Abstract
Dengue virus (DENV) affects millions of people, causing more than 20,000 deaths annually. No effective treatment for the disease caused by DENV infection is currently available, partially due to the lack of knowledge on the basic aspects of the viral life cycle, including the molecular basis of the interaction between viral components and cellular compartments. Here, we characterized the properties of the interaction between the DENV capsid (C) protein and hepatic lipid droplets (LDs), which was recently shown to be essential for the virus replication cycle. Zeta potential analysis revealed a negative surface charge of LDs, with an average surface charge of −19 mV. The titration of LDs with C protein led to an increase of the surface charge, which reached a plateau at +13.7 mV, suggesting that the viral protein-LD interaction exposes the protein cationic surface to the aqueous environment. Atomic force microscopy (AFM)-based force spectroscopy measurements were performed by using C protein-functionalized AFM tips. The C protein-LD interaction was found to be strong, with a single (un)binding force of 33.6 pN. This binding was dependent on high intracellular concentrations of potassium ions but not sodium. The inhibition of Na+/K+-ATPase in DENV-infected cells resulted in the dissociation of C protein from LDs and a 50-fold inhibition of infectious virus production but not of RNA replication, indicating a biological relevance for the potassium-dependent interaction. Limited proteolysis of the LD surface impaired the C protein-LD interaction, and force measurements in the presence of specific antibodies indicated that perilipin 3 (TIP47) is the major DENV C protein ligand on the surface of LDs.
INTRODUCTION
Dengue virus (DENV) causes the most important arthropod-borne human viral disease, with 2.5 billion people at risk, 100 million infections, and more than 20,000 deaths annually, primarily in tropical developing countries (20). Four genetically distinct serotypes (DENV1 to DENV4) have been identified, which are transmitted among humans through the bite of an infected mosquito of the genus Aedes. DENV belongs to the family Flaviviridae, together with other important human pathogens, such as yellow fever virus (YFV), West Nile virus (WNV), and hepatitis C virus (HCV). The clinical manifestations of DENV infection range from a mild illness to a severe and potentially life-threatening disease for which no treatment is available so far, due at least in part to the limited understanding of the molecular mechanisms that underlie the interaction between DENV and its host cells.
The DENV genome is a single-stranded positive-sense RNA molecule of approximately 11 kb that is translated from a single open reading frame, generating a polyprotein associated with the endoplasmic reticulum (ER) membrane (34). The polyprotein is cleaved co- and posttranslationally by cellular and viral proteases into three structural proteins (capsid [C], premembrane [prM], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The DENV particle structure consists of an internal nucleocapsid composed of C protein complexed with the viral genome, enveloped by a lipid bilayer with which E and M proteins associate (26).
The DENV C protein is essential for virus assembly, ensuring the specific encapsidation of the viral genome. It has 100 amino acid residues and forms dimers in solution, which are believed to be the building blocks for the nucleocapsid assembly (23). The DENV C protein is a very basic protein due to its 26 Arg or Lys residues and only 3 negatively charged residues. It also contains a 21-amino-acid hydrophobic segment, spanning residues 45 to 65, which is required for the maturation and assembly of the viral particles (30). The three-dimensional structures of the C proteins of DENV and WNV have been solved by nuclear magnetic resonance (NMR) and X-ray crystallography, respectively (16, 28). Both structures are similar, with each monomer containing four α-helices (α1 to α4) connected by short loops. The α1 helix shows different orientations in DENV and WNV proteins, suggesting that this part of the protein is more flexible. Most of the positive charges are concentrated in one face of the dimer, formed by the α4 helices of each monomer (α4-α4′), while the opposite face, formed by α1-α1′ and α2-α2′ helices, is composed largely of apolar residues. This asymmetric charge distribution in the DENV C protein led other authors to propose that the α4-α4′ region would interact with viral RNA, whereas the α2-α2′ region would bind to the viral membrane (28).
For several RNA viruses, the genome replicates in association with cellular membranes, which are generally modified during infection (33). In the case of DENV, clusters of double-membrane vesicles, probably originating from the ER, have been detected in infected hepatic cells (47). Different viral nonstructural proteins as well as double-stranded RNA, a marker of virus replication, were found to colocalize with these structures. On the other hand, the DENV C protein was found to associate with lipid droplets (LDs), intracellular organelles that originate from the ER (39). Interestingly, the interaction between the C protein and LDs was shown to be essential for virus replication (39).
LDs occur in virtually all cell types. They were initially considered lipid deposits, but they are now regarded as dynamic structures involved in different cellular processes, some of them related to human diseases such as obesity, inflammation, cancer, and several infectious diseases (6, 31). LDs have a globular shape, and their size can vary in most mammalian cells from 0.1 to 2 μm (15). They are composed of a hydrophobic core containing neutral lipids, mainly triglycerides and cholesterol esters, surrounded by a phospholipid monolayer (43). Several proteome analyses of LDs allowed the identification of a large number of proteins with functions beyond the involvement in lipid metabolism, reinforcing the multiple biological roles of these organelles (21). Some pathogens promote the formation of LDs in their host cells, including DENV (2, 39) and HCV (3). Besides inducing LD accumulation in infected cells, the replication of both HCV and DENV was found to be dependent on the association of their capsid proteins with these organelles (5, 39, 42). The hydrophobic D2 domain of the HCV core protein was shown to be responsible for the targeting to LDs (4, 22). This domain is missing in the DENV C protein, suggesting that the C protein-LD interaction occurs in a different manner. Interestingly, mutations in two leucine residues located in the DENV C protein α2 helix impaired the association of the protein with LDs, suggesting the involvement of this region (39). However, the molecular basis of the C protein-LD interaction and the properties of this binding remained to be characterized.
In this study, we investigated the interaction between the DENV C protein and LDs using atomic force microscopy (AFM)-based force spectroscopy, zeta potential, and cell biology approaches. The results indicate that the C protein-LD interaction is strong and specific and reveal the requirement of LD surface proteins, mainly perilipin 3 (tail-interacting protein of 47 kDa [TIP47]), for the binding, which is dependent on a high intracellular potassium concentration.
MATERIALS AND METHODS
DENV C protein expression and purification.
The purification protocol used for the DENV C protein was previously described by Jones et al. (23). The protein was expressed in Escherichia coli (strain Codon Plus) cells transformed with the DENV serotype 2 C protein gene (encoding residues 1 to 100) cloned into plasmid pET21a. Cells were grown at 37°C and at 200 rpm in LB medium in the presence of 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. One-fifth of the culture grown overnight was transferred into a freshly prepared LB culture in the presence of the same antibiotics. Protein expression was induced at an optical density at 600 nm (OD600) of between 0.8 and 1 with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG), and the cell culture was grown overnight at 18°C and at 200 rpm. Next, the cell culture was centrifuged at 7,000 × g for 20 min at 4°C, and the supernatant was discarded. The pellet was resuspended in 70 ml buffer A (25 mM HEPES [pH 7.4], 200 mM NaCl, 1 mM EDTA, 5% glycerol) and 10 μM protease inhibitor mix (phenylmethylsulfonyl fluoride [PMSF], pepstatin, leupeptin, E64, and bestatin). Cells were lysed by heating and thawing (liquid nitrogen and 42°C water bath) for a total of 10 cycles, followed by 20 cycles of sonication. The cell lysate was then centrifuged at 27,000 × g for 30 min at 4°C. A precipitation of 30% and 60% ammonium sulfate was carried out, followed by centrifugation at 42,000 × g for 20 min at 4°C. The pellets were resuspended with buffer A, combined, and injected onto a MonoS 10/100 GL column (GE Healthcare) coupled to an Akta purifier system (GE Healthcare) at a 0.5-ml/min flow rate. The DENV C protein was eluted with an increasing NaCl concentration gradient (0.1 to 2 M) at a 2-ml/min flow rate. The 3-ml fractions containing the DENV C protein were confirmed by 18% SDS-PAGE. The fractions were then pooled and dialyzed against 50 mM NaH2PO4–1 M NaCl (pH 6). The dialysis step was repeated 3 times with decreasing NaCl concentrations (0.5 M and 0.2 M). The DENV C protein was concentrated with a Centriprep instrument (cutoff, 3,000 Da; Millipore) and stored at −80°C.
Isolation of LDs from HepG2 cells.
