGATTACACGGCGATCTTTCCGCCC |
| Nf | Sense | ACGGGATCCATGAGCACCCTCCAAGAATTG |
| Nr | Antisense | ACGGAATTCCTACAACTTAAGTGGCTCTTGG |
| NSsGSTf | Sense | ACGGGATCCATGCCGAGAAGGCAGTGGAGG |
| NSsGSTr | Antisense | ACGGAATTCTTAGATGATCATCAGGCTCAAGC |
| NGSTf | Sense | ACGGGATCCATGAGCACCCTCCAAGAATTGC |
| NGSTr | Antisense | ACGGAATTCCTACAACTTAAGTGGCTCTTGG |
| NSsOCf | Sense | GGATCCATGCCCAGAAGGCAGTGGAGG |
| NSsOCr | Antisense | GAATTCTTAGATGATCATCAGGCTCAAGC |
| F-HantaS | Sense | ACACGAACAACAGCTCGTGAC |
| R-HantaR | Antisense | AGGCTCAAGCCCTGTTGGATC |
| FlucS | Sense | ACTTCGAAATGTCCGTTCGG |
| FLucAS | Antisense | GCAACTCCGATAAATAACGCG |
| GAPDHS | Sense | TCCACCACCCTGTTGCTGTAG |
| GAPDHAS | Antisense | ACCCACTCCTCCACCTTTGAC |
Mutations are indicated in lowercase.
The A in position +4 with respect to the ATG of N was replaced with G.
The ATG of N was replaced with CTC.
The T in position +4 with respect to the ATG of NSs was replaced with G.
The ATG of NSs was replaced with CTC.
Table 2.
Plasmids used to evaluate the initiation codon contexta
| Plasmid name | Primer | AUG in frame with FLuc |
|---|---|---|
| N-DNA | 1AUG-W/2AUGn-W | AUGN |
| W/W | 1AUG-W/2AUGnss-W | AUGNSs |
| X/W | 1AUG-X/2AUGnss-W | AUGNSs |
| X/K | 1AUG-X/2AUGnss-K | AUGNSs |
| W/K | 1AUG-W/2AUGnss-K | AUGNSs |
| K/W | 1AUG-K/2AUGnss-W | AUGNSs |
| K/K | 1AUG-K/2AUGnss-K | AUGNSs |
| W/X | 1AUG-W/2AUG-Xnss | AUGNSs |
| X/X | 1AUG-X/2AUG-Xnss | AUGNSs |
W, wild-type nucleotide context of AUGN (A in position +4) and/or AUGNSs (U in position +4). K, Kozak context (G in position +4). X, replacement of AUG with CUC.
Protein expression and purification.
The GST, GST-NSs, and GST-N proteins were expressed in the Escherichia coli BL21-CodonPlus (DE3)-RIPL bacterial strain (Stratagene, La Jolla, CA). Bacteria expressing the recombinant proteins, which were previously induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h, were lysed on ice using lysis buffer (3 mg of lysozyme in 50 mM Tris, pH 8.0, 200 mM NaCl, 1 mM EDTA, 5 mM dithiothreitol [DTT], 1.5 mM N-lauryl sarcosine, and protease inhibitors [Complete protease inhibitor cocktail tablets; Roche Applied Science, Mannheim, Germany]). Lysates were sonicated and clarified by centrifugation. The supernatant was combined with 1 ml of fresh glutathione-Sepharose 4B beads (GE Healthcare) and rotated at 4°C for 3 h. The beads then were washed three times with washing buffer (50 mM Tris, pH 8.0, 1 mM EDTA, 1% Triton X-100, and 200 mM NaCl). Elutions were obtained in 50 mM Tris, pH 8.0, 10 mM glutathione. Protein concentration and integrity were determined by a Bio-Rad protein assay (Bio-Rad Laboratories Inc., Hercules, CA) and SDS-PAGE, respectively.
In vitro transcription.
Plasmids N-DNA, NSs-DNA, or their variants were linearized with XbaI (Fermentas, Vilnius, Lithuania). Linear DNA (1 μg) was used to program in vitro transcription reactions using 3 mM DTT, 0.5 mM MgCl2, 10 mM rATP, 10 mM recombinant CTP (rCTP), 10 mM rUTP, 2 mM rGTP, Ribomax buffer (Promega), 20 U of RNasin (Promega), 1.28 mM 7mGpppG cap analogue (Promega), and 10 U of T7 RNA polymerase (Fermentas) at 37°C for 2 h. Template DNA was digested with DNase RQ1 (Promega), and the RNA was precipitated with 2.8 M LiCl. RNA was resuspended in nuclease-free water and its concentration determined spectrophotometrically by a Nanodrop (Nanodrop Technology, Wilmington, DE). RNA integrity was confirmed by electrophoresis on denaturing agarose gels.
In vitro translation.
Nuclease-treated Flexi rabbit reticulocyte lysate (RRL) (Promega) (50%, vol/vol, supplemented with 20 μM amino acids [Promega] and 8 U Ribolock RNase inhibitor [Fermentas, Vilnius, Lithuania]) were programmed with 0.06 pmol RNA, and reactions were performed for 90 min at 30°C using previously described salt conditions (44). For assays containing edeine, a kind gift from Ian Brierley (Division of Virology, Department of Pathology, University of Cambridge, United Kingdom), nuclease-treated RRL was incubated in the presence or absence of different concentrations of the drug at 30°C for 10 min prior to use for in vitro translations assays. Luciferase, FLuc, and RLuc activities were measured using the DLR assay system (Promega) on a Sirius single-tube luminometer (Lumat 9507; Berthold Detection Systems GmbH, Pforzheim, Germany) as previously described (1, 49).
DNA transfection.
Cells were seeded at 55 × 103 cells/well in 48-well culture plates or at 110 × 103 cells/well in 12-well culture plates. DNA transfection was performed at 60% confluence by the JetPEI system (Polyplus-Transfection SA, Illkirch, France) according to the manufacturer's protocols. After 24 h the culture medium was removed, and the cells were directly mixed with the passive lysis buffer supplied with the DLR assay system (Promega). The protein concentration was determined by a Bradford assay using the Bio-Rad protein assay (Bio-Rad Laboratories, Inc.). FLuc and RLuc activities were measured as described above. For DNA transfection assays, 100 ng of plasmid N-DNA or NSs-DNA plus 30 ng of a plasmid expressing RLuc (as a transfection normalizer) were transfected. For DNA transfection assays to evaluate the effect of the context of the AUGN or AUGNSs codon, 200 ng of each plasmid together with 30 ng of a plasmid expressing RLuc were transfected as described above. For DNA transfection assays with the plasmid expressing ANDV N or NSs protein, 100 ng of plasmid N-DNA or NSs-DNA plus 30 ng of plasmid expressing RLuc were transfected as described above. The total amount of transfected DNA was constant. When needed, pcDNA4/HisMaxTopo (empty vector) was used to complete the total DNA content. Luciferase activities were determined as indicated above.
