Abstract
Most of an intravenous dose of species C adenovirus serotype 5 (Ad5) is destroyed by liver Kupffer cells. In contrast, another species C virus, Ad6, evades these cells to mediate more efficient liver gene delivery. Given that this difference in Kupffer cell interaction is mediated by the hypervariable (HVR) loops of the virus hexon protein, we genetically modified each of the seven HVRs of Ad5 with a cysteine residue to enable conditional blocking of these sites with polyethylene glycol (PEG). We show that these modifications do not affect in vitro virus transduction. In contrast, after intravenous injection, targeted PEGylation at HVRs 1, 2, 5, and 7 increased viral liver transduction up to 20-fold. Elimination or saturation of liver Kupffer cells did not significantly affect this increase in the liver transduction. In vitro, PEGylation blocked uptake of viruses via the Kupffer cell scavenger receptor SRA-II. These data suggest that HVRs 1, 2, 5, and 7 of Ad5 may be involved in Kupffer cell recognition and subsequent destruction. These data also demonstrate that this conditional genetic-chemical mutation strategy is a useful tool for investigating the interactions of viruses with host tissues.
INTRODUCTION
Adenovirus serotype 5 (Ad5) has proven to be one of the most potent in vivo gene delivery vectors for liver-directed gene therapy. As much as 95 to 98% of an intravenously (i.v.) injected dose of Ad5 is trafficked to the liver (12). The adenovirus capsid is comprised of three major proteins: hexon, penton base, and fiber (reviewed in reference 7). Based on numerous in vitro studies, adenovirus fiber and penton base have been shown to be cellular attachment proteins. The fiber of Ad5 binds coxsackie and adenovirus receptor (CAR) (3), which triggers binding of penton base to αv integrins via an RGD motif (35, 36) and results in viral cell internalization. Although necessary for providing specificity of the virus in vitro, modifications to the fiber have little effect on vector tropism in vivo (25). Instead, increasing evidence suggests that hexon plays a large role in the natural liver tropism of Ad5.
Hexon is the most abundant viral capsid protein with 720 monomers per virion. Hexon organizes into trimers so that three hexon monomers and their loops wrap tightly around each other to create a tower-like structure with a central depression (20, 28). Each hexon monomer has seven flexible, serotype-specific loops, named hypervariable regions (HVRs) (23), that are predicted to be located on the surface of the hexon trimer and the virion (29). This location allows the HVRs of hexon to interact with neutralizing antibodies, receptors, proteins, and cells. Considering there are 5,040 (720 × 7 = 5,040) HVRs per virion, these represent a complex, exposed surface for many interactions.
After i.v. injection, Ad5 exhibits the greatest transduction within liver hepatocytes (12). Despite this robust in vivo gene delivery, ∼90% of the injected dose is sequestered and destroyed by resident liver macrophages called Kupffer cells (1). These antigen-presenting cells not only destroy the virus but are themselves destroyed. This cellular necrosis plays a role in inflammation and triggers innate and adaptive immune responses to the virus (21). Previous studies indicate that Kupffer cells take up Ad5 via scavenger receptors (13, 37). It is hypothesized that scavenger receptors on these cells recognize the virus by interacting with the highly charged HVRs on Ad5 (1, 37). In particular, HVR1 is thought to be a good ligand for scavenger receptors (1), since it is the largest HVR and also has a number of negatively charged amino acids (18). These results indicate that hexon and its HVRs are important surfaces involved in Kupffer cell recognition. By evading such cells, adenoviruses should be able to more readily transduce downstream sites.
After evading the reticuloendothelial system (which includes both Kupffer cells and liver sinusoidal endothelial cells), Ad5 enters the liver parenchyma through fenestrations in the vessel walls (32). If adenovirus can enter the parenchyma, robust hepatocyte infection is dependent not only on cell binding ligands evolved by the virus but also on host blood proteins. Evidence shows that Ad5 hexon binds to vitamin K-dependent coagulation factors, such as factor X (FX), with high affinity, and this interaction markedly increases hepatocyte infection (26, 31). Structural and mutational analysis has revealed that FX binds to the top of the hexon trimer depression, with predicted interactions at HVR5, HVR7, and possibly HVR3 (16, 33). These data implicate the HVRs of the Ad5 hexon in two pharmacologic bottlenecks after i.v. injection of the virus: (i) binding or evasion of Kupffer cells and (ii) binding of FX and retargeting to hepatocytes.
We recently compared the biology of Ad5 with another species C adenovirus, Ad6 (18, 34). In these studies, we found that native Ad6 mediates higher liver transduction than Ad5 after i.v. injection (34). To identify the underlying molecular basis for this difference, the HVRs of Ad5 were replaced with those of Ad6 producing a virus called Ad5/6 (Fig. 1A) (18). When Ad5 and Ad5/6 were compared, the Ad5/6 virus mediated 10-fold increases in hepatocyte transduction after i.v. injection. This effect appeared to be due to reduced interactions of Ad5/6 with macrophages and Kupffer cells.
Fig 1.

The Ad5 hexon HVR domain is critical for scavenger receptor recognition. (A) BALB/c mice were predosed i.v. with PBS or 100 μg of poly(I). They were injected i.v. 5 min later with 1010 vp of each vector. After 24 h, the mice were imaged with a Lumazone imager for 3-min, 2 × 2 binning, and the luciferase expression was quantified. (B) CHO cells expressing the coxsackie and adenovirus receptor (CAR) or scavenger receptors (SRA-II and SREC) were infected with 1,000 vp of Ad5 or a previously created Ad5 vector containing the Ad6 HVR cassette (Ad5/6)/cell for 36 h and then assessed by luciferase assay (n = 10). *, P < 0.05; **, P < 0.0001; ns, not significant.
These data suggest that particular surfaces of the Ad5 hexon are involved in Kupffer cell recognition. Given this and the pivotal role of Kupffer cell depletion during systemic therapy, we evaluated the roles of each of the seven HVRs by conditionally mutating them using genetic and chemical engineering techniques (18, 19). Single cysteine residues were inserted into each of the seven HVRs of Ad5 hexon individually. These “silent” mutations were then “activated” by specifically modifying the site by reaction with cysteine-reactive polyethylene glycol (PEG) to conditionally block interactions with each HVR. By this approach, we probed the role of each HVR in Kupffer cell recognition.