The human hepatocellular liver carcinoma cell line (HepG2) was maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM) with 0.01% sodium pyruvate and 4 mM l-glutamine, supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were grown at 37°C in a humidified 5% carbon dioxide incubator. Once approximately 80% confluence was reached, the cells were treated with 0.1 mM oleic acid (Sigma-Aldrich Co., St. Louis, MO). After 24 h, LDs were isolated by use of sucrose gradients. Cells were washed twice and resuspended in 3 ml TEE buffer (20 mM Tris-HCl, 100 mM KCl, 1 mM EDTA, and 1 mM EGTA [pH 7.4]) (39) with a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). Cells were disrupted by nitrogen cavitation at 700 lb/in2 for 20 min at 4°C by using a cell disruption vessel (model 4639; Parr Instrument Company, Moline, IL). The cell lysate was centrifuged at 1,500 × g for 10 min to remove the nuclei, and the supernatant was collected and mixed with an equal volume of TEE buffer containing 1.08 M sucrose. The sample was transferred into a 12-ml ultracentrifugation tube and sequentially overlaid with 2 ml each of 0.270 M sucrose in TEE buffer, 0.135 mM sucrose in TEE buffer, and TEE buffer without sucrose. Following centrifugation at 250,000 × g for 70 min at 4°C, eight fractions of about 1 ml each were collected from the top to the bottom of the gradient. To evaluate cytosolic contamination in the LD fraction, the activity of lactate dehydrogenase (LDH) was measured by using the CytoTox 96 kit (Promega, Madison, WI). Additionally, Western blot analysis for adipose differentiation-related protein (ADRP) or perilipin 2, one of the major proteins on the LD structure (7), was performed on all gradient fractions. The gradient fractions containing LDs were kept at 4°C before use. The Western blot analysis was performed as previously described (39).
LD preparation for AFM measurements.
Fifty microliters of the HepG2 cell LD suspension was placed onto thin freshly cleaved muscovite mica and allowed to deposit for 30 min at room temperature. Nonadherent LDs were removed by 5 sequential washes with TEE buffer. The sample was loaded into the AFM instrument and allowed to equilibrate in TEE buffer for 10 min before imaging or force spectroscopy measurements.
Labeling of LDs with di-8-ANEPPS.
For fluorescent labeling, LDs were incubated with 10 μM di-8-ANEPPS {4-[2-[6-(dioctylamino)-2-naphthalenyl]ethenyl]-1-(3-sulfopropyl)-pyridinium}, Molecular Probes, Invitrogen, Carlsbad, CA] for 2 h at room temperature, without light exposure. After that, samples were deposited onto mica as described above.
AFM imaging.
A NanoWizard II atomic force microscope (JPK Instruments, Berlin, Germany) mounted onto the top of an Axiovert 200 inverted optical microscope (Zeiss, Jena, Germany) was used for imaging and force spectroscopy experiments. The AFM head is equipped with a 15-μm z-range-linearized piezoelectric scanner and an infrared laser. The imaging of LDs was performed with buffer in the tapping mode. Oxidized sharpened silicon tips with a tip radius of 15 nm, a resonant frequency of approximately 73 kHz, and a spring constant of 0.57 N/m were used for imaging. Imaging parameters were adjusted to minimize the force applied onto the topography scanning of the complexes. The scanning speed was optimized to 0.8 Hz, and acquisition points were 512 by 512. Set points on the region between 350 and 390 mV were chosen as an optimum scanning parameter. Imaging data were analyzed with JPK image processing software, v.3 (JPK Instruments, Berlin, Germany).
Functionalization of AFM tips.
Force spectroscopy measurements were performed by using DENV C protein-functionalized tips. For the functionalization, OMCL TR-400-type silicon nitride tips (Olympus, Japan) were cleaned with an intense UV light source and silanized in a vacuum chamber with 3-aminopropyl-triethoxysilane (APTES) (30 μl) and N,N-diisopropylethylamine (10 μl) for 1 h under an argon atmosphere to be coated with a self-assembled monolayer of amines. After this, the probes were rinsed with fresh chloroform and dried with nitrogen gas. The silanization process results in a uniformly distributed self-assembled monolayer of amino-terminated APTES molecules on the AFM tips, which were then placed into a 2.5% (vol/vol) glutaraldehyde solution for 20 min and washed 3 times with TEE buffer (pH 7.4). Finally, the tips were placed into a 167 μM DENV C protein solution during 30 min to covalently bind the protein. The functionalized C protein tips were immediately mounted onto the AFM instrument and used for the force spectroscopy measurements.
Force spectroscopy measurements.
Force spectroscopy measurements were performed by using the softest triangular cantilevers, with a tip radius of 15 nm and a resonance frequency of 11 kHz. Spring constants of the tips were calibrated by the thermal fluctuation method, resulting in values of 22 ± 5 mN/m. For each contact between LDs and the cantilever, the distance between the cantilever and the LD was adjusted in order to maintain an applied force of 200 pN before retraction. Molecular recognition was searched for by pressing the cantilevers intermittently onto different points of LDs adsorbed to the mica surface. Data were collected for each force-distance cycle at 2 μm/s, leading to a loading rate of 4 nN/s. Force curves were analyzed by using JPK image processing software, v.3. For any given experiment, approximately 5,000 force-distance curves were collected, analyzed, and fitted to the worm-like-chain (WLC) model (38). Each experiment was performed at least three times, each time on different samples and with different functionalized tips. Histograms of the (un)binding forces of each studied protein-LD complex were constructed, choosing the ideal bin size to achieve the best-fitted Gaussian model peak forces. The selected binning size was 6 pN. Force rupture values ranging between 0 and 10 pN were considered to represent noise, artifacts, or nonspecific interactions. According to this, values up to 10 pN were neglected for data presentation and analysis. From each histogram, the most likely single C protein molecule rupture force can be determined by fitting the distributions of the rupture forces with the Gaussian model. The maximum values of the Gaussian peaks represent a single-molecule-based statistical measure of the strength of the molecular bond. The experiments were performed at room temperature. The AFM laboratory is strictly maintained at temperatures between 23°C and 25°C. Small variations in temperature at this range did not affect our force spectroscopy measurements, even after a long period.
Measurements were conducted using TEE buffer (pH 7.4) with 10, 100, or 400 mM KCl. To study the effect of the sodium ions, 100 mM NaCl was used in the TEE buffer instead of KCl. The influence of calcium ions was analyzed by removing EDTA and EGTA from the buffer and adding 1.2 mM CaCl2 to the TEE buffer with 100 mM KCl. The effect of phosphate ions was evaluated by changing the TEE buffer to 50 mM phosphate buffer or by incubating the LD suspension with 0.5 or 2 mM sodium tripolyphosphate (TPP) in TEE buffer (with 100 mM KCl) for 15 min before analysis.
Limited proteolysis of LDs was performed by incubating the LD sample with 1, 5, or 10 μM trypsin in TEE buffer (with 100 mM KCl) for 15 min at room temperature. The reaction was stopped by the addition of 1 mM PMSF to the mixture. After 5 min of incubation and subsequent washes with TEE buffer, force spectroscopy experiments were carried out.
As controls for tip modification, measurements with tips at different steps of their functionalization process (including nonfunctionalized tips) were conducted on glass slides, on mica, and on LD samples. Additionally, tips functionalized with LDH from bovine heart at a concentration of 1 mg/ml (Sigma) were used as the experimental control for a protein that does not bind to LDs.
In order to identify the specific protein(s) on the LD surface involved in the binding to the DENV C protein, we performed identical measurements in the presence of three specific guinea pig anti-human antibodies (Progen, Heidelberg, Germany): anti-perilipin (binding residues 507 to 519 of the C terminus), anti-adipophilin/ADRP (adipose differentiation-related protein) (binding C-terminal residues 423 to 437), and anti-TIP47 (tail-interacting protein of 47 kDa) (binding C-terminal residues 420 to 431) antibodies. Perilipin, adipophilin/ADRP, and TIP47 are the three major proteins on the LD surface, belonging to the PAT (for perilipin, ADRP, and TIP47) family of proteins. The adoption of perilipin as a unifying nomenclature for the mammalian PAT family of LD proteins was recently established (25), renaming perilipin, ADRP, and TIP47 as perilipin 1, perilipin 2, and perilipin 3, respectively. The effects of the antibodies on each of these PAT family proteins were evaluated by force spectroscopy after incubating the LD samples with different concentrations of the antibodies (from 0.1 to 100 μg/liter) for 1 h at room temperature. As a negative control for the antibody-blocking experiments, the effect of the presence of an unspecific isotype antibody was also tested (normal guinea pig IgG serum; Santa Cruz Biotechnology, Santa Cruz, CA).
Zeta potential measurements.