RNA extraction and RT-qPCR.
Total RNA was extracted from cells 24 h posttransfection and used in a real-time quantitative PCR (RT-qPCR) to establish the content of FLuc and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA by following well-established protocols (38). Cells were trypsinized (Gibco BRL) and resuspended in DMEM (Gibco BRL) containing 10% bovine fetal serum (HyClone). Cells were collected by centrifugation at 710 × g for 5 min and washed three times in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 × 7H2O, 1.4 mM KH2PO4, pH 7.4) at 4°C. For RNA extraction, cells were resuspended in 200 μl of cold RLNa buffer (10 mM Tris-HCl, pH 8, 10 mM NaCl, 3 mM MgCl2, 1 mM DTT, 0.5% NP-40, and 10 U/ml of Riboblock [Fermentas]). After a 5-min incubation on ice, lysed cells were centrifuged for 2 min at 400 × g at 4°C, and the supernatant then was recovered. One milliliter of TRIzol (Invitrogen) then was added to the supernatant, and RNAs were extracted by following the protocol provided by the manufacturer. Cytoplasmic RNAs were treated with RQ1 DNase (Promega) to avoid DNA contamination. For RT-qPCR, cDNAs first were synthesized using 200 ng of cytoplasmic RNAs and the qScript master mix (Quanta Biosciences, Inc., Gaithersburg, MD) by following the supplier's instructions. A 20-μl reaction mixture was prepared with 5 μl of template cDNA (1/10 diluted), 10 μl of Faststart universal SYBR green kit with Rox (Roche Diagnostics Corporation, Indianapolis, IN), and 0.2 mM each primer, and then it was subjected to amplification using a fluorescence thermocycler (Step-One-Plus; Applied Biosystems, Foster City, CA) under the following conditions: 5 min at 95°C for initial denaturation, followed by 40 cycles of denaturation at 95°C for 15 s and annealing and elongation at 60°C for 30 s. This program was followed by a melting curve analysis to verify the specificity of the PCR product. Firefly luciferase was amplified in parallel with the housekeeping gene GAPDH, and relative copy numbers of firefly cDNAs were compared to those of GAPDH using x − ΔCT (where x corresponds to the experimentally calculated amplification efficiency of each primer couple and CT is the cyclic threshold). The primer sequences used to amplify FLuc (FlucS and FlucAS) (Table 1) and GAPDH (GAPDHS and GAPDHAS) (Table 1) were designed using Beacon designer software (Premier Biosoft International, Palo Alto, CA) and were validated in a previous study (38).
For ANDV S-RNAdetection, total RNA was extracted using the High Pure viral nucleic acid kit (Roche Applied Science) by following the manufacturer's protocol. Recovered RNA was resuspended in 25 μl of elution buffer (Roche Applied Science), and its concentration was determined spectrophotometrically (Nanodrop Technology). About 50 ng of total RNA (3 μl of resuspended RNA) was amplified with the SuperScript III one-step RT-PCR and platinum Taq polymerase (Invitrogen) using primers F-HantaS and R-HantaR (Table 1), which target the nucleocapsid coding region (S genomic RNA) as previously described (11, 30).
Detection of ANDV proteins by Western blotting and immunofluorescence (IF).
For Western blot analysis, cells were collected by centrifugation and lysed with radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 0.5% NP-40, and protease inhibitors [Roche Applied Science]). The protein concentration was determined by a Bradford assay as described above, and 20 μg of total protein was resolved on a 15% tricine–SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Hybond-P; GE Healthcare). The membrane was blocked, washed, and incubated with an anti-polyhistidine monoclonal antibody (H1029; Sigma-Aldrich, St. Louis, MO) at a 1:3,000 dilution. An anti-mouse horseradish peroxidase (HRP)-conjugated antibody (074-1806; KPL Inc., Gaithersburg, MD) was used as a secondary antibody at a dilution of 1:10,000. For protein detection, the SuperSignal West Pico chemiluminescent kit (Thermo Fisher Scientific, Rockford, IL) was used.
Vero E6 cells grown on coverslips were transfected (2 μg of DNA/well in a 6-well plate) with ANDV protein expressing a plasmid (NSs-DNA or N-DNA) or infected with ANDV strain CHI-7913 as described above. At given times according to the experiment, cells were washed with 1× PBS–0.05% Tween 20 and fixed with 4% paraformaldehyde (PFA) at room temperature for 15 min, washed, blocked for 1 h at room temperature in PBS containing 1% bovine serum albumin (BSA), and permeabilized with 1% BSA and 0.3% Triton X-100 for intracellular staining. In these assays, an anti-His monoclonal antibody (H1029; Sigma-Aldrich) and an anti-His polyclonal antibody (CAPPEL 59257; MP Biomedicals, Solon, OH) were used. The anti-His antibodies were used at a 1:200 dilution and were incubated for 2 h, while the anti-NSs monoclonal antibody at a 1:200 dilution was incubated overnight. The detection of the ANDV N proteins was conducted using a well-characterized monoclonal anti-N antibody at a 1:300 dilution (11, 30, 48) or a previously described polyclonal anti-N antibody at a 1:800 dilution (kindly provided by M. Ferres, Laboratorio de Infectología, Departamento de Pediatría, Facultad de Medicina, Pontificia Universidad Católica de Chile) (10). The cells then were washed three times with 1× PBS and incubated for 30 min with the secondary antibodies using a goat anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody (172-1806; KPL Inc.) and a goat anti-rabbit Alexa Fluor 555-conjugated antibody (A-21429; Invitrogen) at a 1:200 dilution in PBS. Cell nuclear staining with DAPI (710301; KPL Inc.) was performed for 7 min at room temperature (1 μg/ml final concentration). Slides were mounted (S3023; fluorescent mounting medium; Dako Denmark A/S) prior to analysis. Photographs were taken using a high-resolution IEEE 1394 FireWire digital charge-coupled device color camera with high-speed real-time viewing and a MicroPublisher 3.3 RTV camera in conjunction with an Olympus microscope at 40× and digitally analyzed using QCapture Pro 6 software (QImaging, Surrey, British Columbia, Canada) as previously described (11).
Bioinformatic analysis.
The nucleotide sequences of the ANDV smRNA (GenBank NC_003466) were analyzed using the Bioedit shareware program (version 7.0.9; Ibis Bioscience, Carlsbad, CA).
RESULTS
Expression and detection of a recombinant tagged ANDV NSs protein.