MATERIALS AND METHODS
Cell culture.
Human embryonic kidney 293 cells (HEK293) were purchased from Microbix. A549 human lung carcinoma cells (American Type Culture Collection [ATCC] CCL-185), Chinese hamster ovary cells (ATCC CCL-61), and RAW 264.7 murine monocyte/macrophage cells (ATCC CRL-2278) were purchased from the ATCC. All cells were maintained in Dulbecco modified Eagle medium containing 10% heat-inactivated fetal calf serum (Gibco) and penicillin-streptomycin at 100 U/ml (Gibco). CHO-CAR cells were developed by selecting stable transfectants of CMV-CAR plasmids with G418. CHO-SREC and CHO-SRA-II cells were kindly provided by Brent Berwin (Dartmouth, NH) and Monty Krieger (Massachusetts Institute of Technology), respectively. The latter was cultured in Ham F-12 medium supplemented with 10% lipid reduced fetal bovine serum (HyClone), penicillin-streptomycin at 100 U/ml, 250 μM mevalonic acid, 3 μg of acLDL/ml, 40 μM mevinolin, and 300 μg of G418/ml. Normal medium was used during comparative experiments.
Adenoviruses.
(i) For unmodified control adenovirus, first-generation, replication-defective (i.e., with E1 and E3 deleted) Ad5 vector carrying enhanced green fluorescent protein (GFP) and luciferase fusion gene (GL) was produced with the AdEasy system (Qbiogene/MP Biomedicals) in 293 cells as described in Mok et al. (24). (ii) For cysteine-modified adenovirus, Ad5 HVR sequence segments with cysteine insertions were purchased from GenScript. Using flanking ApaI and AvrII restriction sites, each HVR segment was cloned into a shuttle plasmid and recombined into the adenovirus backbone with red recombinase cells. Viruses were purified by two consecutive CsCl gradient centrifugations; in order to prevent disulfide bond formation, the first gradient was supplemented with 10 mM dithiothreitol (DTT), and the second was supplemented with 1 mM DTT according to a previously published protocol. Vectors were desalted using Econopac 10-DG chromatography columns (Bio-Rad), resuspended in degassed 0.5 M sucrose in potassium phosphate-buffered saline (KPBS) buffer and stored at −80°C. The virus particle (vp) concentration was determined by A260 measurements. Infectious unit titers were determined from the 50% tissue culture infective dose (TCID50), where viruses were serially diluted on 293 cells in a 96-well plate. Wells were scored for cytopathic effect after 2 weeks.
PEGylation.
Linear, heterobifunctional PEG with biotin and maleimide (mal-PEG-biotin) or with methyl and maleimide (PEG without biotin cap) linkers was purchased in 5-, 20-, and 40-kDa lengths (JenKem Technology). We purchased 0.5-kDa Peg2–biotin from Thermo Scientific and 5-kDa methyl and succinimide heterobifunctional PEG from Sunbright. Upon use, all PEGs were overlaid with nonreactive nitrogen gas and stored at −20°C in desiccant. For the PEGylation of cysteine-modified vectors, 1 mg of PEG was added per 1011 vp and rotated at room temperature for 1.5 h. After incubation, the virus mixtures were dialyzed at room temperature into degassed and N2-overlaid 0.5 M sucrose–KPBS buffer for 2 h and then in fresh buffer at 4°C overnight. After dialysis, vector suspensions were injected into mice within 3 h. In some cases, genome copies after dialysis were determined by real-time PCR to confirm amounts of virus being injected.
Quantification of PEGylation.
Equivalent amounts of Cys-modified and control viruses were PEGylated. Excess PEG was removed by three sequential buffer exchanges each into 0.5 M sucrose buffer using Bio-Spin 30 Tris columns (Bio-Rad). In three wells, 109 vp of each virus were bound in a 96-well high-binding enzyme-linked immunosorbent assay (ELISA) plate. A protein standard of fully biotinylated MBP-AviTag protein (Avidity) was also plated. After 2 h at room temperature, the wells were blocked with 3% bovine serum albumin overnight. Avidin-horseradish peroxidase (HRP) at a 1:1,000 dilution was used as a probe for 1 h at room temperature. TMB substrate (100 μl/well) was used for detection, followed by 25 μl of 2 M sulfuric acid after 15 min to stop the reaction. The plate was read at 405 nm using a Beckman Coulter DTX 880 multimode detector system. An inverse assay to quantify unconjugated cysteines was also used to assess the number of PEGylation events per virion. Cys-modified virus was mock treated or PEGylated at room temperature for 1 h. Excess IR800-maleimide (LI-COR) was added to each virus suspension for 1 h at room temperature. Mock-treated or PEGylated vectors (1010 vp) were run on a 7.5% polyacrylamide gel. The Kodak Image Station In Vivo FX was used to detect fluorescence for 80 min (755-nm excitation, 830-nm emission).
Western blotting.
PEGylated (biotin–5-kDa PEG–maleimide) or mock-PEGylated vectors were loaded at 1010 vp onto a 7.5% polyacrylamide gel. The proteins were transferred to a nitrocellulose membrane and blocked overnight in 5% milk. The membrane was probed with a 1:2,000 dilution of neutravidin-HRP. SuperSignal West Pico chemiluminescent substrate (Pierce) and Kodak Image Station In Vivo FX were used for detection.
Animals.
Female BALB/c mice (6 to 8 weeks old) were purchased from Harlan Sprague-Dawley (Indianapolis, IN) and housed in the Mayo Clinic Animal Facility. All animal experiments were performed according to the policies and procedures of Mayo Clinic.
Adenovirus injections.
All viruses were diluted in phosphate-buffered saline (PBS) and mice were injected via the tail vein (i.v.) with a total viral dose of 1010 vp in 100 μl of PBS, except when otherwise specified. For adenovirus predosing, mice were injected i.v. with 3 × 1010 vp of Ad5-dsRed in 100 μl of PBS 4 h prior to i.v. injection of vectors. For in vivo scavenger receptor experiments, mice were injected i.v. with 100 μg of poly(I) in a total of 100 μl of PBS for 5 min prior to adenovirus vector injection.
Bioluminescence imaging in vivo.