Zeta potential (ζ) measurements were carried out with Malvern Zetasizer Nano ZS dynamic light scattering and zeta potential equipment (Malvern, United Kingdom) equipped with an He-Ne laser (λ = 632.8 nm). The zeta potential of LDs was determined at 37°C from the mean of 15 measurements (120 runs each) in the absence and presence of different concentrations of the DENV C protein by phase analysis light scattering (PALS), using disposable zeta cells with platinum gold-coated electrodes (Malvern). The viscosity value and the refractive index were 0.8872 cP and 1.330, respectively. The electrophoretic mobility obtained was used for the zeta potential calculation with the help of the Smoluchowski equation (17):
| (1) |
where u is the electrophoretic mobility, η is the viscosity of the solvent, and ε is its dielectric constant. The variation of the zeta potential (Δζ) for each sample was calculated by subtracting the initial value in the absence of added protein from the zeta potential value of the sample. These differences can be plotted as a function of the DENV C protein concentration, and the experimental data were fitted by using the following equation (8, 10, 24):
| (2) |
where Δζmax is the maximum amplitude of variation of the zeta potential induced by the interaction with the DENV C protein and C1/2 corresponds to the value of the C protein concentration at Δζmax/2, or half of the protein concentration needed for LD saturation.
Confocal microscopy experiments.
HepG2 cells were seeded into 24-well culture dishes containing glass coverslips and grown until they reached a density of 8 × 104 cells/well, when cells were either mock infected or infected with DENV serotype 2 Asiatic strain 16681 (multiplicity of infection [MOI] of 1.0 PFU/cell). After 1 h, cells were washed to remove nonadherent viruses and cultured in DMEM supplemented with 5% fetal bovine serum at 37°C in a humidified 5% carbon dioxide incubator. After 16 h of infection, mock-infected and infected cells were treated with different concentrations of ouabain (Sigma) during 5 h. Just after ouabain treatment, the cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.4) at room temperature for 20 min. Cells were then permeated with 0.1% Triton X-100 for 4 min at room temperature and stained by sequential incubation with an anti-C protein antibody (kindly given by A. Gamarnik, Instituto Leloir, Argentina) at a 1:2,000 dilution, Alexa Fluor 594-conjugated goat anti-rabbit IgG secondary antibody (Molecular probes) at a 1:500 dilution, and Bodipy 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene) (Molecular Probes) at a 1:500 dilution. Cells were mounted onto glass slides, and images were obtained by using a Leica TCS SP5 confocal microscope.
DENV replication and HepG2 cell viability after treatment with ouabain.
The infection of HepG2 cells was performed as described above for confocal microscopy experiments, except that the MOI used was 0.5 PFU/cell. After ouabain treatment, cells were washed and incubated with fresh medium until completing 72 h of infection. HepG2 cell viability was determined by using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. Infectious viral particles released into the culture medium were quantified by a plaque assay on BHK-21 cells (12). For viral RNA quantification, cellular extracts were collected in TRIzol reagent (Invitrogen Life Technologies, CA) according to the manufacturer's instructions. First-strand cDNA was synthesized by using a High-Capacity cDNA Archive kit (Applied Biosystems, CA) according to the manufacturer's instructions, and real-time PCR assays were performed by using TaqMan Master Mix reagents (Applied Biosystems, CA), forward primer ATTAGAGAGCAGATCTCTG, reverse primer TGACACGCGGTTTC, and probe TCAATATGCTGAAACGCG (11).
RESULTS
Isolation of LDs from HepG2 cells.
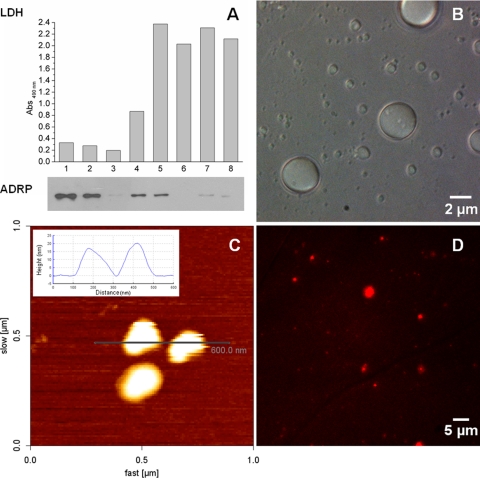
In order to characterize the interaction between LDs and the DENV C protein, it was necessary to obtain isolated LD samples. For this, LD accumulation in a human lineage of hepatic cells, HepG2, was induced by treatment with oleic acid. LDs were subsequently isolated via sucrose gradients after cell disruption. Figure 1A shows the analysis of the eight isolated sucrose gradient fractions by Western blotting for the LD major protein component, adipose differentiation-related protein (ADRP), and by lactate dehydrogenase (LDH) activity. Fractions 1 and 2 were enriched in ADRP, indicating that they correspond to the LD fractions. This was expected, since these were the fractions with the lowest densities. Additionally, these two fractions showed the lowest LDH activity, indicating that no contamination with cytosolic contents occurred. The absence of both ADRP and LDH activity and the low-density property of fraction 3 indicate that it corresponded to the microsomal fraction. The presence of low ADRP levels but high levels of LDH activity in fractions 4 and 5 may indicate that these fractions contain an endoplasmic reticulum membrane, where the synthesis of ADRP occurs and also where LDs biogenesis takes place. All the other isolated fractions (fractions 6 to 8) presented higher levels of LDH activity and no ADRP colocalization, which indicate that the LD-enriched fractions were only the top lowest-density fractions (fractions 1 and 2). Based on these results, the subsequent experiments were performed using fraction 2 to ensure consistent LD structural properties.
Fig 1.
Identification and characterization of isolated HepG2 cell LDs. (A) Western blot analysis of ADRP (perilipin 2) and quantification of the LDH activities in the eight isolated sucrose gradient fractions (see Materials and Methods). (B) Phase-contrast image of LDs of different sizes. (C) Atomic force microscopy scanning image of LDs deposited onto freshly cleaved muscovite mica. The inset shows a cross-section profile of two LDs revealing their diameter (approximately 200 nm) and height (15 to 20 nm). (D) Fluorescence microscopy image of LDs labeled with di-8-ANEPPS.
LDs of HepG2 cells were deposited onto a thin layer of mica and observed by phase-contrast optical microscopy imaging (Fig. 1B), AFM scanning (Fig. 1C), and fluorescence microscopy after di-8-ANEPPS labeling (Fig. 1D). The isolated LDs were round, with a broad size distribution, ranging from 0.1 μm to a few micrometers diameter, in agreement with data from previous reports (45). The inset in Fig. 1C shows a cross-section profile of an AFM scanning image of an LD with a diameter of approximately 200 nm. The lipidic nature of the isolated LDs could be confirmed by successful labeling with di-8-ANEPPS, a lipophilic probe (Fig. 1D).
Assessment of the DENV C protein-LD interaction by force spectroscopy.
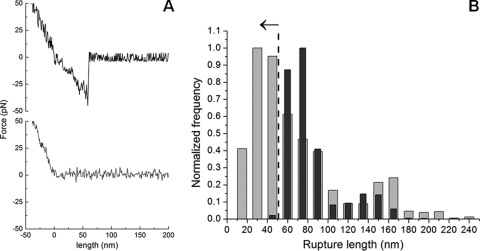
By use of the AFM-based force spectroscopy methodology, it was possible to measure specific interactions between LDs and the DENV C protein by the adhesion profiles obtained for each force curve. Figure 2A shows examples of force curve profiles of a control sample (a nonfunctionalized AFM tip interacting with the mica) (bottom) and of the DENV C protein attached to the AFM tip and LDs (top). By the deflection of the AFM tip, it was possible to measure the (un)binding force between LDs and the DENV C protein. The comparison of the rupture length histograms obtained for the LD interaction with C protein-functionalized AFM tips or for the LD interaction with nonfunctionalized tips showed a partial overlay of the histograms for rupture lengths above 50 nm (Fig. 2B). This can be explained by the fact that with the movement of the AFM tip, some LDs occasionally detach from the mica and adhere to the tip, resulting in binding (and subsequent unbinding) between the LDs attached to the tip and those on the mica surface, which could indeed be observed with the optical microscope (data not shown). To avoid any interference from these events with the force spectroscopy data, ensuring that only the direct interactions between the C protein on the tip and the LDs firmly adherent on mica are considered, only the force-distance events that occurred at a rupture length below 50 nm were taken into consideration for all force spectroscopy data analyses (i.e., LD-LD interactions were therefore excluded).
Fig 2.
Force spectroscopy results for the DENV C protein interaction with LDs. (A) Typical force-extension curves of a control experiment with an unmodified silicon nitride tip interacting with the mica surface (gray line), where adhesion forces were undetectable, and a DENV C protein-functionalized AFM tip and HepG2 cell LDs (black line). (B) Rupture length histograms comparing the unbinding events achieved when a DENV C protein-functionalized AFM tip (lighter gray) or a nonfunctionalized AFM tip (darker gray) interacts with a LD. Histograms were obtained from approximately 5,000 single-event force measurements, carried out with an applied force of 0.2 nN, a pulling speed of 2 μm/s, and a loading rate of 4 nN/s. Once the tip is in contact with the sample surface, the tip-sample separation remains at zero while the force is increased by the cantilever pushing toward the surface. Thus, the zero of length was defined as the point where the tip is no longer in contact with the sample during the retraction of the cantilever.