Several reports, based exclusively on sequence analysis, predicted the expression of the NSs protein in TULA, PUUV, and ANDV from the SmRNA (17, 31, 35). Interestingly, these predictions have been partially confirmed, as the SmRNA of TULA and PUUV have been shown to express the NSs protein (16, 51). Intrigued by the possible expression of NSs in ANDV, we undertook an analysis of ANDV SmRNA. In agreement with a previous report, the analysis of the ANDV SmRNA (GenBank accession no. NC_003466) revealed an (+1) ORF initiating 76 nucleotides downstream from the N protein initiation codon, designated AUGN (nt +3 of the AUGN to nt +1 of the putative AUGNSs) (35). However, one major obstacle to demonstrate the expression of the ANDV NSs was the lack of commercially available or published antibodies against this putative viral protein. For this purpose, anti-ANDV NSs monoclonal antibodies were developed as detailed in Materials and Methods. Two NSs antibodies, NS2-2B11/C2 and NS2-5E7/D9, were selected for this study. The first antibody detected denatured recombinant GST-NSs protein, while the second one identified the native recombinant GST-NSs protein (Materials and Methods and data not shown).
Before testing the anti-ANDV NSs antibody in the context of a viral infection, we evaluated its ability to specifically detect a recombinant ANDV NSs protein in Vero E6 cells. The selection of the cell type was based on the fact that it is commonly used for ANDV propagation (10, 11, 30). For this experiment, an expression plasmid developed to encode a His-tagged version of the putative ANDV NSs protein was used (pNSs-ANDV in Fig. 1A). The plasmid was transfected into Vero E6 cells, and the presence of the recombinant version of the putative viral protein was analyzed by Western blotting using an anti-His antibody (Fig. 1B, left) or the newly developed anti-NSs monoclonal antibody (Fig. 1B, right), followed by an anti-mouse HRP-conjugated antibody as indicated in Materials and Methods. Results showed that the recombinant version of the ANDV NSs protein was expressed and that it could be readily detected by both the anti-His (Fig. 1B, left) or the newly developed anti-NSs monoclonal antibody (Fig. 1B, right). Having established an expression system for a recombinant version of the putative ANDV NSs protein, we next sought to evaluate if the second anti-NSs antibody was suitable for detecting the recombinant ANDV NSs protein in an indirect immunofluorescence (IF) assay.
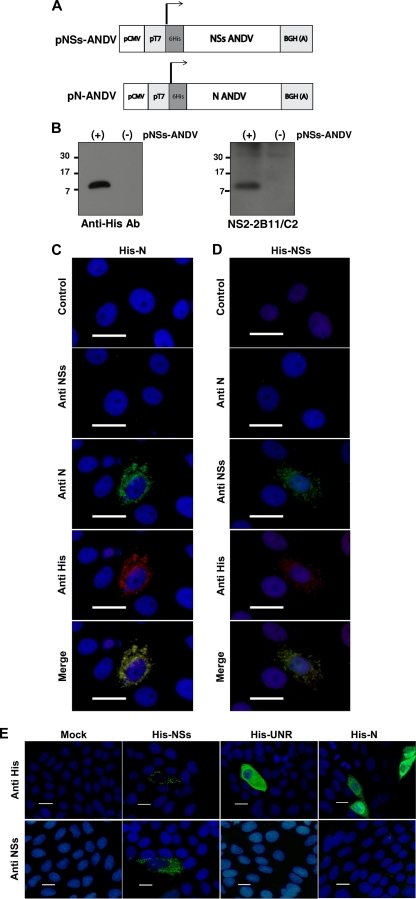
Fig 1.
Detection of a transiently expressed recombinant His-ANDV NSs protein in Vero E6 cells. (A) Schematic representation of the plasmids used to express the recombinant tagged ADNV NSs and ANDV N proteins. The arrows indicate the initiation codon used to generate the recombinant proteins. BGH(A) corresponds to the bovine growth hormone polyadenylation sequences. (B) Plasmid pNSs-ANDV, expressing the recombinant His-ANDV NSs protein, was transfected in Vero E6 cells, and 48 h posttransfection total proteins were extracted. Shown is the expression of the recombinant His-ANDV NSs protein detected by Western blotting using a commercial anti-His monoclonal antibody (left) or the anti-NSs monoclonal antibody NS2-2B11/C2 (right). (C and D) Vero E6 cells transfected with plasmid pN-ANDV (C) or plasmid pNSs-ANDV (D) were fixed, and the expression of the His-tagged protein was detected by immunofluorescence using a commercial polyclonal anti-His antibody, anti-NSs monoclonal antibody NS2-5E7/D9, or a well-characterized anti-ANDV N monoclonal antibody (11, 30, 48). A goat anti-mouse fluorescein isothiocyanate (FITC)-conjugated antibody (green staining) and a goat anti-rabbit Alexa Fluor 555-conjugated antibody (red staining) were used as secondary antibodies. Cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; blue staining). (E) Vero E6 cells transfected with pNSs-ANDV, pN-ANDV, and the His-tagged irrelevant tagged protein upstream of N-ras (His-UNR) were fixed, and the expression of the His-tagged proteins was detected by immunofluorescence using a commercial anti-His monoclonal antibody, followed by an FITC-labeled anti-mouse antibody (green staining). The recombinant His-tagged ANDV NSs protein was specifically detected by the anti-NSs monoclonal antibody NS2-5E7/D9, followed by an FITC-labeled anti-mouse antibody (green staining). Cell nuclei were stained with DAPI (blue staining). For all panels in the figure, size bars corresponds to 20 μm.
As an experimental control, Vero E6 cells were transfected with plasmid pN-ANDV (Fig. 1A), which encodes a His-tagged version of the ANDV N protein. Transfected cells were fixed, and the expression of the recombinant version of the ANDV N protein was evaluated using a commercially available polyclonal anti-His antibody and a well-characterized monoclonal anti-ANDV N antibody (11, 30, 48) as described in Materials and Methods (Fig. 1C). As expected, the ANDV N protein was detected by both the polyclonal anti-His antibody and the monoclonal anti-ANDV N antibody but not by NS2-5E7/D9, the monoclonal anti-ANDV NSs antibody (Fig. 1C). Additionally, stains of the polyclonal anti-His antibody and the monoclonal anti-ANDV N antibody colocalized, suggesting that antibodies recognize the same protein (Fig. 1C, merged image). Based on these findings, a new set of Vero E6 cells was transfected with plasmid pNSs-ANDV (Fig. 1A). Cells were fixed and the expression of the recombinant version of the putative ANDV NSs protein was evaluated using the same commercially available polyclonal anti-His antibody and the NS2-5E7/D9 monoclonal antibody (Fig. 1D). The recombinant version of the putative ANDV NSs protein was detected by both the polyclonal anti-His and the monoclonal NS2-5E7/D9 antibodies but not by the anti-ANDV N monoclonal antibody (Fig. 1D). The fluorescence of the polyclonal anti-His and monoclonal NS2-5E7/D9 antibodies colocalized (Fig. 1D, merged image), suggesting that both recognize the same protein. A granular staining of recombinant His-tagged ANDV NSs was observed in the cytoplasm of transiently transfected cells when detected with either the polyclonal anti-His antibody or with the anti-NSs monoclonal antibody (Fig. 1D).