Mice were imaged 24 h after adenovirus injection. All mice were anesthetized with ketamine-xylazine, injected intraperitoneally (i.p.) with 100 μl of d-luciferin (20 mg/ml; Molecular Imaging Products), and imaged on the Lumazone imaging system (Photometrics, Roper Scientific) for 1 to 10 min with 1 × 1 or 2 × 2 binning, using no filters or photomultiplication. Lumazone imaging software was used to determine the total light intensity per mouse (photons) for data analysis.
Luciferase assay in vitro.
Cell lines were plated to ∼80 to 90% confluence in 96-well plates and infected with adenovirus vectors for 24 h at 37°C. For the FX experiments, FX was added at 10 μg/ml, cells were lysed with 1× passive lysis buffer (Promega), and 50 μl of luciferase assay reagent was added to each well. The relative luminescence units (RLU) were determined with a Beckman Coulter DTX 880 multimode detector system.
Quantitative real-time PCR.
The concentration of the DNA samples was determined based on the A260 and then diluted to 4 ng/μl. Real-time PCR was performed on the DNA using the Applied Biosystems Prism 7900HT sequence detection system with SDS 2.3 software. Each well contained 10 μl of Sybr green (Applied Biosystems), 1 μl of H2O, 2 μl of 3 μM F primer, 2 μl of 3 μM R primer, and 5 μl of sample (i.e., 20 ng of DNA/well). The parameters used were 1 cycle of 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 55°C for 1 min, and 72°C for 30 s. To normalize the data, PCR was performed for both the cellular housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase), using the previously published F primer (5′-CGTGTTCCTACCCCCAATGT-3′ and R primer 5′-TGTCATCATACTTGGCAGGTTTCT-3′), and the cytomegalovirus (CMV) gene, using F primer (5′-CAAGTGTATCATATGCCAAGTACGCCCC-3′ and R primer 5′-CCCCGTGAGTCAAACCGCTATCCACGCC-3′). Viral DNA was normalized to cellular DNA by dividing the CMV genomes by the GAPDH genomes.
Data analysis.
Graphs and statistical analyses were performed using Prism graphical software. Analysis between two groups (such as virus alone or virus with PEG) was determined by using an unpaired t test. Analysis between three or more groups was done by one-way analysis of variance (ANOVA; Bonferroni post test). Graphs show means plus the standard deviations.
RESULTS
Ad5/6 mediates increased liver transduction and evades scavenger receptors.
The Ad5/6 virus mediates 10-fold increases in hepatocyte transduction after i.v. injection (Fig. 1A) (18). This effect appeared to be due to reduced interactions of Ad6 HVRs with macrophages and Kupffer cells (18).
Previous data suggest that Ad5 is phagocytosed by Kupffer cells via scavenger receptors (14). Broadly specific scavenger receptors recognize negative charge clusters, such as those that are present in adenovirus HVRs (Fig. 2A). To compare virus interactions with these receptors, Ad5 and Ad5/6 were incubated with CHO cells that were modified to express CAR (the cognate receptor for Ad5), SRA-II (the Kupffer cell scavenger receptor), or SREC (the endothelial cell scavenger receptor) (2, 4, 18).
Fig 2.
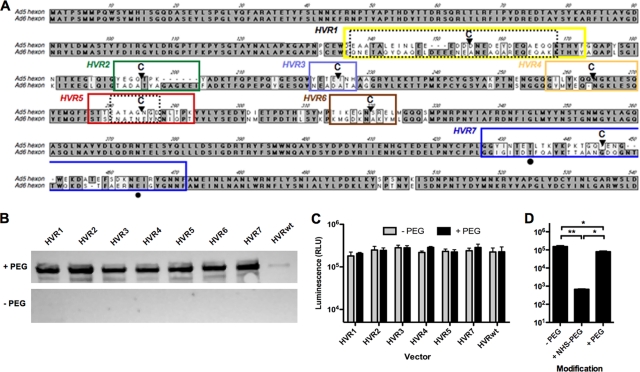
Cysteine-modified vectors can be specifically PEGylated and are still infectious in vitro. (A) Gene sequence alignment of Ad5 and Ad6 hexon. Boxes indicate HVR domains as follows: yellow, HVR1; green, HVR2; light blue, HVR3; orange, HVR4; red, HVR5; brown, HVR6; blue, HVR7. C's indicate the positions of cysteine (Cys) insertions at amino acids 151, 191, 215, 254, 275, 310, and 435. Natural Cys occurs at positions 132, 236, 420, and 523. Black circles indicate potentially critical FX binding residues. (B) Mock-treated (control) or cysteine-modified vectors were conjugated to maleimide–5 kDa PEG-biotin (PEG) or were not conjugated. Samples were subjected to SDS-PAGE, and the PEG-biotin was detected by Western blotting with streptavidin-HRP. (C) A549 cells were transduced with control or PEGylated cysteine-modified vectors at an MOI of 1,000 vp/cell. After 24 h, the cells were lysed, and the luciferase transgene expression was determined by using a spectrophotometer (n = 3). (D) Luciferase expression was also measured in A549 cells transduced with Ad-HVR5 PEGylated with either 5-kDa succinimide-PEG (NHS-PEG) or PEG at an MOI of 1,000 vp/cell. *, P < 0.05; **, P < 0.01.
CHO cells normally express low levels of CAR and are relatively nonpermissive for Ad5 infection (3, 22) (Fig. 1B). The addition of CAR to CHO cells increased transduction 10-fold for both Ad5 and Ad5/6 (Fig. 1B). When CHO cells expressed SRA-II in the absence of CAR, transduction by Ad5 was also increased. In contrast, SRA-II expression caused no increase in transduction by Ad5/6. On CHO-SRA-II cells, transduction by Ad5 was 20-fold higher than transduction by Ad5/6 (P < 0.0001 [two-tailed t test]). SREC increased Ad5 transduction only slightly, but this was higher than transduction for Ad5/6 (P < 0.01 [two-tailed t test]).
Insertion of cysteine residues into single HVRs for targeted PEGylation.
The data in Fig. 1 suggested that Ad5 hexon interacts more strongly with the Kupffer cell scavenger receptor than Ad6 hexon. Given that Ad5 and Ad5/6 differ only in their HVRs, these data suggested that the interaction of Ad5 with SRA-II occurs at the surface of one or more of its HVRs.