The distribution of the length of the rupture adhesion events between the DENV C protein and LDs was analyzed by fitting the obtained histogram with the Gaussian model. A maximum at 35 nm was obtained. No background adhesion force between an unmodified silicon nitride tip and the LDs was detected. When a nonfunctionalized tip or a C protein-functionalized tip made contact with the mica, only weak interactions were detected. For the control experiment with LDH functionalized on the AFM tip, only events with a force lower than 20 pN and a frequency of (un)binding events of 17% were obtained (i.e., binding was detected 17% of the time when the DENV C protein-derivatized AFM tip “touched” an LD, requiring an additional force on the pulling of the tip in order to unbind it). These force values correspond to the unspecific binding between LDH and LDs, and it has been taken into consideration during the evaluation of the results achieved for the interaction between the DENV C protein and LDs.
Binding force measurements and effects of ionic strength/potassium concentration.
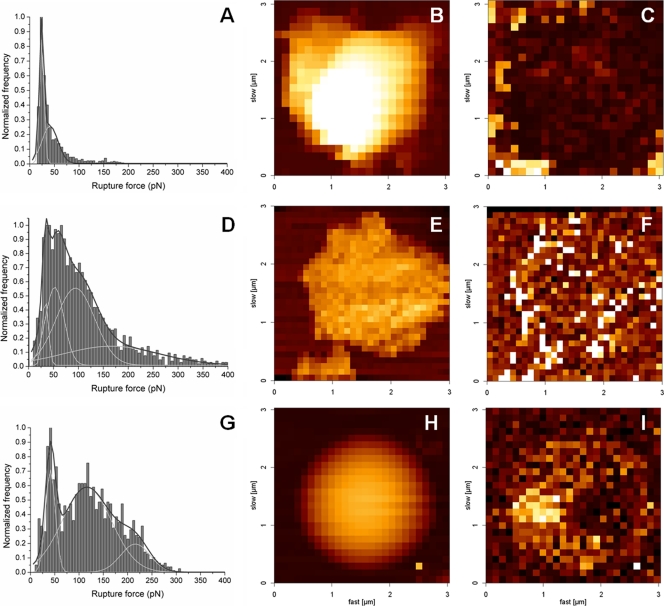
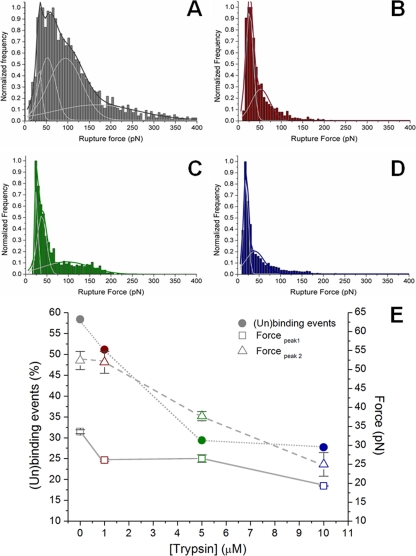
Figure 3 presents the force rupture histogram obtained for the normalized frequency of the forces of the (un)binding events between the C protein-functionalized AFM tip and LDs in the presence of different potassium chloride concentrations. For 100 mM KCl, after applying the Gaussian model to the histogram profile, a force rupture value of 33.6 ± 0.5 pN was obtained, corresponding to the force necessary to break the bond between a DENV C protein dimer and an LD (Fig. 3D). The three other peaks with greater interaction forces (Table 1) correspond to the rupture of multiple bonds due to the interaction of more than one protein dimer attached to the tip with an LD (Fig. 3D). The frequency of the adhesion-rupture events was 58%. Of these adhesion events, 22% corresponded to the force value of the first peak (centered at 33.6 pN), and the remaining 78% were from stronger binding forces or from double- or multiple-step (un)binding events.
Fig 3.
Force rupture histograms and force mapping images at different KCl concentrations. Shown are force rupture histograms of the DENV C protein and LDs in TEE buffer supplemented with 10 mM (A), 100 mM (D), or 400 mM (G) KCl. Each histogram was constructed with the results from approximately 5,000 single rupture force measurements, done with an applied force of 0.2 nN, a pulling speed of 2 μm/s, and a loading rate of 4 nN/s. Peaks were obtained after choosing the best rupture force scale bin and fitting the histograms with the Gaussian model. Shown are force mapping images (height and adhesion) for the three tested ionic strength/KCl concentration conditions, 10 mM (B and C), 100 mM (E and F), or 400 mM (H and I) KCl. These force maps were also obtained with an atomic force microscope by using the force mapping methodology. For all maps, the image is 3 by 3 μm2. Height color scales were adjusted to 800 nm on the height maps (B, E, and H), and force color scales were adjusted to 100 pN on the adhesion maps (C, F, and I). Brighter-colored squares represent regions on the image with higher values of height or adhesion force.
Table 1.
Rupture forces and percentage of (un)binding events under diverse experimental conditionsa
| Experimental condition | % (un)binding events | Rupture force (pN) |
|
|---|---|---|---|
| 1st peak | 2nd peak | ||
| Potassium ([KCl]) | |||
| 10 mM | 34.7 | 24.2 | 40.9 |
| 100 mM | 58.4 | 33.6 | 52.4 |
| 400 mM | 61.0 | 41.0 | 115.8 |
| Trypsin pretreatment ([trypsin] in 100 mM KCl) | |||
| 1 μM | 51.1 | 26.1 | 51.9 |
| 5 μM | 29.4 | 26.6 | 37.7 |
| 10 μM | 27.7 | 19.3 | 25.5 |
| Sodium ([NaCl]), 100 mM | 19.5 | 26.1 | |
| Calcium ([CaCl2] in 100 mM KCl), 1.2 mM | 56.4 | 30.2 | 58.9 |
| Phosphate ([Na2HPO4]), 50 mM | 11.1 | 25.5 | |
| Tripolyphosphate ([Na5P3O10] in 100 mM KCl), 2 mM | 9.6 | 33.4 | |
Average force rupture values obtained for interactions between the DENV C protein and HepG2 cell LDs under the different conditions studied (all with TEE buffer).
In order to evaluate the types of forces involved in the binding between the C protein and LDs, a detailed analysis of the interaction was obtained by electrophoretic mobility and force spectroscopy measurements (Fig. 3 and 4 and Table 1). The percentage of (un)binding events decreased significantly with decreases of the ionic strength of the buffer (from 58% in TEE buffer with 100 mM KCl to 35% in TEE buffer with 10 mM KCl; P < 0.0001), while it slightly increased with TEE buffer with 400 mM KCl (61%; P < 0.05). Also, at a low ionic strength, the force values of the first peak on the histogram decreased significantly (to 24 pN; P < 0.001) (Fig. 3A and Table 1), leading to a significant increase (from 22% to 63%; P < 0.001) in the ratio between the percentage of events of the first peak and the percentage of events of the other peaks (i.e., the ratio between the area of the first peak and the area of the remaining ones). This means that the probability of occurrence of strong binding decreases significantly, leaving only the weaker bindings, characteristic of unspecific interactions. Under conditions of high ionic strength (Fig. 3G), the (un)binding force values of all peaks increased significantly (from 33.6 to 41.0 pN for the first peak; P < 0.01) (Table 1), but the ratio between the percentage of the first peak and that of the other peaks remained similar (26%).
Fig 4.
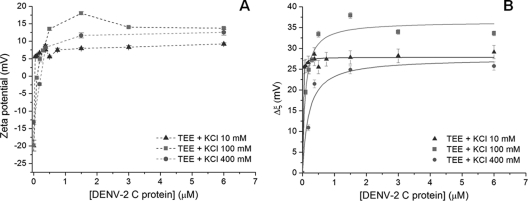
Zeta potential analysis. (A) LD zeta potential values were determined in the absence and in the presence of distinct DENV C protein concentrations, at different KCl concentrations. Dashed lines are a guide for the eye. (B) Variation of the zeta potential (Δζ) values with the DENV C protein concentration. Solid lines were obtained by fitting the experimental data with equation 2. Zeta potential and Δζ values are presented as means ± standard errors.