To further our results, a new series of experiments were conducted using a different anti-His antibody. For this, Vero E6 cells were transfected with the pNSs-ANDV vector, cells were fixed, and the expression of the recombinant version of the putative ANDV NSs protein was evaluated using a commercially available monoclonal anti-His antibody or the NS2-5E7/D9 antibody, followed by an anti-mouse FITC-conjugated antibody (Materials and Methods). As shown in Fig. 1E, the recombinant version of the putative ANDV NSs protein again was detected with both antibodies. In agreement with what was observed when using the polyclonal anti-His antibody (Fig. 1C), a granular staining of the recombinant His-tagged ANDV NSs was observed in the cytoplasm of transiently transfected cells when detected with either the monoclonal anti-His-antibody or with the newly developed anti-NSs monoclonal antibody (Fig. 1E). As an additional control, cells were transfected with plasmid pN-ANDV (Fig. 1A) or a plasmid encoding the irrelevant His-tagged protein upstream of N-ras (His-UNR). In accordance with what was previously observed (Fig. 1D), antibody NS2-5E7/D9 was shown to be specific for the NSs recombinant protein and to have no cross-reactivity with the recombinant His-ANDV N protein or the irrelevant tagged protein (Fig. 1E).
Taken together, these results indicate that when expressed in cells, the tagged NSs protein can be detected using the developed anti-NSs monoclonal antibodies (Fig. 1B, D, and E).
Detection of the ANDV proteins and RNA in infected cells.
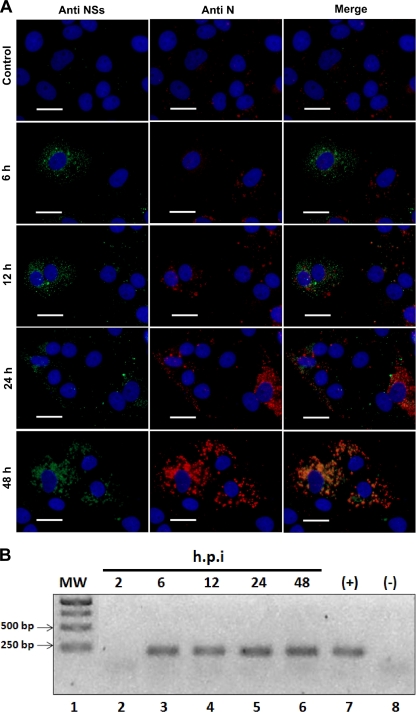
We sought to determine if the NSs protein is expressed in the context of an ANDV infection. A recent report documents the expression of the TULA NSs protein in infected cells (51). Interestingly, the expression of the TULA NSs protein was observed in early stages of viral infection (51). Based on these observations, we decided to evaluate the early expression of the ANDV NSs protein. For this, Vero E6 cells were infected with the ANDV CHI-7913 strain as previously described (10, 11, 30). At 6, 12, 24, and 48 h postinfection (hpi), cells were fixed and the presence of the ANDV NSs protein was evaluated by IF using antibody NS2-5E7/D9, followed by an FITC-labeled anti-mouse secondary antibody as described above (Fig. 2A). At 6, 12, 24, and 48 hpi, ANDV NSs protein was readily detected (Fig. 2A). As when transiently expressed (Fig. 1C), a granular staining of ANDV NSs was observed in the cytoplasm of ANDV-infected Vero E6 cells (Fig. 2A). As an additional control for virus infection, we confirmed the expression of the ADNV-N protein at the same time points (Fig. 2A) and evaluated the presence of the viral S-RNAs (genomic S-RNA, SmRNA, and the positive-sense antigenome small cytoplasmic RNA [scRNA]) in cells at 2, 6, 12, 24, and 48 hpi (Fig. 2B). As shown in Fig. 2A, the ADNV N protein also was detected at all time points. Consistently with the IF data (Fig. 2A), the presence of ANDV S-RNAs in infected Vero E6 cells was confirmed by an ANDV-specific RT-PCR (Fig. 2B) (11, 30). Taken together, these results strongly suggest that the ANDV NSs protein is indeed expressed during ANDV infection.
Fig 2.
Detection of the ANDV NSs protein in infected Vero E6 cells. Vero E6 cells were infected with the ANDV CHI-7913 strain as previously described (10, 11, 30). At 6, 12, 24, and 48 h postinfection (p.i.), cells were fixed and the presence of the ANDV NSs protein was evaluated by IF using antibody NS2-5E7/D9, followed by an FITC-labeled anti-mouse secondary antibody (green staining). In parallel, the ANDV N protein was detected using a well-characterized polyclonal antibody (10) followed by an Alexa Fluor 555-labeled anti-rabbit secondary antibody (red staining). The cellular nucleus was stained with DAPI (blue staining). Size bars correspond to 20 μm. (B) Total RNA was extracted from ANDV-infected cells 2 (lane 2), 6 (lane 3), 12 (lane 4), and 24 h (lane 5) postinfection (h.p.i) and used as the template in an RT-PCR designed to specifically amplify the viral S-RNAs (genomic S-RNA, smRNA, and the positive-sense antigenome scRNA) (11, 30). This assay also included a positive RT-PCR control that consisted of RNA extracted from the supernatant initially used to infect Vero E6 cells (viral stock, lane 7) and a negative RT-PCR control which corresponds to water (lane 8). MW indicates the molecular size marker (1 kb; lane 1; Fermentas).
The putative AUGNSs present in the ANDV S mRNA is recognized as an initiation codon in the context of virus-like mRNA.