To investigate which Ad5 HVRs are critical for these interactions, single cysteines were inserted into each of the seven HVRs of Ad5 by red recombination (Fig. 2A). These modifications were applied in the context of a replication-defective Ad5 viral genome expressing the GFP-luciferase (GFPLuc) fusion transgene. Each virus was rescued in 293 cells and amplified in cell factories. Virus particles were purified by double CsCl banding in the presence of the reducing agent DTT in order to maintain the inserted cysteines in their nonoxidized form, thereby preventing disulfide formation and virion clumping (19). All viruses grew to similar titers of 2 × 1013 to 6 × 1013 vp except for HVR1, which produced ∼10-fold fewer vp (viral harvest repeated four times). When the viruses were assayed for their TCID50, most of the vectors had vp/infectious unit (vp/IU) values within 2- to 18-fold of the unmodified virus (Table 1). In contrast, the HVR1 virus had vp/IU values that were 50-fold higher than those for Ad5. While the IU measurements were somewhat reduced, the transduction efficiency by the modified viruses was largely unchanged compared to unmodified Ad5 (Fig. 2C and Table 1).
Table 1.
Bioactivity of HVR-modified vectorsa
| Virus | vp/IU − PEG |
vp/luciferase − PEG |
vp/luciferase + PEG |
|||
|---|---|---|---|---|---|---|
| vp/IU | vp/IU: vP/IU wt | vp/RLU | vp/RLU: vp/RLU wt | vp/RLU | vp/RLU: vp/RLU wt | |
| HVRwt | 32 | 178 | 177 | |||
| HVR1 | 1,778 | 56 | 221 | 1.2 | 193 | 1.1 |
| HVR2 | 316 | 10 | 160 | 0.9 | 164 | 0.9 |
| HVR3 | 316 | 10 | 141 | 0.8 | 142 | 0.8 |
| HVR4 | 562 | 18 | 183 | 1.0 | 139 | 0.8 |
| HVR5 | 56 | 2 | 172 | 1.0 | 178 | 1.0 |
| HVR6 | 56 | 2 | NA | NA | NA | NA |
| HVR7 | 316 | 10 | 166 | 0.9 | 140 | 0.8 |
IU, infectious units; vp, virus particles; RLU, relative luminescence units; PEG, polyethylene glycol; wt, wild type; NA, not applicable.
To conditionally block the HVRs, each of the cysteine-modified vectors was reacted with maleimide-activated linear 5-kDa PEG, capped with or without a terminal biotin (for detection). Vectors were suspended in a 0.5 M sucrose buffer to maintain virion activity during these reactions (8). Excess PEG was removed by dialysis against 0.5 M sucrose. Mock-PEGylated viruses were treated in parallel without the addition of PEG.
Attachment of PEG to the virion was determined by detecting the biotin head group on each PEG with streptavidin-HRP by Western blotting (Fig. 2B). Faint background staining was shown for control adenoviruses, while all PEGylated Cys-modified viruses were strongly detected by streptavidin-HRP, indicating that each had been conjugated with 5-kDa PEG. To quantify the number of cysteine residues that were conjugated by PEGylation, an unmodified adenovirus and a representative Cys-modified virus, HVR5, were PEGylated with biotin–5-kDa PEG–maleimide. Their biotins were quantified by ELISA. This method demonstrated that there were ∼540 PEG molecules added to the HVR5 virus (data not shown). As a reciprocal assay to detect unreacted cysteines, unmodified and PEGylated HVR5 was reacted with the small fluorophore IR800-maleimide, and labeling was analyzed by fluorescence after SDS-PAGE. By this assay, 67% of available cysteines were protected by PEG (data not shown). These assays suggest that approximately two-thirds of the inserted cysteines are being protected by the 5-kDa PEG. This modification fraction translates to approximately 2 PEG molecules per hexon trimer. Given that PEGylation levels were similar by Western blotting for all of the HVR-modified viruses (Fig. 2B), these data suggest that they all may have similar numbers of PEGs as HVR5.
During recombination, HVR6-modified adenovirus lost its GFPLuc transgene. Since HVR6 is relatively buried and has not been implicated in Ad5 interactions, analysis of this HVR modification was not deemed to be crucial, and testing proceeded without this vector.
Targeted PEGylation of hexon preserves viral infectivity in vitro.
We and others have previously shown that Ad5 can be nonspecifically PEGylated with 5-kDa N-hydroxy-succinimide-activated PEG (NHS-PEG). This reagent reacts with up to 15,000 lysines on the surface of the virus and is useful for protecting the virus from preexisting neutralizing antibodies (9) and reducing side effects (15, 24). While NHS-PEG is potent for shielding, when it is applied under saturating conditions it inactivates the virus in vitro but not in vivo (10, 24).
We hypothesized that targeting PEGylation to specific sites on the virion would only inactivate virus functions specific to that site. Therefore, we predicted that the PEGylated cysteine-modified viruses should retain in vitro activity unless a critical HVR was involved in virus entry events. Previous data targeting PEGylation to HVR5 of Ad5 has shown that viral activity is retained (27), supporting the concept that targeted modification of all seven HVRs may similarly retain in vitro activity.
To investigate the in vitro activity of specifically PEGylated vectors, A549 cells were infected with 1,000 vp/cell of control and cysteine-modified vectors with or without conjugation to PEG. Their transduction activity was compared 24 h later by luciferase assay (Fig. 2C). There was no significant difference in expression between the cysteine-modified vectors and the unmodified control virus with or without PEG modification (Table 1). To contrast this with NHS-PEGylation, the HVR5 modified virus was nonspecifically PEGylated with NHS-PEG or was specifically PEGylated at its targeted cysteine with PEG-maleimide. Consistent with previous results, NHS-PEG reduced transduction >100-fold compared to both mock-treated and specifically PEGylated adenovirus (Fig. 2D, P < 0.01 [one-way ANOVA]). In contrast, targeting PEGylation to HVR5 preserved in vitro virus activity. When each of the viruses was tested for FX-mediated transduction, FX increased the in vitro transduction of all of the viruses, with or without PEG (see Fig. S1 in the supplemental material).
PEGylation of select HVRs increases liver transduction.