Figure 3B, C, E, F, H, and I shows three pairs of atomic force microscopy force mappings (height and adhesion images), corresponding to the three tested ionic strength/potassium concentration conditions. In high-ionic-strength buffer, there was an increase of the adhesion forces at the surface of the LDs. The higher adhesion forces were observed at the LD lateral borders (Fig. 3I), due to the higher surface area of the AFM tip-LD contact at these regions. For the adhesion map for the lower-ionic-strength buffer (Fig. 3C), an opposite situation was detected: the pixels on the map revealed adhesion force over the LDs lower than that at the mica surface. This fact further strengthens the result already observed for the force rupture histograms, in which the interaction between the DENV C protein and LDs was inhibited at low KCl concentrations.
To better understand the relevance of the LD surface charges in the interaction with the C protein, zeta potential analysis, a technique based on dynamic light scattering data for charged particles under electrophoretic conditions, was performed (Fig. 4). In the absence of the C protein, LDs presented zeta potential values of −19 mV in buffer with 100 mM KCl (Fig. 4A). The addition of the C protein induced a progressive increase in the scattering-particle charge, stabilizing at positive values (13.7 mV). Identical titrations of LDs with the DENV C protein were also conducted with buffers with 10 mM or 400 mM KCl (Fig. 4A). After fitting the experimental data with equation 2, we obtained values for the maximum amplitude of variation of the zeta potential induced by the interaction with the C protein, Δζmax, and C1/2 (the DENV C protein concentration at Δζmax/2) under each experimental condition studied (Table 2). Fitted curves obtained under different-ionic-strength conditions are shown in Fig. 4B. At 10 mM KCl, a C1/2 of 5.9 nM was obtained. A value 14.5-fold higher was obtained for 100 mM KCl (85.7 nM). As the concentrations of the salt increased to 400 mM, an increase of the C1/2 to 188.4 nM was observed.
Table 2.
Parameters obtained by zeta potential analysisa
| [KCl] (mM) | Mean C1/2 (nM) ± SE | Mean Δζmax (mV) ± SE |
|---|---|---|
| 10 | 5.9 ± 2.2 | 27.9 ± 0.4 |
| 100 | 85.7 ± 17.6 | 34.4 ± 1.3 |
| 400 | 188.4 ± 68.8 | 27.6 ± 2.3 |
Shown are values of the maximum amplitude of variation of the LD zeta potential (Δζmax) induced by the interaction with the DENV C protein and the DENV C protein concentration at Δζmax/2 (C1/2) in TEE buffer with different potassium concentrations.
When force spectroscopy measurements were performed with the potassium ions replaced by sodium ions in the buffer, a dramatic change in the results was observed. The use of TEE buffer with 100 mM NaCl instead of 100 mM KCl promoted a significant decrease in both the percentage of (un)binding events (to 19.5%) and the rupture force (persisting for only weak interactions with an [un]binding force of 26 pN; P < 0.0001) (Table 1), showing that the DENV C protein-LD interaction is K+ specific rather than an effect of the ionic strength.
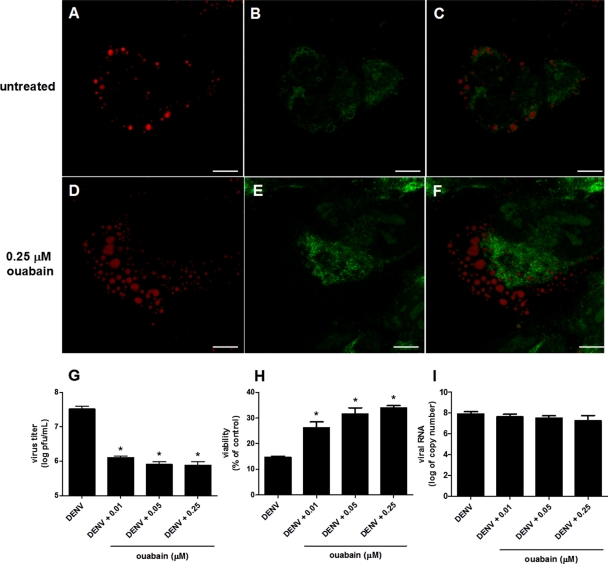
To evaluate the biological relevance of these findings, we investigated whether the inhibition of cellular Na+/K+-ATPase would interfere with C protein binding to LDs and with viral replication. Na+/K+-ATPase is a plasma membrane ion transporter that catalyzes the ATP-dependent transport of K+ into the cell in exchange for Na+, maintaining the intracellular concentration of K+ high and that of Na+ low in relation to the extracellular environment. Ouabain was used to inhibit Na+/K+-ATPase in HepG2 cells infected under experimental conditions similar to those for which it was demonstrated previously that the DENV C protein associates with LDs, an event essential for virus replication (39). After 16 h of infection, HepG2 cells were incubated, during a period of 5 h, with ouabain at concentrations that inhibit Na+/K+-ATPase in HepG2 cells (50). Just after treatment with 0.25 μM ouabain, the cells were observed by confocal microscopy for the detection of both C protein and LDs. In untreated cells, the C protein was clearly localized at the surface of LDs, as previously described (39) (Fig. 5A to C). On the other hand, in cells treated with ouabain, a redistribution of the C protein to a diffuse pattern in the cytosol was observed (Fig. 5D to F). In order to evaluate whether the C protein detachment of LDs interferes with the viral replication cycle, viral particles released from the infected cells were quantified after 72 h of infection (Fig. 5). Treatment with 0.01, 0.05, and 0.25 μM ouabain resulted in 29-, 43-, and 50-fold inhibitions of infectious virus particle production, respectively (Fig. 5G), which was followed by a recovery from the loss of viability caused by infection (Fig. 5H). However, despite the decrease in particle release, no difference was found in the DENV RNA content at 72 h postinfection (Fig. 5I), indicating that the alterations in the intracellular K+ concentration that induce DENV C protein release from LDs inhibit virus particle assembly without interfering with RNA replication. It is important to mention that control experiments were conducted to ensure that the treatment with ouabain at this concentration range did not affect the viability of noninfected HepG2 cells.
Fig 5.
Effects of Na+/K+-ATPase inhibition on different steps of the DENV virus replication cycle. HepG2 cells were infected with DENV and treated 16 h after infection with 0.25 μM ouabain for 5 h. Cells were then fixed with 4% paraformaldehyde and permeated with 0.1% Triton X-100. (A to F) Untreated infected cells (A to C) and ouabain-treated infected cells (D to F) were analyzed by confocal fluorescence microscopy using Bodipy for LD staining (red) (A and D) and a polyclonal anti-C protein antibody (green) (B and E). Merged images are shown in panels C and F. Scale bars correspond to 7.5 μm. The images correspond to representative results from three independent experiments. (G to I) After 72 h of infection, DENV infectious particles released into the culture medium (G), cell viability (H), and DENV RNA (I) were determined by plaque assay, MTT assay, and real-time PCR, respectively. For these experiments, different concentrations of ouabain were used, as indicted in the figure, following the same protocol of treatment as that described above for panels A to F. *, P < 0.05.
Effects of the presence of calcium, phosphate, or tripolyphosphate ions.
The addition of 1.2 mM calcium chloride to TEE buffer led to a nonsignificant decrease in the percentage of (un)binding events, from 58.4% to 56.4%. The values for the force necessary to break the bond between LDs and the C protein did not change under this condition (Table 1). The presence of phosphate ions led to a decrease of the percentage of (un)binding events to 11.1% and a decrease of the forces to only minor values (25 pN; P < 0.0001), while with the addition of 0.5 mM or 2 mM tripolyphosphate (TPP) ions to TEE buffer, the forces necessary to break the bond between the DENV C protein and LDs remained the same as those for the experiment without TPP but with a considerably lower percentage of occurrences (only 9.6%) (Table 1).
Trypsin treatment effects.
To test whether the DENV C protein interacts with lipid or protein components of LDs, force rupture values and the percentage of (un)binding events were determined for LDs preincubated in the absence or in the presence of different concentrations of trypsin (1, 5, or 10 μM) (Fig. 6E) in TEE buffer with 100 mM KCl. The force rupture histograms for the four tested conditions are shown in Fig. 6A to D. Incubation with increasing trypsin concentrations led to a significant decrease of the percentage of force rupture (un)binding events (Table 1). All the values of the (un)binding force decreased significantly compared to those of the experiment without preincubation with trypsin (for the first peak of each histogram, 26 pN for trypsin for 1 μM and 5 μM trypsin [P < 0.01 relative to the absence of trypsin treatment] and 19 pN for 10 μM trypsin [P < 0.0001]) (Table 1). Additionally, all the stronger binding events tended to disappear with increasing trypsin concentrations, leaving only the weaker interactions.
Fig 6.