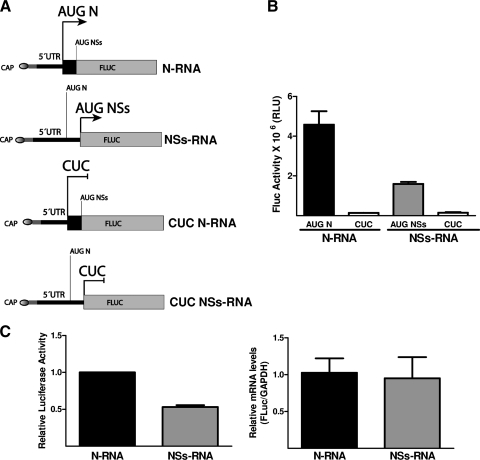
The data presented above suggest that the ANDV NSs is expressed in the context of a viral infection (Fig. 2). Prompted by these observations, we were interested in understanding the molecular mechanism by which the NSs initiation codon is recognized by the host translational machinery in order to drive NSs protein expression. To this end, we constructed a series of plasmids encoding virus-like mRNAs designed to mimic the ANDV SmRNA (Fig. 3A). In these constructs, we took advantage of the natural (+1) shift in the reading frame that exists between the N and the putative NSs ORF (35). In the N RNA, the 5′UTR of the SmRNA from nt +1 to the AUGNSs codon was inserted upstream of the firefly luciferase reporter gene (FLuc), and the reporter was set in frame with the initiation codon of the N ORF, AUGN (depicted by the thick arrow in Fig. 3A). FLuc expression therefore is dependent on the recognition of AUGN as a functional initiation codon. In the NSs RNA, the FLuc reporter was set in frame with the putative initiation codon of the NSs ORF, AUGNSs (depicted by the thick arrow in Fig. 3A). In the NSs RNA, the AUGN codon is out of frame (+1) with respect to the reporter gene; therefore, the expression of FLuc is dependent on the recognition of AUGNSs as a functional initiation codon. In both RNAs the expression of firefly luciferase (FLuc) is expected to be exclusively dependent on the AUGN or AUGNSs initiation codon.
Fig 3.
NSs ORF can be translated in the context of virus-like smRNAs. (A) Schematic representation of the capped monocistronic RNA constructs that mimic the ANDV S mRNA. To faithfully mimic the viral mRNA, these RNAs contain 21 additional nucleotides (5′-GGGAGACCCAAGCTGGCTAGC-3′; gray region) between the 5′ cap structure (oval at the 5′ end of the RNA) and the first nucleotide of the ANDV smRNA sequence (black line) (50). Additionally, the viral NSs ORF has been replaced by the firefly luciferase (FLuc) reporter gene. The arrow indicates the initiation codon used. In the N RNA, the N initiation codon is in frame with FLuc. In the NSs RNA, the putative NSs initiation codon is in frame with FLuc. In the CUC RNAs, the AUGN or AUGNSs codons have been mutated to CUC. (B) The N RNA (black bar) and NSs RNA (gray bar) were translated in a commercial in vitro translation system as indicated in Materials and Methods. FLuc activity (in relative light units [RLU]) was measured as indicated in Materials and Methods. Unshaded bars correspond to FLuc activity obtained with CUC mutations. Values are the means ± standard deviations (SD) from three independent experiments, each conducted in triplicate. (C) DNA plasmids coding for the RNAs depicted in panel A plus an RLuc-expressing plasmid (transfection efficiency control) were transfected into HeLa cells. Total RNA and protein were extracted and analyzed. RNA was used to assess the total amount of the virus-like RNAs in cells by an RT-qPCR assay (right panel). The quantity of N RNA was arbitrarily set to 1, and the total amount of NSs RNA recovered from cells was expressed relative to this value (right panel). FLuc and RLuc activities were determined, and FLuc activity was normalized to RLuc activity (left panel). Normalized N RNA FLuc activity was arbitrarily set to 1 (left panel). Values shown in both panels are the means ± SD from three independent experiments each conducted in triplicate.
To evaluate if the AUGNSs codon can be recognized as an initiation codon within the context of a virus-like mRNA, rabbit reticulocyte lysate (RRL) was programmed using in vitro-transcribed capped N and NSs RNAs, and an in vitro translation reaction was performed (Fig. 3B). Results show that FLuc was expressed from the N RNA, confirming that the in vitro-generated RNAs were functional. Additionally, data show that the NSs initiation codon (AUGNSs) can be recognized, allowing the expression of the FLuc reporter (Fig. 3B), albeit to a lesser extent than that of the AUGN codon (Fig. 3B). In both N and NSs RNAs, where the AUGN and AUGNSs initiation codons were mutated to CUC (Fig. 3A), no FLuc activity was observed, thus confirming the absence of nonspecific reporter protein synthesis (Fig. 3B).
To further confirm the in vitro data, DNA plasmids coding for each tested RNA were transfected together with a constant quantity of a plasmid coding for RLuc into HeLa cells. Total RNA was extracted and used to assess the expression of the virus-like RNAs. Results showed that the N and NSs RNAs were expressed at equivalent levels in HeLa cells, as confirmed by RT-qPCR (Fig. 3C, right). Total proteins were extracted and luciferase activities determined. RLuc activity was used to normalize the assay for variability in plasmid transfection efficiencies. As shown in Fig. 3C (left), the AUGN initiation codon is recognized and leads to the expression of FLuc in HeLa cells. In agreement with the in vitro data shown above, in cells FLuc encoded by the NSs RNA was expressed, albeit to a lesser extent than that from the upstream AUGN codon (Fig. 3C). It should be noted that due to the experimental design, the 5′UTR of the NSs-expressing plasmid is longer than the virus mRNA or the RNA used in the in vitro assay. However, this difference apparently does not affect the ratio of expression from the AUGN and AUGNSs codons (Fig. 3B and C). Similar results were obtained when DNAs encoding the N and NSs RNAs were transfected into Vero E6, 293T, and NIH 3T3 cell lines (data not shown). These results show that in the context of the ANDV-like SmRNAs, the AUGNSs codon is recognized as a functional initiation codon leading to the expression of the FLuc reporter.
Translation from the AUGN and AUGNSs codons displays high sensitivity to edeine.
Edeine, a strongly basic, linear oligopeptide antibiotic, interferes with AUG initiation codon recognition by scanning 40S-eIF2-GTP/Met-tRNAi complexes, precluding the attachment of 60S subunits and therefore the formation of the 80S ribosome (7, 14, 25). At low concentrations, edeine inhibits translation initiation in RRL with no effect on polypeptide elongation (13). We reasoned that the use of this drug in in vitro translation assays would shed some light on the possible mechanism used by 40S ribosomal subunits to recognize the AUGNSs codon. As a positive control for the activity of edeine in our experimental setup and to establish a suitable dose for the drug, we employed RRL programmed with bicistronic dual luciferase (dl) mRNAs containing the internal ribosome entry site (IRES) elements from poliovirus (PV) or the hepatitis C virus (HCV) in the intercistronic spacer region (Fig. 4A). As predicted, the cap-dependent translation of the first cistrons of both dl PV IRES (Fig. 4B) and dl HCV IRES (Fig. 4C) mRNAs was inhibited in the presence of 250 and 500 nM edeine. The expression of the downstream cistron mediated by the PV IRES also was inhibited (Fig. 4B), which is consistent with existing evidence that this IRES requires AUG start codon recognition by scanning ternary complexes (43). In contrast, translation initiation mediated by the HCV IRES remained poorly affected at 250 nM edeine (Fig. 4C), which is consistent with the reported ability of this IRES to directly recruit 40S ribosomal subunits to the initiation codon independently of initiation factors or canonical scanning (26). The partial inhibition of HCV IRES activity at 500 nM edeine probably reflects an effect on processes other than translation initiation (26). Taken together, these observations confirm edeine's efficacy in our experimental system.