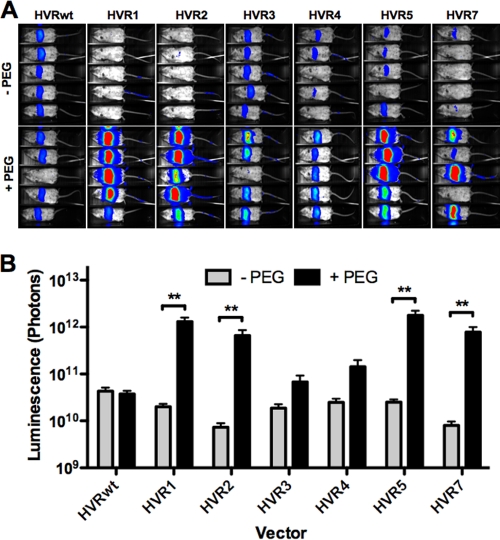
Since targeted PEGylation did not affect in vitro transduction, 1010 vp of each PEGylated and mock-PEGylated vector was injected i.v. into groups of female BALB/c mice to determine whether these modifications altered in vivo transduction. After 24 h, liver transduction was assessed by luciferase imaging (Fig. 3). The wild-type HVR (HVRwt) control virus mediated robust and similar liver transduction whether mock treated or treated with PEG. The transduction levels of mock-PEGylated cysteine-modified vectors were similar or slightly lower than those for HVRwt. Viruses that were PEGylated on HVRs 1, 2, 5, and 7 mediated up to 10- to 40-fold increases in liver transduction over HVRwt expression (P < 0.01 [one-way ANOVA]). In contrast, PEGylation of HVR3 and HVR4 mediated smaller increases in transduction that were not statistically significant. These data suggest that HVRs 1, 2, 5, and 7 may play a role in virus sequestration in vivo.
Fig 3.
PEGylation at HVRs 1, 2, 5, and 7 results in dramatically increased liver expression. BALB/c mice were injected i.v. with 1010 vp of mock-treated or PEGylated virus. (A) They were imaged 24 h later using a Lumazone imager for 3 min 1 × 1 or 2 × 2 binning. Shown are representative white-light images overlaid with luciferase expression in pseudocolor. (B) Quantification of luciferase expression from multiple experimental repeats. Sample sizes (n): HVRwt, 21; HVR1, 20; HVR2, 7 × 10; HVR3, 9; HVR4, 8; HVR5, 18. **, P < 0.01.
Kupffer cell depletion does not affect transduction by vectors after HVR-targeted PEGylation.
Eliminating Kupffer cells by predosing increases liver transduction by unmodified Ad5 10- to 40-fold (18). As found previously with Ad5/6, we hypothesized that HVR PEGylation might reduce uptake of adenovirus by liver Kupffer cells. If so, PEGylated vectors should be largely unaffected by the presence or absence of liver Kupffer cells. To test this, Kupffer cells were eliminated by predosing mice with irrelevant Ad5-dsRed 4 h before HVR vector injection as described previously (18). Luciferase imaging 24 h later showed that predosing increased unmodified Ad5 transduction 10-fold, as expected (Fig. 4A). HVR3-PEGylated vector transduction was also significantly increased by predosing (P < 0.01). In contrast, predosing did not increase transduction by PEGylated HVR 1, 4, or 5, suggesting that Kupffer cell elimination may be dependent on these HVR regions.
Fig 4.
Saturation of Kupffer cells does not boost expression of select Cys-modified vectors. (A) Mice were predosed i.v. with 3 × 1010 vp in 100 μl of PBS. After 4 h, the mice were injected i.v. with 1010 vp in PBS of PEGylated control or HVR-modified vectors (n = 3 to 8). Mice were imaged 24 h later for 3-min, 1 × 1 binning. (B) Mice were injected i.v. with a high dose of 3 × 1011 vp in 100 μl of PBS and then imaged after 24 h for 10-s, 1 × 1 binning. *, P < 0.05; **, P < 0.01.
High dose injection increases liver transduction more for unmodified Ad5 than for PEGylated Ad5.
Kupffer cells are a major viral sink for i.v. injected adenovirus at doses below its “threshold dose” (6). At doses of Ad5 that saturate and exceed this threshold, transduction becomes linear with dose. If HVR-PEGylated vectors avoid Kupffer cells, they should also avoid this threshold effect.
To test this, unmodified Ad5 or HVR5 virus were PEGylated with PEG-maleimide, and mice were injected i.v. with 30-fold-higher doses (3 × 1011 vp) than before. Under these conditions, increasing the Ad5 dose 30-fold increased liver transduction 10-fold (Fig. 4B). In contrast, Ad-HVR5 only increased its expression 2.5-fold. These data suggest that PEGylation of HVR5 avoids the threshold effect that normally impacts Ad5.
Blocking scavenger receptor in vivo does not boost transduction of PEGylated HVR1.
Negatively charged poly(I) is a ligand for scavenger receptors and has been shown to block Kupffer phagocytosis of Ad5 in vivo (14). Mice were injected with poly(I) for 5 min prior to i.v. injection of 1010 vp of Ad5 or Ad5/6 and transgene expression was determined 24 h later by luciferase imaging. Injection of poly(I) before Ad5 increased Ad5 transduction more than 10-fold but had little effect on Ad5/6 (Fig. 1A).
HVR1 has the largest number of charged amino acids (Fig. 2A) and has been implicated as a potential scavenger receptor domain (1) so this vector was used to test the effect of poly(I) on PEGylated adenoviruses. Injection of poly(I) prior to injection of unmodified Ad5 again boosted transduction 10-fold (P < 0.01 [two-tailed t test]) (Fig. 5). Poly(I) also increased the transduction of unPEGylated Ad-HVR1 to a lesser degree. In contrast, poly(I) did not increase the transduction of PEGylated Ad-HVR1 but, surprisingly, decreased transduction by the modified virus (P < 0.01 [two-tailed t test]).
Fig 5.
Poly(I) predosing does not boost the expression of PEGylated Ad-HVR1. BALB/c mice were injected i.v. with PBS or 100 μg of poly(I). After 5 min, 1010 vp of Ad or Ad-HVR1 with or without PEG were injected i.v. Mice were imaged for luciferase expression after 24 h for 3-min, 2 × 2 binning. (A) White-light images overlaid with luciferase expression in pseudocolor from 800 to 3,500 gray values; (B) quantification of luciferase expression (n = 3). **, P < 0.01; ns, not significant.