Effect of pretreatment with trypsin on binding between DENV C protein and LDs. (A to E) Force-rupture histograms in the absence of trypsin treatment (A) and after treatment with 1 μM (B), 5 μM (C), or 10 μM (D) trypsin in TEE buffer with 100 mM KCl. Peaks were obtained after choosing the best rupture force scale bin and fitting the histograms with the Gaussian model. (E) Percentage of (un)binding events and force adhesion values of the first and the second fitted peaks for the DENV C protein-functionalized AFM tips binding to LDs treated with increasing trypsin concentrations (from 0 to 10 μM). Values are presented as means ± standard errors. The percentage of (un)binding events was defined by the ratio between the number of force spectroscopy curves with adhesion events and the total number of curves acquired under each experimental condition.
Identification of LD proteins involved in binding to the DENV C protein.
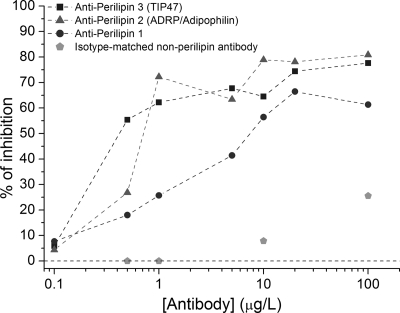
In order to identify the specific protein(s) on the surface of the LDs responsible for the binding to the DENV C protein, we measured the antibody-induced inhibition of the C protein-LD interaction by force spectroscopy. Specific antibodies against the three major PAT family proteins, perilipin 1 (formerly perilipin), perilipin 2 (ADRP/adipophilin), and perilipin 3 (TIP47), were tested. The antibody concentrations varied from 0.1 to 100 μg/liter. As shown in Fig. 7, at an antibody concentration of 0.5 μg/liter, the greater inhibitory effect of anti-perilipin 3 antibody than that of the other two specific antibodies became especially evident: a rate of DENV C protein-LD binding inhibition of 43% was observed for anti-perilipin 3, while 21% and 14% inhibitions were detected for the antibodies against perilipin 2 and perilipin 1, respectively. At higher antibody concentrations, the differences between the levels of inhibition of DENV C protein-LD binding observed for the three LD-specific antibodies became less evident. It is important to bear in mind that the binding inhibition observed occurred mostly at the level of the suppression of the stronger binding events, leaving mainly those with lower (un)binding forces. As a negative control for this experiment, we used an isotype-matched nonperilipin antibody. No detectable binding inhibition was observed at concentrations of IgG serum up to 1 μg/liter (the concentration range used to determined the specificity of the inhibition), and very slight reductions in binding were observed with concentrations of 10 μg/liter (7.9%) and 100 μg/liter (26%), confirming the specificity of the binding between LD perilipins and the DENV C protein (Fig. 7).
Fig 7.
Inhibition of binding between the DENV C protein and LDs by antibodies against different PAT family proteins. Shown are percentages of DENV C protein-LD binding inhibition in the presence of specific antibodies against perilipin 1, perilipin 2 (ADRP or adipophilin), and perilipin 3 (TIP47) as well as for a negative control with an isotype-matched nonperilipin antibody. The antibody concentrations varied from 0.1 to 100 μg/liter (please note the log scale on the x axis) in TEE buffer with 100 mM KCl.
DISCUSSION
The association between the DENV C protein and LDs in different cell types was shown previously to be essential for the virus replication cycle (39). In this study, we characterized for the first time the properties of the interaction between recombinant DENV C protein and purified LDs from a human lineage of hepatic cells (HepG2) by use of force spectroscopy and zeta potential measurements.
The covalent coupling of biomolecules to AFM tips has already proven to be an important nanotool for the measurement of both inter- and intramolecular forces (9, 10, 35, 40, 48). In this study, the use of glutaraldehyde as a flexible cross-linker to covalently couple the C protein to the tip gave this protein the flexibility to diffuse freely in a volume of buffer, restricted only by the dynamic length of the spacer (13, 48). With this cross-linker, the binding sites on the protein were reachable, and bonds between protein and LDs could be formed. The number of DENV C protein molecules bound to each tip is not crucial for the force measurements, since the technique enables the identification of single-molecule detachments from the characteristic stretching curves before rupture (9, 18, 44). Since the C protein has several accessible primary amines (28), its binding site cannot be defined a priori, and the part of the protein that is exposed for binding may vary from molecule to molecule. This is important in order to avoid possible artifacts arising from the immobilization of the protein in an orientation that would impair its correct functionality.
The single DENV C protein-LD interaction was found to be strong and characteristic of specific binding, with a (un)binding force of 33.6 ± 0.5 pN (Table 1), which is of the same order of magnitude as several specific protein-protein, protein-ligand, and protein-phospholipid bilayer interactions previously studied by force spectroscopy (14, 27). The validation of the obtained force measurements for specific molecular recognition events is a key issue for all force spectroscopy studies. This is especially relevant when measuring the interaction of a molecule on the AFM tip with a cell or cellular component, such as LDs, instead of a purified molecule attached to the substrate. For this reason, it was important to ensure that LDs were isolated from HepG2 cells without cytoplasmic contaminants or other cellular organelles. The absence of LDH activity, an enzyme known to be soluble in cytoplasm, and the detection of ADRP (one of the major protein components of LDs) proved that cell cavitation and sucrose gradient procedures allowed the successful isolation of LDs. Additionally, we performed all the controls needed to distinguish the specific interactions between LDs and the DENV C protein from those between one LD on the mica and another LD attached to the tip (after detaching the LD from the mica with the movement of the AFM tip in solution). This distinction could be made by measuring the differences in the AFM tip and the LD separation lengths at the moment of occurrence of the bond break. The value of the difference in the lengths of the tip and sample separation is dependent on the stretching of the C protein and on the elasticity of LDs. The DENV C protein is a symmetric dimer with a total of 100 amino acid residues and with a maximum length of approximately 8.3 nm in the native folded state (28). When the AFM tip pulls away from the LD surface after C protein binding, the protein stays attached to the LD surface and may begin to change conformation until the point when the bond breaks. The mean value of the length of the rupture events calculated by the force spectroscopy technique was 34.6 nm, which means that the extension was 4-fold longer than the DENV C protein native length. Obviously, this extension should not be attributed solely to the capsid protein but also to the LDs component(s) involved in binding.
The zeta potential measurements of LDs at pH 7.4 and 100 mM KCl revealed a negative surface charge, with an average value of −19 mV. This negative charge was expected, due to the phospholipid monolayer at the LD surface (17) and the negatively charged protein content. After an interaction with the C protein, this negative surface charge becomes positive, reaching a maximum plateau at +13.7 mV. The C protein dimer net charge at a neutral pH is +46 mV (assuming solution-state pKa values), with a remarkably nonuniform spatial charge distribution (28). The region with the highest density of positive charges is the α4-α4′ helices, in which 22 of the 52 basic residues lie (28). This suggests that this face of the protein becomes exposed to the solvent after the interaction with LDs. It is important to bear in mind that if the interaction between the C protein and LDs would render all cationic groups attached to the anionic surface of the LD and no hydrophobic/entropic factors were involved, the expected titration curve obtained by zeta potential measurements would tend to a horizontal asymptotic line close to zero, corresponding to electrostatic neutralization (1), which was not the case. The horizontal asymptotic line at higher positive values indicates that the interaction between the C protein and LDs may leave the polycationic domain exposed to the aqueous environment. This hypothesis is in agreement with the model proposed previously by Ma et al., based on the C protein NMR structure, in which the α4-α4′ region interacts with RNA and the apolar α2-α2′ region interacts with the viral membrane (28). A similar model was proposed previously for the M1 protein of influenza virus, in which the membrane and RNA interact with opposite faces of the protein (41). DENV C protein binding to LD can be of key importance to RNA recognition, since extensive aggregation and precipitation occur when this protein is merely added to RNA in solution. It is possible that this aggregation is prevented in vivo by the binding to LDs. This binding would enable the exposure of the positive charges of the protein cationic domain to the aqueous environment and their binding to the viral RNA, which is necessary for the assembly of new virions.
In our experiments with different KCl concentrations, the DENV C protein-LD interaction was shown to be highly dependent on either the ionic strength or the potassium chloride concentration. At a low ionic strength or a low KCl concentration, a significant decrease in the number of force spectroscopy binding events was observed, while at values of ionic strength or KCl concentrations above the physiologic range, significant increases in all the forces and also in the percentages of binding events were obtained. This behavior is the opposite of the commonly observed increase in protein-protein and protein-lipid interactions at lower ionic strengths and decreased interactions at high ionic strengths.
As shown in Fig. 4, at 400 mM KCl, the value of the surface charge of LDs in the absence of the C protein (−13.2 mV) is different from that under other potassium chloride conditions studied (100 mM and 10 mM KCl). This result is expected at high ionic strengths due to the shielding of the LD charges by a large quantity of ions interacting with the particle surface, leading to a decrease of the maximum variation amplitude of the zeta potential (Δζmax), as shown in Table 2. Using buffer with 10 mM KCl, a dependence of zeta potential values on the C protein concentration was not observed, suggesting that the interaction between LDs and the DENV C protein is not specific. This is in accordance with the force spectroscopy results obtained under this condition.