Fig 4.
Translation initiation from the N and NSs initiation codons is sensitive to edeine. (A) Schematic representation of a bicistronic mRNA similar to those used in this study. RNAs correspond to the dual-luciferase (dl) reporter construct containing an upstream Renilla luciferase gene (RLuc) and a downstream firefly luciferase gene (FLuc). IRES activity was monitored using the FLuc activity as the readout, while the RLuc reporter gene served as a control for cap-dependent translation initiation. Two dl RNAs were used in this study, one harboring the poliovirus (PV) IRES (37) and the other harboring the hepatitis C virus (HCV) IRES (1). dl PV (B) or dl HCV (C) bicistronic RNA was translated in RRL in the presence of the translation initiation inhibitor edeine, which interferes with AUG start codon recognition by scanning eIF2-GTP/Met-tRNAiMet complexes (7, 14, 25, 26). The relative RLuc and FLuc activity for each RNA in the absence of edeine was arbitrarily set to 1 (± SD). Values are the means ± SD from three independent experiments each conducted in triplicate. (D) Monocistronic ANDV S-like mRNAs depicted in Fig. 1A were translated in RRL in the presence of edeine. The relative FLuc activity for each RNA in the absence of edeine was arbitrarily set to 1 (± SD). Values are the means ± SD from at least three independent experiments, each conducted in triplicate.
We next sought to evaluate the effect of edeine on translation driven by the ANDV SmRNA 5′UTR. Monocistronic mRNAs that contained the 5′UTR of ANDV (Fig. 3A) were incubated in RRL with increasing amounts of edeine. Results show that translation from both initiation codons, AUGN and AUGNSs, was sensitive to edeine (Fig. 4D), suggesting that the recognition of both initiation codons requires scanning ternary complexes.
Translation from the AUGNSs codon proceeds by leaky scanning from the upstream AUGN initiation codon.
For eukaryotic mRNAs, one of the most crucial elements for the optimal recognition of an initiation codon by a scanning 40S ribosomal subunit is the AUG context (21, 22). The ideal initiation context is defined by a purine at position −3 and guanine at position +4 (21, 22). The analysis of the nucleotides surrounding the AUGN (GGA AUGN A) and AUGNSs (AGG AUGNSs C) codons (underlining indicates initiation codons) in the ANDV smRNA (GenBank accession no. NC_003466) reveals that neither is found in an ideal initiation context (21, 22). Thus, to determine the role of sequence surrounding the AUG codon on translation from both the N and NSs ORFs, a series of point mutations were designed to optimize the AUG context and introduced into the NSs RNA (Table 2). In these constructs K stands for the optimal context (Kozak; A/GNNAUGG, where N is any nucleotide) (21, 22), W stands for the wild-type viral context, and in X the initiation codon has been suppressed by mutating AUG to CUC. In vitro-transcribed RNAs generated from these constructs were used to program the RRL. FLuc activity was analyzed relative to that of W/W NSs RNA, which was arbitrarily defined as 1 (Fig. 5A). Results show that the recognition and translation of FLuc from AUGNSs was increased when the upstream initiation codon was mutated to CUC (Fig. 5A, compare W/W to X/W); thus, the wild-type AUGN codon negatively affects 40S complex recruitment from AUGNSs. In support of this observation, the improvement of AUGN from a W to a K context had a negative effect on initiation from the AUGNSs codon (Fig. 5A, compare W/W to K/W and K/K). It is noteworthy that even though the AUGN codon was in a theoretically optimal K context, translation from the downstream AUGNSs initiation codon was not totally abolished (Fig. 5A, K/W). Optimizing the context of the AUGNSs codon from W to K, however, did not increase activity from the AUGNSs codon (Fig. 5A, compare W/W to W/K, K/W to K/K, and X/W to X/K). As expected, NSs RNAs containing the AUGNSs-CUC mutation exhibited no luciferase activity, confirming the absence of nonspecific FLuc protein synthesis (Fig. 5A, W/X and X/X). Taken together, these observations suggest that the AUGNSs initiation codon is recognized mostly by ribosomes that leak from the upstream AUGN codon.
Fig 5.
Sequence context of the 5′-proximal N initiation codon modulates the recognition of the putative NSs initiation codon. RNAs corresponding to different mutants of the NSs RNA (Table 2) were translated in RRL (A), or plasmid DNA coding for them was transfected into HeLa cells together with an RLuc-expressing plasmid (as a control for the transfection efficiency) (B). W stands for the wild-type context as found in the ADNV smRNA, K stands for the optimal context (purine in position −3 and a G in position +4), and in X the AUG initiation codon has been mutated to CUC. FLuc values were normalized to RLuc activity. The relative FLuc activity for the wild-type (W/W) virus-like RNA was arbitrarily set to 1 (± SD). Values are the means ± SD from three independent experiments each conducted in triplicate.
To extend these observations, plasmids carrying the RNAs were transfected into HeLa cells together with a constant amount of a control plasmid coding for RLuc. Total proteins were extracted and luciferase activities determined. RLuc activity was used to normalize the assay for plasmid transfection efficiency. Normalized data are presented in Fig. 5B. As described above, the FLuc activity exhibited by the W/W NSs RNA was arbitrarily set to 1 (Fig. 5B). Results obtained in cells fully support conclusions drawn from the in vitro assay and show that the upstream AUGN codon negatively affects initiation from the downstream AUGNSs (Fig. 5B, compare W/W, W/K, and K/K to X/W and X/K). Taken together, these sets of in vitro (Fig. 5A) and ex vivo (Fig. 5B) data support the conclusion that translation initiation from the AUGNSs codon proceeds by leaky scanning.
Thus, we propose that initiation from the AUGNSs codon relies primarily on scanning ribosomes that have bypassed the upstream AUGN by a leaky scanning mechanism.