PEGylated HVR vectors are inefficiently recognized by scavenger receptors in vitro.
These data suggested that PEGylated-HVR1 virus bypasses Kupffer cell scavenger receptor uptake. To test this more specifically, CHO cells expressing CAR or the scavenger receptors SRA-II or SREC were incubated with selected HVR-modified adenoviruses at 1,000 vg/cell with or without PEGylation, and transduction was measured 36 h later (Fig. 6). The addition of CAR to the CHO cells increased transduction by the unmodified and PEGylated vectors ∼10-fold. Transduction by unPEGylated HVR vectors was increased by expression of SRA-II. In contrast, PEGylation of all of the tested HVR-modified vectors reduced this SRA-II-mediated transduction. SREC on CHO cells did not mediate increased transduction vector expression, but PEGylation of HVR7 and HVR3 reduced the background transduction on these cells.
Fig 6.
Targeted PEGylation decreases scavenger receptor mediated viral transduction. Control (None), coxsackie and adenovirus receptor (CAR), scavenger receptor AII (SRA-II) or scavenger receptor from endothelial cells (SREC) were stably expressed on CHO cells. These cells were infected with 1,000 viral genomes of HVR wild-type (HVRwt), 1, 2, 3, 5, and 7 vectors/cell that had been mock treated or PEGylated. Expression was determined 36 h later by luciferase assay (n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
DISCUSSION
Ad5 has potential for use as a vector for liver gene transfer due to its natural tropism for hepatocytes. However, sequestration of most of the injected dose of this virus by liver Kupffer cells necessitates the use of extremely large doses of vector that can trigger widespread toxicity. To maximize the effective dose of therapeutic adenoviruses for liver gene therapy, optimal vectors for liver gene delivery must retain their ability to bind blood factors (i.e., retain liver tropism) but minimize cellular sequestration (i.e., evade Kupffer cells) (18). In contrast, increasing adenovirus delivery beyond the liver will likely require evading Kupffer cells and hepatocytes.
Previous work implicated the hexon protein of Ad5 as a target for Kupffer cell sequestration (1). In the present study, we probed the Ad5 hexon in an effort to detect regions involved in its interactions with these cells. This work builds on our recent observations that Ad5 and Ad6, two highly related species C adenoviruses, have profoundly different interactions with liver Kupffer cells (18). We previously demonstrated that the Ad5 hexon is strongly recognized by Kupffer cells and macrophages, whereas Ad5 displaying the Ad6 hexon was not (18). Given that the only molecular differences between Ad5 and Ad5/6 are the HVRs of their hexons, we hypothesized that it was these hypervariable loops that are the key surfaces for interaction with Kupffer cells. We demonstrate here that the Ad5 HVR domain is indeed recognized by the Kupffer cell scavenger receptor SRA-II, whereas the Ad6 HVR domain is not.
To probe HVR-specific interactions with SRA-II and Kupffer cells, we applied targeted PEGylation to all seven of the Ad5 HVRs using the approach that previously was used to target PEG to HVR5 (11, 27). By this systematic approach, targeted PEGylation of HVRs 1, 2, 5, and 7 produced marked increases in liver hepatocyte transduction in vivo after intravenous injection. In contrast, PEGylation of HVRs 3 and 4 produced weak effects that did not reach statistical significance. These data implicate HVRs 1, 2, 5, and 7 as important regions that affect Ad5 pharmacology in vivo.
As shown previously with Ad5/6, the increased in vivo transduction by the PEGylated adenoviruses was not further increased by eliminating Kupffer cells prior to injection of these vectors. Nor was it increased by blocking scavenger receptors in vivo with poly(I). These PEGylated vectors also did not suffer the same threshold effect as unmodified Ad5. These data suggest that the in vivo liver transduction increases that were observed for HVRs 1, 2, 5, and 7 are mediated at least in part by protecting the virus from Kupffer cell sequestration.
Kupffer cells likely play a predominant role in the observed in vivo effects of Ad5/6 and HVR-PEGylated Ad5. However, we cannot exclude that some of these effects may occur by reducing interactions with other cells and proteins. For example, interactions with blood cells, endothelial cells, and blood proteins may be also be reduced by PEGylation (reviewed in reference 17). In the context of Ad5 (which suffers from strong Kupffer cell depletion), targeted PEGylation may affect this by modulating sequestration. If this bottleneck is blunted, other downstream interactions may be secondarily reduced by PEGylation. For other viruses that may natively avoid Kupffer cells, targeted HVR PEGylation may probe only downstream events. This study begins probing these interactions at this first bottleneck and lays the foundation for probing other downstream events.
Since HVR1 is the most negatively charged HVR (with a net negative charge of −13 amino acids [18]), it is reasonable to implicate HVR1 as a ligand for scavenger receptor recognition (1). Consistent with this, targeted PEGylation of HVR1 markedly increased in vivo transduction and reduced in vitro interactions with SRA-II, as expected. However, targeted PEGylation of HVRs 2, 5, and 7 also reduced these interactions and appeared to modulate interactions with Kupffer cells. HVR3 PEGylation also displays reduced interactions with SRA-II in vitro but showed weaker effects in vivo.
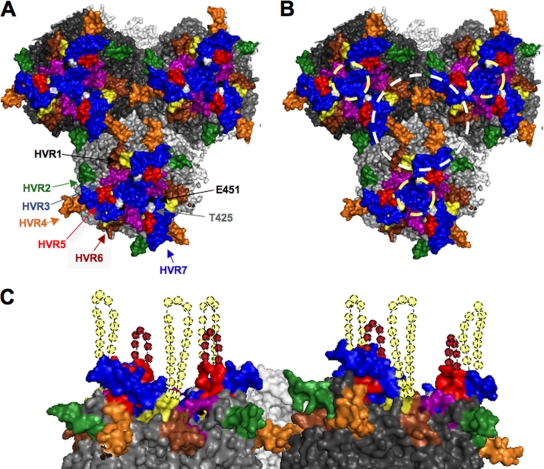
The X-ray structure of the hexon trimer shows that HVRs 1, 2, 5, and 7 (i.e., the four sites at which PEGylation causes >10-fold increased liver expression) are clustered on the surface of the hexon trimer (Fig. 7). In the context of the entire virion, two regions that may contribute to binding to Kupffer cells are: (i) the depression at the center of each hexon trimer and (ii) a depression at the center of three hexon trimers (Fig. 7B).