The force spectroscopy approach was also used to probe the influence of different ions (sodium, calcium, phosphate, and tripolyphosphate) on the binding between the DENV C protein and LDs, as shown on Table 1. In all cases except for the presence of both Ca2+ and K+ ions in solution, drastic decreases in the percentages of events and in the (un)binding forces were observed. The replacement of potassium by the same concentration of sodium, another monovalent cation with a smaller size, decreased the binding between the C protein and LDs significantly, demonstrating that the interaction is K+ dependent and that the decreased level of binding observed at low potassium concentrations (Fig. 3 and Table 1) is not a consequence of the decreased ionic strength but a consequence of a decrease of the potassium concentration itself.
It is important to bear in mind that K+ is the main intracellular cation, and its concentration is low in extracellular medium, where NaCl is mainly responsible for osmolarity. Our data strongly suggest that the K+-dependent interaction between the DENV C protein and LDs may be a means for regulating C protein release in the cytosol. We found that the inhibition of Na+/K+-ATPase promoted the release of the C protein from LDs in infected cells, resulting in a significant decrease in infectious viral particle release from the cells. The inhibition of particle assembly was not due to an interference with viral RNA replication, in agreement with previous results obtained by using C protein mutants that do not bind to LDs (39). Viruses commonly modulate the biochemistry and physiology of infected cells in order to optimize the conditions for different steps of the viral life cycle. This fine-tuning of the ideal cellular conditions for viral replication occurs often at the level of specific intracellular ion concentrations. Viroporins are a class of viral proteins that enhance cell membrane permeability in order to favor viral replication (for a review, see reference 46). HCV, like DENV, belongs to the family Flaviviridae and depends on LDs for replication (for a review, see reference 32). p7 is an HCV viroporin that promotes membrane permeability to potassium and other cations (19), and although it seems essential for virus infection, its role in the viral life cycle is still partially unexplained (32, 46). Additionally, HCV is also known to modulate the function of Kv2.1, a K+-specific channel, through the action of the viral nonstructural protein NS5A (29). It is tempting to speculate that (i) the interaction of the HCV core protein with LDs is also K+ specific, being modulated at different stages of the viral life cycle by the viral proteins p7 and/or NS5A and (ii) equivalent DENV proteins influence intracellular potassium concentrations, therefore promoting the binding of the C protein to LDs and/or its release at a later stage of infection. The C-terminal domain of the DENV M protein may also play a role in this process, as it presents ion channel activity, with a higher permeability for K+ than for other ions (37).
By increasing the number of negative phosphate ions or polyphosphate concentrations, a remarkable decrease in the binding between the C protein and LDs was observed. These results can also be related to the poorly understood DENV assembly process: as proposed above, the association of the DENV C protein with the LD surface would constitute the proper scaffold for a correct protein-RNA interaction upon the initiation of the assembly process. The protein-RNA complex would then be released from the LD surface. As higher phosphate or polyphosphate concentrations may partially mimic the interaction between the positively charged amino acid residues of the exposed domain of the C protein and the negatively charged phosphate groups of RNA, this could lead to a shift of the molecular equilibrium toward the detachment of the protein from the LD surface.
LDs contain several proteins on their surface, namely, the proteins of the PAT family, perilipin 1, perilipin 2 (ADRP), and perilipin 3 (TIP47), and other proteins in small quantities (36). To investigate a possible role of these proteins as well as to distinguish a situation of a protein-protein interaction from a protein-lipid interaction, we performed identical force spectroscopy experiments after the pretreatment of the LDs with different concentrations of trypsin. Force spectroscopy data showed very significant decreases in the force values and in the frequency of binding with increasing trypsin concentrations. We can conclude from these results that the weaker interactions become more frequent than the stronger, specific ones after the removal of the proteins from the LD surface, indicating that the DENV C protein binds to a protein component on the surface of the LD. At this level, it is worth mentioning that PAT proteins cover 15 to 20% of the surface of LDs (45) and are negatively charged at the studied pH (the theoretical isoelectric points of perilipin 1, perilipin 2, and perilipin 3 are 6.03, 6.34, and 5.30, respectively), contributing to the negative charge registered for our zeta potential results for LDs before the addition of the DENV C protein.
Based on the LD-DENV C protein binding inhibition studies, conducted using specific antibodies against perilipin 1, perilipin 2 (ADRP), and perilipin 3 (TIP47), we can conclude that these proteins have distinguishable affinities for the DENV C protein. Perilipin 3 is the main protein at the LD surface involved in LD-DENV C protein binding, while perilipin 2 and perilipin 1 seem to have lower affinities for the viral protein and a minor influence on its interaction with LDs. The stronger affinity of perilipin 3 for the DENV C protein than that of the other two perilipins becomes especially evident at lower specific antibody concentrations (e.g., 0.5 μg/liter), where the greater magnitude of binding inhibition with anti-perilipin 3 becomes evident (Fig. 7). The isotype-matched nonperilipin control ruled out any unspecific effect at lower antibody concentrations. These results led to the conclusion that perilipin 3 is the main protein on the LD surface responsible for binding to the DENV C protein.
The perilipin 3 amino acid sequence has a high degree of similarity to the sequences of the other two major LD-associated proteins, perilipin 1 and perilipin 2. It is 43% identical to perilipin 2 throughout the length of the sequence, with a higher level of homology in the N-terminal region (60% identity and 80% similarity), which is shared by perilipin 1, but differs in the amino acid sequences of the C termini (49). These similarities may account for the binding of the DENV C protein to perilipin 2 and perilipin 1, but to a lesser extent than the binding to perilipin 3, following a general trend of DENV C protein affinity of perilipin 3 > perilipin 2 > perilipin 1, in agreement with the degree of similarity between these three proteins.
The inhibition results at antibody concentrations above 10 μg/liter may result from the saturation of the surface of the LDs with any of the antibodies tested. With these high antibody concentrations, the surface crowding effect would make it difficult to distinguish between the effects of targeting each specific protein.
It could also be reasoned that the DENV C protein ligand on the LD surface could be one of its minor components, such as perilipin 4 (S3-12) or perilipin 5 (PAT1, LSDP5, OXPAT, or MLDP) (25). However, this would not be possible, as binding to a minor component of LDs would not account for the high percentage of (un)binding events observed by force spectroscopy in the presence of K+ (DENV C protein-LD binding occurred in almost 80% of the AFM tip approach-retraction cycles).
In summary, based on our findings, we propose that (i) the DENV C protein binds to proteins on the surface of intracellular LDs, primarily perilipin 3 (TIP47); (ii) the interaction is not electrostatic driven but depends on a high intracellular potassium concentration; and (iii) the interaction between the C protein and perilipin 3 on the LD surface may regulate the availability of the C protein in the cytosol or another cellular compartment, allowing its proper participation in the virus replication cycle. Furthermore, an understanding of these molecular interactions during the viral replication cycle may allow the identification of new targets for the treatment of DENV infection.
ACKNOWLEDGMENTS
We thank Teresa Freitas, Marco M. Domingues (Instituto de Medicina Molecular, Faculdade de Medicina da Universidade de Lisboa, Portugal), and Ricardo Vilela (Laboratório de Biologia, Instituto Nacional de Metrologia, Qualidade e Tecnologia-INMETRO, Rio de Janeiro, Brazil) for technical assistance and for the help with the zeta potential measurements and confocal microscopy experiments, respectively.
This work was supported by FP7-PEOPLE IRSES (International Research Staff Exchange Scheme) project MEMPEPACROSS (European Union), by the Fundação para a Ciência e a Tecnologia—Ministério da Educação e Ciência (FCT-MEC, Portugal) (projects PTDC/QUI-BIQ/112929/2009 and PTDC/QUI/69937/2006), by Fundação Calouste Gulbenkian (Portugal), by the FCT-CAPES Portugal-Brazil joint cooperation projects, and by the Brazilian funding agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Financiadora de Estudos e Projetos (FINEP), and National Institute of Science and Technology in Dengue (INCT-Dengue). I. C. Martins also acknowledges consecutive postdoctoral funding from a Marie Curie International Outgoing Fellowship (MC-IOF-237373) and FCT-MEC postdoctoral fellowships (SFRH/BPD/46324/2008 and SFRH/BPD/74287/2010). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
F. A. Carvalho, F. A. Carneiro, I. Assunção-Miranda, M. A. R. B. Castanho, R. Mohana-Borges, A. T. Da Poian, and N. C. Santos conceived and designed experiments; F. A. Carvalho, F. A. Carneiro, I. C. Martins, I. Assunção-Miranda, and A. F. Faustino performed experiments; F. A. Carvalho, F. A. Carneiro, I. C. Martins, I. Assunção-Miranda, A. F. Faustino, A. T. Da Poian, and N. C. Santos analyzed the data; R. M. Pereira and R. Mohana-Borges contributed with reagents, materials, and/or analysis tools; and F. A. Carvalho, F. A. Carneiro, I. C. Martins, I. Assunção-Miranda, P. T. Bozza, M. A. R. B. Castanho, R. Mohana-Borges, A. T. Da Poian, and N. C. Santos wrote the manuscript.