DISCUSSION
Sequence analysis of the SmRNA segment reveals that some hantaviruses exhibit an overlapping (+1) ORF for the putative NSs (35) in addition to the ORF for the N protein. Intrigued by these observations and by the fact that several studies have predicted, but not demonstrated, the expression of the NSs protein in ANDV (17, 31, 35, 47), we generated ANDV NSs antibodies to determine if the putative NSs protein could be detected during the ANDV replication cycle. These anti-NSs antibodies were evaluated using a recombinant version of the putative ANDV NSs (Fig. 1). Using these new tools, we showed that the ANDV NSs is expressed in the context of a viral infection (Fig. 2). Having established that the ANDV NSs protein is expressed during ANDV infection, we were interested in understanding the molecular mechanism by which the initiation codon driving the ANDV NSs protein was recognized during mRNA translation. The ANDV SmRNA exhibits an overlapping (+1) ORF for the putative NSs in addition to the ORF for the N protein; thus, to facilitate experiments and their interpretation, we decided to replace the NSs coding region with a firefly luciferase reporter gene (FLuc) placed in frame with the NSs or N initiation codon (Fig. 3A). This experimental approach enables the independent evaluation of the N and NSs initiation codons. Using this strategy, we studied the translation properties of the N and NSs 5′ untranslated regions (5′UTRs) of the ANDV SmRNA. Both in vitro and in cells, we found that the NSs initiation codon was recognized by the translation initiation machinery and that the ORF corresponding to NSs, replaced by FLuc, is indeed expressed (Fig. 3). The sensitivity of translation initiation to edeine (Fig. 4) suggests that the AUGNSs codon is recognized by scanning ribosomes (26, 33, 54). Further analysis based on the optimization of the nucleotide context surrounding the N and NSs initiation codon (Fig. 5) indicates that the recognition of the NSs initiation codon occurs mainly by 40S ribosomal subunits that have bypassed the 5′-proximal N initiation codon by a mechanism known as leaky scanning.
The scanning model of translation initiation predicts that ribosomes should initiate at the first AUG codon encountered by a scanning 40S subunit (23). For the vast majority of mRNAs, initiation indeed usually occurs at the 5′-proximal AUG codon in a favorable context (22). However, the first encountered AUG codon can be bypassed if it is present in a nonoptimal context, and leaky scanning is said to account for initiation by the 40S subunit at a downstream AUG (23). Leaky scanning is widely encountered in viral RNAs, where it presumably helps optimize the coding capacity (5, 23, 39, 40, 42, 45, 55, 56). In support of a mechanism of leaky scanning, the context optimization of the upstream AUGN initiation codon had a negative effect on translation from the downstream AUGNSs (Fig. 5, K/W and K/K). Taken together, these observations suggest that scanning 40S ribosomal subunits that bypass the AUGN codon can be recruited at the AUGNSs. This conclusion is further supported by increased translation from the NSs ORF when the AUGN codon is abolished altogether by the CUC mutation (Fig. 5, X/W and X/K). Interestingly, translation initiation from AUGNSs was not enhanced when its sequence context was optimized (Fig. 5, compare X/W to X/K and W/W to W/K), suggesting that in our experimental setting the viral wild-type context is recognized as efficiently as an optimal context by scanning 40S ribosomal subunits. Moreover, results presented in Fig. 5 suggest that not all 40S ribosomal subunits loaded onto the virus-like RNA efficiently recognize the first initiation codon, even when the AUGN codon is in an optimal context. This leakiness from the N initiation codon in the face of a favorable context cannot be readily explained, although it may be related to a 5′UTR of less than optimal length (24).
Further studies are needed to fully understand the relevance of the ANDV NSs protein during the viral replication cycle. Its role is hard to predict, as the reported function of known NSs proteins differs greatly among the different members of the Bunyaviridae family of viruses (6, 15, 19, 27, 51, 53). Nonetheless, this report describing the expression of a new viral protein is expected to further propel research regarding the function of ANDV proteins during the viral replication cycle.
ACKNOWLEDGMENTS
We thank M. Rau (Oxford, United Kingdom) for the critical reading and editing of the manuscript. We are grateful to Ian Brierley (Division of Virology, Department of Pathology, University of Cambridge, United Kingdom) for kindly providing edeine. We thank Stephen C. St. Jeor (Department of Microbiology, University of Nevada) for kindly providing the plasmid containing the full-length ANDV S segment. We thank Nahum Sonenberg (Department of Biochemistry, McGill University, Canada) for providing the bicistronic construct harboring the poliovirus IRES.
This study was supported by the Proyecto Instituto Milenio P09/016-F IMII, Financiado con Fondos Programa ICM, and PHS grant 2U01AI045452-11 to M.L.-L.; by CONICYT through grant FONDECYT 1100756 to N.T. and M.L.-L.; and by a travel grant from MECESUP-USACH to J.V.-O. J.V.-O. was supported by an MECESUP-USACH doctoral fellowship, and L.S. is a recipient of a Ph.D. fellowship from CONICYT, Chile. J.V.-O. and L.S. conducted this work as part of their Ph.D. theses. R.S.-R. was supported by an M2/doctoral fellowship from CONICYT and the French Embassy in Chile. E.P.R. was supported by grants from Fondation pour la Recherche Medicale (FRM) and the French Ministry (MENRT).
Footnotes
Published ahead of print 7 December 2011
REFERENCES
- 1. Barría MI, et al. 2009. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 37:957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Billecocq A, et al. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798–9806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brasey A, et al. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 77:3939–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bridgen A, Weber F, Fazakerley JK, Elliott RM. 2001. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 98:664–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chenik M, Chebli K, Blondel D. 1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 69:707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colón-Ramos DA, et al. 2003. Inhibition of translation and induction of apoptosis by bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell 14:4162–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dinos G, et al. 2004. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site RNA binding. Mol. Cell 13:113–124 [DOI] [PubMed] [Google Scholar]
- 8. Dunn EF, Pritlove DC, Elliott RM. 1994. The S RNA genome segments of Batai, Cache Valley, Guaroa, Kairi, Lumbo, Main Drain and Northway bunyaviruses: sequence determination and analysis. J. Gen. Virol. 75:597–608 [DOI] [PubMed] [Google Scholar]
- 9. Fuller F, Bhown AS, Bishop DH. 1983. Bunyavirus nucleoprotein, N, and a non-structural protein, NSS, are coded by overlapping reading frames in the S RNA. J. Gen. Virol. 64:1705–1714 [DOI] [PubMed] [Google Scholar]
- 10. Galeno H, et al. 2002. First human isolate of Hantavirus (Andes virus) in the Americas. Emerg. Infect. Dis. 8:657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Godoy P, et al. 2009. Andes virus antigens are shed in urine of patients with acute hantavirus cardiopulmonary syndrome. J. Virol. 83:5046–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hart TJ, Kohl A, Elliott RM. 2008. Role of the NSs protein in the zoonotic capacity of orthobunyaviruses. Zoonoses Public Health 56:285–296 [DOI] [PubMed] [Google Scholar]