Fig 7.
Model of HVR domain interactions with host components. A hexon trimer of trimers, based on the recent X-ray structure of adenovirus, with structured HVR domains in color as follows: yellow, HVR1; green, HVR2; light blue, HVR3; orange, HVR4; red, HVR5; brown, HVR6; blue, HVR7. (A and B) Top-down view of hexon nonamer (trimer of trimers) with HVRs highlighted. (B) Potential FX and scavenger receptor binding surfaces. The light yellow dashed circle indicates the hexon trimer central depression and putative FX binding region; the white dashed circle indicates the hexon nonamer central depression and alternate recognition surfaces. (C) Side view, with a simulation of unstructured residues. Speculated locations for the unstructured HVR1 and HVR5 loops are shown as dashed loops of dashed spheres.
The binding site of FX has been localized by cryo-electron microscopy and mutagenesis studies to the center depression of the hexon trimer (indicated by a yellow dashed circle, Fig. 7B) (5). Several pieces of data suggest that FX and scavenger receptor may not bind the same surface on hexon. First, FX binding does not preclude Kupffer cells phagocytosis. Second, PEGylation of the HVRs inhibited SRA-II binding but did not inhibit interactions with FX in vitro (see Fig. S1 in the supplemental material). Nor did this PEGylation affect liver transduction in vivo. Indeed, hepatocyte transduction was markedly enhanced by shielding the HVRs, an effect opposite to what one would predict if FX binding was at the same sites. Given that PEGylation of HVRs 1, 2, 5, and 7 lines the rim of the depression formed by three hexon trimers, this region may be a binding surface or SRA-II and scavenger receptors (white circle, Fig. 7B) rather than in the center of each hexon trimer where FX binds (yellow circle, Fig. 7B). Interestingly, HVR4 is situated on the bottom surface of this cavity between hexon trimers and HVR3 projects more into this depression than into the depression within each trimer (Fig. 7B). Their close proximity to this depression may explain why PEGylation of HVR3 and HVR4 mediated detectable, but weaker effects on liver expression or SRA-II binding than PEGylation of HVRs 1, 2, 5, and 7, which are more accessible on the surface of the virion.
If this depression between trimers or the rim of this depression is involved in scavenger receptor interactions, Kupffer cell interactions could also occur at different locations on the virion. The 12 hexon trimers on each facet of the virion essentially all face the same direction on this surface. Therefore, the depressions between trimers within the facet are chemically identical with the exception of where the minor coat proteins IIIa and pIX protrude (28). In addition, a wider depression between trimers is also present at the edge of each facet. Therefore, it is possible that scavenger receptor binding may also occur on the face of the facet, at the facet's edge, or at sites where minor capsid proteins are localized.
Both FX and SRA-II have an affinity for anionic surfaces. FX binds anions via chelated calcium on its GLA domain (30) and SRA-II binds polyanions through its scavenger receptor cysteine-rich domain (18). It is interesting that adenoviruses that do not appear to be taken up by Kupffer cells (species B Ad11 and Ad35, species D Ad26 and Ad48 [11, 27; unpublished observations]) are not thought to bind FX (4). However, Ad6 (of species C) breaks this concordance. The Ad6 hexon appears to have reduced interactions with SRA-II and Kupffer cells but still binds FX, albeit at 10-fold-lower affinity than Ad5 hexon (18). This suggests that FX and scavenger receptor binding are separable, but more direct studies are needed to clarify this biology.
In summary, targeted PEGylation of each HVR domain appears to be a useful strategy to investigate the specific surfaces on hexon that mediate interactions with a variety of host components. This conditional shielding approach may have utility for probing the interactions of HVRs and other viral surfaces with other proteins in and outside of cells. Beyond its use in probing virus biology, the targeted PEGylation approach may also have practical value for liver gene transfer. In this case, targeted PEGylation of HVRs 1, 2, 5, or 7 generated viruses with 10- to 40-fold-enhanced in vivo hepatocyte transduction. Unlike previous efforts to randomly PEGylate adenovirus with NHS-PEG, this approach and previous targeted modification of HVR5 (27) preserves virus binding to CAR and FX and retains full virus function in vitro. Therefore, targeted PEGylation may provide insight into the interactions of adenovirus with the host and may be useful in optimizing vectors for purposes such as liver gene transfer and beyond.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brent Berwin and Monty Krieger for supplying scavenger receptor cell lines. We also thank Mary Barry for helpful technical assistance.
This study was supported by National Institutes of Health (NIH) grant R01-CA136945 to M.A.B. and NIH grant RO1-AI070771 to V.S.R.