Footnotes
Published ahead of print 30 November 2011
REFERENCES
- 1. Alves CS, et al. 2010. Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J. Biol. Chem. 285:27536–27544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Assunção-Miranda I, et al. 2010. Contribution of macrophage migration inhibitory factor to the pathogenesis of dengue virus infection. FASEB J. 24:218–228 [DOI] [PubMed] [Google Scholar]
- 3. Barba G, et al. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. U. S. A. 94:1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boulant S, et al. 2006. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281:22236–22247 [DOI] [PubMed] [Google Scholar]
- 5. Boulant S, Targett-Adams P, McLauchlan J. 2007. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 88:2204–2213 [DOI] [PubMed] [Google Scholar]
- 6. Bozza PT, Magalhães KG, Weller PF. 2009. Leukocyte lipid bodies—biogenesis and functions in inflammation. Biochim. Biophys. Acta 1791:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brasaemle DL, et al. 1997. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38:2249–2263 [PubMed] [Google Scholar]
- 8. Brewer SH, Glomm WR, Johnson MC, Knag MK, Franzen S. 2005. Probing BSA binding to citrate-coated gold nanoparticles and surfaces. Langmuir 21:9303–9307 [DOI] [PubMed] [Google Scholar]
- 9. Carvalho FA, et al. 2010. Atomic force microscopy-based molecular recognition of a fibrinogen receptor on human erythrocytes. ACS Nano 4:4609–4620 [DOI] [PubMed] [Google Scholar]
- 10. Carvalho FA, de Oliveira S, Freitas T, Gonçalves S, Santos NC. 2011. Variations on fibrinogen-erythrocyte interactions during cell aging. PLoS One 6:e18167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conceição TM, Da Poian AT, Sorgine MH. 2010. A real-time PCR procedure for detection of dengue virus serotypes 1, 2, and 3, and their quantitation in clinical and laboratory samples. J. Virol. Methods 163:1–9 [DOI] [PubMed] [Google Scholar]
- 12. Conceição TM, et al. 2010. Gene expression analysis during dengue virus infection in HepG2 cells reveals virus control of innate immune response. J. Infect. 60:65–75 [DOI] [PubMed] [Google Scholar]
- 13. de Odrowaz Piramowicz M, Czuba P, Targosz M, Burda K, Szymonski M. 2006. Dynamic force measurements of avidin-biotin and streptavidin-biotin interactions using AFM. Acta Biochim. Pol. 53:93–100 [PubMed] [Google Scholar]
- 14. Desmeules P, Grandbois M, Bondarenko VA, Yamazaki A, Salesse C. 2002. Measurement of membrane binding between recoverin, a calcium-myristoyl switch protein, and lipid bilayers by AFM-based force spectroscopy. Biophys. J. 82:3343–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Digel M, Ehehalt R, Fullekrug J. 2010. Lipid droplets lighting up: insights from live microscopy. FEBS Lett. 584:2168–2175 [DOI] [PubMed] [Google Scholar]
- 16. Dokland T, et al. 2004. West Nile virus core protein; tetramer structure and ribbon formation. Structure 12:1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Domingues MM, Castanho MARB, Santos NC. 2009. rBPI21 promotes lipopolysaccharide aggregation and exerts its antimicrobial effects by (hemi)fusion of PG-containing membranes. PLoS One 4:e8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Francius G, et al. 2009. Stretching polysaccharides on live cells using single molecule force spectroscopy. Nat. Protoc. 4:939–946 [DOI] [PubMed] [Google Scholar]
- 19. Griffin SD, et al. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34–38 [DOI] [PubMed] [Google Scholar]
- 20. Guzman A, Isturiz RE. 2010. Update on the global spread of dengue. Int. J. Antimicrob. Agents 36(Suppl 1):S40–S42 [DOI] [PubMed] [Google Scholar]
- 21. Hodges BD, Wu CC. 2010. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 51:262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hope RG, McLauchlan J. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913–1925 [DOI] [PubMed] [Google Scholar]
- 23. Jones CT, et al. 2003. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 77:7143–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaufman ED, et al. 2007. Probing protein adsorption onto mercaptoundecanoic acid stabilized gold nanoparticles and surfaces by quartz crystal microbalance and zeta-potential measurements. Langmuir 23:6053–6062 [DOI] [PubMed] [Google Scholar]
- 25. Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. 2010. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J. Lipid Res. 51:468–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuhn RJ, et al. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee CK, Wang YM, Huang LS, Lin S. 2007. Atomic force microscopy: determination of unbinding force, off rate and energy barrier for protein-ligand interaction. Micron 38:446–461 [DOI] [PubMed] [Google Scholar]
- 28. Ma L, Jones CT, Groesch TD, Kuhn RJ, Post CB. 2004. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. U. S. A. 101:3414–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mankouri J, et al. 2009. Suppression of a pro-apoptotic K+ channel as a mechanism for hepatitis C virus persistence. Proc. Natl. Acad. Sci. U. S. A. 106:15903–15908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Markoff L, Falgout B, Chang A. 1997. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 233:105–117 [DOI] [PubMed] [Google Scholar]
- 31. Martin S, Parton RG. 2006. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 7:373–378 [DOI] [PubMed] [Google Scholar]
- 32. McLauchlan J. 2009. Lipid droplets and hepatitis C virus infection. Biochim. Biophys. Acta 1791:552–559 [DOI] [PubMed] [Google Scholar]
- 33. Miller S, Krijnse-Locker J. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mukhopadhyay S, Kuhn RJ, Rossmann MG. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22 [DOI] [PubMed] [Google Scholar]
- 35. Muller DJ. 2008. AFM: a nanotool in membrane biology. Biochemistry 47:7986–7998 [DOI] [PubMed] [Google Scholar]
- 36. Olofsson SO, et al. 2009. Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim. Biophys. Acta 1791:448–458 [DOI] [PubMed] [Google Scholar]
- 37. Premkumar A, Horan CR, Gage PW. 2005. Dengue virus M protein C-terminal peptide (DVM-C) forms ion channels. J. Membr. Biol. 204:33–38 [DOI] [PubMed] [Google Scholar]
- 38. Ratto TV, Rudd RE, Langry KC, Balhorn RL, McElfresh MW. 2006. Nonlinearly additive forces in multivalent ligand binding to a single protein revealed with force spectroscopy. Langmuir 22:1749–1757 [DOI] [PubMed] [Google Scholar]
- 39. Samsa MM, et al. 2009. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Santos NC, Castanho MARB. 2004. An overview of the biophysical applications of atomic force microscopy. Biophys. Chem. 107:133–149 [DOI] [PubMed] [Google Scholar]
- 41. Sha B, Luo M. 1997. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat. Struct. Biol. 4:239–244 [DOI] [PubMed] [Google Scholar]
- 42. Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169 [DOI] [PubMed] [Google Scholar]
- 43. Thiele C, Spandl J. 2008. Cell biology of lipid droplets. Curr. Opin. Cell Biol. 20:378–385 [DOI] [PubMed] [Google Scholar]
- 44. Tsapikouni TS, Missirlis YF. 2009. Measuring the force of single protein molecule detachment from surfaces with AFM. Colloids Surf. B Biointerfaces 75:252–259 [DOI] [PubMed] [Google Scholar]
- 45. Walther TC, Farese RV., Jr 2009. The life of lipid droplets. Biochim. Biophys. Acta. 1791:459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang K, Xie S, Sun B. 2011. Viral proteins function as ion channels. Biochim. Biophys. Acta. 1808:510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Welsch S, et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Willemsen OH, et al. 2000. Biomolecular interactions measured by atomic force microscopy. Biophys. J. 79:3267–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolins NE, Rubin B, Brasaemle DL. 2001. TIP47 associates with lipid droplets. J. Biol. Chem. 276:5101–5108 [DOI] [PubMed] [Google Scholar]
- 50. Xu ZW, et al. 2010. Targeting the Na(+)/K(+)-ATPase alpha1 subunit of hepatoma HepG2 cell line to induce apoptosis and cell cycle arresting. Biol. Pharm. Bull. 33:743–751 [DOI] [PubMed] [Google Scholar]