- 13. Hunt T. 1974. The control of globin synthesis in rabbit reticulocytes. Ann. N. Y. Acad. Sci. 241:223–231 [DOI] [PubMed] [Google Scholar]
- 14. Hunter AR, Jackson RJ, Hunt T. 1977. The role of complexes between the 40-S ribosomal subunit and Met-tRNA-Met-f in the initiation of protein synthesis in the wheat-germ system. Eur. J. Biochem. 75:159–170 [DOI] [PubMed] [Google Scholar]
- 15. Ikegami T, Peters CJ, Makino S. 2005. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J. Virol. 79:5606–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jääskeläinen KM, et al. 2007. Tula and Puumala hantavirus NSs ORFs are functional and the products inhibit activation of the interferon-beta promoter. J. Med. Virol. 79:1527–1536 [DOI] [PubMed] [Google Scholar]
- 17. JÄäskeläinen KM, Plyusnina A, Lundkvist A, Vaheri A, Plyusnin A. 2008. Tula hantavirus isolate with the full-length ORF for nonstructural protein NSs survives for more consequent passages in interferon-competent cells than the isolate having truncated NSs ORF. Virol. J. 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khaiboullina SF, Morzunov SP, St. Jeor SC. 2005. Hantaviruses: molecular biology, evolution and pathogenesis. Curr. Mol. Med. 5:773–790 [DOI] [PubMed] [Google Scholar]
- 19. Kohl A, et al. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999–8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Köhler G, Milstein C. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495–497 [DOI] [PubMed] [Google Scholar]
- 21. Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kozak M. 1981. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 9:5233–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozak M. 2002. Pushing the limits of the scanning mechanism for initiation of translation. Gene 299:1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozak M. 1991. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1:111–115 [PMC free article] [PubMed] [Google Scholar]
- 25. Kozak M, Shatkin AJ. 1978. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 253:6568–6577 [PubMed] [Google Scholar]
- 26. Lancaster AM, Jan E, Sarnow P. 2006. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12:894–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Le May N, et al. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550 [DOI] [PubMed] [Google Scholar]
- 28. Le May N, et al. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Léonard VH, Kohl A, Hart TJ, Elliott RM. 2006. Interaction of Bunyamwera orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J. Virol. 80:9667–9675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marsac D, et al. 2011. Infection of human monocyte-derived dendritic cells by ANDES hantavirus enhances pro-inflammatory state, the secretion of active MMP-9 and indirectly enhances endothelial permeability. Virol. J. 8:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meissner JD, Rowe JE, Borucki MK, St. Jeor SC. 2002. Complete nucleotide sequence of a Chilean hantavirus. Virus Res. 89:131–143 [DOI] [PubMed] [Google Scholar]
- 32. Mohamed M, McLees A, Elliott RM. 2009. Viruses in the Anopheles A, Anopheles B, and Tete serogroups in the Orthobunyavirus genus (family Bunyaviridae) do not encode an NSs protein. J. Virol. 83:7612–7618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Odon OW, Kramer G, Henderson AB, Pinphanichakarn P, Hardesty B. 1978. GTP hydrolysis during methionyl-tRNAf binding to 40 S ribosomal subunits and the site of edeine inhibition. J. Biol. Chem. 253:1807–1816 [PubMed] [Google Scholar]
- 34. Pini N. 2004. Hantavirus pulmonary syndrome in Latin America. Curr. Opin. Infect. Dis. 17:427–431 [DOI] [PubMed] [Google Scholar]
- 35. Plyusnin A. 2002. Genetics of hantaviruses: implications to taxonomy. Arch. Virol. 147:665–682 [DOI] [PubMed] [Google Scholar]
- 36. Plyusnin A, Vapalahti O, Vaheri A. 1996. Hantaviruses: genome structure, expression and evolution. J. Gen. Virol. 77:2677–2687 [DOI] [PubMed] [Google Scholar]
- 37. Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 273:14002–14007 [DOI] [PubMed] [Google Scholar]
- 38. Ricci EP, et al. 2009. Translation of intronless RNAs is strongly stimulated by the Epstein-Barr virus mRNA export factor EB2. Nucleic Acids Res. 37:4932–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ryabova LA, Pooggin MM, Hohn T. 2006. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 119:52–62 [DOI] [PubMed] [Google Scholar]
- 40. Schaecher SR, Mackenzie JM, Pekosz A. 2007. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 81:718–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmaljohn CS, Jennings GB, Hay J, Dalrymple JM. 1986. Coding strategy of the S genome segment of Hantaan virus. Virology 155:633–643 [DOI] [PubMed] [Google Scholar]
- 42. Schwartz S, Felber BK, Fenyo EM, Pavlakis GN. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J. Virol. 64:5448–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonenberg N, Meerovitch K. 1990. Translation of poliovirus mRNA. Enzyme 44:278–291 [DOI] [PubMed] [Google Scholar]
- 44. Soto Rifo R, Ricci EP, Decimo D, Moncorge O, Ohlmann T. 2007. Back to basics: the untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 35:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stacey SN, et al. 2000. Leaky scanning is the predominant mechanism for translation of human papillomavirus type 16 E7 oncoprotein from E6/E7 bicistronic mRNA. J. Virol. 74:7284–7297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas D, et al. 2004. Inhibition of RNA polymerase II phosphorylation by a viral interferon antagonist. J. Biol. Chem. 279:31471–31477 [DOI] [PubMed] [Google Scholar]
- 47. Tischler ND, et al. 2003. Complete sequence of the genome of the human isolate of Andes virus CHI-7913: comparative sequence and protein structure analysis. Biol. Res. 36:201–210 [DOI] [PubMed] [Google Scholar]
- 48. Tischler ND, Rosemblatt M, Valenzuela PD. 2008. Characterization of cross-reactive and serotype-specific epitopes on the nucleocapsid proteins of hantaviruses. Virus Res. 135:1–9 [DOI] [PubMed] [Google Scholar]
- 49. Vallejos M, et al. 2010. The 5′-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Res. 38:618–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vera-Otarola J, et al. 2010. The 3′ untranslated region of the Andes hantavirus small mRNA functionally replaces the poly(A) tail and stimulates cap-dependent translation initiation from the viral mRNA. J. Virol. 84:10420–10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Virtanen JO, Jaaskelainen KM, Djupsjobacka J, Vaheri A, Plyusnin A. 2010. Tula hantavirus NSs protein accumulates in the perinuclear area in infected and transfected cells. Arch. Virol. 155:117–121 [DOI] [PubMed] [Google Scholar]
- 52. Weber F, et al. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Weber F, Dunn EF, Bridgen A, Elliott RM. 2001. The Bunyamwera virus nonstructural protein NSs inhibits viral RNA synthesis in a minireplicon system. Virology 281:67–74 [DOI] [PubMed] [Google Scholar]
- 54. Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102:511–520 [DOI] [PubMed] [Google Scholar]
- 55. Xu K, et al. 2009. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology 388:279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Y, Hussain S, Wang H, Ke M, Guo D. 2009. Translational control of the subgenomic RNAs of severe acute respiratory syndrome coronavirus. Virus Genes 39:10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]