Footnotes
Published ahead of print 7 December 2011
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Alemany R, Suzuki K, Curiel DT. 2000. Blood clearance rates of adenovirus type 5 in mice. J. Gen. Virol. 81:2605–2609 [DOI] [PubMed] [Google Scholar]
- 2. Ashkenas J, et al. 1993. Structures and high and low affinity ligand binding properties of murine type I and type II macrophage scavenger receptors. J. Lipid Res. 34:983–1000 [PubMed] [Google Scholar]
- 3. Bergelson JM, et al. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320–1323 [DOI] [PubMed] [Google Scholar]
- 4. Berwin B, Delneste Y, Lovingood RV, Post SR, Pizzo SV. 2004. SREC-I, a type F scavenger receptor, is an endocytic receptor for calreticulin. J. Biol. Chem. 279:51250–51257 [DOI] [PubMed] [Google Scholar]
- 5. Bowdish DM, Gordon S. 2009. Conserved domains of the class A scavenger receptors: evolution and function. Immunol. Rev. 227:19–31 [DOI] [PubMed] [Google Scholar]
- 6. Bristol JA, Shirley P, Idamakanti-N, Kaleko M, Connelly S. 2000. In vivo dose threshold effect of adenovirus-mediated factor VIII gene therapy in hemophiliac mice. Mol. Ther. 2:223–232 [DOI] [PubMed] [Google Scholar]
- 7. Campos SK, Barry MA. 2007. Current advances and future challenges in adenoviral vector biology and targeting. Curr. Gene Ther. 7:189–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Croyle MA, Cheng X, Wilson JM. 2001. Development of formulations that enhance physical stability of viral vectors for gene therapy. Gene Ther. 8:1281–1290 [DOI] [PubMed] [Google Scholar]
- 9. Croyle MA, Chirmule N, Zhang Y, Wilson JM. 2001. “Stealth” adenoviruses blunt cell-mediated and humoral immune responses against the virus and allow for significant gene expression upon readministration in the lung. J. Virol. 75:4792–4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doronin K, Shashkova EV, May SM, Hofherr SE, Barry MA. 2009. Chemical modification with high molecular weight polyethylene glycol reduces transduction of hepatocytes and increases efficacy of intravenously delivered oncolytic adenovirus. Hum. Gene Ther. 20:975–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espenlaub S, et al. 2010. Capsomer-specific fluorescent labeling of adenoviral vector particles allows for detailed analysis of intracellular particle trafficking and the performance of bioresponsive bonds for vector capsid modifications. Hum. Gene Ther. 21:1155–1167 [DOI] [PubMed] [Google Scholar]
- 12. Green NK, et al. 2004. Extended plasma circulation time and decreased toxicity of polymer-coated adenovirus. Gene Ther. 11:1256–1263 [DOI] [PubMed] [Google Scholar]
- 13. Haisma HJ, et al. 2009. Scavenger receptor A: a new route for adenovirus 5. Mol. Pharm. 6:366–374 [DOI] [PubMed] [Google Scholar]
- 14. Haisma HJ, et al. 2008. Polyinosinic acid enhances delivery of adenovirus vectors in vivo by preventing sequestration in liver macrophages. J. Gen. Virol. 89:1097–1105 [DOI] [PubMed] [Google Scholar]
- 15. Hofherr SE, Mok H, Gushiken FC, Lopez JA, Barry MA. 2007. Polyethylene glycol modification of adenovirus reduces platelet activation, endothelial cell activation, and thrombocytopenia. Hum. Gene Ther. 18:837–848 [DOI] [PubMed] [Google Scholar]
- 16. Kalyuzhniy O, et al. 2008. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. U. S. A. 105:5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khare R, Chen CY, Weaver EA, Barry MA. 2011. Advances and future challenges in adenoviral vector pharmacology and targeting. Curr. Gene Ther. 11:241–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khare R, et al. 2011. Generation of a Kupffer cell-evading adenovirus for systemic and liver-directed gene transfer. Mol. Ther. 19:1254–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kreppel F, Gackowski J, Schmidt E, Kochanek S. 2005. Combined genetic and chemical capsid modifications enable flexible and efficient de- and retargeting of adenovirus vectors. Mol. Ther. 12:107–117 [DOI] [PubMed] [Google Scholar]
- 20. Liu H, et al. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 329:1038–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Q, Muruve DA. 2003. Molecular basis of the inflammatory response to adenovirus vectors. Gene Ther. 10:935–940 [DOI] [PubMed] [Google Scholar]
- 22. McDonald D, Stockwin L, Matzow T, Blair Zajdel ME, Blair GE. 1999. Coxsackie and adenovirus receptor (CAR)-dependent and major histocompatibility complex (MHC) class I-independent uptake of recombinant adenoviruses into human tumor cells. Gene Ther. 6:1512–1519 [DOI] [PubMed] [Google Scholar]
- 23. Mizuta K, et al. 2009. Stability of the seven hexon hypervariable region sequences of adenovirus types 1–6 isolated in Yamagata, Japan between 1988 and 2007. Virus Res. 140:32–39 [DOI] [PubMed] [Google Scholar]
- 24. Mok H, Palmer DJ, Ng P, Barry MA. 2005. Evaluation of polyethylene glycol modification of first-generation and helper-dependent adenoviral vectors to reduce innate immune responses. Mol. Ther. 11:66–79 [DOI] [PubMed] [Google Scholar]
- 25. Nicol CG, et al. 2004. Effect of adenovirus serotype 5 fiber and penton modifications on in vivo tropism in rats. Mol. Ther. 10:344–354 [DOI] [PubMed] [Google Scholar]
- 26. Parker AL, et al. 2006. Multiple vitamin K-dependent coagulation zymogens promote adenovirus-mediated gene delivery to hepatocytes. Blood 108:2554–2561 [DOI] [PubMed] [Google Scholar]
- 27. Prill JM, et al. 2010. Modifications of adenovirus hexon allow for either hepatocyte detargeting or targeting with potential evasion from Kupffer cells. Mol. Ther. 19:83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reddy VS, Natchiar SK, Stewart PL, Nemerow GR. 2010. Crystal structure of human adenovirus at 3.5 Å resolution. Science 329:1071–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rux JJ, Burnett RM. 2000. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol. Ther. 1:18–30 [DOI] [PubMed] [Google Scholar]
- 30. Shashkova EV, May SM, Barry MA. 2009. Characterization of human adenovirus serotypes 5, 6, 11, and 35 as anticancer agents. Virology [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shayakhmetov DM, Gaggar A, Ni S, Li ZY, Lieber A. 2005. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol. 79:7478–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Snoeys J, et al. 2007. Species differences in transgene DNA uptake in hepatocytes after adenoviral transfer correlate with the size of endothelial fenestrae. Gene Ther. 14:604–612 [DOI] [PubMed] [Google Scholar]
- 33. Waddington SN, et al. 2008. Adenovirus serotype 5 hexon mediates liver gene transfer. Cell 132:397–409 [DOI] [PubMed] [Google Scholar]
- 34. Weaver EA, et al. 2011. Characterization of species C human adenovirus serotype 6 (Ad6). Virology 412:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wickham TJ, Mathias P, Cheresh DA, Nemerow GR. 1993. Integrins avb3 or avb5 promote adenovirus internalization but not virus attachment. Cell 73:309–319 [DOI] [PubMed] [Google Scholar]
- 36. Wu E, Nemerow GR. 2004. Virus yoga: the role of flexibility in virus host cell recognition. Trends Microbiol. 12:162–169 [DOI] [PubMed] [Google Scholar]
- 37. Xu Z, Tian J, Smith JS, Byrnes AP. 2008. Clearance of adenovirus by Kupffer cells is mediated by scavenger receptors, natural antibodies, and complement. J. Virol. 82:11705–11713 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.